Abstract
Anaplasma phagocytophilum is a zoonotic obligate intracellular bacterium known to be transmitted by ticks belonging to the Ixodes persulcatus complex. This bacterium can infect several mammalian species, and is known to cause diseases with variable symptoms in many domestic animals. Specifically, it is the causative agent of tick-borne fever (TBF), a disease of important economic impact in European domestic ruminants, and human granulocytic anaplasmosis (HGA), an emerging zoonotic disease in Asia, USA and Europe. A. phagocytophilum epidemiological cycles are complex and involve different ecotypes, vectors, and mammalian host species. Moreover, the epidemiology of A. phagocytophilum infection differs greatly between Europe and the USA. These different epidemiological contexts are associated with considerable variations in bacterial strains. Until recently, few A. phagocytophilum molecular typing tools were available, generating difficulties in completely elucidating the epidemiological cycles of this bacterium. Over the last few years, many A. phagocytophilum typing techniques have been developed, permitting in-depth epidemiological exploration. Here, we review the current knowledge and future perspectives regarding A. phagocytophilum epidemiology and phylogeny, and then focus on the molecular typing tools available for studying A. phagocytophilum genetic diversity.
Keywords: Anaplasma phagocytophilum, diversity, epidemiology, granulocytic anaplasmosis, phylogeny, typing technique, tick-borne fever
Introduction
Anaplasma phagocytophilum is a tick-borne intragranulocytic alpha-proteobacterium, transmitted by hard ticks of the Ixodes persulcatus complex. It infects a large range of hosts worldwide, and is the causative agent of tick-borne fever (TBF) in ruminants (also known as bovine or ovine granulocytic anaplasmosis), and of human granulocytic anaplasmosis (HGA), two diseases which are becoming increasingly recognized in both Europe and the USA. A. phagocytophilum epidemiological cycles are complex, and involve different ecotypes circulating in various host species, as vector ticks feed on a large variety of vertebrates. To date, these epidemiological cycles are poorly understood, especially in Europe, as European reservoir hosts of the bacterium have not yet been identified.
In this review, we first present current knowledge concerning A. phagocytophilum phylogeny and epidemiology, with a particular focus on Europe and the USA where the vast majority of studies have been undertaken. We then review molecular typing tools that are currently available for studying A. phagocytophilum.
Taxonomy
Veterinary medicine has been aware of the existence of A. phagocytophilum for decades. In 1932, during a study of louping-ill virus in sheep (Ovis aries) from Scotland, Gordon et al. discovered a new infectious disease caused by an unknown agent, but which was also transmitted by ticks (Gordon et al., 1932). The disease was named TBF, and in 1951 the causative agent was first described as Rickettsia phagocytophila (Foggie, 1951). However, the exact taxonomic position of A. phagocytophilum has been modified several times since 1951. The bacterium was reclassified as Cytoecetes phagocytophila in 1962 (Foggie, 1962), and as Ehrlichia phagocytophila in 1974 (Philip, 1974). Phylogenetic studies based on 16S RNA locus and groESL operon sequences drove Dumler et al. in 2001 to propose the unification of E. phagocytophila, Ehrlichia equi [first described in 1969 as the causative agent of equine granulocytic ehrlichiosis (Gribble, 1969)], and the agent of human granulocytic ehrlichiosis [first described in 1994 (Chen et al., 1994)] into one single species, A. phagocytophilum (Dumler et al., 2001). Using gltA gene sequences, Inokuma et al. also postulated that these three species were phylogenetically close, but did not question their placement within the Ehrlichia genus (Inokuma et al., 2001). In 2006 Dunning Hotopp et al. generated a phylogenetic tree based on the amino-acid sequences of 31 housekeeping proteins, which unified A. phagocytophilum and Anaplasma marginale into the same cluster, supporting the hypothesis of Dumler et al. (Dunning Hotopp et al., 2006). Even though their study provided strong arguments which supported the current classification of A. phagocytophilum, its taxonomy remains unclear, as only one A. phagocytophilum genome was compared. Considerable genetic variation has been consistently observed between different A. phagocytophilum strains (Barbet et al., 2013). For this reason, future phylogenomic analysis with the numerous A. phagocytophilum genomes (Dunning Hotopp et al., 2006; Barbet et al., 2013; Dugat et al., 2014b) and other newly available Anaplasmataceae genomic sequences will aid in refining the correct taxonomic position for A. phagocytophilum, possibly leading to further reclassification.
Because A. phagocytophilum has been reclassified several times since its discovery, the disease caused by this bacterium has multiple denominations: TBF, granulocytic ehrlichiosis, granulocytic anaplasmosis. In order to use a nomenclature common to all these diseases and to clearly designate the main host cell in which A. phagocytophilum multiplies, the most universal disease name will be used in this review: granulocytic anaplasmosis. For further clarification, the disease will be specified depending on the affected host species: HGA for humans, bovine granulocytic anaplasmosis (BGA) for cows (Bos taurus), ovine granulocytic anaplasmosis (OGA) for sheep, equine granulocytic anaplasmosis (EGA) for horses (Equus caballus) feline granulocytic anaplasmosis for cats (Felis silvestris catus), canine granulocytic anaplasmosis for dogs (Canis lupus familiaris).
Granulocytic anaplasmosis symptoms
GA symptoms in humans
HGA cases have been described throughout Asia, Europe, and North America (Jin et al., 2012; Ohashi et al., 2013; Kim et al., 2014). Clinical cases are both more serious and more frequent in the USA, where 2389 cases were reported by the Centers for Disease Control (CDC) in 2012 (Adams et al., 2014), as compared to the whole of Europe, where fewer than 100 confirmed cases have been described since its initial identification in Slovenia in 1997 (Petrovec et al., 1997). However, the number of reported European cases has risen over recent years, probably linked in part to improved surveillance (Cochez et al., 2011; Edouard et al., 2012). In Asia, HGA cases have been described in China, Japan, and South Korea (Ohashi et al., 2013; Zhang et al., 2013; Kim et al., 2014). In China, 46 confirmed and 16 probable cases were diagnosed from 2009 to 2010 with a 8.1% fatality rate (Zhang et al., 2013). However, seroprevalence studies suggest that the number of reported cases is underestimated in this country (Zhang et al., 2012, 2014).
HGA symptoms are variable and non-specific. Generally, patients present influenza-like symptoms with fever, headache, myalgia, and malaise (Ismail et al., 2010), and neurological symptoms have also been described (Ismail et al., 2010). Symptoms can be associated with decreased blood cell counts, thrombocytopenia, leukopenia, and anemia (Ismail et al., 2010). HGA can be fatal, but more than 60% of patients only present with moderate symptoms. However, HGA is not a benign disease, at least in the USA. In 2010, the CDC reported that 36% of HGA cases required hospitalization, of which 7% of these patients were referred to intensive care units (Centers for Disease Control and Prevention, Statistics and Epidemiology of Anaplasmosis)1.
A recent study reported severe clinical manifestations in Chinese patients, which have never been described to date in the USA and Europe: a multiple organ dysfunction syndrome (41.2%), gastrointestinal complications (22.6%), hemorrhagic complications (12.9%), a slow pulse (24.2%), facial edema (32.3%), and mental confusion (16.1%) (Zhang et al., 2013).
GA symptoms in domestic ruminants
To date, confirmed GA cases in domestic ruminants have only ever been diagnosed in Europe. Even if A. phagocytophilum infection has been suggested by the presence of DNA and/or serological conversion in domestic ruminants from other continents, no actual clinical cases have been described outside Europe.
The most frequent GA symptoms in domestic ruminants are fever (> 41°C), weakness, anorexia, and abortion storms, occasionally resulting in death (Stuen et al., 2013a). In dairy cattle, a significant drop in milk production is a prominent clinical sign, and respiratory distress and leg edema have also been observed (Woldehiwet, 2006, 2010). As A. phagocytophilum also has immunosuppressive effects, secondary infection represents a common indication of GA in cattle herds (Woldehiwet, 2006, 2010). In addition, up to 30% of lambs (O. aries) infected with A. phagocytophilum developed tick pyaemia due to Staphylococcus aureus infection, which is considered the most common and serious GA complication in sheep. Pasteurellosis and septicemic listeriosis have also been described in this animal species (Woldehiwet, 2006).
Even though limited data has been collected in the field, it is commonly accepted that ruminant GA has an important economic impact in Europe. In the UK, approximately 300,000 lambs develop tick pyaemia annually, secondary to A. phagocytophilum infection. Most of them die or are subsequently of no economic value. Economic losses associated with cattle abortions and reduced milk production can also be significant (Brodie et al., 1986).
GA symptoms in other domestic animals
In horses, EGA cases have been reported in both the USA and Europe. Many symptoms are similar to those observed in humans, and include fever, weakness, anorexia, ataxia, icterus leg edemas, thrombocytopenia, anemia, and leucopenia (Bermann et al., 2002; Butler et al., 2008; Jahn et al., 2010). Neurological symptoms have also been reported (Gussmann et al., 2014).
In dogs, malaise, lethargy, fever, inappetence or anorexia, weakness, indisposition, lymphadenomegaly, hepatomegaly, and splenomegaly have all been described (Carrade et al., 2009; Silveira et al., 2015).
In cats, a recent study reported lethargy in all animals, and fever and anorexia in 15/16 studied cats (Savidge et al., 2015). Hepatosplenomegaly, ataxia, conjunctivitis, and elevation of the nictitating membranes were also observed in some animals, associated with different blood cell count disorders, such as lymphopenia (7/11 cats), thrombocytopenia (7/11), neutropenia (3/11), and leukopenia (2/11) (Savidge et al., 2015).
A. phagocytophilum epidemiological cycles
As Stuen et al. reviewed the distribution of A. phagocytophilum according to continents and host species in 2013 (Stuen et al., 2013a), our review will first focus on the roles of the different actors involved in A. phagocytophilum epidemiological cycles, and then on the spread of A. phagocytophilum infection in both hosts and populations.
One bacterium, but multiple variants
The A. phagocytophilum species can be divided into several genetic variants2 likely involved in different epidemiological cycles. Here we will describe the most studied variants defined using the nucleotide sequence of the 16S RNA locus. Variants defined using other loci will be discussed in the paragraph “Tools for studying the genetic diversity of A. phagocytophilum.”
In the USA, variations in the 16S RNA locus distinguishes at least two major variants: Ap-V1 and Ap-ha (Chen et al., 1994). Other 16S RNA variants have also been detected, but as few epidemiological and biological data are currently available concerning these variants (Belongia et al., 1997; Massung et al., 2002; Michalski et al., 2006), we will focus on the major variants, Ap-V1 and Ap-ha. These two variants can coexist in the same geographical areas, and could also be transmitted by the same vectors (Courtney et al., 2003). These observations suggest that Ap-V1 and Ap-ha do not segregate according to geography, but rather according to the vertebrate hosts that they can or cannot infect. Indeed, the white-tailed deer (Odocoileus virginianus) is a major reservoir host for Ap-V1, but not Ap-ha (Massung et al., 2005; Reichard et al., 2009). Conversely, the white-footed mouse (Peromyscus leucopus) is a principle reservoir host for Ap-ha, but not Ap-V1 (Massung et al., 2003). Moreover, Ap-ha is pathogenic for humans, dogs and horses, whereas Ap-V1 has never been implicated in human infections. The tropism observed at the host level can also be observed at the cellular level: whereas Ap-ha can infect and multiply in both Ixodes (ISE6, IDE8) (Munderloh et al., 1999) and human (HL-60) cell lines (Goodman et al., 1996; Horowitz et al., 1998), Ap-V1 has only been cultivated in Ixodes cell lines (Woldehiwet et al., 2002; Massung et al., 2007).
A. phagocytophilum epidemiology in Europe is poorly understood compared to the USA. In contrast to what has been observed in the USA, European variants carrying 16S RNA sequences which correspond to both Ap-V1 and Ap-ha variants have been detected in several wild ruminant species.
Vectors
Granulocytic anaplasmosis is not a contagious disease, as A. phagocytophilum is only transmitted by vectors3. Known arthropod vectors of A. phagocytophilum are ticks belonging to the Ixodes genus: I. ricinus in Europe, I. scapularis in Eastern USA, I. pacificus and I. spinipalpis in Western USA, and I. persulcatus in Asia and Russia (MacLeod and Gordon, 1933; Telford et al., 1996; Des Vignes et al., 1999; Zeidner et al., 2000; Burkot et al., 2001; Stuen et al., 2013a). A. phagocytophilum DNA has also been detected in many other tick genera and species, but their vector competence has not yet been proven (Stuen et al., 2013a). Some other Ixodes species, such as I. trianguliceps (Bown et al., 2003, 2008, 2009) or I. hexagonus (Silaghi et al., 2012a) appear to be involved in epidemiological cycles distinct from those involving I. ricinus. This will be further discussed in the next section. Moreover, A. phagocytophilum DNA has been detected in areas where I. ricinus is rare or even absent. For example, several elements suggest that Rhipicephalus sp could be the vector involved in A. phagocytophilum transmission in the French Camargue region (Leblond et al., 2005; Moulin, 2009; Chastagner et al., 2013; Dugat et al., 2014a). However, as the vector competence of these ticks is unknown, the next paragraph focuses on confirmed A. phagocytophilum vectors.
Ixodes spp are telotropic species, meaning that they are able to feed on a large variety of host species (Stanek, 2009). True trophic “preferences” are difficult to identify, and are currently under discussions. Nevertheless, it has been suggested that larvae preferentially parasitize small mammals (such as rodents), nymphs parasitize bigger mammals (such as rabbits), and adult ticks target large mammals (such as wild and domestic ruminants) (Chauvet, 2004; Sonenshine and Roe, 2014).
