Background: TGF-β1 produced by regulatory T lymphocytes suppresses excessive immune responses.
Results: Lysosomal-associated transmembrane protein 4B (LAPTM4B) was found to inhibit TGF-β1 production in Tregs.
Conclusion: LAPTM4B, known to exert oncogenic functions in tumor cells, also plays a role in the immune system.
Significance: LAPTM4B may represent a new therapeutic target to modulate immunosuppression by Tregs.
Keywords: immunosuppression, protein-protein interaction, T cell biology, TGF-β, yeast two-hybrid, glycoprotein A repetition predominant (GARP), leucine-rich repeat-containing protein 32 (LRRC32), regulatory T cells (Tregs)
Abstract
Production of active TGF-β1 is one mechanism by which human regulatory T cells (Tregs) suppress immune responses. This production is regulated by glycoprotein A repetitions predominant (GARP), a transmembrane protein present on stimulated Tregs but not on other T lymphocytes (Th and CTLs). GARP forms disulfide bonds with proTGF-β1, favors its cleavage into latent inactive TGF-β1, induces the secretion and surface presentation of GARP·latent TGF-β1 complexes, and is required for activation of the cytokine in Tregs. We explored whether additional Treg-specific protein(s) associated with GARP·TGF-β1 complexes regulate TGF-β1 production in Tregs. We searched for such proteins by yeast two-hybrid assay, using GARP as a bait to screen a human Treg cDNA library. We identified lysosomal-associated transmembrane protein 4B (LAPTM4B), which interacts with GARP in mammalian cells and is expressed at higher levels in Tregs than in Th cells. LAPTM4B decreases cleavage of proTGF-β1, secretion of soluble latent TGF-β1, and surface presentation of GARP·TGF-β1 complexes by Tregs but does not contribute to TGF-β1 activation. Therefore, LAPTM4B binds to GARP and is a negative regulator of TGF-β1 production in human Tregs. It may play a role in the control of immune responses by decreasing Treg immunosuppression.
Introduction
Regulatory T lymphocytes (Tregs)4 are a subset of CD4+ T cells that maintain immune tolerance by suppressing autoreactive T cells (1, 2). Their development and function requires transcription factor FOXP3, as illustrated by the severe autoimmune syndrome that affects mice and humans carrying a mutated FOXP3 gene (3–5). Foxp3 expression is a specific marker of Tregs in mice. This is not true in humans, where non-regulatory CD4+ or CD8+ T lymphocytes transiently express FOXP3 upon T cell receptor (TCR) stimulation (2). Stable FOXP3 expression, a hallmark of Tregs in mice and humans, is ensured by the demethylation of a conserved non-coding region of gene FOXP3, called FOXP3i1, TSDR, or CNS2 (6–10). Demethylated FOXP3i1 can serve to identify and quantify Tregs in human blood or cell samples (11–13).
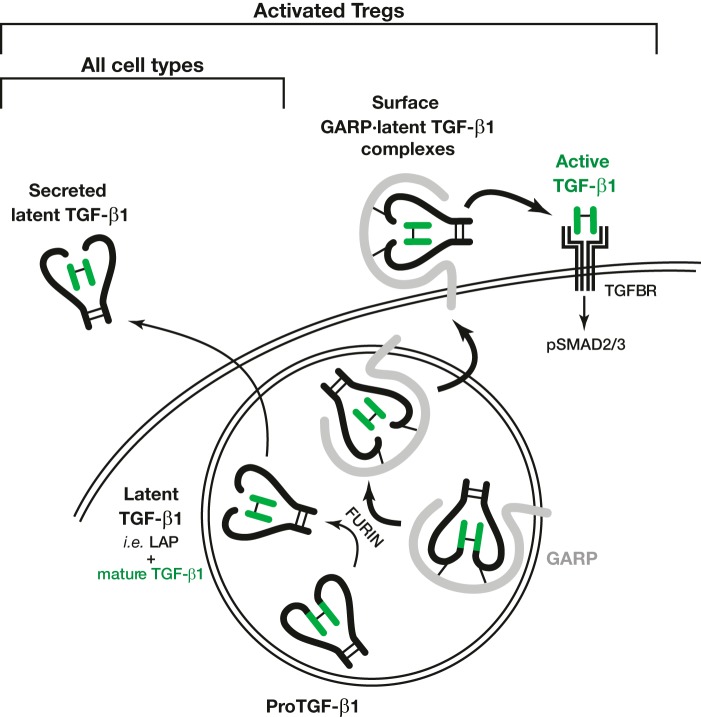
Depending on the context or the cell type to suppress, Tregs use various mechanisms of immune suppression. One mechanism implies the production of the potent immunosuppressive cytokine TGF-β1 (1, 14). Its production by Tregs is regulated by GARP, a surface protein expressed on stimulated Tregs but no other T lymphocytes (11, 15–17). TGF-β1 is synthesized in all cell types as a homodimeric proTGF-β1 precursor (Fig. 1) (18, 19). FURIN cleaves proTGF-β1 to generate a C-terminal fragment or mature TGF-β1, which remains non-covalently bound to the N-terminal fragment known as latency-associated peptide (LAP). This complex, called latent TGF-β1, is inactive because LAP prevents mature TGF-β1 from binding to its receptor. Latent TGF-β1 is secreted by most cell types as a soluble form. In the secretory pathway of stimulated human Tregs, GARP forms disulfide bonds with the proTGF-β1 precursor and favors its FURIN-dependent cleavage into latent TGF-β1 (11, 20). GARP·latent TGF-β1 complexes are then presented on the Treg surface (11, 15, 16). Stimulated Tregs release mature TGF-β1 from surface GARP·latent TGF-β1 complexes, a process called “latent TGF-β1 activation.” This allows binding of active TGF-β1 to its receptor, leading to autocrine and paracrine signaling followed by phosphorylation of SMAD transcription factors (8). GARP is necessary for TGF-β1 activation by Tregs because some anti-GARP monoclonal antibodies are able to block this process (21). GARP is therefore a regulator of TGF-β1 production by human Tregs.
FIGURE 1.
Production and processing of TGF-β1 in cells expressing (Tregs) or not expressing (other cell types) GARP. Double curves represent cell membranes, with the secretory pathway shown as a circle. In latent TGF-β1, the LAP is represented as thick black lines, and the mature TGF-β1 as green lines. Thin black lines represent disulfide bonds. Although not shown in this figure, in Tregs, latent TGF-β1 can also be secreted as a soluble form, in which it is covalently associated with GARP (11).
We postulated that additional proteins expressed in Tregs but not in other T lymphocytes cooperate with GARP to regulate TGF-β1 production. Here we sought to identify proteins that bind to GARP and are expressed at higher levels in Tregs than in Th cells to identify new regulators of TGF-β1 production in human Tregs. As a source of human Tregs, we used Treg clones described previously, i.e. pure populations of cells with a demethylated FOXP3i1 allele, or blood CD4+CD25+CD127lo cells shortly expanded in vitro, i.e. polyclonal populations enriched in cells with a demethylated FOXP3i1 allele. Both cell populations are suppressive in vitro, express FOXP3, and, upon TCR stimulation, induce GARP expression and produce active TGF-β1 (8, 11, 15, 22–24). On the basis of their suppressive activity and mRNA, protein, and epigenetic profile, we termed these human cells “Tregs” (Treg clones or polyclonal Tregs), as proposed by Abbas et al. (25).
