Abstract
Fucose is an L-configuration sugar found abundantly in the mammalian gut. It has long been known to be induced there by the presence of bacteria, but only recently have some of the molecular mechanisms behind this process been uncovered. New work suggests that fucose can have a protective role in both gut-centered and systemic infection and inflammation. This review highlights recent studies showing that in addition to acting as a food source for beneficial gut symbionts, host fucose can also suppress the virulence of pathogens and pathobionts. The relevance of gut fucosylation to human diseases will also be discussed.
Introduction
L-fucose is a six-carbon sugar originally discovered in seaweed of the genus Fucus, but found in all branches of life. It is one of the few sugars in nature with L stereochemistry. Like all carbohydrates it has many potential functions, whether as a monosaccharide or as part of a more complex glycan structure, but this review focuses on its presence in the mammalian gut, and how it may moderate the host-microbiota relationship there.
Types of fucosylation
Mammals incorporate fucose into glycans in several ways. The sugar can be attached directly to serine or threonine in proteins (O-fucosylation). It can also be attached to sugars at the base of glycan chains in the α(1,6) configuration (known as core fucosylation). Lastly, it can be displayed near the terminal end of glycans in the α(1,2), (1,3) or (1,4) configurations, where it is well positioned to engage in interactions with other cells. Each of these structures is synthesized by a distinct fucosyltransferase enzyme, and can only be cleaved by a specific fucosidase.
Developmental and environmental control of fucosylation in the gut
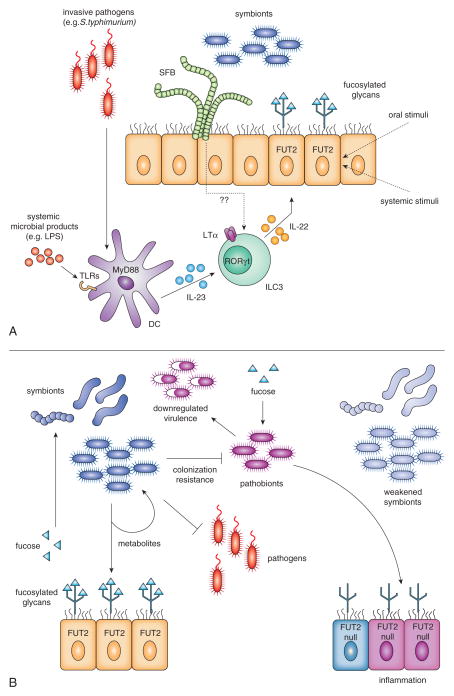
In the mammalian gastrointestinal tract, fucose is an abundant component of glycans decorating proteins and lipids, especially on the epithelial surface facing the lumen and in mucosal secretions. This is predominantly in the form of α(1,2)fucosylation (1, 2), which can be detected in stomach, distal small intestine (ileum), and large intestine, using the lectin UEA-1 (Ulex europaeus Agglutinin-1). Of the two functional mammalian α(1,2)fucosyltransferases, the Fut2 protein is responsible for the majority of fucosylation in the mouse and human gut (1, 3–6). In Fut2-negative mice, other types of fucosylation were not found by a broadly reactive fucose-specific lectin (1). Fut1 expression is restricted to M cells, a rare intestinal epithelial population (3). In rodents, the stomach and parts of large intestine are constitutively fucosylated, while that in the small intestine is variable and can be induced by a variety of environmental factors. This concentration of fucose in tissues that are in intimate contact with the highest density of microbes immediately suggests a connection. Indeed, fucosylation of the distal small intestine increases in rats and mice around weaning (3–4 weeks) when a shift in bacterial populations occurs (7, 8). Germ-free (GF) mice do not maintain ileal fucosylation after weaning, but colonization with bacteria from conventionally-housed mice restores it (8, 9). Not all bacteria are capable of causing fucosylation, nor can it be recapitulated by oral administration of a bacterial TLR ligand such as LPS to GF mice (10). Monocolonization of mice with segmented filamentous bacteria (SFB)(11) or Bacteroides thetaiotaomicron (8) is effective, while Lactobacillus murinus is not (12)). Conversely, certain oral antibiotics can abolish fucosylation in conventional mice (12). Clearly some bacteria have the ability to cause fucosylation, while others do not. How do gut bacteria cause fucosylation? The model human gut symbiont B. thetaiotaomicron provides one interesting example. When monocolonizing mice at sufficiently high densities it triggers fucosylation of the ileum (8), and since it resides in the lumen this must be done via a soluble signal secreted by the bacteria. Genetic experiments suggest that the putative signaling molecule is controlled by fucose availability. That is, the fucose-inducing signal is regulated by the same mechanisms as the bacteria’s fucose catabolism genes, via a fucose-responsive transactivator (13). This co-regulation strongly suggests that host-derived fucose is energetically useful for this symbiont. The identity of the putative signal, and how it activates fucosylation on the host, remains unknown.