The presence of Ixodes spp within a territory mainly depends on climatic conditions (between 10 and 30°C, and >80% relative humidity) and on the availability of hosts. In Europe, I. ricinus territories have expanded over the past few years (Materna et al., 2005; Jore et al., 2011; Jaenson et al., 2012), probably due to climatic and ecological modifications linked to both anthropic factors (Medlock et al., 2013; Jore et al., 2014) and forest extension. The low host specificity of I. ricinus, and its tolerance to various environments, can explain its broad spread across Europe.
Ticks belonging to the Ixodes genus have a three-host triphasic lifecycle. Each stage requires one blood meal (except for males which do not feed) before moving to the next stage. Ticks may acquire A. phagocytophilum while feeding on an infected animal host or through transstadial transmission (from one stage to the next). For Ixodes species, transovarial transmission (from the female adult to the eggs) does not occur according to current knowledge. Two experimental studies have reported A. phagocytophilum transmission between ticks via co-feeding4, without the need for an infected animal, but this mechanism appears to be rare (Levin and Fish, 2000; Ogden et al., 2003). Infected ticks can then transmit A. phagocytophilum to new hosts during the blood meal of its following stage. To summarize, Ixodes spp can be infected by A. phagocytophilum at each stage (except as eggs), but only nymphs and adult females can transmit this bacterium. For this reason, and because transovarial transmission has never been described in these ticks, Ixodes spp are unable to support persistent A. phagocytophilum infection, and therefore cannot be considered as reservoir hosts for this pathogen.
Reservoir and/or spillover hosts
A reservoir is a biotic or abiotic environment which permits a pathogen to persist in a sustainable manner, under given conditions. As A. phagocytophilum is an obligate intracellular bacterium, its reservoirs should be animal hosts which permit its survival, particularly (but not exclusively) outside the activity period of its vectors. For tick-borne pathogens, if the microorganism is transovary transmitted, the tick can also be a reservoir host, as has been demonstrated for Rickettsia helvetica and for the tick-borne encephalitis virus in I. ricinus (Nuttall and Labuda, 2003; Sprong et al., 2009; Rizzoli et al., 2014). As mentioned above for A. phagocytophilum, such transmission has never been described in Ixodes spp, indicating that they are not reservoir hosts for this bacterium. A. phagocytophilum transovarial transmission has been shown to occur at high frequency (up to 40%) for Dermacentor albipictus in the USA. However, under experimental conditions, infected F1 larval ticks which were reared to maturity failed to transmit the bacteria to their F2 larval progeny (Baldridge et al., 2009). Thus, even though A. phagocytophilum transovarial transmission in D. albipictus has been proven, its impact on bacterial persistence remains highly questionable. Therefore, according to currently available data, we assume that A. phagocytophilum reservoir hosts must be vertebrate organisms.
For some wild species, their precise role in A. phagocytophilum epidemiological cycles remains to be determined. Some species, rather than being reservoir hosts, could be spillover hosts, thus facilitating the transmission of A. phagocytophilum between reservoir hosts and susceptible hosts, particularly but not only when contacts between these categories of hosts are infrequent. As A. phagocytophilum epidemiological cycles are complex and not completely elucidated, it is important to keep in mind that some of the host species listed below that are supposed or suspected to be reservoir hosts, could in fact be spillover hosts.
Wild ruminants
In the USA, the white-tailed deer is the main reservoir host for A. phagocytophilum variant Ap-V1. The role of other wild ruminant species has not been investigated in depth to date. In California, one study has reported high infection prevalence5 in mule deer (Odocoileus hemonius hemonius) and Tule elks (Cervus elaphus nannodes) (Foley et al., 1998), suggesting that these two wild ruminants could also be reservoir hosts for A. phagocytophilum.
In Europe, A. phagocytophilum DNA has been detected in many wild ruminant species. In particular, red deer (Cervus elaphus) and roe deer (Capreolus capreolus) have been suspected to be A. phagocytophilum reservoir hosts (Alberdi et al., 2000) for several reasons. Firstly, A. phagocytophilum infection can persist for up to 3 months in red deer (Stuen et al., 2001). Secondly, both red and roe deer have high prevalences of A. phagocytophilum infection, as indicated by the presence of A. phagocytophilum DNA: up to 87.5 and 98.9% for red and roe deer, respectively (Stuen et al., 2013a). However, several recent studies strongly suggest that roe deer are not reservoir hosts for human, dog, or horse variants, nor for domestic ruminant variants (Rymaszewska, 2008; Scharf et al., 2011; Chastagner et al., 2014; Dugat et al., 2014a; Huhn et al., 2014). For this reason, we and other authors have hypothesized that roe deer could be reservoir hosts for their own specific A. phagocytophilum variants. Conversely, further data also indicate that red deer could be reservoir hosts for domestic ruminant variants (Petrovec et al., 2002; Majazki et al., 2013; Chastagner et al., 2014; Dugat et al., 2014a; Huhn et al., 2014). However, in contrast to what has been previously proposed (Rymaszewska, 2008), red deer are not likely to be reservoir hosts for human, dog, or horse variants (Chastagner et al., 2014; Dugat et al., 2014a; Huhn et al., 2014).
A. phagocytophilum is also highly prevalent in other European wild ruminants, such as elks (Alces alces), alpine ibex (Capra ibex), sika deer (Cervus Nippon), fallow deer (Dama dama), and chamois (Rupicapra rupicapra). However, the role of these animals in A. phagocytophilum epidemiological cycles has not yet been clarified.
In Asia, A. phagocytophilum DNA has been detected at high prevalence rates in Korean water deer (Hydropotes inermis argyropus) and in sika deer (C. nippon) up to 46 and 63.6%, respectively (Stuen et al., 2013a), suggesting that these animals could be reservoir hosts. Two studies reported that the variants obtained from sika deer in Japan are genetically distant from those infecting wild ruminants and other animal species in Asia, Europe and the USA (Kawahara et al., 2006; Masuzawa et al., 2011). However, the number of studies that have been conducted on wild ruminants in Asia is too limited to draw any definitive conclusion about the role played by these animals in A. phagocytophilum epidemiological cycles.
Rodents
Rodents are major hosts for ticks, mainly the larval and nymph stages. In the USA, A. phagocytophilum infection prevalence varies from 1.8 to 88.4%, depending on the study and the rodent species (Stuen et al., 2013a). In the eastern part of the country, the white-footed mouse, and possibly the gray squirrel (Sciurus carolinensis), are reservoir hosts for the Ap-ha variant (Levin et al., 2002; Massung et al., 2003). Dusky-footed woodrats (Neotoma fuscipes) and yellow-cheeked chipmunks (Tamias ochrogenys) are also considered as reservoir hosts for this variant, particularly in the western part of the USA, from where the white-footed mouse is absent (Nicholson et al., 1999; Levin et al., 2002; Foley et al., 2008, 2011; Nieto and Foley, 2009). Finally, in Colorado, the high prevalence of A. phagocytophilum infection observed in deer mouse (Peromyscus maniculatus) and in mexican woodrat (Neotoma mexicana), have led some authors to conclude that these rodents are reservoir hosts for this pathogen. They also suggested that these animals could be involved in an alternative epidemiological cycle, in which I. spinipalpis could be the vector (Zeidner et al., 2000).
In Europe, A. phagocytophilum DNA has been detected in at least nine different rodent species (Stuen et al., 2013a). However, the prevalence of A. phagocytophilum infection is lower in rodents than in wild ruminants (Barandika et al., 2007; Silaghi et al., 2012b; Stuen et al., 2013a; Baráková et al., 2014). Phylogenetic analysis based on groEL (Jahfari et al., 2014), msp4 (Baráková et al., 2014) and ankA (Majazki et al., 2013) loci have revealed that rodent strains belong to a different cluster than other A. phagocytophilum strains, making it unlikely that these rodent strains circulate in A. phagocytophilum epidemiological cycles involving other mammals. According to recent studies, rodents could be A. phagocytophilum reservoir hosts in an independent epidemiological cycle, involving only rodents as mammalian hosts (Bown et al., 2009; Blaòarová et al., 2014). In the UK and Central Europe, at least two independent epidemiological cycles have been described: the first may involve rodents as reservoir hosts and I. trianguliceps as vectors, whereas the second might involve wild ruminants as reservoir hosts and I. ricinus as vectors (Bown et al., 2003, 2008, 2009; Blaòarová et al., 2014).
In Asia, A. phagocytophilum DNA has been detected in at least 16 different rodent species (Stuen et al., 2013a). High prevalences of infection have been observed in several species in this continent-up to 55.5 and 23.6% in brown house rats (Rattus norvegicus) and black-striped field mice (Apodemus agrarius), respectively (Stuen et al., 2013a)-suggesting that they could be reservoir hosts for A. phagocytophilum. In particular, due to its high rate of infection and its wide geographical distribution in Asia, some authors consider that the black-striped field mice is the most important reservoir of A. phagocytophilum in this continent (Zhan et al., 2010; Jin et al., 2012; Yang et al., 2013). However, its reservoir competence has not been yet experimentally demonstrated (Zhan et al., 2008); thus, more studies are needed before drawing any definitive conclusion regarding the role of rodents as reservoir hosts of A. phagocytophilum in Asia.
Other wild species
A. phagocytophilum DNA has been detected in 24.6% of raccoons (Procyon lotor) tested in Connecticut (Levin et al., 2002). Raccoons can be experimentally infected by both Ap-ha and Ap-V1 variants, but only appear to be reservoir hosts for Ap-ha (Levin et al., 2002; Yabsley et al., 2008).
On Nantucket island (Massachusetts), the cottontail rabbit (Sylvilagus floridanus) could be involved in an alternative epidemiological cycle as a reservoir host, with Ixodes dentatus as the vector (Goethert and Telford, 2003).
In Europe, hedgehogs (Erinaceus europaeus) and wild boars (Sus scrofa) are also suspected to be reservoir hosts for human A. phagocytophilum variants for the following reasons (Skuballa et al., 2010; Silaghi et al., 2012a, 2014; Huhn et al., 2014). Firstly, high A. phagocytophilum infection prevalence has been observed in these animals (up to 85.4 and 12%, respectively) (Stuen et al., 2013a). Moreover, variants infecting hedgehogs and wild boars have identical 16S RNA sequences to those variants infecting humans (Silaghi et al., 2012a, 2014). In addition, a recent MLST study has revealed that variants infecting hedgehogs, wild boars and humans cluster together (Huhn et al., 2014). However, the immune system of wild boars quickly eliminates their infection (Galindo et al., 2012) and infection duration in hedgehogs also appears to be short (Silaghi et al., 2012a). Thus, their role as reservoir hosts has been recently questioned (Galindo et al., 2012; Silaghi et al., 2012a).
Hedgehogs could also be involved in an alternative epidemiological cycle in which I. hexagonus could be the vector (Silaghi et al., 2012a). Hedgehogs are often heavily parasitized by ticks, particularly I. hexagonus and I. ricinus, (Pfäffle et al., 2011; Silaghi et al., 2012a). Silaghi et al. have shown that hedgehogs, as well as the I. ricinus and I. hexagonus that feed on them, are all infected with A. phagocytophilum (Silaghi et al., 2012a). All variants reported in that study were able to infect both Ixodes species and hedgehogs (Skuballa et al., 2010; Silaghi et al., 2012a), but one variant (human pathogenic variant A) was more often associated with hedgehogs and I. ricinus (Silaghi et al., 2012a). Although the 16S RNA locus is not a highly discriminatory marker, this study suggested that variants infecting I. hexagonus could also infect I. ricinus, therefore the epidemiological cycles involving these two vectors may be interconnected. Moreover, the coexistence of variants involved in different epidemiological cycles in these hedgehogs could lead to genetic recombinations between variants, contributing to A. phagocytophilum genetic diversity. If this hypothesis of interconnected epidemiological cycles is validated, the hedgehog situation could be completely different from the independent epidemiological cycles described in UK rodents.
To date, only a few groups have studied the role of birds in A. phagocytophilum epidemiological cycles (Stuen et al., 2013a). One such study did detect high levels of A. phagocytophilum DNA in several different bird species. However, only a few birds from each species were tested (often only one), and for this reason it is difficult to conclude if birds play a significant role in A. phagocytophilum epidemiological cycles (De la Fuente et al., 2005). Contradictorily, a second study reported that only 1/11 common blackbirds (Turdus merula), was infected with A. phagocytophilum, and that no infection was observed in any of the eight other bird species examined. In this study, the authors used groEL sequences to demonstrate that birds and bird-fed ticks harbored specific A. phagocytophilum variants, suggesting that these animals could be involved in an independent avian epidemiological cycle (Jahfari et al., 2014). Studying tick feeding on an animal species can provide indirect clues as to its role in a pathogen's epidemiological cycle. Prevalence of A. phagocytophilum infection in bird-fed ticks is low, suggesting that birds are unlikely to be reservoir hosts (Skotarczak et al., 2006; Hildebrandt et al., 2010; Palomar et al., 2012; Geller et al., 2013; Lommano et al., 2014). Nevertheless, migrating birds can travel long distances and could be important for the spread of A. phagocytophilum and its vector(s) (Hildebrandt et al., 2010; Palomar et al., 2012; Dingler et al., 2014).
Domestic ruminants
In Europe, sheep are one of the main hosts of A. phagocytophilum. Intra-herd infection prevalence can reach up to 37% (Norway) (Stuen et al., 2013b), and at least 300,000 OGA cases are diagnosed every year in both Norway and the UK (Woldehiwet, 2006). During experimental infection of sheep, A. phagocytophilum DNA was detected by qPCR at 358 days post-infection. The authors did not confirm the presence of viable bacteria, yet this result suggests that sheep can maintain A. phagocytophilum infection for at least a year (Thomas et al., 2012). Thus, even if sheep develop OGA symptoms during the acute phase of infection, they could potentially also be reservoir hosts for their own A. phagocytophilum, as they can maintain constant infection with certain periods of high bacteremia. This dual role could contribute to the high number of OGA cases observed every year in some European countries.
