Background: The survival motor neuron (SMN) protein forms oligomeric complexes involved in ribonucleoprotein (RNP) biogenesis.
Results: SMN forms stable dimers, which in turn self-associate to form tetramers and octamers.
Conclusion: SMN complexes form discrete oligomers with unusually large hydrodynamic sizes.
Significance: Understanding the oligomeric nature of SMN provides an important foundation for exploring the biochemical bases of RNP assembly and spinal muscular atrophy.
Keywords: analytical ultracentrifugation, neurodegenerative disease, oligomerization, ribonuclear protein (RNP), RNA metabolism, Gemin2, snRNP assembly, spinal muscular atrophy, survival motor neuron
Abstract
The survival motor neuron (SMN) protein forms the oligomeric core of a multiprotein complex required for the assembly of spliceosomal small nuclear ribonucleoproteins. Deletions and mutations in the SMN1 gene are associated with spinal muscular atrophy (SMA), a devastating neurodegenerative disease that is the leading heritable cause of infant mortality. Oligomerization of SMN is required for its function, and some SMA patient mutations disrupt the ability of SMN to self-associate. Here, we investigate the oligomeric nature of the SMN·Gemin2 complexes from humans and fission yeast (hSMN·Gemin2 and ySMN·Gemin2). We find that hSMN·Gemin2 forms oligomers spanning the dimer to octamer range. The YG box oligomerization domain of SMN is both necessary and sufficient to form these oligomers. ySMN·Gemin2 exists as a dimer-tetramer equilibrium with Kd = 1.0 ± 0.9 μm. A 1.9 Å crystal structure of the ySMN YG box confirms a high level of structural conservation with the human ortholog in this important region of SMN. Disulfide cross-linking experiments indicate that SMN tetramers are formed by self-association of stable, non-dissociating dimers. Thus, SMN tetramers do not form symmetric helical bundles such as those found in glycine zipper transmembrane oligomers. The dimer-tetramer nature of SMN complexes and the dimer of dimers organization of the SMN tetramer provide an important foundation for ongoing studies to understand the mechanism of SMN-assisted small nuclear ribonucleoprotein assembly and the underlying causes of SMA.
Introduction
The survival motor neuron (SMN)2 protein forms the oligomeric core of a multiprotein complex that functions in the biogenesis of spliceosomal snRNPs and other RNP complexes (1, 2). Additional potential functions of SMN include modulating apoptosis (3), mRNA localization (4), and translational regulation (5). SMN binds tightly to the Gemin2 protein in vivo and in vitro and interacts with additional “Gemin” proteins in the cells of most higher eukaryotes (6–8). Both SMN and Gemin2 are conserved from fission yeast to humans and have been shown to be essential in each organism that has been examined (9).
Deletions and mutations in the SMN1 gene are associated with spinal muscular atrophy (SMA), an autosomal recessive neurodegenerative disorder that affects one in 6,000 births and is a leading genetic cause of infant mortality (10, 11). A second gene, SMN2, is found only in humans and is the sole source of wild-type SMN in most SMA patients. However, the majority of SMN2 pre-mRNA transcripts undergo alternative splicing that removes the seventh exon, resulting in a truncated protein (12, 13). In the most severe form of SMA, patients typically do not live past 2 years of age; in the mildest form, they reach adulthood with only minor motor function defects. The clinical severity of SMA is strongly correlated with a reduction of functional SMN (for reviews, see Refs. 9 and 14–16).
Nearly half of the missense mutations found in SMA patients map to the C-terminal region of SMN (9), within a highly conserved oligomerization domain termed the YG box (Fig. 1A). SMN complexes purified from cells are large particles that are composed of SMN oligomers and other SMN-interacting proteins (6, 7, 17). Some SMA patient YG box mutations have been shown to result in a decrease in the ability of SMN to self-interact and to interact with other proteins (18, 19). However, the size and nature of the oligomers formed by SMN are not known. Furthermore, a specific mechanistic role for the YG box or for SMN oligomerization in RNP biogenesis that could explain why this region is so sensitive to mutation has not yet been established.
FIGURE 1.