Another common gut resident, SFB, also cause increased fucosylation in the ileum that they populate (11). SFB came to prominence based on their induction of IL-17 and IL-22 production in the small intestine lamina propria, and the effects of this on the immune system (14). Ileal fucosylation in SFB-harboring specific pathogen-free (SPF) mice was shown to depend on IL-22 and TNF family member lymphotoxin alpha (LTα)(12). IL-22 is a cytokine of the IL-10 family whose expression can be induced by commensal and pathogenic bacteria (15). Its receptor is expressed on epithelial cells, and can activate defense and tissue repair mechanisms via STAT3 signaling (16). LTα is involved in lymphoid tissue organogenesis, and has been implicated in maintenance of IL-22 production (17). In contrast to IL-22, its expression was not affected by antibiotic treatment. To cause fucosylation, IL-22 and LTα both needed to be produced by innate lymphoid cells (ILCs), which require the transcription factors RORγt and Id2 for development. Acute blockade of IL-22 or the lymphotoxin beta receptor (which LTα signals through) abolished fucosylation in the ileum (12). Unlike most gut bacteria, SFB is in intimate physical contact with epithelial cells, but the molecular mechanisms that allow it to activate IL-22 and fucosylation are not yet clear.
The innate and adaptive arms of the immune system combine to sequester most bacteria in the lumen of the ileum and large intestine. In several mouse strains that lack components of the adaptive immune system (RAG −/−, scid, nude, pIgR −/−), there is increased small and large intestine fucosylation (12, 18). One explanation is that certain members of the gut community (such as SFB (19)) that are normally limited by secreted IgA antibodies could trigger excess fucosylation when uncontrolled (either by expanding in numbers, invading, or altering their behavior). Fucosylation in this case could be part of an increased innate immune response compensating for the absence of adaptive immunity (20). A mouse model of cystic fibrosis also results in increased fucosylation of small intestine mucins (21), which could be secondary to dysbiosis.
Although the commensal-derived molecules that normally cause fucosylation are not yet known, some microbial products are sufficient to do it. Polyamines are produced by bacteria and found in food, and their concentration in the gut increases at weaning concurrent with the increase in fucosylation. Feeding polyamines to young rats results in early development of fucosylation (22). Gavage of the cAMP-activating toxin from a pathogen, Vibrio cholerae, is also effective (3). Cecal and colonic fucosylation, although present in GF animals, is further induced by bacterial colonization (23). Although the upstream sensor is not known, this may occur via the kinases JNK and ERK: feeding of small-molecule inhibitors of these kinases reduced fucosylation (24). IL-22, which is induced by microbes, can also enhance cecum and colon fucosylation, but it is not absolutely required, as some fucosylation is still found in these tissues in IL-22 receptor knockout mice (25). Finally, injection of substances as diverse as indomethacin (3), glucocorticoids (26), the protein synthesis inhibitors cycloheximide or emetine, or the DNA synthesis inhibitor cytosine arabinoside (27) also trigger small intestine fucosylation.
Thus, fucosylation can be activated by a variety of signals originating from both inside the gut lumen and from the systemic circulation (summarized in Figure 1A). Some of these phenomena may be physiologically relevant, and some may be non-specific effects. How many unique fucosylation-inducing pathways exist and whether any are activated directly in the gut epithelium or all of them require mediators produced by hematopoeitic cells, are outstanding questions.
Figure 1. Inducible α(1,2)fucosylation of the epithelium in the small intestine.
A) Fucosylation inducing pathways.
Fut2, the enzyme primarily responsible for fucosylation in the gut, is activated by IL-22 and requires LTα, both produced by RORγt-dependent ILC3 cells. Systemic invasion of pathogens like S. typhimurium, or microbial products in the circulation, activate DCs via TLRs and the adaptor MyD88 to produce IL-23, which also acts upon ILC3 cells. ILC3 cells can be activated by epithelia-associated SFB and lumen-resident bacteria through unknown mediators. Other compounds either administered orally (polyamines, cholera toxin) or injected systemically (glucocorticoids, and inhibitors of cyclooxygenase, protein or DNA synthesis) induce Fut2 through unknown pathways. It is possible that they directly stimulate epithelial cells.