Conclusions: hypothetical epidemiological cycles of A. phagocytophilum
In the USA (Figure 1, Table 1)
Figure 1.
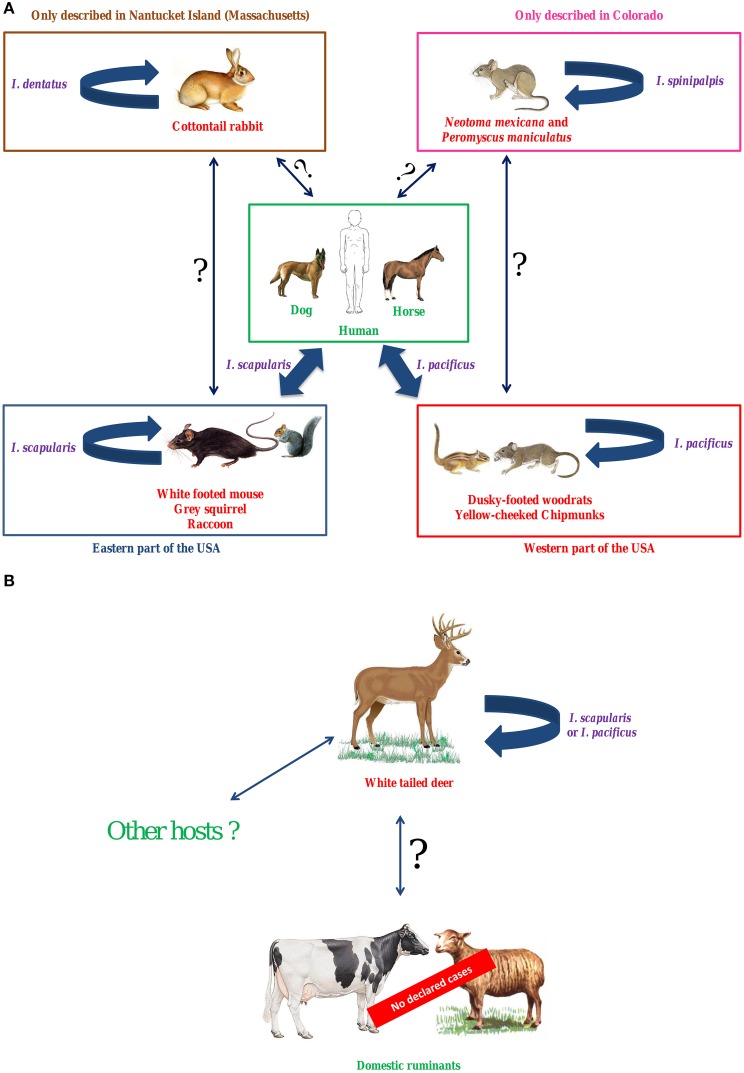
Hypothetical epidemiological cycles of A. phagocytophilum variants Ap-ha (A) and Ap-V1 (B) in the USA. In the USA, I. scapularis (Eastern states) and I. pacificus (Western states) are the main vectors of A. phagocytophilum. To date, two predominant independent epidemiological cycles, involving the two different variants Ap-ha and Ap-V1, have been identified in this country. White-footed mice, raccoons, gray squirrels (Eastern USA) and other rodents (Western USA) are reservoir hosts for the first variant, Ap-ha, which also infects humans, dogs, and horses. Variant Ap-V1 circulates in a secondary epidemiological cycle, which involves white-tailed deer as reservoir hosts. Domestic ruminants can be experimentally infected by this variant, however to date, no BGA cases have been reported in the USA. Thus, under natural conditions, variant Ap-ha does not appear capable of infecting ruminants, whereas variant Ap-V1 cannot infect either rodents or humans. Finally, two potential alternative A. phagocytophilum epidemiological cycles have been described in the USA: the first involves N. mexicana and P. maniculatus as reservoir hosts and the tick I. spinipalpis as vector, whereas the second involves the cottontail rabbit as a reservoir host and I. dentatus as vector. In purple, vectors; in red, reservoir hosts; in green, dead-end hosts (or clinical hosts); Large solid arrow, demonstrated transmission; Solid arrow with question mark, unknown transmission.
Table 1.
Main variants, reservoir hosts, and vectors of A. phagocytophilum in the USA.
| Variant | Reservoir host(s) | Vector(s) | Geographical area | Reference(s) |
|---|---|---|---|---|
| Ap-ha | Cottontail rabbit (S. floridanus) | I. dentatus | Nantucket Island (Massachusetts) | Goethert and Telford, 2003 |
| Mexican woodrat (N. mexicana)Deer mouse (P. maniculatus) | I. spinipalpis | Colorado | Zeidner et al., 2000 | |
| White-footed mouse (P. leucopus)Gray squirrel (S. carolinensis)Raccoon (P. lotor) | I. scapularis | Eastern USA | Levin et al., 2002; Massung et al., 2003 | |
| Dusky-footed woodrats (N. fuscipes)Yellow-cheeked chipmunks (T. ochrogenys) | I. pacificus | Western USA | Nicholson et al., 1999; Levin et al., 2002; Foley et al., 2008, 2011; Nieto and Foley, 2009 | |
| Ap-V1 | White-tailed deer (Odocoileus virginianus) |
I. scapularis I. pacificus |
Probably the whole country | Massung et al., 2005; Reichard et al., 2009 |
In the USA, I. scapularis (Eastern states) and I. pacificus (Western states) are the main vectors of A. phagocytophilum. To date, two predominant independent epidemiological cycles, involving the two different variants Ap-ha and Ap-V1, have been identified in this country. White-footed mice, raccoons, gray squirrels (Eastern USA), and other rodents (Western USA) are reservoir hosts for the first variant, Ap-ha, which also infects humans, dogs and horses (Figure 1A). Variant Ap-V1 circulates in a secondary epidemiological cycle, which involves white-tailed deer as reservoir hosts (Figure 1B). Domestic ruminants can be experimentally infected by this variant, however to date, no BGA cases have been reported in the USA. Thus, under natural conditions, variant Ap-ha does not appear capable of infecting ruminants, whereas variant Ap-V1 cannot infect either rodents or humans. Finally, two potential alternative A. phagocytophilum epidemiological cycles have been described in the USA: the first involves N. mexicana and P. maniculatus as reservoir hosts and the tick I. spinipalpis as vector, whereas the second involves the cottontail rabbit as a reservoir host and I. dentatus as vector.
In Europe (Figure 2, Table 2)
Figure 2.
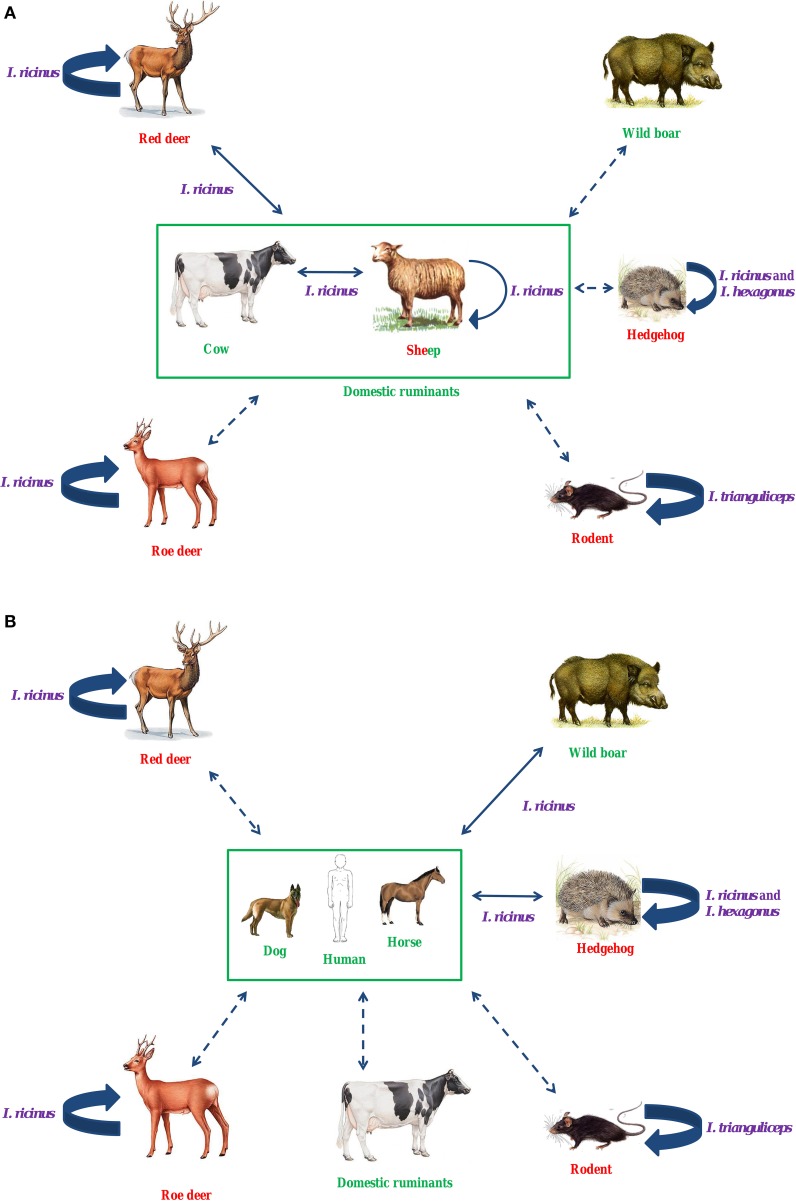
Hypothetical epidemiological cycles of A. phagocytophilum in ruminants (A) and in humans, dogs, and horses (B) in Europe. I. ricinus is the main European vector of A. phagocytophilum. Several studies suggest that red deer could be reservoir hosts for domestic ruminant variants. Roe deer are unlikely to be reservoir hosts for human, horse or pet variants, nor for domestic ruminant variants. Roe deer may be involved in another epidemiological cycle, and could maintain their “own” specific variant(s). Hedgehogs have been suspected as reservoir hosts for human variants, but this remains unproven. Similar to roe deer, hedgehogs could also be involved in an alternative epidemiological cycle, in which I. hexagonus could be the vector. Different rodent species could be involved in an independent epidemiological cycle involving I. trianguliceps as vector. In purple, vectors; in red, reservoir hosts; in green, dead-end hosts (or clinical hosts); Large solid arrow, known transmission; Solid arrow, unknown, but possible transmission; Dotted arrow, unknown, but unexpected transmission.
Table 2.
Major theoretical reservoir hosts, dead-end hosts, and vectors of A. phagocytophilum in Europe.
| Theoretical reservoir host | Vector(s) | Theoretical dead-end host(s) | Geographical area | Reference(s) |
|---|---|---|---|---|
| Roe deer (C. capreolus) | I. ricinus | Unknown | Europe | Alberdi et al., 2000; Rymaszewska, 2008; Scharf et al., 2011; Chastagner et al., 2014; Dugat et al., 2014a; Huhn et al., 2014 |
| Red deer (C. elaphus) | I. ricinus | Dometic ruminants | Europe | Stuen et al., 2001; Petrovec et al., 2002; Majazki et al., 2013; Chastagner et al., 2014; Dugat et al., 2014a; Huhn et al., 2014 |
| Hedgehog (E. europaeus) | I. ricinus and I. hexagonus | Human | Germany probably other countries | Skuballa et al., 2010; Pfäffle et al., 2011; Silaghi et al., 2012a; Huhn et al., 2014 |
| Different rodent species | I. trianguliceps | Unknown | United Kingdom and Slovakia | Bown et al., 2008, 2009; Majazki et al., 2013; Baráková et al., 2014; Blaòarová et al., 2014; Jahfari et al., 2014 |
| Sheep (O. aries) | I. ricinus | Dometic ruminants | Europe | Thomas et al., 2012; Stuen et al., 2013b |
I. ricinus is the main European vector of A. phagocytophilum, but to date, reservoir hosts for this bacterium have not been clearly identified. However, several studies suggest that red deer could be reservoir hosts for domestic ruminant variants (Figure 2A). Roe deer are unlikely to be reservoir hosts for human, horse or pet variants, nor for domestic ruminant variants (Figure 2). Roe deer may be involved in another epidemiological cycle, and could maintain their “own” specific variant(s). Hedgehogs have been suspected as reservoir hosts for human variants, but this remains unproven (Figure 2B). Similar to roe deer, hedgehogs could also be involved in an alternative epidemiological cycle, in which I. hexagonus could be the vector. Different rodent species could be involved in an independent epidemiological cycle involving I. trianguliceps as vector.
Dynamics of the infection
A. phagocytophilum transmission between vertebrate and invertebrate hosts
A. phagocytophilum transmission from the vector to the vertebrate host
Experimental transmission of A. phagocytophilum via I. scapularis to mice occurs from between 24 and 48 h after tick attachment (Hodzic et al., 1998; des Vignes et al., 2001). The median infectious dose at which morulae are observed in mice is between 104 and 105 A. phagocytophilum cells (Hodzic et al., 1998). In sheep, intravenously injecting an average of 1.3 infected neutrophil polynuclear cells was sufficient to induce TBF symptoms, but initial bacterial number per polynuclear cell was not assessed (Stuen and Artursson, 2000). As the very processes of tick attachment and feeding stimulate A. phagocytophilum multiplication, it is not possible to infer naturally occurring infection rates from these experimental conditions (Hodzic et al., 1998, 2001; Foley and Nieto, 2007). Additionally, incubation period is inversely dose-dependent: the higher the infectious dose, the shorter the incubation period (Stuen and Artursson, 2000), but according to Stuen et al., infectious dose is not correlated with severity of symptoms (Stuen and Artursson, 2000).