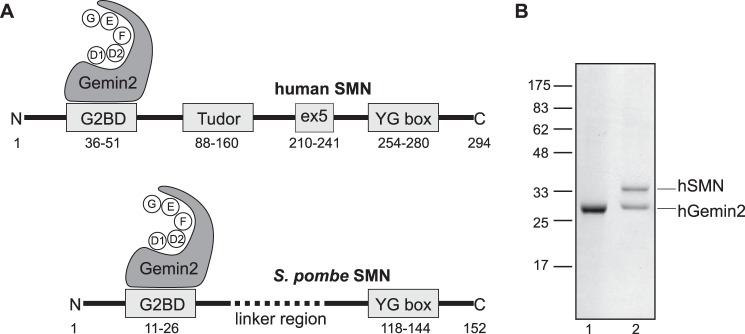
Domain structures of human and yeast SMN·Gemin2 complexes. A, the Gemin2-binding domain (G2BD) and YG box regions of SMN are highly conserved, but fungal SMNs lack the Tudor domain found in metazoans. Human and fly Gemin2 have been shown to bind a pentamer of Sm proteins (D1, D2, F, E, and G) during snRNP assembly (37, 38); strong conservation of Gemin2 suggests that fungal Gemin2 has a similar function. B, SDS-PAGE of purified hGemin2 (lane 1) and hSMN·Gemin2 (lane 2).
Here, we describe experiments aimed toward understanding the oligomeric nature of the human and fission yeast SMN·Gemin2 complexes (hSMN·Gemin2 and ySMN·Gemin2). We find that the hSMN·Gemin2 complex forms an equilibrium mixture of oligomers in the dimer to octamer range. In this paper, an oligomer of SMN·Gemin2 refers to a species with stoichiometry (SMN·Gemin2)n, where n = 2 for a dimer, n = 4 for a tetramer, etc. hGemin2 is monomeric at micromolar concentration, and neither hGemin2 nor regions upstream of the YG box in hSMN strongly influences the oligomeric state of the hSMN·Gemin2 complex.
The ySMN·Gemin2 complex forms an equilibrium mixture of dimers and tetramers but does not form higher order oligomers even at elevated concentrations. A crystal structure of the ySMN YG box reveals remarkable conservation of structure with the previously reported hSMN YG box (20) and extends that structural model in both directions from the highly conserved core sequence. We have exploited the simplicity and favorable biochemistry of the yeast system to further show that the SMN tetramer is formed by self-association of stable SMN·Gemin2 dimers and is not a symmetrical bundle of glycine zipper helices similar to membrane ion channels with related oligomerization motifs (21).
These findings provide answers to key questions about the core SMN·Gemin2 complex that have gone unanswered for nearly 20 years. The results also provide an important foundation for investigating the biochemical defects in SMA patients and for dissecting the functions and mechanisms of action of SMN.
Experimental Procedures
Plasmid Construction
Plasmids were constructed using standard restriction enzyme cloning methods (22). Mutants were generated using inverse PCR and overlap extension PCR methods (22). Maltose-binding protein (MBP) fusions were constructed using a pETDuet (Novagen) construct containing Escherichia coli MalE residues Lys27–Thr395 and included no more than two plasmid-derived linker residues. Internal laboratory database numbers for expression plasmids are given in Table 1 or are noted below.
TABLE 1.
SEC-MALS and SV analyses of SMN·Gemin2 complexes
Plasmid numbers refer to internal laboratory database entries for the plasmids used for expression or co-expression. Calculated complex molecular masses are for a 1:1 stoichiometry. SEC-MALS concentration is at the UV absorbance peak, as determined by refractive index. MA, apparent molecular mass based on calibration of the SEC column with globular standards; M̄w, weight-average molecular mass from MALS; ND, not determined.