Experimental Procedures
Split-ubiquitin System in Yeast
We used the split-ubiquitin system (Dualsystems Biotech AG) according to the instructions of the manufacturer. Briefly, the GARP ORF was cloned into SfiI sites of the bait plasmid pBT3-SUC. The resulting plasmid was used to transform and select GARP-expressing NMY51 yeasts. A prey cDNA library was synthesized in the pPR3N plasmid, starting from 2 μg of total RNA isolated from three different human Treg clones stimulated during 24 h with anti-CD3 and anti-CD28, as described previously (8). The Treg cDNA library contained 5.2 × 106 independent bacterial colonies, which were collected and used to purify plasmid DNA with the PureLink Plasmid Maxiprep kit (Life Technologies). Library plasmid DNA (28 μg) was transformed in GARP-expressing yeasts. An aliquot of transformed yeasts were selected on synthetic defined Leu−Trp− medium to assess transformation efficiency. The remaining yeasts were selected on synthetic defined Leu−Trp−His−Ade− medium to isolate clones transformed with potential GARP interactants.
RT-PCR and RT-qPCR Analyses of LAPTM4B Expression in Human Tregs
Total RNA was extracted, reverse-transcribed, and submitted to PCR or qPCR as described previously (8). qPCR amplifications were done with the ABI Prism 7300 real-time PCR system (Applied Biosystems) under standard conditions: 95 °C for 10 min, 45 cycles of 95 °C for 15 s, and 60 °C for 1 min.
The sequences of primers used to amplify the LAPTM4B mRNA variants shown in Fig. 3 (5′ to 3′) were as follows: primer A1, ATTAACAAGGATCCGCGATGACGTCACGGACTCGG; primer A2, ATTAACAAGGATCCGCGATGAAGATGGTCGCGCCC; primer B, GCTCTATGGTGCCTGGGCCA; and primer R, AACTGATTCTCGAGCGCAGACACGTAAGGTGGCGG. The primers used for RT-qPCR analysis of LAPTM4B Va expression (5′ to 3′) were as follows: sense, ACCATCCTGCTCGGCGTCTG; antisense, CGGATCAGCCAGGGCACTCAAT. The primers used for RT-qPCR analysis of LAPTM4B Vb expression (5′ to 3′) were as follows: sense, GCTCTATGGTGCCTGGGCCA; antisense, CGGATCAGCCAGGGCACTCAAT; and FAM-TAMRA TaqMan probe, GTACCACAGCATTGATGATCTCATTCCCC.
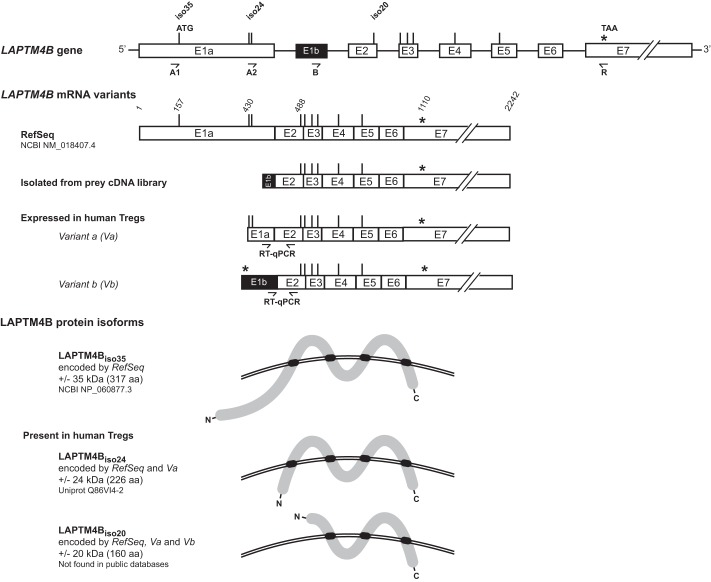
FIGURE 3.
LAPTM4B mRNA variants expressed in human Tregs encode two protein isoforms that differ at their N termini. Shown are schematics of the LAPTM4B gene, mRNA variants, and proteins isoforms. Exons are represented as boxes and introns as thin horizontal lines. All start and stop codons in-frame with the reported start of the RefSeq mRNA are indicated by vertical lines and asterisks, respectively. Primers used for RT-PCR are indicated by arrows. LAPTM4B protein isoforms are represented as thick gray lines, with transmembrane domains highlighted in black. Double curved lines represent the plasma membrane. Start codons for the translation of the various protein isoforms are indicated above the representation of the gene.
Electroporation of siRNA in Tregs
Human polyclonal cell populations enriched in Tregs (CD4+CD25+CD127lo cells) were isolated and expanded from hemochromatosis donors as described previously (11). Expanded Tregs were mixed with siRNAs (3–80 pmol/106 cells, as indicated in the figure legends) and electroporated using unstimulated human T cells 4D-Nucleofector solution and a 4D-Nucleofector instrument (Lonza). Immediately after transfection, cells were restimulated with anti-CD3/CD28-coated beads (Dynabeads human T-activator CD3/CD28, Life Technologies) in Iscove's modified Dulbecco's medium supplemented with 10% human serum, l-arginine, l-asparagine, l-glutamine, β-mercaptoethanol (5 × 10−5 m), and methyl-tryptophan (200 μm).
The sequences of siRNAs (Silencer Select siRNAs from Life Technologies, 5′ to 3′) were as follows: siLAPTM4B #1 (catalog no. S30812), GGAUCAGUAUAACUUUUCAtt (sense) and UGAAAAGUUAUACUGAUCCgg (antisense); siLAPTM4B #2 (catalog no. s502845, UCAAUGCUGUGGUACUGUUtt (sense) and AACAGUACCACAGCAUUGAtg (antisense).
Cell Transfections
293T cells or a 293T cell clone stably expressing human GARP and human TGF-β1 were transiently transfected with the plasmids indicated in the figures using TransIT-LT1 transfection reagent (Mirus Bio). Cells were analyzed 24 h after transfection. Luciferase activity was measured with the Britelite Plus reporter gene assay system (PerkinElmer Life Sciences).
Western Blot Analysis
Cells were lysed in Laemmli buffer supplemented with 5% β-mercaptoethanol as described previously (8) and submitted to SDS-PAGE and Western blot analysis with the following primary antibodies, as indicated in the figures: anti-GARP (Enzo Life Sciences, catalog no. ALX-804-867), anti-TGF-β1 (BD Biosciences, catalog no. 555052), biotinylated anti-LAP (R&D Systems, catalog no. BAF246), anti-Myc (Roche, catalog no. 11-667-149-001), anti-PSMAD2 (Cell Signaling Technology, catalog no. 3108), anti-SMAD2 (Cell Signaling Technology, catalog no. 3122), anti-IGF1Rβ (Cell Signaling Technology, catalog no. 3027), anti-HA (Eurogentec, catalog no. MMS-101R), or anti-β-Actin (Sigma, catalog no. A5441).
FACS Analyses
Cells were stained with mouse monoclonal biotinylated anti-GARP antibody (clone MHG-6 (21)) followed by streptavidin coupled to phycoerythrin (BD Biosciences, catalog no. 554061), anti-LAP antibody coupled to allophycocyanin (R&D Systems, catalog no. 27232), anti-CD9 antibody coupled to phycoerythrin (BioLegend, catalog no. 312105), or anti-HLA-A2 antibody coupled to FITC (BioLegend, catalog no. 343304). Data were collected on a FACS LSR Fortessa (BD Biosciences) and analyzed with FlowJo software (Tree Star).