B) Consequences of inducible fucosylation of the epithelial cells
Upon activation of Fut2, fucosylated proteins (and lipids?) produced by small intestine epithelium are shed into the lumen and become available for consumption by the downstream microbiota. Fucose is liberated by microbial fucosidases. It can then enhance the beneficial activity of symbionts, including their production of metabolites for the host or other bacteria, and improve colonization resistance against pathogens and pathobionts. Free fucose may directly suppress bacterial virulence.
In the absence of functional Fut2, an acute or chronic stress weakens beneficial symbionts (pale blue), and/or reduces their abundance while activating genes encoding virulence factors. Such activation of pathobionts then would sustain the host’s inflammatory responses. This could contribute to the development of conditions such as Crohn’s disease, which has been genetically linked to FUT2 null alleles. Importantly, other genes required for inducible fucosylation, IL23 and STAT3, have been also linked to Crohn’s disease.
Fucose use by gut symbionts
The available evidence suggests that rodents do not themselves metabolize fucose for energy (28, 29), while other mammals (pigs (30) and humans (31)) may be able to do so. On the other hand, diverse bacterial species possess the enzymes necessary to metabolize fucose (13, 32, 33), and the gut bacteria do so actively (34). To obtain fucose in the gut, bacteria must first liberate it from host or dietary glycans with an α-fucosidase enzyme. Since these enzymes are generally secreted, fucose that they liberate will be available to other members of the community. B. thetaiotaomicron, for example, is adept at liberating and utilizing many host sugars, including fucose. It upregulates its fucose cleavage and metabolism genes when living in mice compared to growth in vitro (35), and is under selective pressure to maintain the capacity to metabolize fucose (13). Importantly, it also triggers host fucosylation, likely to use as a food source (13). Similarly, non-pathogenic E. coli is impaired in mouse colonization when its fucose metabolism is disabled (36).
Besides its use as an energy source, some bacteria also display fucose on their own proteins, lipids and polysaccharides. Several members of the Bacteroides genus are able to incorporate exogenous fucose into their own glycans (37), in addition to making it de novo from other sugars. For B. fragilis, at least, the ability to fucosylate its proteins is crucial for its fitness in the gut (37, 38). Proper glycosylation is likely required for the function of numerous glycoproteins. Another possibility is that Bacteroides species mimic host glycans in order to evade the immune system. There is precedence for this kind of host mimicry in Helicobacter pylori, for instance (39). This idea could be tested by colonizing immunodeficient mice or mice lacking fucosylation: any competitive advantage of fucose “camouflage” should disappear in these mice.
How important is host fucose then to the resident bacteria in the gut without an external stress? Mice that lack the Fut2 enzyme and thus lose almost all gut fucosylation grow normally and are healthy in SPF conditions. Some measurable differences in their gut communities have been reported (40), although the extent of the changes is smaller than the effect of diet manipulation or cohousing (10).
Besides shifting bacterial populations in the gut, the presence or absence of fucose could also affect bacterial gene expression. In vitro effects of fucose on gene expression have been shown for at least one species, the human gut symbiont R. inulinivorans (32). At the very least, provision of fucose in the gut will alter the metabolites produced by bacteria that feed on it (33), which could subsequently affect the host. Analyzing the downstream products of fucose catabolism with techniques such as metabolomics, or stable isotope probing (41) to identify the key fucose users in the gut, could be helpful for understanding better the functions of fucose in the healthy gut ecosystem.
Fucosylation in response to infection
In healthy SPF mice, α(1,2)fucosylation is found in stomach, cecum and colon, but little is found in the small intestine (except the ileum, which is also inducible by microbes). In contrast, during systemic infection or after injection of microbial products (such as intraperitoneal injection of LPS or other TLR ligands), robust fucosylation of the entire small intestine occurs (10). The mechanism of this rapid small intestine fucosylation is through MyD88-dependent sensing of microbial products by dendritic cells (DCs), which produce IL-23, triggering IL-22 production in RORγt-dependent ILCs. IL-22 then causes upregulation of the Fut2 enzyme in small intestine epithelial cells, much as it does in response to normal bacterial colonization, except that it is more intense and widespread. This not only affects the small intestine, but perhaps more importantly results in fucosylated glycans travelling “downstream” to the densely-populated cecum and colon, where they are harvested by the resident bacteria. This is then translated into an improvement in host health (10).