| Complex | Plasmid | Mcalc | SEC-MALS (20 °C) |
SV (20 °C) |
SV (4 °C) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Concentration | MA | M̄w | Concentration | s20,w | Concentration | s20,w | |||
| kDa | μm | kDa | kDa | μm | μm | ||||
| hGemin2 | 371 | 30.8 | 2.3 | 56.3 | 27.7 ± 0.1 | ND | ND | ||
| hSMN(26–51)·Gemin2 | 1449/371 | 33.7 | 1.9 | 54.4 | 31.7 ± 0.3 | ND | ND | ||
| hSMN(14–156)·Gemin2 | 384 | 46.6 | 4.7 | 74.6 | 44.4 ± 0.2 | ND | 21 | 1.9 | |
| hSMN(14–209)·Gemin2 | 2150/371 | 52.3 | 2.8 | 86.8 | 62.1 ± 2.4 | 32 | 3.1 | ND | |
| hSMN·Gemin2 | 400 | 62.3 | 0.14 | > 670 | 200–600 | 4 | 11–12a | 4 | 5–6 |
| SMNΔ5·Gemin2 | 408 | 58.4 | 0.7 | > 670 | 200–454 | 4 | 11–12a | 4 | 5–6 |
| 0.5 | > 670 | 236–368 | ND | ND | |||||
| 0.4 | > 670 | 175–280 | ND | ND | |||||
| SMNΔ7·Gemin2 | 401 | 61.1 | 0.2 | 100 | 66.0 ± 7.9 | ND | ND | ||
| ySMN·Gemin2 | 1173/414 | 44.7 | 1.3 | 305 | 110–180 | 10.3 | 5–8 | 24 | 4–5 |
| 0.4 | 256 | 51–160 | ND | ND | |||||
| 0.13 | 164 | 54–95 | ND | ND | |||||
| MBP-hSMN(229–294) | 1736 | 48.3 | 0.9 | 128 | 200–410 | 10.2 | 10–11 | 10.2 | 7–8 |
| MBP-ySMN(39–152) | 1774 | 53.7 | 0.5 | 337 | 189.1 ± 13.6 | ND | ND | ||
a Measurements performed at 25 °C.
Expression and Purification
hSMN·Gemin2, hSMNΔ5·Gemin2, hSMNΔ7·Gemin2, and hSMN(14–156)·Gemin2 were produced by co-expression of hSMN with Gemin2(12–280) fused to a C-terminal Mxe intein (New England Biolabs) containing a chitin-binding domain and hexahistidine tags in pETDuet. Soluble hSMN·Gemin2 complex was obtained following induction in BL21(DE3) cells for 5–16 h at 20 °C. The complex was purified by nickel-nitrilotriacetic acid (Ni-NTA; Qiagen) and chitin (New England Biolabs) chromatography at 4 °C followed by intein cleavage and release from the chitin resin with 50 mm 2-mercaptoethanol. The complex was further purified on a Superdex 200 16/60 column (GE Healthcare) at 20 °C using 20 mm Tris-HCl, pH 7.5, 400 mm NaCl, 5 mm DTT. hSMN(14–209)·Gemin2 was produced using a similar protocol, except that SMN was expressed from pCDFDuet (Novagen) and hGemin2-Mxe-His6 was produced from pETDuet. hGemin2 alone was produced using the same intein construct by induction of BL21(DE3) cells for 16 h at 15 °C. After the Ni-NTA and chitin steps, hGemin2 was further purified using a MonoQ ion exchange column (GE Healthcare). The hSMN(26–51)·Gemin2 complex was reconstituted from purified hGemin2 and purified hSMN(26–51) peptide as described (23). MBP-hSMN(229–294) was expressed as a C-terminal Mxe-His6 fusion in pETDuet at 37 °C and purified on Ni-NTA, chitin, and MonoQ resins followed by Superdex-200 sizing.
ySMN (Yab8p) and yGemin2 (Yip1p) were co-expressed from pCDFDuet and pETDuet vectors, respectively, in BL21(DE3) cells at 20 °C for 5 h. yGemin2 was expressed as a C-terminal Mxe-His6 fusion, as described above for hGemin2. The complex was purified using Ni-NTA and chitin resins at 4 °C, followed by MonoQ ion exchange and Superdex-200 columns at 20 °C with sizing buffer composed of 20 mm Tris-HCl, pH 7.4, 300 mm NaCl, and 1 mm dithiothreitol (DTT). The MBP-ySMN(39–152) fusion protein was expressed at 37 °C in BL21(DE3) cells for 2.5 h and purified on amylose beads (New England Biolabs) followed by Superdex-200 sizing as described for ySMN·Gemin2. The MBP-SMN(119–152) fusion used to crystallize the YG box (plasmid 2169) was purified on amylose beads, followed by MonoQ, and Superdex-200 columns.
Biophysical analyses of hSMN·Gemin2 and SMNΔ7·Gemin2 were carried out in buffer composed of 20 mm Tris-HCl, pH 7.5, 400 mm NaCl, and 1–10 mm DTT. Analyses of SMNΔ5·Gemin2 were carried out in 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5% glycerol, and 5 mm DTT. Analyses of ySMN·Gemin2 were carried out in 10 mm Na2HPO4, 2 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, pH 7.4, and 2 mm DTT. All other analyses were carried out in 10 mm sodium-potassium phosphate, pH 7.0, 100 mm NaCl, 1–10 mm DTT. All buffers were 0.1 μm-filtered. Protein concentrations were determined using absorbance at 280 nm (A280) and extinction coefficients calculated from amino acid composition.