Latent TGF-β1 Concentrations in Supernatants
Supernatants treated with acid or left untreated were analyzed by ELISA according to the instructions of the manufacturer (human TGF-β1 Duoset, R&D Systems).
Fluorogenic Assay for FURIN-specific Activity
Assays were performed as described by Bourne and Grainger (26). Briefly, transfected 293T cells were lysed in 5× lysis buffer (500 mm HEPES (pH 7.0), 2.5% Triton X-100, 5 mm calcium chloride, and 5 mm β-mercaptoethanol). Lysates containing 3.3 × 105 cell equivalents were seeded in black opaque 96-well plates precoated with a goat polyclonal anti-FURIN antibody (R&D Systems, catalog no. AF1503). Where indicated, the FURIN inhibitor I Dec-RVKR-CMK (decanoyl-Arg-Val-Lys-Arg-chloromethylketone; Calbiochem) was added at a final concentration of 50 μm. The FURIN fluorogenic peptide substrate pERTKR-AMC (R&D Systems) was added at a final concentration of 100 μm. Fluorescence intensities were measured in duplicate wells every 3 min after the addition of substrate on a Victor X2 plate reader (PerkinElmer Life Sciences), with excitation at 380 nm and emission at 460 nm.
Gaussia Luciferase Complementation Assay
Protein complementation assays were performed as described by Remy and Michnick (27). Briefly, 293T cells were transfected with Lipofectamine 2000 transfection reagent (Life Technologies) in triplicate wells (96-well plates) with 0.05 μg of each plasmid. The medium was changed to DMEM without phenol red (Life Technologies), and native coelenterazine (Nano Technologies, catalog no. 303) was added to live cells 24 h after transfection. Luciferase activities were measured on live cells in a Victor X Light device (PerkinElmer Life Sciences).
Ethics Statement
Experiments with human cells were approved by the ethics committee of our institution's (Commission d'Ethique Biomédicale Hospitalo-Facultaire de l'Université catholique de Louvain) under registration number B403201110966.
Results
Identification of GARP Binding Proteins in a Yeast Two-hybrid System
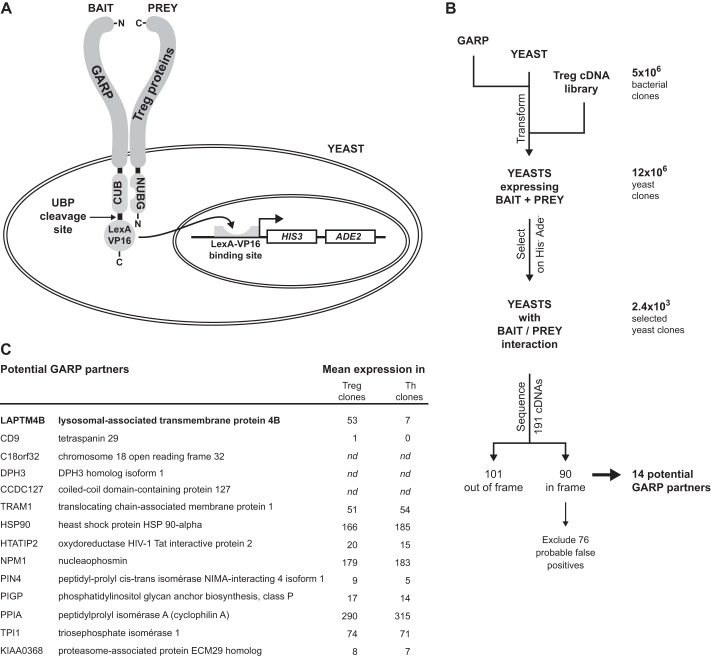
To identify proteins binding to GARP in human Tregs, we used the yeast two-hybrid system known as split-ubiquitin. As shown in Fig. 2A, the bait is a transmembrane protein fused to the C-terminal moiety of Ubiquitin (CUB) and to the LexA-VP16 hybrid transcription factor. Because of the fusion, LexA-VP16 is retained in the cytoplasm and cannot exert transcriptional activity. The prey cDNA library encodes proteins fused to the N-terminal moiety of Ubiquitin (NUBG), which harbors a mutation that prevents spontaneous association with CUB. In yeasts transfected with bait and prey, reconstitution of a functional Ubiquitin by reassembly of CUB and NUBG occurs only when bait and prey interact. Reconstituted Ubiquitin is recognized by Ubiquitin-specific proteases that cleave off LexA-VP16 from the bait, allowing its migration to the nucleus, transcription of HIS3 and ADE2 reporter genes, and growth of yeast on media lacking histidine and adenine (Fig. 2A). It must be noted that productive bait/prey interactions occur only when CUB and NUBG are both located in the cytosol. Therefore, when NUBG is fused to the N-terminal extremity of prey, only cytosolic proteins or type II membrane proteins can be identified using this approach. Here we used GARP-CUB-LexA-VP16 as a bait and a prey cDNA library constructed from three different human Treg clones stimulated for 24 h with anti-CD3 and anti-CD28 antibodies (8).
FIGURE 2.
Identification of potential GARP binding partners using a two-hybrid split-ubiquitin system in yeast. A, the yeast two-hybrid split-ubiquitin system. B, screening of a Treg cDNA library to identify GARP binding partners using the split-ubiquitin system in yeast. C, list of 14 potential GARP partners and their expression levels in TCR-stimulated human Treg and Th clones (means of three or two clones, respectively), as measured with Affymetrix expression microarrays (8). nd, not determined.
We transformed the Treg cDNA library in yeasts expressing GARP-CUB-LexA-VP16 (Fig. 2B). Selection on His−Ade− medium yielded 2371 yeast clones in which the bait interacted with a prey. We sequenced prey cDNAs from 191 selected clones. Of these, 101 encoded out-of-frame fusions or contained non-coding genomic DNA, whereas 90 encoded proteins fused in-frame with the NUBG coding sequence. The list of these cDNAs was compared with that of cDNAs frequently isolated in unrelated split-ubiquitin screens (Dualsystems)5 to exclude 45 cDNAs that were likely to be false positives. We further excluded 31 other cDNAs encoding proteins involved in protein translation or posttranslational modifications or proteins that do not have a predicted N terminus in the cytosol. This left us with 14 cDNAs encoding proteins that potentially interact with GARP.
We examined the expression profiles of the corresponding mRNAs in stimulated human Treg and Th clones using available expression microarray data (8). Two mRNAs, LAPTM4B and CD9, were expressed at >7-fold higher levels in Treg versus Th clones (Fig. 2C). Because CD9 expression in Tregs was low and could not be confirmed to be higher than in Th by RT-qPCR, we focused our analyses on LAPTM4B.
Two mRNA Variants of LAPTM4B Are Expressed in Tregs
According to public databases, the transmembrane protein LAPTM4B is encoded by a gene reported to comprise seven exons: E1a and E2-E7. The mRNA variant isolated from our prey cDNA library contained an alternative first exon, which we named E1b (Fig. 3).