Since fucosylated glycans shed from the small intestine can have an impact on the bacteria of the large intestine, and the host, could feeding of exogenous glycans be therapeutically useful? Mice lacking IL-22 signaling are known to be more susceptible to gut inflammation such as that caused by dextran sodium sulfate or the attaching-effacing pathogen Citrobacter rodentium (42). Recent work showed that one cause of this is not the pathogen or inflammation per se, but expansion and invasion of a pathobiont, Enterococcus faecalis, from the inflamed gut to systemic tissues (25). Since one function of IL-22 signaling was to enhance fucosylation of the gut epithelium, the authors gavaged fucosylated glycans to the susceptible mice and found that it improved animal survival. Fucosyl-glycan supplementation reduced the E. faecalis numbers in the gut and systemic organs, and shifted the overall bacterial community structure, but did not affect the numbers of the inciting pathogen, C. rodentium (25). In a different model, the capacity to fucosylate the small intestine after LPS injection correlated with a reduction of pathology caused by C. rodentium without reducing its numbers (10). In both of these cases, fucosylated glycans could be improving host health in several ways:
Fucose could feed into specific bacterial metabolic pathways, resulting in production of metabolites useful to the host. Short-chain fatty acids, for instance, are major bacterial metabolites and a significant energy source for mammals that can result from fucose catabolism.
Fucose could cause changes in bacterial gene expression that result in favorable effects on the host, by suppressing virulence genes in pathobionts like E. faecalis. The phenomenon of reduced virulence due to exposure to fucose has been demonstrated for pathogenic E. coli (43).
Fucose could bolster the beneficial members of the gut community, like the Ruminococcaceae and Bacteroides species that expanded in fucose-treated mice (25), thus maintaining their beneficial functions, such as colonization resistance.
Community-wide bacterial RNA expression (transcriptome) analysis in mice that could or could not produce α(1,2)fucosylation(10) upon systemic treatment with LPS revealed that bacteria in fucose-producing mice increased their expression of genes coding for the fucose transporter, indicating that they were taking up more fucose. They also showed signs of increased metabolic activity (protein synthesis, ATP synthesis and use) compared to bacteria in mice lacking fucose. Conversely, in fucosylation-deficient mice, bacteria showed increased expression of virulence-related genes. These results support the idea that fucose, especially when induced during severe sickness of the host, supports the beneficial activity of resident bacteria while suppressing their potential virulence (Figure 1B). In other words: rapid mobilization of an internal resource (available fucose) serves to maintain host-commensal symbiosis.
Pathogens and fucosylation
Although many symbiotic gut bacteria can and do utilize fucose, and it is protective in several disease models, it can also be taken advantage of by harmful microbes. Useful nutrients in the gut, like fucose, are normally rapidly scavenged by resident bacteria. This forms one important cornerstone of colonization resistance in the gut: the filling of nutritional niches. If this stable state is disrupted, for example when antibiotics are given, these niches are no longer filled, allowing pathogens to gain a foothold or pathobionts to realize their virulent potential. The common foodborne pathogen S. typhimurium, for example, is normally kept at bay by colonization resistance in mice, requiring pre-treatment with antibiotics for effective colonization of the gut (44, 45). Soon after infection, S. typhimurium upregulates its fucose catabolism genes (46, 47), indicating that it is metabolizing this sugar. Crucially, S. typhimurium can’t access fucose from complex glycans by itself, as it lacks an α-fucosidase enzyme--it must rely on other bacteria to free the sugar. Is fucose an important nutrient for pathogens in the gut? For S. typhimurium, the answer is not clear: in different experimental settings (different bacterial strains and mouse models) bacteria that couldn’t metabolize fucose had either a slight reduction in fecal levels compared to wild type (46), or not (47). For another enteric pathogen, Campylobacter jejuni, fucose metabolism seems more important for survival in the gut (48).
In addition to their potential use as a carbon and energy source, fucosylated glycans can serve as adhesion sites or receptors for pathogens, including S. typhimurium (49), H. pylori (50), enterotoxigenic E. coli (51), and norovirus (52, 53). The high levels of fucose in the stomach and large intestine but usually low levels in the small intestine, as well as local gradients in the crypt/villus axis, could also make fucose a useful location marker for bacteria. C. jejuni, for example, chemotaxes toward fucose (54).
In Fut2-deficient mice, early S. typhimurium invasion into the cecal tissue was ~2-fold higher (12). The reason for this is unclear, but probably does not result from a difference in growth since bacterial numbers in the cecal lumen were the same. An effect on chemotaxis is also unlikely (55). The increased invasion may be due to the loss of fucosylation’s indirect effects on the resident bacteria, resulting in reduced colonization resistance. Interestingly, S. typhimurium induces robust fucosylation of the small intestine when infecting GF or antibiotic-treated mice, and this is dependent on MyD88 and RORγt (10, 12). S. typhimurium is known to exploit host inflammatory responses (56, 57), so determining whether fucosylation is protective for the host or taken advantage of by this or other pathogens will require further examination of both sides of the interaction.