A. phagocytophilum transmission from vertebrate host to vector
Experimental A. phagocytophilum transmission from infected mice to I. scapularis occurs between 24 and 48 h after the initiation of blood meals (Hodzic et al., 1998). Subsequently A. phagocytophilum migrates to the gut and the salivary glands of the tick, with positive detection at these locations from 24 h after the start of the blood meal (Sukumaran et al., 2006).
The probability of A. phagocytophilum being transmitted from an infected sheep to I. ricinus is highest during the acute phase of infection, which then decreases with time as persistent infection is established and bacteremia decreases (post-acute phase) (Ogden et al., 2003). The risk of transmission also increases with the number of ticks parasitizing the animal (Ogden et al., 2003), and nymphs also have a higher likelihood of acquiring A. phagocytophilum than larvae (Ogden et al., 2003).
In order to more efficiently control GA, it is important to understand the dynamics of A. phagocytophilum infection, but this is generally an aspect that has been poorly studied. However, some important data have been obtained using sheep and mouse models. Nevertheless, as various A. phagocytophilum ecotypes appear to circulate in differing epidemiological cycles, the results presented here cannot be generalized across all species.
Infection spread within vertebrate host populations
A. phagocytophilum spread within host populations depends of the presence of: (1) the bacteria itself; (2) A. phagocytophilum-receptive hosts; (3) A. phagocytophilum-susceptible hosts; (4) competent vectors for A. phagocytophilum; and (5) the ability of these vectors to transmit A. phagocytophilum. In addition, increases in host and/or vector population size and/or their distribution areas can promote the geographical spread of the bacterium.
Roe deer can have high prevalence levels of A. phagocytophilum infection (Stuen et al., 2013a), and are often parasitized by many ticks (Vor et al., 2010). Deer can travel long distances (Vor et al., 2010), aiding the spread of both the bacteria and its vector(s) (Overzier et al., 2013). The same phenomenon could also occur with migratory birds (Alekseev et al., 2001; Bjöersdorff et al., 2001). However, the very low prevalence of A. phagocytophilum infection in such birds (Stuen et al., 2013a) suggests that their involvement in A. phagocytophilum expansion is limited.
A. phagocytophilum transplacental transmission has been described under natural conditions in cows (Henniger et al., 2013), and observed under experimental conditions in sheep (Reppert et al., 2013). However, evaluating the role of this transmission route in the spread of A. phagocytophilum infection has been difficult due to lack of data. In France, one study reported the presence of A. phagocytophilum DNA in 52/84 aborted bovine fetuses (5.2%), and in 484 placental swabs or 357 vaginal swabs from cows which had aborted (Guatteo et al., 2014). The presence of DNA alone does not conclusively confirm either the existence of viable bacteria, or their direct involvement in abortions, therefore additional studies are needed before drawing any definitive conclusions. Several human cases of A. phagocytophilum transmission occurring via blood transfusion have also been reported (Annen et al., 2012; Alhumaidan et al., 2013; Townsend et al., 2014). Finally, Zhang et al., reported nosocomial transmission of HGA in a Chinese hospital, through direct contact with blood or respiratory secretions (Zhang et al., 2008).
To summarize this section, many gaps still exist in current A. phagocytophilum epidemiology knowledge. This could be at least partly due to the fact that until very recently, few effective typing techniques were available. The second part of this review will be devoted to describing these tools.
Molecular tools for studying A. phagocytophilum
Historically, phenotypic approaches (such as serotyping) were used to study bacterial diversity. Apart from a study performed by Inokuma et al. (2003), A. phagocytophilum serotyping methods have rapidly been replaced by more efficient molecular biology-based typing methods.
A. phagocytophilum genome features
The A. phagocytophilum genome is a double-stranded chromosome from 1.47 to 1.48 Mb (only fully sequenced genomes were considered here) without any associated plasmids (Dunning Hotopp et al., 2006; Barbet et al., 2013). Complete sequenced genomes contain between 1140 and 1411 genes (including protein coding sequences, rRNA, tRNA, and pseudogenes) (Dunning Hotopp et al., 2006; Barbet et al., 2013; Dugat et al., 2014b). The HZ genome reference strain encodes 1369 proteins, of which 747 (55%) had known functions at the time of sequencing (Dunning Hotopp et al., 2006). Following infection of HL-60 cells, the expression of 1212 proteins was detected, corresponding to 89.3% of predicted proteins from the HZ strain genome (Lin et al., 2011). The vast majority of proteins with known function (99%) are produced, vs. only 80% of proteins with unknown function. The A. phagocytophilum genome also includes many repetitive elements (12.7% of the entire HZ genome sequence), containing 113 copies of the msp2/p44 gene and many variable number tandem repeat (VNTR) sequences (Dunning Hotopp et al., 2006; Barbet et al., 2013; Dugat et al., 2014a,b).
Tools for studying the genetic diversity of A. phagocytophilum
Pulsed-field gel electrophoresis (PFGE)
For decades, PFGE was considered the gold standard for epidemiological studies of clinically important bacteria, which is still the case for some species (Sabat et al., 2013). For this method, restriction enzymes cleave total genomic DNA at infrequently occurring restriction sites, and the resulting DNA fragments are then separated on an agarose gel using an alternating electric field.
Dumler et al. developed a PFGE approach for A. phagocytophilum (Dumler et al., 2003). However, their method could not discriminate between strains isolated from different animal species, and consequently is not appropriate for epidemiological studies. This technique is laborious, time consuming, and requires specific expertise, and because molecular tools have simultaneously been improved and diversified, becoming both faster and more user friendly, these new approaches have become widely adopted.
Single locus sequence typing (SLST)
SLST was the most common molecular approach for studying A. phagocytophilum genetic diversity, and is still often used. The groESL operon, 16S RNA locus, ankA, and msp2 genes are the most frequently utilized markers.
16S RNA locus
As previously stated, Massung et al. described six A. phagocytophilum variants (including Ap-V1 and Ap-ha) in the USA, by sequencing the 5′ region of the 16S RNA locus (Massung et al., 2002), and in 2011, 15 variants were defined using this sequence (Rar and Golovljova, 2011). This method can discriminate the human pathogenic variant Ap-ha from other non-pathogenic variants (Massung et al., 2002). However, the authors emphasized this marker's poor resolution, and recommended using other genes to study A. phagocytophilum genetic diversity.
In Europe, using the 16S RNA locus, several authors clearly distinguished variants infecting red deer from the those infecting roe deer (Petrovec et al., 2002; Zeman and Pecha, 2008). However, this conclusion was challenged by other studies, where results with this marker did not confirm any host species segregation for A. phagocytophilum variants, even including those variants infecting roe deer and red deer (Scharf et al., 2011; Silaghi et al., 2011a; Majazki et al., 2013). Some 16S RNA variants seem to infect wild and domestic ruminants more frequently, whereas other variants have been detected more often in humans, horses and dogs (Silaghi et al., 2011a,b). For this reason, in contrast to the USA, this locus does not seem appropriate for determining if one or many ecotype(s)6 circulate in European wild ruminants vs. other host species (Bown et al., 2007; Silaghi et al., 2011a; Overzier et al., 2013). Finally, 16S RNA data is unable to distinguish variants according to their geographical origins (Casey et al., 2004).
Even if some conclusions can be drawn using the 16S RNA locus, it does not have the required level of discrimination, such that markers generating more detailed information are essential for studying A. phagocytophilum diversity (Huhn et al., 2014).
The groESL operon
The groESL operon spans a region covering two genes encoding the GroES and GroEL chaperone proteins, separated by an intergenic region. In Europe, variations in groESL can differentiate variants infecting roe deer from those infecting others animals (Rymaszewska, 2008; Silaghi et al., 2011a,b). However, in Austria, other groups detected only one groESL variant infecting both red and roe deer (Polin et al., 2004). Using groEL sequences, Jahfari et al. described four A. phagocytophilum clusters circulating in Europe (Jahfari et al., 2014). The first included all 34 human variants and 209 A. phagocytophilum variants obtained from animals (including both wild and domestic animals). The second cluster grouped the majority of roe deer variants (66/72), several sheep variants (5/29), red deer (2/47), and rodent variants (3/30). The third cluster exclusively included rodent variants (27/30), whereas the fourth was only composed of eight A. phagocytophilum variants obtained from birds. According to this study, groESL could possibly discriminate between those variants isolated from birds and rodents, but not variants originating from other animals.
Also using the groESL operon, Sumner et al. identified variants circulating in the USA that originated from Europe (Sumner et al., 1997). However, this conclusion could not be generalized for all variants, as only three European strains and five American strains were screened. In another study, Alberti et al. described two clusters: the first grouped pathogenic variants and variants of unknown pathogenicity circulating in both the USA and Europe, whereas the second only included European variants of unknown pathogenicity (Alberti et al., 2005). Within the first cluster, two subclusters were also identified, grouping all variants isolated from the USA and Sardinia on one hand, and a subset of European variants on the other. However, Jahfari et al. could not discriminate variants originating from different European areas using the groESL operon (Jahfari et al., 2014).
Thus, the groESL operon can distinguish variants of different pathogenicity or geographic origin better than the 16S RNA locus, but further information is required for clearer discrimination of variants and ecotypes.
ankA gene
ankA encodes an ankyrin repeat protein involved in host cell transcription regulation, which, when sequenced, is able to distinguish five A. phagocytophilum variant clusters, relating to the animal host. It must be noted that the majority of samples for the two studies cited here were collected in Europe, except for human variants originating from the USA (Scharf et al., 2011; Majazki et al., 2013). The first cluster mostly contained variants obtained from humans, dogs, cats, and horses, with several variants obtained from domestic and wild ruminants. Within this group, European variants and American human variants belonged to separate subgroups (Rar and Golovljova, 2011). The second cluster included variants isolated from roe and red deer. The third contained variants from domestic (cattle and sheep) as well as wild ruminants (including red and roe deer). The fourth cluster exclusively grouped those variants isolated from roe deer, whereas the fifth was exclusively composed of rodent variants.
Sequencing ankA distinguishes variants according to their animal hosts, therefore although this method could be used for phylogenic analysis, it may not have the required level of discrimination for epidemiological studies, such that its applicability remains to be determined.
Msp2 gene
Msp2 is an A. phagocytophilum surface protein, belonging to the OMP-1/MSP2/P44 superfamily. msp2 comprises two conserved sequences flanking a hypervariable region, which is sequenced for A. phagocytophilum phylogenetic analysis. Using this hypervariable region, one study distinguished European from American variants (Silaghi et al., 2011b). However, horses were the only animal hosts common to these two continents. For this reason, the observed clustering could be linked to the animal host species, rather than to the geographical origin, as suggested by De La Fuente et al. (2005). Before drawing any definite conclusions, larger sample numbers obtained from various American and European animal host species will have to be tested.
Thus, the SLST method can potentially discriminate variants according to their geographical or animal host origins. However, current markers cannot reveal the full genetic diversity of A. phagocytophilum. Moreover, depending on the locus used, contradictory results have been obtained. For example, isolates from roe deer and domestic ruminants (sheep and cattle) belonged to different clusters when based on ankA gene phylogeny (Scharf et al., 2011), whereas examining the groEL locus showed that isolates from domestic ruminants (goats), belonged to the same cluster as those from roe deer (Silaghi et al., 2011a). In order to at least partly solve these problems, multilocus strategies should be employed.
Multilocus sequence analysis
Multilocus sequence typing (MLST)
The MLST method is based on the PCR amplification and sequencing of several loci. These loci generally consist of seven housekeeping genes, but their functions and numbers can vary according to the bacterial species for which the techniques are developed. For a given locus, each nucleotide sequence obtained corresponds to an allele. The combination of alleles for each locus, also named as a profile, can determine the sequence type (ST) of the microorganism in question. Because housekeeping genes have a rather slow molecular clock, and thus can represent excellent phylogenetic markers if they have been properly selected, MLST has been increasingly recognized as the gold standard phylogenetic method, at least for bacteria with clonal evolution. Various MLST techniques have been developed for A. phagocytophilum. Here, we will focus on the two most recent, both developed in 2014 (Chastagner et al., 2014; Huhn et al., 2014). These two techniques have produced results consistent with phylogenies built using 16S RNA and ankA loci, and have improved discriminatory power, as could be expected from a technique based on several genes.
The MLST method developed by Chastagner et al. was used to type 160 A. phagocytophilum-infected samples obtained from France: 104 cattle samples, 40 roe deer, 13 horse, and 3 dog samples (Chastagner et al., 2014). Interpretable results were obtained for 88 samples (55% typeability). Three clonal complexes were observed. The first only included cattle variants, the second comprised variants isolated from cattle, horses, and dogs, and the third comprised all variants sampled from roe deer and three A. phagocytophilum obtained from cattle. The authors concluded that three A. phagocytophilum epidemiological cycles exist in French cattle. However, as some potentially important host species, such as red deer, were not included in their analysis, additional studies are needed before drawing definite conclusions.
The second MLST technique, developed by Huhn et al., was used for typing 383 A. phagocytophilum-positive samples, obtained from various animal species (Huhn et al., 2014). Interpretable results were obtained for 284 samples (74% typeability). For other samples, double peaks were observed, probably due to sample co-infection by different A. phagocytophilum variants. One major clonal complex was detected, which contained the majority of European A. phagocytophilum obtained from humans, dogs, and horses. Wild boar and hedgehog variants were also included within this clonal complex, suggesting that they may serve as reservoir hosts for A. phagocytophilum capable of infecting humans, horses and dogs in Europe. Variants infecting roe deer and voles were located in another clonal complex, indicating that these animals are involved in (an)other A. phagocytophilum epidemiological cycle(s). However, 54% of obtained sequence types did not belong to any of the defined clonal complexes, indicating that the A. phagocytophilum population structure could be semi-clonal, raising questions about the reliability of MLST results for this bacterium (Huhn et al., 2014).