Size Exclusion Chromatography and Multiangle Light Scattering (SEC-MALS)
Absolute molecular masses were determined by multiangle light scattering coupled with refractive interferometric detection (Wyatt Technology Corp.) and a Superdex 200 10/300 GL column (GE Healthcare) at 20 °C, as described previously (24).
Sedimentation Equilibrium (SE) and Sedimentation Velocity (SV)
Analytical ultracentrifugation experiments were performed with an XL-A analytical ultracentrifuge (Beckman-Coulter) and a TiAn60 rotor with six-channel (for SE) or two-channel (for SV) charcoal-filled epon centerpieces and quartz windows. SE data were collected at 4 °C with detection at 280 nm for 1–3 sample concentrations. SE analyses were carried out using global fits to data acquired at multiple speeds for each concentration with strict mass conservation using the program SEDPHAT (25). Error estimates for equilibrium constants were determined from a 1,000-iteration Monte Carlo simulation. Complete SV profiles were recorded every 30 s for 200 boundaries at 45,000 rpm. Data were fit using the c(S) distribution model of the Lamm equation as implemented in SEDFIT (26). After optimizing meniscus position and fitting limits, sedimentation coefficients (S) and frictional ratios (f/f0) were determined by interative least squares fitting of the Lamm equation, with all fit root mean square deviations less than 0.01. The partial specific volume (¯υ), solvent density (ρ), and viscosity (η) were derived from chemical composition by SEDNTERP (27).
Small Angle X-ray Scattering
X-ray scattering data were measured at three different synchrotron sources: beam line F2 at the Cornell University High Energy Synchrotron Source (Ithaca, NY) (28), beam line X9 at the National Synchrotron Light Source (Upton, NY) (29), and the SIBYLS beam line at the Advanced Light Source (Berkeley, CA) (30). Data were also recorded using a rotating anode SAXS instrument as described previously (31). In all cases, the forward scattering from the samples studied was recorded on a CCD or multiwire detector and circularly averaged to yield one-dimensional intensity profiles as a function of q (q = 4πsinθ/λ, where 2θ is the scattering angle). Samples were centrifuged at 10,000 × g for 10 min at 4 °C before 0.5–20-s exposures were taken at 4 °C. Scattering from a matching buffer solution was subtracted from the data and corrected for the incident intensity of x-rays. Replicate exposures were examined carefully for evidence of radiation damage by Guinier analysis and Kratky plot analysis. Silver behenate powder was used to locate the beam center and to calibrate the sample-to-detector distance. All of the preparations analyzed were monodisperse, as evidenced by linearity in the Guinier region of the scattering data and agreement of the I(0) and Rg values determined with inverse Fourier transform analyses using the program GNOM (32). Molecular masses were derived from I(0) measurements using the forward scatter of a protein standard of known mass and concentration as a control.
Structure Determination
To obtain crystals of the ySMN YG box, we used the same strategy of screening MBP-YG box fusions that proved successful in earlier work (20). Only one fusion led to diffraction quality crystals, resulting in a structural model for residues 119–149 (residues 150–152 are poorly ordered). Crystals of MBP-YG(119–152) were grown at 21 °C by hanging drop vapor diffusion in 0.1 m sodium malonate, pH 5.6, 3.0 m ammonium sulfate, 20% sucrose. Rodlike crystals in space group P212121 grew to a maximum of 0.3 × 0.04 × 0.02 mm and appeared after 2 days. The crystals were flash-frozen and stored in liquid nitrogen prior to diffraction experiments. High resolution diffraction data were collected at the National Synchrotron Light Source X29 beam line and processed using HKL2000 (33). Initial phases were determined to 3 Å by molecular replacement using the domains of MBP as search models (Protein Data Bank code 1OMP), and the YG box dimer was fit into clear, unambiguous electron density during subsequent model building with COOT (34). Iterative rounds of refinement with REFMAC (35) yielded a final refined model at 1.7 Å with Rwork and Rfree values of 0.169 and 0.191, respectively. The final structure was consistent with a composite omit map generated using CNS (36). Four residues (0.4%) located in MBP loops are Ramachandran outliers.