Applications to human disease
Humans display Fut2-dependent α(1,2)fucosylation in the stomach, small and large intestine similarly to rodents (4–6). It is likely to be inducible and controlled by the signals that have been found in rodents in the small intestine in humans (but this has not been definitively shown). However, large intestine fucosylation was shown to increase during local inflammation (58). Interestingly, ~20% of humans are homozygous loss-of-function mutants for Fut2(59, 60) and thus completely lack α(1,2)fucosylated glycans in the gastrointestinal tract. This gene has evidently been subject to balancing selection, indicating that it can enhance or reduce fitness depending on the circumstances (61). Susceptibility to certain pathogens like norovirus that use α(1,2)fucosylated glycans as receptors is a clear downside to expressing it (52, 53), and could have helped select for its loss of function. On the other hand, provision of fucose to the gut could protect the host by supporting beneficial gut bacteria. As in mice, there does seem to be an effect of Fut2 expression on the human gut bacterial community structure (62, 63). This could explain why Fut2-negative humans are more susceptible to various infections and death from sepsis as infants (64–67). Fucosylated glycans provided in milk (also Fut2-dependent) could protect from some infections as well (68). Fut2 has also been linked by genome-wide association to several diseases with potential connections to the gut microbiota (69–71), most notably Crohn’s disease (72). Interestingly, other genes in the Fut2-inducing pathway (IL23 and STAT3) are also associated with this disease (73). Examining the activity of the gut bacteria in humans with or without fucosylation, especially in the context of disease, would help in understanding these connections. If fucose acts to suppress virulence in pathobionts or pathogens, as seen in mice, it might prevent an at-risk individual from progressing to full inflammatory disease.
Conclusions
α(1,2)fucosylation displayed on host glycans is ideally positioned to interact with the many bacteria inhabiting the gut. It is induced by the normal resident bacteria, and produced even more abundantly during infection and inflammation. For bacteria, it can function as a food source, an adhesion site, or a modifier of gene expression. Fucosylated glycans in the gut are protective in several models of systemic and intestinal inflammation and infection. However, like many host immune mechanisms, it can also be taken advantage of by some pathogens. Humans, which are polymorphic at the Fut2 locus, offer a natural model of fucosylation deficiency. It does not seem to have profound effects on the microbiota composition or the host’s health, until a stress (acute or chronic) occurs. Then the ability to provide fucose to one’s own microbiota becomes important. We would like to hypothesize that such is the mechanism linking Fut2 deficiency with Crohn’s disease (Figure 1B).
Although fucose plays an important role in the gut, there are undoubtedly other glycans that have similar effects on gut bacteria and could be manipulated to enhance health (74). Understanding the functions of the large spectrum of glycan structures will require analysis of the glycosylation profile of the host, along with high-throughput measurements of bacterial activity, including their transcriptomes and metabolomes. The other unknown is the role of fucosylation (or other inducible glycosylation events) of the mucosal surfaces outside of the gut in maintenance of the host-microbial symbiosis. Better understanding how fucose in the gut (or lungs, or urogenital tract) protects from or predisposes to disease will be a challenging but rewarding endeavor, with clear clinical benefits.
Footnotes
The authors’ work is supported by the Digestive Disease Research Core Center grant DK42086, T32 AI007090 NIH training grant and a Kenneth Rainin Foundation grant to AVC.