Multiple locus variable number tandem repeat (VNTR) analysis (MLVA)
The MLVA method is based on PCR amplification of different VNTR. VNTR are mini- or micro-satellite DNA sequences repeated in tandem. The principle of the MLVA method is that depending on the isolate, each VNTR may have a variable number of repeats, thus permitting isolate discrimination. After PCR amplification of a given VNTR, its length is determined by agarose gel migration, or by capillary electrophoresis, and the exact number of repeats is then calculated. Using multiple VNTR increases the discriminatory power of the technique.
Two MLVA techniques for A. phagocytophilum typing have been developed to date, the first was developed by Bown et al. (2007). It was based on intergenic microsatellite VNTR, but paradoxically, was too discriminatory for epidemiological studies. The 20 strains studied only shared a few alleles at each of the four loci. For this reason, their technique does not seem well adapted to studying either A. phagocytophilum transmission between species, or the circulation of isolates within host populations such as domestic ruminant herds.
The second MLVA technique was developed by Dugat et al. (2014a). In contrast to the first system, this method is based on intragenic minisatellite VNTR, which are theoretically less variable than intergenic microsatellite VNTR. In this study, A. phagocytophilum sampled from different cattle herds showed higher diversity than those variants sampled from the same herd, emphasizing the concordance of the information provided by this technique with epidemiological contexts. This technique also identified at least two separate A. phagocytophilum clusters based on their hosts: red deer and domestic ruminant variants on one hand, vs. roe deer variants on the other. Epidemiologically unlinked horse variants also grouped together in their own cluster, but only two samples were tested. The authors conclude that at least two A. phagocytophilum epidemiological cycles exist in France: the first involving red deer as reservoir hosts, and domestic ruminants as either accidental or longer-term hosts, whereas the second might involve roe deer as reservoir hosts.
A. phagocytophilum genetic diversity studies using whole genome sequencing
The dramatic cost reduction of whole genome sequencing technologies has enabled their use across a wide range of bacterial genome sequencing projects (Schuster, 2008; MacLean et al., 2009; Fournier et al., 2014). They can now even be used for routine medical investigations in bacteriology (Bertelli and Greub, 2013; Firth and Lipkin, 2013; Fournier et al., 2014). Here we review the current state of the art and future directions for A. phagocytophilum genomic studies.
Current state of the art
Twenty A. phagocytophilum genomes have been sequenced to date, 10 of which were released in March 2015 but without reference to any publication at the time of writing (Dunning Hotopp et al., 2006; Barbet et al., 2013; Dugat et al., 2014b; National Center for Biotechnology Information, 2015). Indeed, the few currently available genomes are far from being representative of global A. phagocytophilum diversity. Sixteen genomes are from American strains, whereas only four are from European strains (Table 3). Moreover, with the exception of human variants, only between one and three genomes are available per sampled host species. Thus, neither can these genomes represent the diversity of strains infecting these hosts. Finally, genomes from strains infecting hosts that could play key roles in A. phagocytophilum epidemiological cycles, such as roe or red deer, have not yet been released.
Table 3.
A. phagocytophilum genomes currently available in public databases.
| Strain/variant | Host | Geographical area | Size (Mb) | Number of genes* | Level of assembly | References |
|---|---|---|---|---|---|---|
| HZ | Human (Homo sapiens) | USA, New York state | 1.47 | 1411 | Close genome | Dunning Hotopp et al., 2006 |
| HZ2 | Human (Homo sapiens) | USA, New York state | 1.48 | 1141 | Close genome | Barbet et al., 2013 |
| ApNYW | Human (Homo sapiens) | USA, New York state | 1.5 | 1635 | 16 scaffolds | National Center for Biotechnology Information, 2015 |
| ApWI1 | Human (Homo sapiens) | USA, Wisconsin | 1.5 | 1589 | 1 scaffold | National Center for Biotechnology Information, 2015 |
| HGE1 | Human (Homo sapiens) | USA, Minnesota | 1.47 | 1188 | 2 scaffolds | Barbet et al., 2013 |
| HGE2 | Human (Homo sapiens) | USA, Minnesota | 1.48 | 1548 | 1 scaffold | National Center for Biotechnology Information, 2015 |
| NCH-1 | Human (Homo sapiens) | USA, Nantucket | 1.5 | 1646 | 15 scaffolds | National Center for Biotechnology Information, 2015 |
| Webster | Human (Homo sapiens) | USA, Wisconsin | 1.48 | 1570 | 1 scaffold | National Center for Biotechnology Information, 2015 |
| Dog2 | Dog (Canis lupus familiaris) | USA, Minnesota | 1.47 | 1148 | 1 scaffold | Barbet et al., 2013 |
| ApMUC09 | Dog (Canis lupus familiaris) | Europe, Netherlands | 1.52 | 1675 | 1 scaffold | National Center for Biotechnology Information, 2015 |
| ApNP | Dog (Canis lupus familiaris) | Europe, Austria | 1.52 | 1827 | 1 scaffold | National Center for Biotechnology Information, 2015 |
| Annie | Horse (Equus caballus) | USA, Minnesota | 1.52 | 1642 | 15 scaffolds | National Center for Biotechnology Information, 2015 |
| MRK | Horse (Equus caballus) | USA, California | 1.48 | 1155 | 9 scaffolds | Barbet et al., 2013 |
| BOV_10-179 | Cow (Bos taurus) | Europe, France | 1.37 | 1041 | 169 scaffolds | Dugat et al., 2014b |
| Norway Variant 2 | Sheep (Ovis aries) | Europe, Norway | 1.52 | 1174 | 23 scaffolds | Barbet et al., 2013 |
| JM | Zapus hudsonius | USA, Minnesota | 1.48 | 1140 | Close genome | Barbet et al., 2013 |
| CR1007 | Tamias striatus | USA, Minnesota | 1.5 | 1554 | 4 scaffolds | National Center for Biotechnology Information, 2015 |
| CRT38 | Ixodes scapularis | USA, Minnesota | 1.51 | 1203 | 2 scaffolds | Barbet et al., 2013 |
| CRT35 | Ixodes scapularis | USA, Minnesota | 1.45 | 1148 | 25 scaffolds | Barbet et al., 2013 |
| CRT53-1 | Ixodes scapularis | USA, Minnesota | 1.57 | 1655 | 45 scaffolds | National Center for Biotechnology Information, 2015 |
Genes include complete CDS, rRNA, tRNA and pseudogenes.
In summary, whereas whole genome sequencing is a powerful tool for studying A. phagocytophilum genetic diversity, there are too few published genomes to fully exploit their potential, especially when considering the low number of genomes available for each investigated animal species. More genomes sequenced from strains or samples originating from various animal hosts and geographical areas are understandably needed.
Perspectives for A. phagocytophilum genomic studies
The difficulty of cultivating Anaplasma sp. can be a critical barrier for accessing genomic sequences. In order to circumvent this difficulty, some authors have attempted to sequence Anaplasma sp. genomes without the intervening culturing steps (Dark et al., 2012; Dugat et al., 2014b).
In 2012, Dark et al. sequenced the genome of Anaplasma marginale Florida and St. Maries strains directly from infected cattle blood samples (Dark et al., 2012). They performed whole genome amplification using the Phi29 DNA polymerase, followed by A. marginale genome sequencing. This approach was probably successful because A. marginale multiplies in anucleate erythrocytes. In nucleated cells, host DNA would inevitably contaminate the bacterial genomic amplification reaction. In the same study, Dark et al. also successfully sequenced the genome of the A. phagocytophilum HZ strain, cultivated in HL-60 cells (Dark et al., 2012). A bacterial purifying step was still required with this method, and its efficiency for sequencing A. phagocytophilum genomes directly from field samples is yet to be explored.
Another strategy without the need for bacterial isolation or purification, was proposed by Dugat et al. (2014b). Using a whole genome capture approach, they sequenced the genome of A. phagocytophilum directly from an infected cattle blood sample. This method enriched A. phagocytophilum DNA 197-fold in the field sample, permitting successful sequencing. This is the first reported A. phagocytophilum whole genome sequencing performed directly from a field sample. The 1.37 Mb BOV-10_179 A. phagocytophilum genome obtained was composed of 169 scaffolds, compared to 56 in the approach used by Dark et al. (2012), and included 1281 coding DNA sequences. As the study of Dark et al. resequenced the HZ strain genome (first sequenced by Dunning Hotopp et al., 2006) it is not surprising that the number of scaffolds obtained was lower than the de novo sequencing project of the BOV-10_179 genome.
To capture the European bovine BOV-10_179 genome, Dugat et al. designed capture probes using the American human HZ strain genome, which was the only available A. phagocytophilum genome at the time. For this reason, and also due to the differences between human and ruminant strains (and potentially between American and European strains) (Barbet et al., 2013), it is plausible that at least some specific BOV-10_179 sequences have not been captured. However, is it interesting to note that the capture probes also captured flanking regions around target sequences, which could minimize this phenomenon. Using the 20 A. phagocytophilum genomes that are currently available, new capture probes could be designed which would better represent A. phagocytophilum genetic diversity, facilitating the capture of a wider range of A. phagocytophilum genome sequences. Finally, according to their results, the authors concluded that it should be relatively simple to sequence several A. phagocytophilum genomes via multiplexing, without compromising the results in terms of coverage (Dugat et al., 2014b). Thus, whole genome capture is a promising tool for studying A. phagocytophilum genetic diversity.
Conclusion
In conclusion, distinct A. phagocytophilum ecotypes appear to be adapted to different vertebrate hosts and/or vectors. As a result, A. phagocytophilum ecotypes could circulate within several different epidemiological cycles, which could be independent or interconnected. This adds additional complexity to the puzzle of A. phagocytophilum epidemiology, and alongside the lack of sufficiently accurate or well-adapted molecular tools, explains why these epidemiological cycles have not yet been completely elucidated. Indeed, until just a few years ago, single locus sequence typing techniques were used for studying both A. phagocytophilum epidemiology and phylogeny. During that time, various multilocus typing techniques have been developed. These techniques are an impressive stride forward in improving knowledge of A. phagocytophilum diversity. In the near future, whole genome sequencing will enable access to the entire genetic diversity of A. phagocytophilum, and will also facilitate the development of whole genome typing techniques such as single-nucleotide polymorphism approaches or whole genome MLST. These techniques will be invaluable in unlocking the black box of A. phagocytophilum diversity, and promoting continual cross-talk between the laboratory and the field.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was performed within the the Laboratory of Excellence (Labex) of Integrative Biology of Emerging Infectious Diseases (IBEID). This work was supported by the Alfort National Veterinary School (ENVA), by the French National Institute for Agricultural Research (INRA) and by the French Agency for Food, Environmental and Occupational Health and Safety (Anses). TD was funded by Anses.
Glossary
Abbreviations
- BGA
Bovine granulocytic anaplasmosis
- CDC
Centers for Disease Control
- EGA
Equine granulocytic anaplasmosis
- HGA
Human granulocytic anaplasmosis
- MLST
Multilocus sequence typing
- MLVA
Multiple locus VNTR analysis
- OGA
Ovine granulocytic anaplasmosis
- PFGE
Pulsed-field gel electrophoresis
- SLST
Single locus sequence typing
- ST
Sequence type
- TBF
Tick-borne fever
- VNTR
Variable number tandem repeat.
Footnotes
1Centers for Disease Control and Prevention, Statistics and Epidemiology of Anaplasmosis. Available online at: http://www.cdc.gov/anaplasmosis/stats/ (Accessed April 23, 2014).
2A genetic variant is defined here as a population identified within a bacterial species using one or more genetic markers (such as the 16S RNA locus), without any obligatory links to phenotypic variations. This subdivision depends on the marker used, and thus on the technique performed.
3A vector is defined here as an arthropod which transmits a pathogen from a receptive host (i.e., able to be infected by the pathogen and permits pathogen multiplication without developing symptoms) and/or a susceptible host (i.e., which can develop symptoms), to another receptive and/or susceptible host.
4The transmission of a pathogen by co-feeding is defined by Jones et al. as “an exchange of a pathogen between infected and uninfected arthropod vectors feeding simultaneously on a host in the absence of systemic infection” (Jones et al., 1987).
5In this review, for simplification purposes, the term “prevalence” refers to studies in which the prevalence was determined by detection of A.phagocytophilum DNA. The term “seroprevalence” is used when the prevalence was estimated through detection of specific antibodies.
6For A.phagocytophilum, an ecotype is defined as a bacterial population sharing the same ecological niche.
References
- Adams D., Jajosky R., Ajani U., Kriseman J., Sharp P., Onweh D., et al. (2014). Morbidity Mortality Weekly Report Summary of Notifiable Diseases. Available online at: http://www.cdc.gov/mmwr/pdf/wk/mm6153.pdf (Accessed July 21, 2015).