Disulfide Cross-linking
ySMN has no naturally occurring cysteine residues. Four cysteine variants of ySMN were generated for these experiments: A145C (plasmid 2714), S147C (plasmid 2715), HSF-A145C (plasmid 2721), and HSF-S147C (plasmid 2722), where HSF is a 28-residue sequence containing heptahistidine, streptactin-binding, and FLAG tags (MSHHHHHHHASWSHPQFEKDYKDDDDKA). These complexes were expressed and purified as described for wild-type ySMN·Gemin2 but with a marked increase in the amount of SMN proteolysis observed upon lysis of the bacterial cells. The pColADuet vector (Novagen) was used for co-expression of untagged ySMN with the cysteine mutants (plasmid 2803). ySMN·Gemin2 complexes were incubated for 2–18 h at 4 °C in 10 mm sodium potassium phosphate, pH 7.0, 300 mm NaCl in the presence of either 10 mm DTT or 0.1 mm diamide and analyzed for disulfide cross-link formation by non-reducing SDS-PAGE.
The ability of ySMN·Gemin2 complexes to form mixed tetramers by exchange of SMN dimers was performed by Ni-NTA pull-down experiments. ySMN·Gemin2 was mixed with HSF-ySMN·Gemin2 and Ni-NTA beads at 4 °C for 30 min by nutation in 20 mm Tris-HCl, pH 7.4, 300 mm NaCl, 5 mm DTT, and 10 mm imidazole. The beads were washed with 20 column volumes of wash buffer (20 mm Tris, pH 7.4, 300 mm NaCl, 20 mm imidazole) before elution with 20 mm Tris, pH 7.4, 300 mm NaCl, 250 mm imidazole. Eluted proteins were analyzed by SDS-PAGE.
Results
Human Gemin2 Is Monomeric
In our experience, native hGemin2 and most N-terminal tagged hGemin2 constructs are poorly expressed in E. coli. A C-terminal Mxe intein fusion modified to include a hexahistidine tag performed best in our hands and was used both to express hGemin2 alone and to co-express hGemin2 with hSMN variants (Fig. 1B).
We determined the oligomeric state of purified hGemin2 using size exclusion chromatography with in-line multiangle light scattering detection (SEC-MALS). At micromolar concentration, hGemin2 has a gel filtration retention time that suggests a size roughly twice that expected for a 29-kDa monomer when compared with globular standards. However, light scattering analysis of the eluted species indicates that the protein is monomeric, with a weight average molecular mass (Mw) of 28 kDa (Fig. 2A and Table 1).
FIGURE 2.
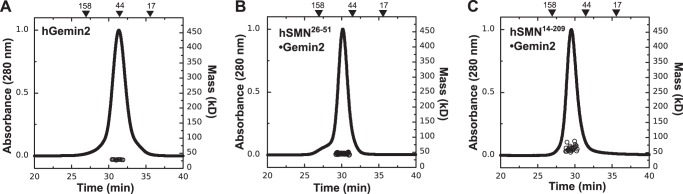
Gemin2 is not oligomeric. SEC-MALS analyses at 20 °C are shown for hGemin2 (A), hSMN(28–51)·Gemin2 (B), and hSMN(14–209)·Gemin2 (C). Although SEC elution times suggest dimeric species, Mw values from light scattering indicate a lack of oligomerization in each case. See also Table 1.
We previously showed using NMR and small angle x-ray scattering (SAXS) methods that the hGemin2 core domain (residues 95–280) adopts a partially unfolded structure in the absence of hSMN but becomes more globular when bound to the hSMN Gemin2-binding domain (23). In addition, crystal structures of hGemin2 bound to a pentamer of Sm proteins indicate that the N-terminal 86 residues of hGemin2 wrap around the partial Sm ring and therefore may not form a globular structure in the absence of Sm proteins (37, 38). These observations could explain the large apparent mass obtained for hGemin2 using gel filtration alone (39).
To examine whether hGemin2 becomes more compact and/or gains the ability to self-associate when bound to hSMN, we purified the complex formed between hGemin2 and the Gemin2-binding domain of hSMN (hG2BD; Fig. 1A) and compared its properties with those of unbound hGemin2. SEC-MALS analysis indicates that at eluted concentrations of 1–2 μm, this complex is also monomeric (Fig. 2B and Table 1). This result is recapitulated in the molecular masses determined using SAXS I(0) measurements at concentrations up to 500 μm (Table 2).
TABLE 2.