To determine which LAPTM4B mRNA variants are expressed in human Tregs, we analyzed stimulated Treg clones by RT-PCR. Sense primer A1, located immediately upstream of the described start codon in the RefSeq sequence, combined to antisense primer R did not yield a PCR amplification product, indicating that the RefSeq LAPTM4B mRNA is not expressed in human Tregs (Fig. 3 and data not shown). Primer A2, straddling a described alternative start codon (28), and R amplified a product of 677 bp, indicating that an mRNA variant that we will refer to as “Variant a” (Va) is expressed in human Tregs. Sequencing confirmed that Va does not contain exon E1b. The precise 5′ extremity of Va is not known. Finally, primers B and R yielded a product of 608 bp, indicating that human Tregs express a second mRNA variant we named “Variant b” (Vb). We used 5′ rapid amplification of cDNA ends to identify the 5′ extremity of Vb (Fig. 3).
LAPTM4B variants have multiple AUG codons in-frame with the longest open reading frame. Although translation of most eukaryotic mRNA initiates at the first AUG, an alternative mechanism, termed leaky scanning, can result in translation initiation at more distal AUG codons and has been reported to lead to the production of at least two LAPTM4B isoforms (28). LAPTM4B variants therefore potentially encode several LAPTM4B isoforms that differ only by the length of their N termini (Fig. 3). LAPTM4Biso35 (35 kDa) is encoded by the RefSeq variant only, whereas LAPTM4Biso24 (24 kDa) is encoded by RefSeq and Va. LAPTM4B Vb can encode a previously unknown 20-kDa isoform, LAPTM4Biso20, that lacks the first 66 amino acids of LAPTM4Biso24. RefSeq and Va also encode LAPTM4Biso20. Because Tregs express Va and Vb but not RefSeq, we hypothesized that LAPTM4Biso24 and LAPTM4Biso20, but not LAPTM4Biso35, are present in Tregs. We could not verify this hypothesis because of the lack of appropriate antibodies.
The Va and Vb mRNA of LAPTM4B Are Expressed at Higher Levels in Tregs Than in Th Cells
We used RT-qPCR to measure the expression levels of LAPTM4B Va and Vb mRNAs in resting or stimulated Treg and Th clones using the primers illustrated in Fig. 3. As shown in Fig. 4, Va is expressed at higher levels than Vb in both types of T cells. Expression of Va was significantly higher in Treg than in Th clones, both at rest and after stimulation (on average 1.9-fold higher at rest and 2.7-fold higher after stimulation). Expression of Vb was detected in stimulated Treg clones only (more than three copies/105 EF-1 copies in five of seven stimulated Treg clones). Together, LAPTM4B Va and Vb are expressed at higher levels in stimulated human Tregs than in resting Tregs and in resting or stimulated Th cells.
FIGURE 4.
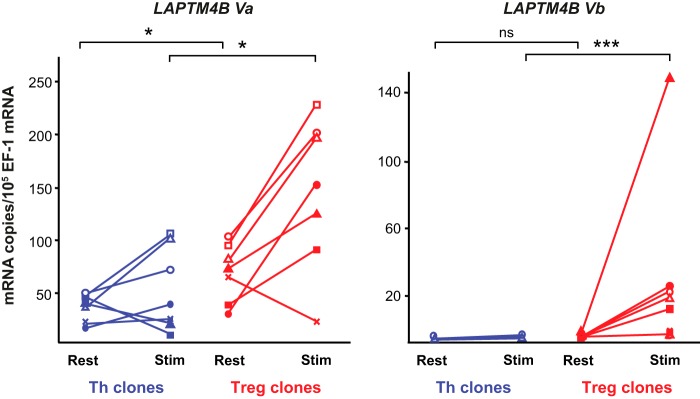
LAPTM4B Va and Vb are expressed at higher levels in human Tregs compared with Th cells. Expression of the indicated LAPTM4B mRNA variants was measured by RT-qPCR in seven Th clones (blue lines) and seven Treg clones (red lines) at rest or 24 h after stimulation with anti-CD3 and anti-CD28 antibodies. *, p < 0,05; ***, p < 0,001; ns, not significant; Mann-Whitney analysis.
Confirmation of the Interaction between GARP and LAPTM4B in Mammalian Cells
To test whether GARP and LAPTM4B interact in mammalian cells, we used a protein complementation assay in which two distinct, inactive fragments of humanized Gaussia luciferase (hGLuc1 and hGLuc2) are fused to the C terminus of candidate proteins and expressed in 293T cells (27). Luciferase activity is recovered only when candidate proteins interact with each other. We cotransfected constructs encoding hGLuc1 fused to N-terminally HA-tagged GARP and hGLuc2 fused to HA-tagged LAPTM4B. Two LAPTM4B constructs were tested, encoding only LAPTM4Biso20 or both LAPTM4Biso20 and LAPTM4Biso24 (i.e. LAPTM4Biso20/24). In the latter construct, only LAPTM4Biso24 is tagged with HA. All HA-tagged proteins were expressed at similar levels, as determined by Western blot analysis (Fig. 5, top panel). High luciferase activity was detected in cells coexpressing GARP and LAPTM4Biso24/20 or GARP and LAPTM4Biso20 (Fig. 5, bottom panel). As expected, no or very low activity was detected in cells expressing any protein alone or in cells coexpressing GLuc fragments fused to GARP and EPOR or to LAPTM4B and EPOR. EPOR was taken as a negative control. These results show that GARP and LAPTM4B isoforms interact in mammalian cells and that the first 66 amino acids of LAPTM4Biso24 (Fig. 3) are not required for this interaction.
FIGURE 5.
GARP interacts with LAPTM4B in mammalian cells. 293T cells were transfected with constructs encoding GARP, LAPTM4B, or negative control EPOR fused to hGLuc1 or hGLuc2 as indicated. Top panel, Western blot analysis of transfected cells. Bottom panel, luciferase activity in live cells 24 h after transfection. One experiment representative of three independent experiments is shown. WB, Western blot; RLU, relative light units.