- Alberdi M. P., Walker A. R., Urquhart K. A. (2000). Field evidence that roe deer (Capreolus capreolus) are a natural host for Ehrlichia phagocytophila. Epidemiol. Infect. 124, 315–323. 10.1017/S0950268899003684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti A., Zobba R., Chessa B., Addis M. F., Sparagano O., Pinna Parpaglia M. L., et al. (2005). Equine and canine Anaplasma phagocytophilum strains isolated on the Island of Sardinia (Italy) are phylogenetically related to pathogenic strains from the United States. Appl. Environ. Microbiol. 71, 6418–6422. 10.1128/AEM.71.10.6418-6422.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseev A. N., Dubinina H. V., Semenov A. V., Bolshakov C. V. (2001). Evidence of ehrlichiosis agents found in ticks (Acari: Ixodidae) collected from migratory birds. J. Med. Entomol. 38, 471–474. 10.1603/0022-2585-38.4.471 [DOI] [PubMed] [Google Scholar]
- Alhumaidan H., Westley B., Esteva C., Berardi V., Young C., Sweeney J. (2013). Transfusion-transmitted anaplasmosis from leukoreduced red blood cells. Transfusion 53, 181–186. 10.1111/j.1537-2995.2012.03685.x [DOI] [PubMed] [Google Scholar]
- Annen K., Friedman K., Eshoa C., Horowitz M., Gottschall J., Straus T. (2012). Two cases of transfusion-transmitted Anaplasma phagocytophilum. Am. J. Clin. Pathol. 137, 562–565. 10.1309/AJCP4E4VQQQOZIAQ [DOI] [PubMed] [Google Scholar]
- Baldridge G. D., Scoles G. A., Burkhardt N. Y., Schloeder B., Kurtti T. J., Munderloh U. G. (2009). Transovarial transmission of Francisella-like endosymbionts and Anaplasma phagocytophilum variants in Dermacentor albipictus (Acari: Ixodidae). J. Med. Entomol. 46, 625–632. 10.1603/033.046.0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baráková I., Derdáková M., Carpi G., Rosso F., Collini M., Tagliapietra V., et al. (2014). Genetic and ecologic variability among Anaplasma phagocytophilum strains, northern Italy. Emerg. Infect. Dis. 20, 1082–1084. 10.3201/eid2006.131023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barandika J. F., Hurtado A., García-Esteban C., Gil H., Escudero R., Barral M., et al. (2007). Tick-borne zoonotic bacteria in wild and domestic small mammals in northern Spain. Appl. Environ. Microbiol. 73, 6166–6171. 10.1128/AEM.00590-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet A. F., Al-Khedery B., Stuen S., Granquist E. G., Felsheim R. F., Munderloh U. G. (2013). An emerging tick-borne disease of humans is caused by a subset of strains with conserved genome structure. Pathogens 2, 544–555. 10.3390/pathogens2030544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belongia E. A., Reed K. D., Mitchell P. D., Kolbert C. P., Persing D. H., Gill J. S., et al. (1997). Prevalence of granulocytic Ehrlichia infection among white-tailed deer in Wisconsin. J. Clin. Microbiol. 35, 1465–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermann F., Davoust B., Fournier P. E., Brisou-Lapointe A. V., Brouqui P. (2002). Ehrlichia equi (Anaplasma phagocytophila) infection in an adult horse in France. Vet. Rec. 150, 787–788. 10.1136/vr.150.25.787 [DOI] [PubMed] [Google Scholar]
- Bertelli C., Greub G. (2013). Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin. Microbiol. Infect. 19, 803–813. 10.1111/1469-0691.12217 [DOI] [PubMed] [Google Scholar]
- Bjöersdorff A., Bergström S., Massung R. F., Haemig P. D., Olsen B. (2001). Ehrlichia-infected ticks on migrating birds. Emerg. Infect. Dis. 7, 877–879. 10.3201/eid0705.017517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaòarová L., Stanko M., Carpi G., Miklisová D., Víchová B., Mošanský L., et al. (2014). Distinct Anaplasma phagocytophilum genotypes associated with Ixodes trianguliceps ticks and rodents in Central Europe. Ticks Tick Borne Dis. 5, 928–938. 10.1016/j.ttbdis.2014.07.012 [DOI] [PubMed] [Google Scholar]
- Bown K. J., Begon M., Bennett M., Woldehiwet Z., Ogden N. H. (2003). Seasonal dynamics of Anaplasma phagocytophila in a rodent-tick (Ixodes trianguliceps) system, United Kingdom. Emerg. Infect. Dis. 9, 63–70. 10.3201/eid0901.020169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown K. J., Lambin X., Ogden N. H., Begon M., Telford G., Woldehiwet Z., et al. (2009). Delineating Anaplasma phagocytophilum ecotypes in coexisting, discrete enzootic cycles. Emerg. Infect. Dis. 15, 1948–1954. 10.3201/eid1512.090178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown K. J., Lambin X., Ogden N. H., Petrovec M., Shaw S. E., Woldehiwet Z., et al. (2007). High-resolution genetic fingerprinting of European strains of Anaplasma phagocytophilum by use of multilocus variable-number tandem-repeat analysis. J. Clin. Microbiol. 45, 1771–1776. 10.1128/JCM.00365-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown K. J., Lambin X., Telford G. R., Ogden N. H., Telfer S., Woldehiwet Z., et al. (2008). Relative importance of Ixodes ricinus and Ixodes trianguliceps as vectors for Anaplasma phagocytophilum and Babesia microti in field vole (Microtus agrestis) populations. Appl. Environ. Microbiol. 74, 7118–7125. 10.1128/AEM.00625-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie T. A., Holmes P. H., Urquhart G. M. (1986). Some aspects of tick-borne diseases of British sheep. Vet. Rec. 118, 415–418. 10.1136/vr.118.15.415 [DOI] [PubMed] [Google Scholar]
- Burkot T. R., Maupin G. O., Schneider B. S., Denatale C., Happ C. M., Rutherford J. S., et al. (2001). Use of a sentinel host system to study the questing behavior of Ixodes spinipalpis and its role in the transmission of Borrelia bissettii, human granulocytic ehrlichiosis, and Babesia microti. Am. J. Trop. Med. Hyg. 65, 293–299. [DOI] [PubMed] [Google Scholar]
- Butler C. M., Nijhof A. M., Jongejan F., van der Kolk J. H. (2008). Anaplasma phagocytophilum infection in horses in the Netherlands. Vet. Rec. 162, 216–217. 10.1136/vr.162.7.216 [DOI] [PubMed] [Google Scholar]
- Carrade D. D., Foley J. E., Borjesson D. L., Sykes J. E. (2009). Canine granulocytic anaplasmosis: a review. J. Vet. Intern. Med. Am. Coll. Vet. Intern. Med. 23, 1129–1141. 10.1111/j.1939-1676.2009.0384.x [DOI] [PubMed] [Google Scholar]
- Casey A. N. J., Birtles R. J., Radford A. D., Bown K. J., French N. P., Woldehiwet Z., et al. (2004). Groupings of highly similar major surface protein (p44)-encoding paralogues: a potential index of genetic diversity amongst isolates of Anaplasma phagocytophilum. Microbiology 150, 727–734. 10.1099/mic.0.26648-0 [DOI] [PubMed] [Google Scholar]
- Chastagner A., Bailly X., Leblond A., Pradier S., Vourc'h G. (2013). Single Genotype of Anaplasma phagocytophilum Identified from Ticks, Camargue, France. Emerg. Infect. Dis. 19, 825–826. 10.3201/eid1905.121003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastagner A., Dugat T., Vourc'h G., Verheyden H., Legrand L., Bachy V., et al. (2014). Multilocus sequence analysis of Anaplasma phagocytophilum reveals three distinct lineages with different host ranges in clinically ill French cattle. Vet. Res. 45, 114. 10.1186/s13567-014-0114-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet S. (2004). Etude Dynamique des Populations de Tiques Dans des Élevages Bovins en Corrèze. DVM thesis, Ecole Nationale Vétérinaire, Agroalimentaire et de l'Alimentation, Nantes.
- Chen S. M., Dumler J. S., Bakken J. S., Walker D. H. (1994). Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochez C., Ducoffre G., Vandenvelde C., Luyasu V., Heyman P. (2011). Human anaplasmosis in Belgium: a 10-year seroepidemiological study. Ticks Tick-Borne Dis. 2, 156–159. 10.1016/j.ttbdis.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Courtney J. W., Dryden R. L., Montgomery J., Schneider B. S., Smith G., Massung R. F. (2003). Molecular characterization of Anaplasma phagocytophilum and Borrelia burgdorferi in Ixodes scapularis ticks from Pennsylvania. J. Clin. Microbiol. 41, 1569–1573. 10.1128/JCM.41.4.1569-1573.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dark M. J., Lundgren A. M., Barbet A. F. (2012). Determining the repertoire of immunodominant proteins via whole-genome amplification of intracellular pathogens. PLoS ONE 7:e36456. 10.1371/journal.pone.0036456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fuente J., Massung R. F., Wong S. J., Chu F. K., Lutz H., Meli M., et al. (2005). Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 43, 1309–1317. 10.1128/JCM.43.3.1309-1317.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente J., Naranjo V., Ruiz-Fons F., Höfle U., Fernández De Mera I. G., Villanúa D., et al. (2005). Potential vertebrate reservoir hosts and invertebrate vectors of Anaplasma marginale and A. phagocytophilum in central Spain. Vector Borne Zoonotic Dis. 5, 390–401. 10.1089/vbz.2005.5.390 [DOI] [PubMed] [Google Scholar]
- Des Vignes F., Levin M. L., Fish D. (1999). Comparative vector competence of Dermacentor variabilis and Ixodes scapularis (Acari: Ixodidae) for the agent of human granulocytic ehrlichiosis. J. Med. Entomol. 36, 182–185. 10.1093/jmedent/36.2.182 [DOI] [PubMed] [Google Scholar]
- des Vignes F., Piesman J., Heffernan R., Schulze T. L., Stafford K. C. S., III, Fish D. (2001). Effect of Tick Removal on Transmission of Borrelia burgdorferi and Ehrlichia phagocytophila by Ixodes scapularis Nymphs. J. Infect. Dis. 183, 773–778. 10.1086/318818 [DOI] [PubMed] [Google Scholar]
- Dingler R. J., Wright S. A., Donohue A. M., Macedo P. A., Foley J. E. (2014). Surveillance for Ixodes pacificus and the tick-borne pathogens Anaplasma phagocytophilum and Borrelia burgdorferi in birds from California's Inner Coast Range. Ticks Tick-Borne Dis. 5, 436–445. 10.1016/j.ttbdis.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Dugat T., Chastagner A., Lagrée A.-C., Petit E., Durand B., Thierry S., et al. (2014a). A new multiple-locus variable-number tandem repeat analysis reveals different clusters for Anaplasma phagocytophilum circulating in domestic and wild ruminants. Parasit. Vectors 7:439. 10.1186/1756-3305-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugat T., Loux V., Marthey S., Moroldo M., Lagrée A.-C., Boulouis H.-J., et al. (2014b). Comparative genomics of first available bovine Anaplasma phagocytophilum genome obtained with targeted sequence capture. BMC Genomics 15:973. 10.1186/1471-2164-15-973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler J. S., Asanovich K. M., Bakken J. S. (2003). Analysis of genetic identity of North American Anaplasma phagocytophilum strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41, 3392–3394. 10.1128/JCM.41.7.3392-3394.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler J. S., Barbet A. F., Bekker C. P., Dasch G. A., Palmer G. H., Ray S. C., et al. (2001). Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51, 2145–2165. 10.1099/00207713-51-6-2145 [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp J. C., Lin M., Madupu R., Crabtree J., Angiuoli S. V., Eisen J., et al. (2006). Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. 10.1371/journal.pgen.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edouard S., Koebel C., Goehringer F., Socolovschi C., Jaulhac B., Raoult D., et al. (2012). Emergence of human granulocytic anaplasmosis in France. Ticks Tick-Borne Dis. 3, 403–405. 10.1016/j.ttbdis.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Firth C., Lipkin W. I. (2013). The genomics of emerging pathogens. Annu. Rev. Genomics Hum. Genet. 14, 281–300. 10.1146/annurev-genom-091212-153446 [DOI] [PubMed] [Google Scholar]
- Foggie A. (1951). Studies on the infectious agent of tick-borne fever in sheep. J. Pathol. Bacteriol. 63, 1–15. 10.1002/path.1700630103 [DOI] [PubMed] [Google Scholar]
- Foggie A. (1962). Studies on Tick Pyaemia and Tick-borne Fever. London: Symposium of the Zoological Society of London. [Google Scholar]
- Foley J. E., Barlough J. E., Kimsey R. B., Madigan J. E., DeRock E., Poland A. (1998). Ehrlichia spp. in cervids from California. J. Wildl. Dis. 34, 731–737. 10.7589/0090-3558-34.4.731 [DOI] [PubMed] [Google Scholar]