SAXS analyses of SMN·Gemin2 complexes
Rg and Dmax were derived from the inverse Fourier transform as implemented in the program GNOM (32). Results are in agreement with classical Guinier analyses.
| Complex | Mcalc | Concentration | Rg | Dmax | Mass by I(0) |
|---|---|---|---|---|---|
| kDa | μm | Å | Å | kDa | |
| hSMN(26–51)·Gemin2 | 33.7 | 504 | 28.1 | 97 | 39.1 |
| 252 | 27.5 | 94 | 38.4 | ||
| 202 | 27.2 | 91 | 37.9 | ||
| 160 | 28.1 | 94 | 38.5 | ||
| hSMN(14–156)·Gemin2 | 46.6 | 275 | 40.2 | 139 | 46.1 |
| 138 | 36.8 | 126 | 48.6 | ||
| 69 | 35.5 | 121 | 58.8 | ||
| hSMN(14–209)·Gemin2 | 52.3 | 176 | 43.7 | 147 | 59.9 |
| 88 | 43.5 | 146 | 53.1 | ||
| 23 | 44.6 | 146 | 59.9 | ||
| hSMNΔ5·Gemin2 | 58.4 | 101 | 89.3 | 293 | 434 |
| 77 | 97.8 | 311 | 438 | ||
| 63 | 99.0 | 300 | 493 | ||
| 50 | 93.4 | 316 | 525 | ||
| ySMN·Gemin2 | 44.7 | 134 | 80.8 | 275 | 208 |
| 67 | 82.8 | 277 | 174 | ||
| 34 | 83.0 | 276 | 141 | ||
| MBP-hSMN(229–294) | 48.3 | 120 | 65.2 | 228 | 491 |
| 107 | 65.9 | 232 | 480 | ||
| 72 | 65.4 | 224 | 345 | ||
| 58 | 67.6 | 237 | 298 | ||
| 47 | 64.2 | 218 | 275 |
The SEC elution time of hG2BD·Gemin2 indicates that the complex has become slightly more globular compared with hGemin2 alone but still has an apparent molecular weight that is larger than that indicated by light scattering (Table 1). Thus, hGemin2 adopts an extended conformation in solution but does not have a strong self-association affinity either alone or when bound to hSMN.
hSMN·Gemin2 Complexes Require the YG Box for Oligomerization
To determine whether regions of hSMN outside of the YG box are capable of mediating self-interaction, we purified and tested hGemin2 bound to hSMN(14–156) and hSMN(14–209). The hSMN(14–156) construct includes the Tudor domain (encoded by exon 3), and the hSMN(14–209) construct includes the region coded by SMN exon 4. Each complex was monomeric based on SEC-MALS and SAXS analyses (Fig. 2C and Tables 1 and 2). However, the Mw determined for hSMN(14–209)·Gemin2 was slightly higher than the calculated molecular mass, supporting the idea that a weak self-interaction motif may be present in the segment coded by exon 4 (40).
We further examined the hSMN(14–156)·Gemin2 complex using SV and SE. The SMN(14–156) construct includes exon 2B and the Tudor domain, both of which have been suggested as possible protein-protein interaction motifs (40, 41). The SV c(S) distribution for this complex at 21 μm gave a single, sharp peak near 2 S, consistent with a 1:1 hSMN·Gemin2 complex (Table 1). SE analysis at 10–20 μm confirmed these conclusions, because global fitting of these data were best described as a single species with a mass of 42 kDa (Table 3). Although weak interactions involving regions upstream of the YG box may exist in an oligomeric complex, we conclude that self-association of hSMN·Gemin2 is driven primarily by the YG box.
TABLE 3.
Sedimentation equilibrium analyses of SMN·Gemin2 complexes
All experiments were performed at 4 °C. Calculated complex molecular masses are for a 1:1 stoichiometry. Mfixed, fixed molecular mass of the smallest species in equilibrium models; Mfit, best fit molecular mass for single species models. S, single species; D, dimer; T, tetramer; O, octamer. NA, not applicable.