Overexpression of LAPTM4B in 293T Cells Decreases Cleavage of proTGF-β1, Secretion of Latent TGF-β1, and Surface Presentation of GARP-latent TGF-β1 Complexes
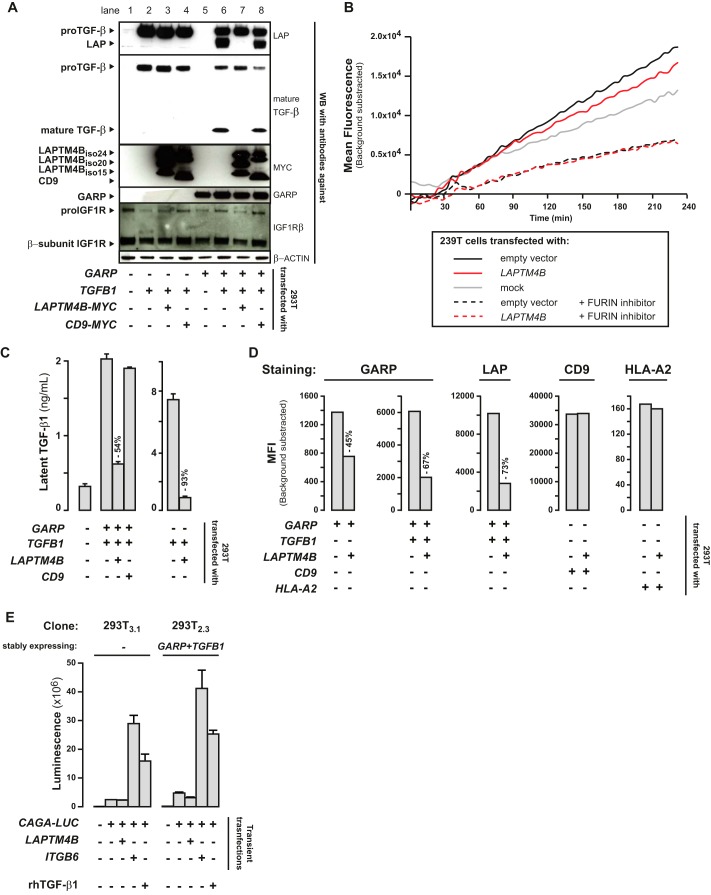
We used constructs encoding both LAPTM4Biso24 and LAPTM4Biso20 to evaluate whether LAPTM4B influences the regulation of TGF-β1 production by GARP in transfected 293T cells. First, we examined, by Western blot analysis, whether LAPTM4B affects the cleavage of proTGF-β1 into latent TGF-β1 (Fig. 6A). In cells transfected with TGFB1 alone (Fig. 6A, lane 2), proTGF-β1 is abundant, but LAP or mature TGF-β1 are not detected, indicating that cleavage of the precursor does not occur at high levels. Cotransfection of TGFB1 with LAPTM4B (Fig. 6A, lane 3) does not increase cleavage. In contrast and as expected, cotransfection of TGFB1 with GARP (Fig. 6A, lane 6) induces cleavage, as evidenced by the detection of abundant LAP and mature TGF-β1. Cotransfection of LAPTM4B (Fig. 6A, lane 7) abolished this GARP-induced cleavage, an effect that was not observed upon cotransfection of CD9 (Fig. 6A, lane 8), taken here as a negative control.
FIGURE 6.
LAPTM4B decreases cleavage of proTGF-β1, surface presentation of GARP·TGF-β1 complexes, and secretion of latent TGF-β1. A, cell lysates of 293T cells transfected as indicated were analyzed by Western blot. B, specific FURIN activity was measured in cell lysates 24 h after transfection by capturing FURIN on immobilized anti-FURIN antibody and then incubating captured proteins with the fluorogenic substrate pERTKR-AMC. Graphs show mean fluorescence intensity (MFI) at the indicated time after addition of the substrate. The FURIN inhibitor Dec-RVKR-CMK was added under some conditions to verify the specificity of the assay. C, concentration of latent TGF-β1 in the acid-treated supernatants was measured by ELISA. D, surface levels of GARP, LAP, CD9, and HLA-A2 in transiently transfected 293T were assessed by FACS. E, clones of 293T cells stably expressing or not expressing human GARP and human TGF-β1 were transfected with LAPTM4B or ITGB6 together with the CAGA-LUC reporter. Luciferase activity was measured 24 h after transfection. Recombinant human (rh) TGF-β1 was added during 6 h at 4 ng/ml under the positive control condition, as indicated. The TGFB1 construct used for transfections contains the full-length TGFB1 ORF coding for the pre-proTGF-β1 precursor, which is processed in 293T cells like in all other cell types.
Because proTGF-β1 cleavage depends on FURIN, decreased proTGF-β1 cleavage in the presence of LAPTM4B could result from a direct effect on FURIN activity. We examined whether the cleavage of endogenous proIGF1R, another known substrate of FURIN, into its β subunit was also reduced in 293T cells transfected with LAPTM4B, but this was not the case (Fig. 6A). We also measured FURIN activity on an exogenous fluorogenic substrate, and the transfection of LAPTM4B did not decrease FURIN activity in this assay either (Fig. 6B). We conclude that LAPTM4B reduces the cleavage of proTGF-β1 in the presence of GARP without reducing overall FURIN activity.
We next examined whether this decreased cleavage of proTGF-β1 resulted in a reduction of latent TGF-β1 secretion. We indeed found 54% less latent TGF-β1 in the supernatants of 293T cells transfected with GARP, TGFB1, and LAPTM4B compared with cells transfected with GARP and TGFB1 only (Fig. 6C). This effect was not observed with a negative control (CD9). Interestingly, the reduction of latent TGF-β1 secretion by LAPTM4B was also observed in the absence of GARP, indicating that the regulatory effect of LAPTM4B on latent TGF-β1 secretion is not dependent on its interaction with GARP (Fig. 6C). Incidentally and as expected, secretion of latent TGF-β1 was lower in the presence of GARP because GARP tethers latent TGF-β1 at the 293T cell surface (11, 20).
We then used flow cytometry to examine whether LAPTM4B influenced surface levels of GARP or GARP·latent TGF-β1 complexes on transfected 293T cells. GARP levels were reduced by 45% upon cotransfection with LAPTM4B (Fig. 6D). In cells transfected with GARP and TGFB1, cotransfection of LAPTM4B reduced surface GARP by 67% and surface LAP by 73%. Surface levels of unrelated proteins such as HLA-A2 or CD9 were not affected by coexpression of LAPTM4B. Therefore, LAPTM4B reduces surface levels of GARP and GARP·latent TGF-β1 complexes.
Finally, we tested whether LAPTM4B regulates latent TGF-β1 activation (Fig. 6E). We used a reporter assay in which the luciferase gene is under the control of a CAGA promoter activated by SMAD2/3 transcription factors in response to TGF-β1 signals (29). We transiently cotransfected the CAGA-LUC reporter with LAPTM4B in clones of 293T cells stably expressing or not expressing GARP and TGFB1. Cotransfection of LAPTM4B did not increase luciferase activity above background (i.e. transfection of the reporter alone), indicating that LAPTM4B expression does not activate latent TGF-β1. This was true also in a clone stably expressing GARP, which is not sufficient to induce active TGF-β1 production (Fig. 6E). As expected, high luciferase activity was induced by transfection of integrin β6, a known activator of TGF-β1 (30), or by addition of recombinant active TGF-β1, both used here as a positive controls.
We conclude that, in transfected 293T cells, LAPTM4B decreases the cleavage of proTGF-β1, the secretion of latent TGF-β1, and the surface presentation of GARP·latent TGF-β1 complexes but does not activate latent TGF-β1 in cooperation with GARP. Therefore, LAPTM4B appears to be a negative regulator of TGF-β1 production in these cells.
LAPTM4B Silencing in Human Polyclonal Tregs Increases the Surface Presentation of GARP·TGF-β1 Complexes and Secretion of Latent TGF-β1
We next wished to evaluate whether LAPTM4B plays a role in human Tregs. Polyclonal Tregs were obtained by a short in vitro amplification of blood CD4+CD25+CD127lo cells. They contained 42–58% of cells with demethylated FOXP3i1. They were transfected with siRNAs and stimulated through their TCR to induce TGF-β1 activation or left unstimulated. siLAPTM4B #1 reduced the expression of LAPTM4B Va and Vb by >85%. It increased by 41–53% the secretion of latent TGF-β1 by resting and stimulated Tregs and by ±100% the surface presentation of GARP and latent TGF-β1 on stimulated Tregs (Fig. 7A). Increases in surface GARP and latent TGF-β1 were detected with amounts of siLAPTM4B #1 as low as 16 pmol/106 cells (Fig. 7B) and were also observed with siLAPTM4B #2, which targets LAPTM4B at a site different from that targeted by siLAPTM4B #1 (Fig. 7C). Neither of the two siLAPTM4B modified surface levels of CD4, used here as an unrelated negative control (Fig. 7C). As expected, siLAPTM4B did not reduce the activation of TGF-β1 by stimulated Tregs (Fig. 7D). These results confirm that LAPTM4B is a negative regulator of TGF-β1 production in human Tregs.