References
- 1.Hurd EA, Holmen JM, Hansson GC, Domino SE. Gastrointestinal mucins of Fut2-null mice lack terminal fucosylation without affecting colonization by Candida albicans. Glycobiology. 2005;15:1002–1007. doi: 10.1093/glycob/cwi089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bry L, Falk PG, Gordon JL. Genetic engineering of carbohydrate biosynthetic pathways in transgenic mice demonstrates cell cycle-associated regulation of glycoconjugate production in small intestinal epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1161–1166. doi: 10.1073/pnas.93.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terahara K, Nochi T, Yoshida M, Takahashi Y, Goto Y, Hatai H, Kurokawa S, Jang MH, Kweon MN, Domino SE, Hiroi T, Yuki Y, Tsunetsugu-Yokota Y, Kobayashi K, Kiyono H. Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochemical and biophysical research communications. 2011;404:822–828. doi: 10.1016/j.bbrc.2010.12.067. [DOI] [PubMed] [Google Scholar]
- 4.Bjork S, Breimer ME, Hansson GC, Karlsson KA, Leffler H. Structures of blood group glycosphingolipids of human small intestine. A relation between the expression of fucolipids of epithelial cells and the ABO, Le and Se phenotype of the donor. The Journal of biological chemistry. 1987;262:6758–6765. [PubMed] [Google Scholar]
- 5.Yuan M, Itzkowitz SH, Palekar A, Shamsuddin AM, Phelps PC, Trump BF, Kim YS. Distribution of blood group antigens A, B, H, Lewisa, and Lewisb in human normal, fetal, and malignant colonic tissue. Cancer research. 1985;45:4499–4511. [PubMed] [Google Scholar]
- 6.Sakamoto S, Watanabe T, Tokumaru T, Takagi H, Nakazato H, Lloyd KO. Expression of Lewisa, Lewisb, Lewisx, Lewisy, siayl-Lewisa, and sialyl-Lewisx blood group antigens in human gastric carcinoma and in normal gastric tissue. Cancer research. 1989;49:745–752. [PubMed] [Google Scholar]
- 7.Srivastava OP, Steele MI, Torres-Pinedo R. Maturational changes in terminal glycosylation of small intestinal microvillar proteins in the rat. Biochimica et biophysica acta. 1987;914:143–151. doi: 10.1016/0167-4838(87)90057-4. [DOI] [PubMed] [Google Scholar]
- 8.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science (New York, NY) 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 9.Umesaki Y, Tohyama K, Mutai M. Appearance of fucolipid after conventionalization of germ-free mice. Journal of biochemistry. 1981;90:559–561. doi: 10.1093/oxfordjournals.jbchem.a133506. [DOI] [PubMed] [Google Scholar]
- 10.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiology and immunology. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 12.Goto Y, Obata T, Kunisawa J, Sato S, Ivanov, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, Setoyama H, Imaoka A, Uematsu S, Akira S, Domino SE, Kulig P, Becher B, Renauld JC, Sasakawa C, Umesaki Y, Benno Y, Kiyono H. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science (New York, NY) 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL–22. Nature immunology. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 17.Tumanov AV, Koroleva EP, Guo X, Wang Y, Kruglov A, Nedospasov S, Fu YX. Lymphotoxin controls the IL–22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwamori M, Iwamori Y, Matsumoto S, Adachi S, Nomura T. Enhanced expression of fucosyl GA1 in the digestive tract of immune-deficient scid, nude and pIgR(−/−) mice. Journal of biochemistry. 2013;154:541–549. doi: 10.1093/jb/mvt087. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Thomsson KA, Hinojosa-Kurtzberg M, Axelsson KA, Domino SE, Lowe JB, Gendler SJ, Hansson GC. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fucalpha1-2 glycosyltransferase. The Biochemical journal. 2002;367:609–616. doi: 10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biol-N’Garagba MC, Greco S, George P, Hugueny I, Louisot P. Polyamine participation in the maturation of glycoprotein fucosylation, but not sialylation, in rat small intestine. Pediatric research. 2002;51:625–634. doi: 10.1203/00006450-200205000-00014. [DOI] [PubMed] [Google Scholar]
- 23.Nanthakumar NN, Meng D, Newburg DS. Glucocorticoids and microbiota regulate ontogeny of intestinal fucosyltransferase 2 requisite for gut homeostasis. Glycobiology. 2013;23:1131–1141. doi: 10.1093/glycob/cwt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng D, Newburg DS, Young C, Baker A, Tonkonogy SL, Sartor RB, Walker WA, Nanthakumar NN. Bacterial symbionts induce a FUT2-dependent fucosylated niche on colonic epithelium via ERK and JNK signaling. American journal of physiology. Gastrointestinal and liver physiology. 2007;293:G780–787. doi: 10.1152/ajpgi.00010.2007. [DOI] [PubMed] [Google Scholar]