| Complex | Mcalc | Concentrations | Rotor speeds | Mfixed | Mfit | Model | Kd |
|---|---|---|---|---|---|---|---|
| kDa | μm | krpm | kDa | kDa | μm | ||
| hSMN(14–156)·Gemin2 | 46.6 | 10.5, 14.8, 19.6 | 18, 22, 25 | NA | 42.4 ± 0.3 | S | NA |
| hSMN·Gemin2 | 62.3 | 2.6, 3.8, 5.2 | 8, 10, 12 | 249.2 | NA | T-O | 0.5 ± 0.9 |
| hSMNΔ5·Gemin2 (20) | 58.4 | 3.0, 4.3 | 8, 10, 12, 16 | 116.8 | NA | D-T-O | 0.4 ± 0.6 (D-T) |
| 3.0 ± 0.6 (T-O) | |||||||
| ySMN·Gemin2 | 44.7 | 3.5, 8.8, 14.0 | 8,10, 12 | 89.4 | NA | D-T | 1.0 ± 0.9 |
| MBP-hSMN(229–294) | 48.3 | 4.9, 7.3, 8.3 | 12, 14, 16 | 193.2 | NA | T-O | 9.7 ± 0.9 |
Oligomeric Properties of Human SMN·Gemin2 Complexes
hSMN forms highly oligomeric structures both when purified from eukaryotic cells (6, 7) and when purified in recombinant forms (19). hSMN is poorly soluble in isolation, but 1:1 hSMN·Gemin2 complexes are sufficiently soluble in high ionic strength buffers to study using biophysical techniques. We therefore co-expressed hSMN and hGemin2 in E. coli and purified the complex using Ni-NTA, chitin, and Superdex-200 chromatography. A consistently higher expression level of hGemin2 using our C-terminal Mxe construct ensured saturation of the Gemin2-binding domains of SMN, leading to a stoichiometric 1:1 complex upon purification based on SDS-PAGE analyses (Fig. 1B).
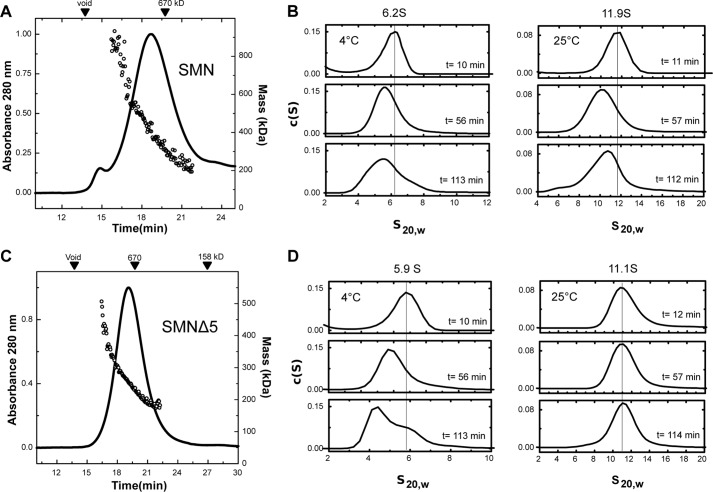
The hSMN·Gemin2 complex elutes near the void volume of Superdex 200 columns, with an apparent molecular mass of >670 kDa based on calibration with globular standards (Fig. 3A). When SEC analysis of this complex is coupled with multiangle light scattering detection, the Mw distribution calculated across the peak indicates that multiple species are present with masses that range from ∼200–550 kDa. Due to limited solubility of the complex, the concentrations of hSMN·Gemin2 eluting from the SEC column in this experiment are ≤140 nm (Table 1). Thus, at submicromolar concentrations, the majority of hSMN·Gemin2 oligomers present in solution are much smaller than what is implied by SEC alone.
FIGURE 3.
Properties of hSMN·Gemin2 complexes. A, SEC-MALS of hSMN·Gemin2 at 20 °C. The Mw values measured across the sizing peak range from ∼200 to 600 kDa. A leading shoulder contains high molecular weight aggregates. B, SV analyses of hSMN·Gemin2. The complex sediments as a 11–12 S peak at 25 °C but a 5–6 S peak at 4 °C. C, SEC-MALS of SMNΔ5·Gemin2 at 20 °C. Mw values range from ∼200–400 kDa. D, SV analyses of SMNΔ5·Gemin2. The complex sediments as a 11 S peak at 25 °C but as 4.5 S and 5.9 S species at 4 °C. See also Table 1.
SV experiments carried out at 25 °C and 4 μm concentration indicate a broad peak at 11–12 S in the c(S) distribution that is relatively stable throughout the 2-h time course of the experiment (Fig. 3B and Table 1). The broad nature of the peak is consistent with a dissociating species (42) and/or a highly flexible species that can adopt an ensemble of different conformations (43). Given the masses indicated by SEC-MALS, this sedimentation behavior implies a relatively high frictional ratio (f/f0; the ratio of the frictional coefficient to that expected for a spherical particle) and therefore an elongated or extended shape for hSMN·Gemin2 oligomers. For comparison, a globular hSMN·Gemin2 octamer (500 kDa) would have a sedimentation coefficient of 14 S under these conditions, assuming f/f0 = 1.2. An octamer with f/f0 = 1.6 would have a sedimentation coefficient of 11 S, close to that observed experimentally.