FIGURE 7.
LAPTM4B decreases GARP surface levels and TGF-β1 secretion in human Tregs. A, polyclonal human Tregs were transfected with the indicated siRNAs (50 pmol/106 cells) and then left resting (Rest) or stimulated (Stim) with anti-CD3/CD28-coated beads for 3 days. Levels of LAPTM4B Va and Vb mRNAs were measured by RT-qPCR. Amounts of total and active TGF-β1 were measured by ELISA in supernatants treated with acid or left untreated. Because no active TGF-β1 was detected in non-acidified supernatants, total TGF-β1 in acidified supernatants corresponded to latent TGF-β1. Surface levels of GARP and latent TGF-β1 were measured by flow cytometry with anti-GARP and anti-LAP antibodies, respectively. The percent increase or decrease compared with the control siRNA is indicated above the bar graphs. One experiment representative of three is shown. B, as A, with the indicated amounts of siRNAs. FI, fluorescence intensity. C, as A, with 80 pmol/106 cells siRNAs and cells stimulated during 4 days. D, Western blot analysis of cells shown in A with anti-pSMAD2, anti-total SMAD2, and anti-β-ACTIN antibodies. Detection of pSMAD2 indicates active TGF-β1 production.
Discussion
LAPTM4B belongs to a family of three glycoproteins with four or five transmembrane domains. The two other members of the family are LAPTM4A and LAPTM5, with 46 and 23% sequence identities with LAPTM4B, respectively. Little is known about the physiological function of LAPTM4B. In tumors, it appears to play an oncogenic role. High LAPTM4B levels are associated with poor prognosis in many types of cancers (31–41), and in vitro and in vivo analyses of cells in which LAPTM4B was overexpressed or silenced indicate that LAPTM4B favors proliferation, migration, invasion, tumorigenesis, and metastasis (42, 43). Two mechanisms by which LAPTM4B plays oncogenic roles have recently been identified. Both imply facilitation of the prosurvival functions of the EGF receptor in cancer cells. In the presence of EGF, LAPTM4B enhances signaling by blocking the lysosomal degradation of activated EGF receptor (44), and in the absence of EGF, it interacts with inactive EGF receptor in the endosomes to initiate cell-protective autophagy (45).
Our results are the first to describe a function for LAPTM4B in the immune system. It decreases the production, secretion, and surface presentation of latent TGF-β1 by Tregs. Down-regulation of surface presentation appears to result from a direct effect of LAPTM4B on surface GARP levels because we show that LAPTM4B interacts with GARP, a receptor for latent TGF-β1, and reduces surface GARP levels in transfected 293T cells in the absence of TGF-β1. Another LAPTM family member has been reported to interact and to down-regulate surface receptors on immune cells. Murine Laptm5 binds to proteins of the T and B cell antigenic receptor complexes (TCR and BCR, respectively), and surface levels of TCR and BCR were higher on activated Laptm5−/− lymphocytes than in activated wild-type lymphocytes (46, 47). C-terminal polyproline-tyrosine (PY) motifs target Laptm5 to lysosomes and are required for the Laptm5-mediated down-modulation of surface TCR levels, suggesting a mechanism by which Laptm5 promotes lysosomal targeting and degradation of receptors to which it is bound (47, 48). PY motifs are also present in the C terminus of LAPTM4B (47, 49). It is therefore possible that LAPTM4B down-regulates GARP surface levels in human Tregs through a similar mechanism. However, we observed in transfected cells that GARP and LAPTM4B colocalize mostly in the median Golgi and that the localization of GARP was not modified in the presence of LAPTM4B (unpublished observations).6 It is worth noting that the down-regulation of latent TGF-β1 secretion by LAPTM4B may occur through another mechanism, independent from GARP, because it was also observed in transfected 293T cells in the absence of GARP.
Two isoforms of LAPTM4B, LAPTM4Biso35 and LAPTM4Biso24, have been described previously (28). Here we identify, in stimulated human Tregs, an mRNA variant coding for a third, shorter isoform, LAPTM4Biso20. Tregs also contain LAPTM4Biso24 but not LAPTM4Biso35. We showed that LAPTM4Biso20 interacts with GARP, indicating that the 66 N-terminal amino acids of LAPTM4Biso24 are not required for the interaction. We could not determine whether LAPTM4Biso24 also interacts with GARP because all of our constructs encoding LAPTM4Biso24 also encode LAPTM4Biso20. We do not know whether LAPTM4Biso24 or LAPTM4Biso20 or both regulate GARP and TGF-β1 production in Tregs because our siRNA approach silenced all of these LAPTM4B mRNA variants.
We report that LAPTM4B expression is increased upon TCR activation of human Tregs and that it is higher in Tregs than in Th cells. Overexpression of LAPTM4B in human or mouse Tregs as well as in non-Tregs transduced with FOXP3 was observed in published expression microarray data sets (50–52), suggesting that gene LAPTM4B might be transcriptionally regulated by FOXP3. Up-regulation of LAPTM4B expression in Tregs after TCR stimulation may represent a negative feedback mechanism that eventually shuts down production of TGF-β1 to ensure return to a resting state.
In conclusion, we uncovered an inhibitory mechanism of GARP and TGF-β1 production mediated by LAPTM4B in human Tregs. Mechanisms of immunosuppression by human Tregs include the production of active TGF-β1, released from GARP·latent TGF-β1 complexes at the Treg surface (15, 16). Tregs are potent inhibitors of immune responses, and excessive or insufficient Treg function is implicated in various human diseases. Pharmacological inhibition of LAPTM4B could sustain or increase TGF-β1 production and, therefore, Treg function, in patients suffering from autoimmune disease or allograft rejection.
Author Contributions
C. H. and S. Lucas designed the study. C. H., S. Liénart, O. D., J. S., and E. G. performed the experiments. C. H., S. Lucas, and P. G. C. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Suzanne Depelchin for editorial help, Nicolas Dauguet for FACS sorting, Maria Panagiotakopoulos and Stéphanie D'Hondt for technical assistance, Vitalina Gryshkova (Ludwig Cancer Research, Brussels, Belgium) for complementation assay vectors, A. Moustakas (Ludwig Cancer Research, Uppsala, Sweden) for the TGF-β signaling reporter construct, and Donatienne Tyteca (de Duve Institute) for help with immunofluorescence and confocal microscopy.
Note Added in Proof
As was brought to our attention by Dr Z. Li, we wish to mention that GARP was also shown to interact with GP96, a protein controlling the folding of several immune-related proteins in the ER (Zhang, Y., Wu, B. X., Metelli, A., Thaxton, J. E., Hong, F., Rachidi, S., Ansa-Addo, E., Sun, S., Vasu, C., Yang, Y., Liu, B., and Li, Z. (2015) GP96 is a GARP chaperone and controls regulatory T cell functions. J. Clin. Invest. 125, 859–869).