- Foley J. E., Nieto N. C., Adjemian J., Dabritz H., Brown R. N. (2008). Anaplasma phagocytophilum infection in small mammal hosts of Ixodes ticks, Western United States. Emerg. Infect. Dis. 14, 1147–1150. 10.3201/eid1407.071599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J., Nieto N. (2007). Anaplasma phagocytophilum subverts tick salivary gland proteins. Trends Parasitol. 23, 3–5. 10.1016/j.pt.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Foley J., Rejmanek D., Fleer K., Nieto N. (2011). Nidicolous ticks of small mammals in Anaplasma phagocytophilum-enzootic sites in northern California. Ticks Tick-Borne Dis. 2, 75–80. 10.1016/j.ttbdis.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P.-E., Dubourg G., Raoult D. (2014). Clinical detection and characterization of bacterial pathogens in the genomics era. Genome Med. 6:114. 10.1186/s13073-014-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo R. C., Ayllon N., Smrdel K. S., Boadella M., Beltrán-Beck B., Mazariegos M., et al. (2012). Gene expression profile suggests that pigs (Sus scrofa) are susceptible to Anaplasma phagocytophilum but control infection. Parasit. Vectors 5:181. 10.1186/1756-3305-5-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller J., Nazarova L., Katargina O., Leivits A., Järvekülg L., Golovljova I. (2013). Tick-borne pathogens in ticks feeding on migratory passerines in Western Part of Estonia. Vector Borne Zoonotic Dis. 13, 443–448. 10.1089/vbz.2012.1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethert H. K., Telford S. R. (2003). Enzootic transmission of the agent of human granulocytic ehrlichiosis among cottontail rabbits. Am. J. Trop. Med. Hyg. 68, 633–637. [PubMed] [Google Scholar]
- Goodman J. L., Nelson C., Vitale B., Madigan J. E., Dumler J. S., Kurtti T. J., et al. (1996). Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334, 209–215. 10.1056/NEJM199601253340401 [DOI] [PubMed] [Google Scholar]
- Gordon W. S., Brownlee A., Wilson D. R., Macleod J. (1932). Tick-borne fever: a hitherto undescribed disease of sheep. J. Comp. Pathol. Ther. 45, 301–307. 10.1016/S0368-1742(32)80025-1 [DOI] [Google Scholar]
- Gribble D. H. (1969). Equine ehrlichiosis. J. Am. Vet. Med. Assoc. 462–469. [PubMed] [Google Scholar]
- Guatteo R., Nicollet P., LeDréan E., Ninio C., Joly A. (2014). Diagnostic Différentiel des Causes Infectieuses D'avortements Chez les Bovins Intérêt D'une Démarche Harmonisée Et D'une Technique PCR Multi-Agents. Reims: Journées Nationales GTV. [Google Scholar]
- Gussmann K., Czech C., Hermann M., Schaarschmidt-Kiener D., von Loewenich F. D. (2014). Anaplasma phagocytophilum infection in a horse from Switzerland with severe neurological symptoms. Schweiz. Arch. Für Tierheilkd. 156, 345–348. 10.1024/0036-7281/a000605 [DOI] [PubMed] [Google Scholar]
- Henniger T., Henniger P., Grossmann T., Distl O., Ganter M., von Loewenich F. D. (2013). Congenital infection with Anaplasma phagocytophilum in a calf in northern Germany. Acta Vet. Scand. 55:38. 10.1186/1751-0147-55-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt A., Franke J., Meier F., Sachse S., Dorn W., Straube E. (2010). The potential role of migratory birds in transmission cycles of Babesia spp., Anaplasma phagocytophilum, and Rickettsia spp. Ticks Tick-Borne Dis. 1, 105–107. 10.1016/j.ttbdis.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Hodzic E., Borjesson D. L., Feng S., Barthold S. W. (2001). Acquisition dynamics of Borrelia burgdorferi and the agent of human granulocytic ehrlichiosis at the host-vector interface. Vector Borne Zoonotic Dis. 1, 149–158. 10.1089/153036601316977750 [DOI] [PubMed] [Google Scholar]
- Hodzic E., Fish D., Maretzki C. M., Silva A. M. D., Feng S., Barthold S. W. (1998). Acquisition and transmission of the agent of human granulocytic ehrlichiosis by Ixodes scapularis ticks. J. Clin. Microbiol. 36, 3574–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H. W., Aguero-Rosenfeld M. E., McKenna D. F., Holmgren D., Hsieh T. C., Varde S. A., et al. (1998). Clinical and laboratory spectrum of culture-proven human granulocytic ehrlichiosis: comparison with culture-negative cases. Clin. Infect. Dis. 27, 1314–1317. 10.1086/515000 [DOI] [PubMed] [Google Scholar]
- Huhn C., Winter C., Wolfsperger T., Wüppenhorst N., Strašek Smrdel K., Skuballa J., et al. (2014). Analysis of the population structure of Anaplasma phagocytophilum using multilocus sequence typing. PLoS ONE 9:e93725. 10.1371/journal.pone.0093725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuma H., Brouqui P., Drancourt M., Raoult D. (2001). Citrate synthase gene sequence: a new tool for phylogenetic analysis and identification of Ehrlichia. J. Clin. Microbiol. 39, 3031–3039. 10.1128/JCM.39.9.3031-3039.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuma H., Brouqui P., Dumler J. S., Raoult D. (2003). Serotyping isolates of Anaplasma phagocytophilum by using monoclonal antibodies. Clin. Diagn. Lab. Immunol. 10, 969–972. 10.1128/cdli.10.5.969-972.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N., Bloch K. C., McBride J. W. (2010). Human ehrlichiosis and anaplasmosis. Clin. Lab. Med. 30, 261–292. 10.1016/j.cll.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenson T. G. T., Jaenson D. G. E., Eisen L., Petersson E., Lindgren E. (2012). Changes in the geographical distribution and abundance of the tick Ixodes ricinus during the past 30 years in Sweden. Parasit. Vectors 5:8. 10.1186/1756-3305-5-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahfari S., Coipan E. C., Fonville M., van Leeuwen A. D., Hengeveld P., Heylen D., et al. (2014). Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasit. Vectors 7:365. 10.1186/1756-3305-7-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn P., Zeman P., Bezdekova B., Praskova I. (2010). Equine granulocytic anaplasmosis in the Czech Republic. Vet. Rec. 166, 646–649. 10.1136/vr.4852 [DOI] [PubMed] [Google Scholar]
- Jin H., Wei F., Liu Q., Qian J. (2012). Epidemiology and control of human granulocytic anaplasmosis: a systematic review. Vector-Borne Zoonotic Dis. 12, 269–274. 10.1089/vbz.2011.0753 [DOI] [PubMed] [Google Scholar]
- Jones L. D., Davies C. R., Steele G. M., Nuttall P. A. (1987). A novel mode of arbovirus transmission involving a nonviremic host. Science 237, 775–777. 10.1126/science.3616608 [DOI] [PubMed] [Google Scholar]
- Jore S., Vanwambeke S. O., Viljugrein H., Isaksen K., Kristoffersen A. B., Woldehiwet Z., et al. (2014). Climate and environmental change drives Ixodes ricinus geographical expansion at the northern range margin. Parasit. Vectors 7:11. 10.1186/1756-3305-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore S., Viljugrein H., Hofshagen M., Brun-Hansen H., Kristoffersen A. B., Nygård K., et al. (2011). Multi-source analysis reveals latitudinal and altitudinal shifts in range of Ixodes ricinus at its northern distribution limit. Parasit. Vectors 4:84. 10.1186/1756-3305-4-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara M., Rikihisa Y., Lin Q., Isogai E., Tahara K., Itagaki A., et al. (2006). Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 72, 1102–1109. 10.1128/AEM.72.2.1102-1109.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-H., Yi J., Oh W. S., Kim N.-H., Choi S. J., Choe P. G., et al. (2014). Human granulocytic anaplasmosis, South Korea, 2013. Emerg. Infect. Dis. 20, 1708–1711. 10.3201/eid2010.131680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond A., Pradier S., Pitel P., Fortier G., Boireau P., Chadoeuf J., et al. (2005). Enquête épidémiologique sur l'anaplasmose équine (Anaplasma phagocytophilum) dans le sud de la France. Rev. Sci. Tech. 24, 899–908. [PubMed] [Google Scholar]
- Levin M. L., Fish D. (2000). Immunity reduces reservoir host competence of Peromyscus leucopus for Ehrlichia phagocytophila. Infect. Immun. 68, 1514–1518. 10.1128/IAI.68.3.1514-1518.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. L., Nicholson W. L., Massung R. F., Sumner J. W., Fish D. (2002). Comparison of the reservoir competence of medium-sized mammals and Peromyscus leucopus for Anaplasma phagocytophilum in Connecticut. Vector Borne Zoonotic Dis. 2, 125–136. 10.1089/15303660260613693 [DOI] [PubMed] [Google Scholar]
- Lin M., Kikuchi T., Brewer H. M., Norbeck A. D., Rikihisa Y. (2011). Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichia chaffeensis. Front. Microbiol. 2:24. 10.3389/fmicb.2011.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommano E., Dvoøák C., Vallotton L., Jenni L., Gern L. (2014). Tick-borne pathogens in ticks collected from breeding and migratory birds in Switzerland. Ticks Tick-Borne Dis. 5, 871–882. 10.1016/j.ttbdis.2014.07.001 [DOI] [PubMed] [Google Scholar]
- MacLean D., Jones J. D. G., Studholme D. J. (2009). Application of “next-generation” sequencing technologies to microbial genetics. Nat. Rev. Microbiol. 7, 287–296. 10.1038/nrmicro2122 [DOI] [PubMed] [Google Scholar]
- MacLeod J., Gordon W. S. (1933). Studies in tick-borne fever of sheep. I. Transmission by the tick, Ixodes ricinus, with a description of the disease produced. Parasitology 25, 273–283. 10.1017/S0031182000019442 [DOI] [Google Scholar]
- Majazki J., Wüppenhorst N., Hartelt K., Birtles R., von Loewenich F. D. (2013). Anaplasma phagocytophilum strains from voles and shrews exhibit specific ankA gene sequences. BMC Vet. Res. 9:235. 10.1186/1746-6148-9-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massung R. F., Courtney J. W., Hiratzka S. L., Pitzer V. E., Smith G., Dryden R. L. (2005). Anaplasma phagocytophilum in white-tailed deer. Emerg. Infect. Dis. 11, 1604–1606. 10.3201/eid1110.041329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massung R. F., Levin M. L., Munderloh U. G., Silverman D. J., Lynch M. J., Gaywee J. K., et al. (2007). Isolation and propagation of the Ap-variant 1 strain of Anaplasma phagocytophilum in a tick cell line. J. Clin. Microbiol. 45, 2138–2143. 10.1128/JCM.00478-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massung R. F., Mauel M. J., Owens J. H., Allan N., Courtney J. W., Stafford K. C., et al. (2002). Genetic variants of Ehrlichia phagocytophila, Rhode Island and Connecticut. Emerg. Infect. Dis. 8, 467–472. 10.3201/eid0805.010251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massung R. F., Priestley R. A., Miller N. J., Mather T. N., Levin M. L. (2003). Inability of a variant strain of Anaplasma phagocytophilum to infect mice. J. Infect. Dis. 188, 1757–1763. 10.1086/379725 [DOI] [PubMed] [Google Scholar]
- Masuzawa T., Uchishima Y., Fukui T., Okamoto Y., Muto M., Koizumi N., et al. (2011). Detection of Anaplasma phagocytophilum from Wild Boars and Deer in Japan. Jpn. J. Infect. Dis. 64, 333–336. [PubMed] [Google Scholar]
- Materna J., Daniel M., Danielová V. (2005). Altitudinal distribution limit of the tick Ixodes ricinus shifted considerably towards higher altitudes in central Europe: results of three years monitoring in the Krkonose Mts. (Czech Republic). Cent. Eur. J. Public Health 13, 24–28. [PubMed] [Google Scholar]
- Medlock J. M., Hansford K. M., Bormane A., Derdakova M., Estrada-Peña A., George J.-C., et al. (2013). Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasit. Vectors 6:1. 10.1186/1756-3305-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski M., Rosenfield C., Erickson M., Selle R., Bates K., Essar D., et al. (2006). Anaplasma phagocytophilum in central and western Wisconsin: a molecular survey. Parasitol. Res. 99, 694–699. 10.1007/s00436-006-0217-9 [DOI] [PubMed] [Google Scholar]
- Moulin E. (2009). Tiques Potentiellement Vectrices De L'anaplasmose Granulocytaire Équine en Camargue. DVM thesis, Université Claude Bernard Lyon 1, Ecole Nationale Vétérinaire de Lyon, Marcy-l'Étoile.
- Munderloh U. G., Jauron S. D., Fingerle V., Leitritz L., Hayes S. F., Hautman J. M., et al. (1999). Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 37, 2518–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (2015). Anaplasma phagocytophilum Genomes, NCBI. Available online at: http://www.ncbi.nlm.nih.gov/genome/genomes/1083 (Accessed July 4, 2015).