- 25.Pham TA, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP, Keane JA, Page AJ, Kumasaka N, Kane L, Mottram L, Harcourt K, Hale C, Arends MJ, Gaffney DJ, Dougan G, Lawley TD. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biol-N’garagba MC, Niepceron E, Mathian B, Louisot P. Glucocorticoid-induced maturation of glycoprotein galactosylation and fucosylation processes in the rat small intestine. The Journal of steroid biochemistry and molecular biology. 2003;84:411–422. doi: 10.1016/s0960-0760(03)00062-1. [DOI] [PubMed] [Google Scholar]
- 27.Umesaki Y, Ohara M. Factors regulating the expression of the neutral glycolipids in the mouse small intestinal mucosa. Biochimica et biophysica acta. 1989;1001:163–168. doi: 10.1016/0005-2760(89)90143-4. [DOI] [PubMed] [Google Scholar]
- 28.Shull KH, Miller ON. Formation in vivo of glycogen by certain intermediates of the lactate-propanediol pathway. The Journal of biological chemistry. 1960;235:551–553. [PubMed] [Google Scholar]
- 29.Coffey JW, Miller ON, Sellinger OZ. THE METABOLISM OF L-FUCOSE IN THE RAT. The Journal of biological chemistry. 1964;239:4011–4017. [PubMed] [Google Scholar]
- 30.Chan JY, Nwokoro NA, Schachter H. L-Fucose metabolism in mammals. The conversion of L-fucose to two moles of L-lactate, of L-galactose to L-lactate and glycerate, and of D-arabinose to L-lactate and glycollate. The Journal of biological chemistry. 1979;254:7060–7068. [PubMed] [Google Scholar]
- 31.Segal S, Topper YJ. On the biosynthesis of L-fucose and L-fucose metabolism in man. Biochimica et biophysica acta. 1960;42:147–151. doi: 10.1016/0006-3002(60)90761-7. [DOI] [PubMed] [Google Scholar]
- 32.Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. Journal of bacteriology. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himelbloom BH, Canale-Parola E. Clostridium methylpentosum sp. nov.: a ring-shaped intestinal bacterium that ferments only methylpentoses and pentoses. Archives of microbiology. 1989;151:287–293. doi: 10.1007/BF00406553. [DOI] [PubMed] [Google Scholar]
- 34.Bocci V, Winzler RJ. Metabolism of L-fucose-1-14C and of fucose glycoproteins in the rat. The American journal of physiology. 1969;216:1337–1342. doi: 10.1152/ajplegacy.1969.216.6.1337. [DOI] [PubMed] [Google Scholar]
- 35.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (New York, NY) 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 36.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. Carbon nutrition of Escherichia coli in the mouse intestine. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science (New York, NY) 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 38.Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 2009;137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pohl MA, Romero-Gallo J, Guruge JL, Tse DB, Gordon JI, Blaser MJ. Host-dependent Lewis (Le) antigen expression in Helicobacter pylori cells recovered from Leb-transgenic mice. The Journal of experimental medicine. 2009;206:3061–3072. doi: 10.1084/jem.20090683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, Loy A, Wagner M. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 43.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohnhoff M, Miller CP. Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. The Journal of infectious diseases. 1962;111:117–127. doi: 10.1093/infdis/111.2.117. [DOI] [PubMed] [Google Scholar]
- 45.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deatherage Kaiser BL, Li J, Sanford JA, Kim YM, Kronewitter SR, Jones MB, Peterson CT, Peterson SN, Frank BC, Purvine SO, Brown JN, Metz TO, Smith RD, Heffron F, Adkins JN. A Multi-Omic View of Host-Pathogen-Commensal Interplay in -Mediated Intestinal Infection. PloS one. 2013;8:e67155. doi: 10.1371/journal.pone.0067155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stahl M, Friis LM, Nothaft H, Liu X, Li J, Szymanski CM, Stintzi A. L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7194–7199. doi: 10.1073/pnas.1014125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chessa D, Winter MG, Jakomin M, Baumler AJ. Salmonella enterica serotype Typhimurium Std fimbriae bind terminal alpha(1,2)fucose residues in the cecal mucosa. Mol Microbiol. 2009;71:864–875. doi: 10.1111/j.1365-2958.2008.06566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boren T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science (New York, NY) 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 51.Coddens A, Diswall M, Angstrom J, Breimer ME, Goddeeris B, Cox E, Teneberg S. Recognition of blood group ABH type 1 determinants by the FedF adhesin of F18-fimbriated Escherichia coli. The Journal of biological chemistry. 2009;284:9713–9726. doi: 10.1074/jbc.M807866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorven M, Grahn A, Hedlund KO, Johansson H, Wahlfrid C, Larson G, Svensson L. A homozygous nonsense mutation (428G-->A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. Journal of virology. 2005;79:15351–15355. doi: 10.1128/JVI.79.24.15351-15355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hutson AM, Airaud F, LePendu J, Estes MK, Atmar RL. Norwalk virus infection associates with secretor status genotyped from sera. Journal of medical virology. 2005;77:116–120. doi: 10.1002/jmv.20423. [DOI] [PubMed] [Google Scholar]