This work was supported by grants from the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming and by grants from the Belgian Cancer Plan (Action 29_024), the Fonds National pour la Recherche Scientifique (Belgium), the Fondation contre le Cancer (Belgium), the Fondation Salus Sanguinis (Belgium), and the Actions de Recherche Concertées ARC 09/14-021 (Belgium) and the Fonds J. Maisin (Belgium). The authors declare that they have no conflict of interest with the contents of the article.
C. Huygens, S. Liénart, O. Dedobbeleer, J. Stockis, E. Gauthy, P. G. Coulie, and S. Lucas, unpublished data.
C. Huygens, D. Tyteca, and S. Lucas, unpublished observations.
- Treg
- regulatory T cell
- GARP
- glycoprotein A repetitions predominant
- LAP
- latency-associated peptide
- Th
- T helper lymphocyte
- CTL
- cytotoxic T lymphocyte
- TSDR
- Treg-specific demethylated region
- TCR
- T cell receptor
- EPOR
- erythropoietin receptor
- BCR
- B cell receptor
- RT-qPCR
- quantitative RT-PCR
- CUB
- C-terminal moiety of ubiquitin
- NUBG
- N-terminal moiety of ubiquitin.
References
- 1. Josefowicz S. Z., Lu L. F., Rudensky A. Y. (2012) Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sakaguchi S., Miyara M., Costantino C. M., Hafler D. A. (2010) FOXP3+ regulatory T cells in the human immune system. Nat. Rev. Immunol. 10, 490–500 [DOI] [PubMed] [Google Scholar]
- 3. Brunkow M. E., Jeffery E. W., Hjerrild K. A., Paeper B., Clark L. B., Yasayko S. A., Wilkinson J. E., Galas D., Ziegler S. F., Ramsdell F. (2001) Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 [DOI] [PubMed] [Google Scholar]
- 4. Bennett C. L., Christie J., Ramsdell F., Brunkow M. E., Ferguson P. J., Whitesell L., Kelly T. E., Saulsbury F. T., Chance P. F., Ochs H. D. (2001) The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 [DOI] [PubMed] [Google Scholar]
- 5. Wildin R. S., Ramsdell F., Peake J., Faravelli F., Casanova J. L., Buist N., Levy-Lahad E., Mazzella M., Goulet O., Perroni L., Bricarelli F. D., Byrne G., McEuen M., Proll S., Appleby M., Brunkow M. E. (2001) X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27, 18–20 [DOI] [PubMed] [Google Scholar]
- 6. Baron U., Floess S., Wieczorek G., Baumann K., Grützkau A., Dong J., Thiel A., Boeld T. J., Hoffmann P., Edinger M., Türbachova I., Hamann A., Olek S., Huehn J. (2007) DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3+ conventional T cells. Eur. J. Immunol. 37, 2378–2389 [DOI] [PubMed] [Google Scholar]
- 7. Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H. D., Bopp T., Schmitt E., Klein-Hessling S., Serfling E., Hamann A., Huehn J. (2007) Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stockis J., Fink W., François V., Connerotte T., de Smet C., Knoops L., van der Bruggen P., Boon T., Coulie P. G., Lucas S. (2009) Comparison of stable human Treg and Th clones by transcriptional profiling. Eur. J. Immunol. 39, 869–882 [DOI] [PubMed] [Google Scholar]
- 9. Zheng Y., Josefowicz S., Chaudhry A., Peng X. P., Forbush K., Rudensky A. Y. (2010) Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Polansky J. K., Kretschmer K., Freyer J., Floess S., Garbe A., Baron U., Olek S., Hamann A., von Boehmer H., Huehn J. (2008) DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38, 1654–1663 [DOI] [PubMed] [Google Scholar]
- 11. Gauthy E., Cuende J., Stockis J., Huygens C., Lethé B., Collet J. F., Bommer G., Coulie P. G., Lucas S. (2013) GARP is regulated by miRNAs and controls latent TGF-β1 production by human regulatory T cells. PLoS ONE 8, e76186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Vries I. J., Castelli C., Huygens C., Jacobs J. F., Stockis J., Schuler-Thurner B., Adema G. J., Punt C. J., Rivoltini L., Schuler G., Coulie P. G., Lucas S. (2011) Frequency of circulating Tregs with demethylated FOXP3 intron 1 in melanoma patients receiving tumor vaccines and potentially Treg-depleting agents. Clin. Cancer Res. 17, 841–848 [DOI] [PubMed] [Google Scholar]
- 13. Wieczorek G., Asemissen A., Model F., Turbachova I., Floess S., Liebenberg V., Baron U., Stauch D., Kotsch K., Pratschke J., Hamann A., Loddenkemper C., Stein H., Volk H. D., Hoffmüller U., Grützkau A., Mustea A., Huehn J., Scheibenbogen C., Olek S. (2009) Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 69, 599–608 [DOI] [PubMed] [Google Scholar]
- 14. Vignali D. A., Collison L. W., Workman C. J. (2008) How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stockis J., Colau D., Coulie P. G., Lucas S. (2009) Membrane protein GARP is a receptor for latent TGF-β on the surface of activated human Treg. Eur. J. Immunol. 39, 3315–3322 [DOI] [PubMed] [Google Scholar]
- 16. Tran D. Q., Andersson J., Wang R., Ramsey H., Unutmaz D., Shevach E. M. (2009) GARP (LRRC32) is essential for the surface expression of latent TGF-β on platelets and activated FOXP3+ regulatory T cells. Proc. Natl. Acad. Sci. U.S.A. 106, 13445–13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang R., Wan Q., Kozhaya L., Fujii H., Unutmaz D. (2008) Identification of a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression. PLoS ONE 3, e2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. ten Dijke P., Arthur H. M. (2007) Extracellular control of TGFβ signalling in vascular development and disease. Nat. Rev. Mol. Cell Biol. 8, 857–869 [DOI] [PubMed] [Google Scholar]
- 19. Travis M. A., Sheppard D. (2014) TGF-β activation and function in immunity. Annu. Rev. Immunol. 32, 51–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang R., Zhu J., Dong X., Shi M., Lu C., Springer T. A. (2012) GARP regulates the bioavailability and activation of TGFβ. Mol. Biol. Cell 23, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cuende J., Liénart S., Dedobbeleer O., van der Woning B., De Boeck G., Stockis J., Huygens C., Colau D., Somja J., Delvenne P., Hannon M., Baron F., Dumoutier L., Renauld J.-C., De Haard H., Saunders M., Coulie P. G., Lucas S. (2015) Monoclonal antibodies against GARP/TGF-β1 complexes inhibit the immunosuppressive activity of human regulatory T cells in vivo. Sci. Transl. Med. 7, 248ra56. [DOI] [PubMed] [Google Scholar]