- Nicholson W. L., Castro M. B., Kramer V. L., Sumner J. W., Childs J. E. (1999). Dusky-footed wood rats (Neotoma fuscipes) as reservoirs of granulocytic Ehrlichiae (Rickettsiales: Ehrlichieae) in Northern California. J. Clin. Microbiol. 37, 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto N. C., Foley J. E. (2009). Reservoir competence of the redwood chipmunk (Tamias ochrogenys) for Anaplasma phagocytophilum. Vector Borne Zoonotic Dis. 9, 573–577. 10.1089/vbz.2008.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall P. A., Labuda M. (2003). Dynamics of infection in tick vectors and at the tick-host interface. Adv. Virus Res. 60, 233–272. 10.1016/S0065-3527(03)60007-2 [DOI] [PubMed] [Google Scholar]
- Ogden N. H., Casey A. N. J., Woldehiwet Z., French N. P. (2003). Transmission of Anaplasma phagocytophilum to Ixodes ricinus ticks from sheep in the acute and post-acute phases of infection. Infect. Immun. 71, 2071–2078. 10.1128/IAI.71.4.2071-2078.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi N., Gaowa Wuritu, Kawamori, F., Wu D., Yoshikawa Y., et al. (2013). Human granulocytic anaplasmosis, Japan. Emerg. Infect. Dis. 19, 289–292. 10.3201/eid1902.120855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overzier E., Pfister K., Herb I., Mahling M., Böck G., Jr., Silaghi C. (2013). Detection of tick-borne pathogens in roe deer (Capreolus capreolus), in questing ticks (Ixodes ricinus), and in ticks infesting roe deer in southern Germany. Ticks Tick-Borne Dis. 4, 320–328. 10.1016/j.ttbdis.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Palomar A. M., Santibáñez P., Mazuelas D., Roncero L., Santibáñez S., Portillo A., et al. (2012). Role of birds in dispersal of etiologic agents of tick-borne zoonoses, Spain, 2009. Emerg. Infect. Dis. 18, 1188–1191. 10.3201/eid1807.111777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovec M., Bidovec A., Sumner J. W., Nicholson W. L., Childs J. E., Avsic-Zupanc T. (2002). Infection with Anaplasma phagocytophila in cervids from Slovenia: evidence of two genotypic lineages. Wien. Klin. Wochenschr. 114, 641–647. [PubMed] [Google Scholar]
- Petrovec M., Lotric Furlan S., Zupanc T. A., Strle F., Brouqui P., Roux V., et al. (1997). Human disease in Europe caused by a granulocytic Ehrlichia species. J. Clin. Microbiol. 35, 1556–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfäffle M., Petney T., Skuballa J., Taraschewski H. (2011). Comparative population dynamics of a generalist (Ixodes ricinus) and specialist tick (I. hexagonus) species from European hedgehogs. Exp. Appl. Acarol. 54, 151–164. 10.1007/s10493-011-9432-x [DOI] [PubMed] [Google Scholar]
- Philip C. B. (1974). Bergey's Manual of Determinative Bacteriology, 8th Edn. Baltimore, MD: Williams and Wilkins Co. [Google Scholar]
- Polin H., Hufnagl P., Haunschmid R., Gruber F., Ladurner G. (2004). Molecular evidence of Anaplasma phagocytophilum in Ixodes ricinus ticks and wild animals in Austria. J. Clin. Microbiol. 42, 2285–2286. 10.1128/JCM.42.5.2285-2286.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rar V., Golovljova I. (2011). Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect. Genet. Evol. 11, 1842–1861. 10.1016/j.meegid.2011.09.019 [DOI] [PubMed] [Google Scholar]
- Reichard M. V., Roman R. M., Kocan K. M., Blouin E. F., de la Fuente J., Snider T. A., et al. (2009). Inoculation of White-Tailed Deer (Odocoileus Virginianus) with Ap-V1 Or NY-18 strains of Anaplasma Phagocytophilum and microscopic demonstration of Ap-V1 In Ixodes Scapularis adults that acquired infection from Deer as Nymphs. Vector-Borne Zoonotic Dis. 9, 565–568. 10.1089/vbz.2008.0106 [DOI] [PubMed] [Google Scholar]
- Reppert E., Galindo R. C., Breshears M. A., Kocan K. M., Blouin E. F., de la Fuente J. (2013). Demonstration of transplacental transmission of a human isolate of Anaplasma phagocytophilum in an experimentally infected sheep. Transbound. Emerg. Dis. 60(Suppl. 2), 93–96. 10.1111/tbed.12120 [DOI] [PubMed] [Google Scholar]
- Rizzoli A., Silaghi C., Obiegala A., Rudolf I., Hubálek Z., Földvári G., et al. (2014). Ixodes ricinus and Its transmitted pathogens in Urban and Peri-urban areas in Europe: new hazards and relevance for public health. Front. Public Health 2:251. 10.3389/fpubh.2014.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymaszewska A. (2008). Divergence within the marker region of the groESL operon in Anaplasma phagocytophilum. Eur. J. Clin. Microbiol. Infect. Dis. 27, 1025–1036. 10.1007/s10096-008-0539-x [DOI] [PubMed] [Google Scholar]
- Sabat A. J., Budimir A., Nashev D., Sá-Leão R., van Dijl J. m, Laurent, F., et al. (2013). Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 18, 20380. [DOI] [PubMed] [Google Scholar]
- Savidge C., Ewing P., Andrews J., Aucoin D., Lappin M. R., Moroff S. (2015). Anaplasma phagocytophilum infection of domestic cats: 16 cases from the northeastern USA. J. Feline Med. Surg. 17, 87–93. 10.1177/1098612x15571148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf W., Schauer S., Freyburger F., Petrovec M., Schaarschmidt-Kiener D., Liebisch G., et al. (2011). Distinct Host Species Correlate with Anaplasma phagocytophilum ankA Gene Clusters∇. J. Clin. Microbiol. 49, 790–796. 10.1128/JCM.02051-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S. C. (2008). Next-generation sequencing transforms today's biology. Nat. Methods 5, 16–18. 10.1038/nmeth1156 [DOI] [PubMed] [Google Scholar]
- Silaghi C., Hamel D., Thiel C., Pfister K., Passos L. M. F., Rehbein S. (2011a). Genetic variants of Anaplasma phagocytophilum in wild caprine and cervid ungulates from the Alps in Tyrol, Austria. Vector Borne Zoonotic Dis. 11, 355–362. 10.1089/vbz.2010.0051 [DOI] [PubMed] [Google Scholar]
- Silaghi C., Liebisch G., Pfister K. (2011b). Genetic variants of Anaplasma phagocytophilum from 14 equine granulocytic anaplasmosis cases. Parasit. Vectors 4:161. 10.1186/1756-3305-4-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silaghi C., Pfister K., Overzier E. (2014). Molecular investigation for bacterial and protozoan tick-borne pathogens in Wild Boars (Sus scrofa) from Southern Germany. Vector Borne Zoonotic Dis. 14, 371–373. 10.1089/vbz.2013.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silaghi C., Skuballa J., Thiel C., Pfister K., Petney T., Pfäffle M., et al. (2012a). The European hedgehog (Erinaceus europaeus)–a suitable reservoir for variants of Anaplasma phagocytophilum? Ticks Tick-Borne Dis. 3, 49–54. 10.1016/j.ttbdis.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Silaghi C., Woll D., Hamel D., Pfister K., Mahling M., Pfeffer M. (2012b). Babesia spp. and Anaplasma phagocytophilum in questing ticks, ticks parasitizing rodents and the parasitized rodents - analyzing the host-pathogen-vector interface in a metropolitan area. Parasit. Vectors 5:191. 10.1186/1756-3305-5-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira J. A. G., Valente P. C. L. G., Paes P. R. O., Vasconcelos A. V., Silvestre B. T., Ribeiro M. F. B. (2015). The first clinical and laboratory evidence of co-infection by Anaplasma phagocytophilum and Ehrlichia canis in a Brazilian dog. Ticks Tick-Borne Dis. 6, 242–245. 10.1016/j.ttbdis.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Skotarczak B., Rymaszewska A., Wodecka B., Sawczuk M., Adamska M., Maciejewska A. (2006). PCR detection of granulocytic Anaplasma and Babesia in Ixodes ricinus ticks and birds in west-central Poland. Ann. Agric. Environ. Med. 13, 21–23. [PubMed] [Google Scholar]
- Skuballa J., Petney T., Pfäffle M., Taraschewski H. (2010). Molecular detection of Anaplasma phagocytophilum in the European hedgehog (Erinaceus europaeus) and its ticks. Vector Borne Zoonotic Dis. 10, 1055–1057. 10.1089/vbz.2009.0150 [DOI] [PubMed] [Google Scholar]
- Sonenshine D. E., Roe R. M. (eds.). (2014). Biology of Ticks Volume 2, Second Edition. Oxford: Oxford University Press. [Google Scholar]
- Sprong H., Wielinga P. R., Fonville M., Reusken C., Brandenburg A. H., Borgsteede F., et al. (2009). Ixodes ricinus ticks are reservoir hosts for Rickettsia helvetica and potentially carry flea-borne Rickettsia species. Parasit. Vectors 2:41. 10.1186/1756-3305-2-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek G. (2009). Büchse der Pandora: krankheitserreger in Ixodes ricinus-Zecken in Mitteleuropa. Wien. Klin. Wochenschr. 121, 673–683. 10.1007/s00508-009-1281-9 [DOI] [PubMed] [Google Scholar]
- Stuen S., Artursson K. (2000). Effects of the dose of Ehrlichia phagocytophila on the severity of experimental infections in lambs. Vet. Rec. 146, 669–672. 10.1136/vr.146.23.669 [DOI] [PubMed] [Google Scholar]
- Stuen S., Granquist E. G., Silaghi C. (2013a). Anaplasma phagocytophilum–a widespread multi-host pathogen with highly adaptive strategies. Front. Cell. Infect. Microbiol. 3:31. 10.3389/fcimb.2013.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuen S., Handeland K., Frammarsvik T., Bergström K. (2001). Experimental Ehrlichia phagocytophila infection in red deer (Cervus elaphus). Vet. Rec. 149, 390–392. 10.1136/vr.149.13.390 [DOI] [PubMed] [Google Scholar]
- Stuen S., Pettersen K. S., Granquist E. G., Bergström K., Bown K. J., Birtles R. J. (2013b). Anaplasma phagocytophilum variants in sympatric red deer (Cervus elaphus) and sheep in southern Norway. Ticks Tick-Borne Dis. 4, 197–201. 10.1016/j.ttbdis.2012.11.014 [DOI] [PubMed] [Google Scholar]
- Sukumaran B., Narasimhan S., Anderson J. F., DePonte K., Marcantonio N., Krishnan M. N., et al. (2006). An Ixodes scapularis protein required for survival of Anaplasma phagocytophilum in tick salivary glands. J. Exp. Med. 203, 1507–1517. 10.1084/jem.20060208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner J. W., Nicholson W. L., Massung R. F. (1997). PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J. Clin. Microbiol. 35, 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford S. R., III., Dawson J. E., Katavolos P., Warner C. K., Kolbert C. P., Persing D. H. (1996). Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc. Natl. Acad. Sci. U.S.A. 93, 6209–6214. 10.1073/pnas.93.12.6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. J., Birtles R. J., Radford A. D., Woldehiwet Z. (2012). Recurrent bacteraemia in sheep infected persistently with Anaplasma phagocytophilum. J. Comp. Pathol. 147, 360–367. 10.1016/j.jcpa.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Townsend R. L., Moritz E. D., Fialkow L. B., Berardi V., Stramer S. L. (2014). Probable transfusion-transmission of Anaplasma phagocytophilum by leukoreduced platelets. Transfusion 54, 2828–2832. 10.1111/trf.12675 [DOI] [PubMed] [Google Scholar]
- Vor T., Kiffner C., Hagedorn P., Niedrig M., Rühe F. (2010). Tick burden on European roe deer (Capreolus capreolus). Exp. Appl. Acarol. 51, 405–417. 10.1007/s10493-010-9337-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldehiwet Z. (2006). Anaplasma phagocytophilum in ruminants in Europe. Ann. N.Y. Acad. Sci. 1078, 446–460. 10.1196/annals.1374.084 [DOI] [PubMed] [Google Scholar]
- Woldehiwet Z. (2010). The natural history of Anaplasma phagocytophilum. Vet. Parasitol. 167, 108–122. 10.1016/j.vetpar.2009.09.013 [DOI] [PubMed] [Google Scholar]
- Woldehiwet Z., Horrocks B. K., Scaife H., Ross G., Munderloh U. G., Bown K., et al. (2002). Cultivation of an ovine strain of Ehrlichia phagocytophila in tick cell cultures. J. Comp. Pathol. 127, 142–149. 10.1053/jcpa.2002.0574 [DOI] [PubMed] [Google Scholar]
- Yabsley M. J., Murphy S. M., Luttrell M. P., Little S. E., Massung R. F., Stallknecht D. E., et al. (2008). Experimental and field studies on the suitability of Raccoons (Procyon lotor) as hosts for tick-borne pathogens. Vector-Borne Zoonotic Dis. 8, 491–504. 10.1089/vbz.2007.0240 [DOI] [PubMed] [Google Scholar]
- Yang J., Liu Z., Guan G., Liu Q., Li Y., Chen Z., et al. (2013). Prevalence of Anaplasma phagocytophilum in ruminants, rodents and ticks in Gansu, north-western China. J. Med. Microbiol. 62, 254–258. 10.1099/jmm.0.046771-0 [DOI] [PubMed] [Google Scholar]
- Zeidner N. S., Burkot T. R., Massung R., Nicholson W. L., Dolan M. C., Rutherford J. S., et al. (2000). Transmission of the agent of human granulocytic ehrlichiosis by Ixodes spinipalpis ticks: evidence of an enzootic cycle of dual infection with Borrelia burgdorferi in Northern Colorado. J. Infect. Dis. 182, 616–619. 10.1086/315715 [DOI] [PubMed] [Google Scholar]
- Zeman P., Pecha M. (2008). Segregation of genetic variants of Anaplasma phagocytophilum circulating among wild ruminants within a Bohemian forest (Czech Republic). Int. J. Med. Microbiol. 298(Suppl. 1), 203–210. 10.1016/j.ijmm.2008.03.003 [DOI] [Google Scholar]
- Zhan L., Cao W.-C., de Vlas S., Xie S.-Y., Zhang P.-H., Wu X.-M., et al. (2008). A newly discovered Anaplasma phagocytophilum variant in rodents from southeastern China. Vector Borne Zoonotic Dis. 8, 369–380. 10.1089/vbz.2007.0211 [DOI] [PubMed] [Google Scholar]
- Zhan L., Cao W.-C., Jiang J.-F., Zhang X.-A., Liu Y.-X., Wu X.-M., et al. (2010). Anaplasma phagocytophilum from Rodents and Sheep, China. Emerg. Infect. Dis. 16, 764–768. 10.3201/eid1605.021293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu H., Xu B., Zhang Z., Jin Y., Li W., et al. (2014). Rural residents in China are at increased risk of exposure to tick-borne pathogens Anaplasma phagocytophilum and Ehrlichia chaffeensis. BioMed Res. Int. 2014:313867. 10.1155/2014/313867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu Y., Ni D., Li Q., Yu Y., Yu X., et al. (2008). Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA 300, 2263–2270. 10.1001/jama.2008.626 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang G., Liu Q., Chen C., Li J., Long B., et al. (2013). Molecular analysis of Anaplasma phagocytophilum isolated from patients with febrile diseases of unknown etiology in China. PLoS ONE 8:e57155. 10.1371/journal.pone.0057155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. C., Zhang L. X., Li W. H., Wang S. W., Sun Y. L., Wang Y. Y., et al. (2012). Ehrlichiosis and zoonotic anaplasmosis in suburban areas of Beijing, China. Vector Borne Zoonotic Dis. 12, 932–937. 10.1089/vbz.2012.0961 [DOI] [PubMed] [Google Scholar]