- 54.Hugdahl MB, Beery JT, Doyle MP. Chemotactic behavior of Campylobacter jejuni. Infection and immunity. 1988;56:1560–1566. doi: 10.1128/iai.56.6.1560-1566.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. Motility allows S. Typhimurium to benefit from the mucosal defence. Cellular microbiology. 2008;10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- 56.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS biology. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyoshi J, Yajima T, Okamoto S, Matsuoka K, Inoue N, Hisamatsu T, Shimamura K, Nakazawa A, Kanai T, Ogata H, Iwao Y, Mukai M, Hibi T. Ectopic expression of blood type antigens in inflamed mucosa with higher incidence of FUT2 secretor status in colonic Crohn’s disease. Journal of gastroenterology. 2011;46:1056–1063. doi: 10.1007/s00535-011-0425-7. [DOI] [PubMed] [Google Scholar]
- 59.Koda Y, Soejima M, Liu Y, Kimura H. Molecular basis for secretor type alpha(1,2)-fucosyltransferase gene deficiency in a Japanese population: a fusion gene generated by unequal crossover responsible for the enzyme deficiency. American journal of human genetics. 1996;59:343–350. [PMC free article] [PubMed] [Google Scholar]
- 60.Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. The Journal of biological chemistry. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 61.Koda Y, Tachida H, Pang H, Liu Y, Soejima M, Ghaderi AA, Takenaka O, Kimura H. Contrasting patterns of polymorphisms at the ABO-secretor gene (FUT2) and plasma alpha(1,3)fucosyltransferase gene (FUT6) in human populations. Genetics. 2001;158:747–756. doi: 10.1093/genetics/158.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rausch P, Rehman A, Kunzel S, Hasler R, Ott SJ, Schreiber S, Rosenstiel P, Franke A, Baines JF. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wacklin P, Makivuokko H, Alakulppi N, Nikkila J, Tenkanen H, Rabina J, Partanen J, Aranko K, Matto J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PloS one. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaudhuri A, DasAdhikary CR. Possible role of blood-group secretory substances in the aetiology of cholera. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1978;72:664–665. doi: 10.1016/0035-9203(78)90031-7. [DOI] [PubMed] [Google Scholar]
- 65.Blackwell CC, Jonsdottir K, Hanson M, Todd WT, Chaudhuri AK, Mathew B, Brettle RP, Weir DM. Non-secretion of ABO antigens predisposing to infection by Neisseria meningitidis and Streptococcus pneumoniae. Lancet. 1986;2:284–285. doi: 10.1016/s0140-6736(86)92103-3. [DOI] [PubMed] [Google Scholar]
- 66.Thom SM, Blackwell CC, MacCallum CJ, Weir DM, Brettle RP, Kinane DF, Wray D. Non-secretion of blood group antigens and susceptibility to infection by Candida species. FEMS microbiology immunology. 1989;1:401–405. doi: 10.1111/j.1574-6968.1989.tb02428.x. [DOI] [PubMed] [Google Scholar]
- 67.Morrow AL, Meinzen-Derr J, Huang P, Schibler KR, Cahill T, Keddache M, Kallapur SG, Newburg DS, Tabangin M, Warner BB, Jiang X. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. The Journal of pediatrics. 2011;158:745–751. doi: 10.1016/j.jpeds.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. The Journal of nutrition. 2005;135:1304–1307. doi: 10.1093/jn/135.5.1304. [DOI] [PubMed] [Google Scholar]
- 69.Smyth DJ, Cooper JD, Howson JM, Clarke P, Downes K, Mistry T, Stevens H, Walker NM, Todd JA. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011;60:3081–3084. doi: 10.2337/db11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parmar AS, Alakulppi N, Paavola-Sakki P, Kurppa K, Halme L, Farkkila M, Turunen U, Lappalainen M, Kontula K, Kaukinen K, Maki M, Lindfors K, Partanen J, Sistonen P, Matto J, Wacklin P, Saavalainen P, Einarsdottir E. Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue antigens. 2012;80:488–493. doi: 10.1111/tan.12016. [DOI] [PubMed] [Google Scholar]
- 71.Xavier JM, Shahram F, Sousa I, Davatchi F, Matos M, Abdollahi BS, Sobral J, Nadji A, Oliveira M, Ghaderibarim F, Shafiee NM, Oliveira SA. FUT2: filling the gap between genes and environment in Behcet’s disease? Annals of the rheumatic diseases. 2013 doi: 10.1136/annrheumdis-2013-204475. [DOI] [PubMed] [Google Scholar]
- 72.McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, Ippoliti A, Vasiliauskas E, Berel D, Derkowski C, Dutridge D, Fleshner P, Shih DQ, Melmed G, Mengesha E, King L, Pressman S, Haritunians T, Guo X, Targan SR, Rotter JI. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Human molecular genetics. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory Bowel Disease as a Model for Translating the Microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lewis AL, Lewis WG. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cellular microbiology. 2012;14:1174–1182. doi: 10.1111/j.1462-5822.2012.01807.x. [DOI] [PubMed] [Google Scholar]