- 22. Liu W., Putnam A. L., Xu-Yu Z., Szot G. L., Lee M. R., Zhu S., Gottlieb P. A., Kapranov P., Gingeras T. R., Fazekas de St Groth B., Clayberger C., Soper D. M., Ziegler S. F., Bluestone J. A. (2006) CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J. Exp. Med. 203, 1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peters J. H., Preijers F. W., Woestenenk R., Hilbrands L. B., Koenen H. J., Joosten I. (2008) Clinical grade Treg: GMP isolation, improvement of purity by CD127 Depletion, Treg expansion, and Treg cryopreservation. PLoS ONE 3, e3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan H., Yang J., Hao J., Ren Y., Chen L., Li G., Xie R., Yang Y., Gao F., Liu M. (2012) Comparative study of regulatory T cells expanded ex vivo from cord blood and adult peripheral blood. Immunology 136, 218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abbas A. K., Benoist C., Bluestone J. A., Campbell D. J., Ghosh S., Hori S., Jiang S., Kuchroo V. K., Mathis D., Roncarolo M. G., Rudensky A., Sakaguchi S., Shevach E. M., Vignali D. A., Ziegler S. F. (2013) Regulatory T cells: recommendations to simplify the nomenclature. Nat. Immunol. 14, 307–308 [DOI] [PubMed] [Google Scholar]
- 26. Bourne G. L., Grainger D. J. (2011) Development and characterisation of an assay for furin activity. J. Immunol. Methods 364, 101–108 [DOI] [PubMed] [Google Scholar]
- 27. Remy I., Michnick S. W. (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3, 977–979 [DOI] [PubMed] [Google Scholar]
- 28. Shao G. Z., Zhou R. L., Zhang Q. Y., Zhang Y., Liu J. J., Rui J. A., Wei X., Ye D. X. (2003) Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene 22, 5060–5069 [DOI] [PubMed] [Google Scholar]
- 29. Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., Sheppard D. (1999) The integrin α v β 6 binds and activates latent TGF β 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 31. Kang Y., Yin M., Jiang W., Zhang H., Xia B., Xue Y., Huang Y. (2012) Overexpression of LAPTM4B-35 is associated with poor prognosis in colorectal carcinoma. Am. J. Surg. 204, 677–683 [DOI] [PubMed] [Google Scholar]
- 32. Tang H., Tian H., Yue W., Li L., Li S., Gao C., Si L., Qi L., Lu M. (2014) Overexpression of LAPTM4B is correlated with tumor angiogenesis and poor prognosis in non-small cell lung cancer. Med. Oncol. 31, 974. [DOI] [PubMed] [Google Scholar]
- 33. Yang H., Xiong F., Qi R., Liu Z., Lin M., Rui J., Su J., Zhou R. (2010) LAPTM4B-35 is a novel prognostic factor of hepatocellular carcinoma. J. Surg. Oncol. 101, 363–369 [DOI] [PubMed] [Google Scholar]
- 34. Yang H., Xiong F. X., Lin M., Yang Y., Nie X., Zhou R. L. (2010) LAPTM4B-35 overexpression is a risk factor for tumor recurrence and poor prognosis in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 136, 275–281 [DOI] [PubMed] [Google Scholar]
- 35. Yang Y., Yang H., McNutt M. A., Xiong F., Nie X., Li L., Zhou R. (2008) LAPTM4B overexpression is an independent prognostic marker in ovarian carcinoma. Oncol. Rep. 20, 1077–1083 [PubMed] [Google Scholar]
- 36. Yin M., Li C., Li X., Lou G., Miao B., Liu X., Meng F., Zhang H., Chen X., Sun M., Ling Q., Zhou R. (2011) Over-expression of LAPTM4B is associated with poor prognosis and chemotherapy resistance in stages III and IV epithelial ovarian cancer. J. Surg. Oncol. 104, 29–36 [DOI] [PubMed] [Google Scholar]
- 37. Yin M., Xu Y., Lou G., Hou Y., Meng F., Zhang H., Li C., Zhou R. (2011) LAPTM4B overexpression is a novel predictor of epithelial ovarian carcinoma metastasis. Int. J. Cancer 129, 629–635 [DOI] [PubMed] [Google Scholar]
- 38. Zhang G., Liang Y., Huang Y., Chen Y., Zhou R. (2012) Elevated lysosome-associated protein transmembrane-4β-35 is an independent prognostic marker in pancreatic carcinoma. J. Int. Med. Res. 40, 1275–1283 [DOI] [PubMed] [Google Scholar]
- 39. Zhang H., Wei Q., Liu R., Qi S., Liang P., Qi C., Wang A., Sheng B., Li L., Xu Y. (2014) Overexpression of LAPTM4B-35: a novel marker of poor prognosis of prostate cancer. PLoS ONE 9, e91069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou L., He X. D., Chen J., Cui Q. C., Qu Q., Rui J. A., Zhao Y. P. (2007) Overexpression of LAPTM4B-35 closely correlated with clinicopathological features and post-resectional survival of gallbladder carcinoma. Eur. J. Cancer 43, 809–815 [DOI] [PubMed] [Google Scholar]
- 41. Zhou L., He X. D., Cui Q. C., Zhou W. X., Qu Q., Zhou R. L., Rui J. A., Yu J. C. (2008) Expression of LAPTM4B-35: a novel marker of progression, invasiveness and poor prognosis of extrahepatic cholangiocarcinoma. Cancer Lett. 264, 209–217 [DOI] [PubMed] [Google Scholar]
- 42. Liu X., Xiong F., Wei X., Yang H., Zhou R. (2009) LAPTM4B-35, a novel tetratransmembrane protein and its PPRP motif play critical roles in proliferation and metastatic potential of hepatocellular carcinoma cells. Cancer Sci. 100, 2335–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang H., Xiong F., Wei X., Yang Y., McNutt M. A., Zhou R. (2010) Overexpression of LAPTM4B-35 promotes growth and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer Lett. 294, 236–244 [DOI] [PubMed] [Google Scholar]
- 44. Tan X., Sun Y., Thapa N., Liao Y., Hedman A. C., Anderson R. A. (2015) LAPTM4B is a PtdIns(4,5)P2 effector that regulates EGFR signaling, lysosomal sorting, and degradation. EMBO J. 34, 475–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan X., Thapa N., Sun Y., Anderson R. A. (2015) A kinase-independent role for EGF receptor in autophagy initiation. Cell 160, 145–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ouchida R., Kurosaki T., Wang J. Y. (2010) A role for lysosomal-associated protein transmembrane 5 in the negative regulation of surface B cell receptor levels and B cell activation. J. Immunol. 185, 294–301 [DOI] [PubMed] [Google Scholar]
- 47. Ouchida R., Yamasaki S., Hikida M., Masuda K., Kawamura K., Wada A., Mochizuki S., Tagawa M., Sakamoto A., Hatano M., Tokuhisa T., Koseki H., Saito T., Kurosaki T., Wang J. Y. (2008) A lysosomal protein negatively regulates surface T cell antigen receptor expression by promoting CD3ζ-chain degradation. Immunity 29, 33–43 [DOI] [PubMed] [Google Scholar]
- 48. Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. (2006) Transport of LAPTM5 to lysosomes requires association with the ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J. Cell Biol. 175, 631–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Milkereit R., Rotin D. (2011) A role for the ubiquitin ligase Nedd4 in membrane sorting of LAPTM4 proteins. PLoS ONE 6, e27478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Herman A. E., Freeman G. J., Mathis D., Benoist C. (2004) CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J. Exp. Med. 199, 1479–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ocklenburg F., Moharregh-Khiabani D., Geffers R., Janke V., Pfoertner S., Garritsen H., Groebe L., Klempnauer J., Dittmar K. E., Weiss S., Buer J., Probst-Kepper M. (2006) UBD, a downstream element of FOXP3, allows the identification of LGALS3, a new marker of human regulatory T cells. Lab. Invest. 86, 724–737 [DOI] [PubMed] [Google Scholar]
- 52. Williams L. M., Rudensky A. Y. (2007) Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 [DOI] [PubMed] [Google Scholar]