When we analyzed the sedimentation behavior of the hSMN·Gemin2 complex at 4 °C, the results were quite different (Fig. 3B and Table 1). The complex sediments over a broad distribution from 4 S to 8 S, with a peak at 5.8 S. This temperature effect is reversible, as demonstrated by sedimentation experiments that cycled between 4 and 25 °C. Our interpretation of this result is that the lower temperature and the large hydrostatic pressure (70–140 bars) present in the SV experiment destabilize the larger hSMN·Gemin2 oligomers (44), leading to a shifted distribution that favors the smaller species. A tetramer of hSMN·Gemin2 (250 kDa) with f/f0 = 1.6 has a calculated sedimentation coefficient of ∼6 S and could explain the smaller species observed at 4 °C. The temperature dependence of hSMN oligomerization implies that the larger hSMN·Gemin2 oligomers are stabilized primarily by hydrophobic interactions.
A simple model for hSMN oligomerization that emerges from SEC-MALS and SV analyses is that of a tetramer-octamer equilibrium, where the octamer is favored at 25 °C but readily dissociates to form tetramers at 4 °C. To further test this model, we performed SE experiments on 3–5 μm hSMN·Gemin2 at 4 °C. Linearized plots and global fits of the radial distributions show Mw values close to that expected for a hSMN·Gemin2 tetramer (Fig. 4A and Table 3).
FIGURE 4.
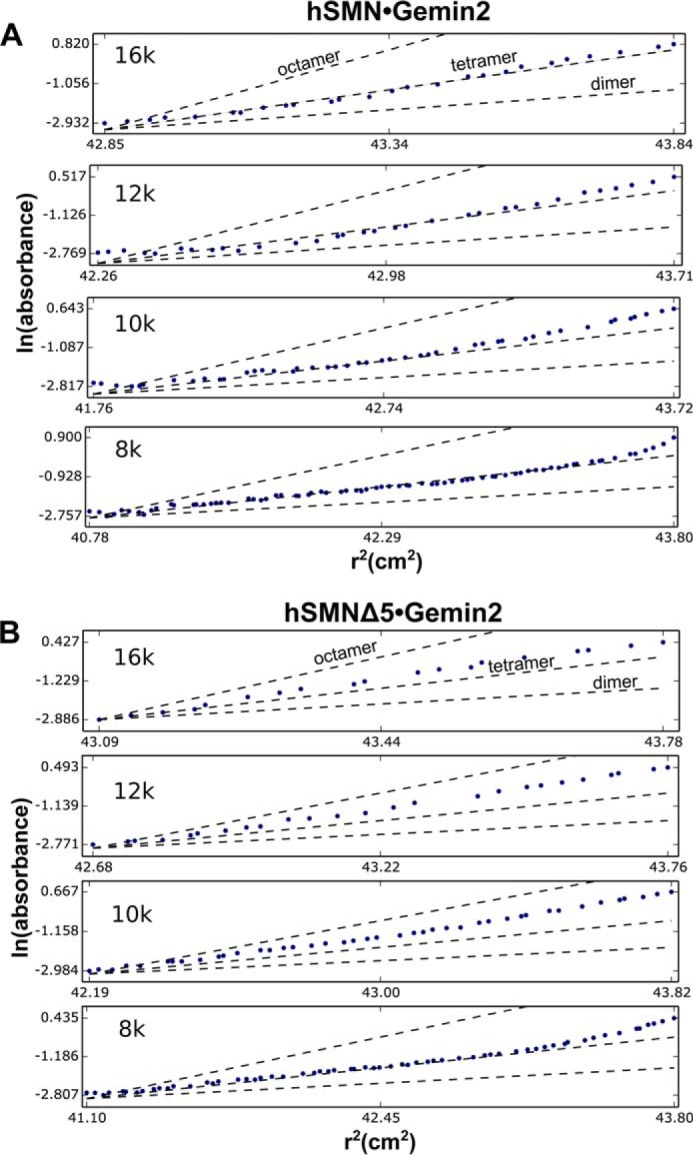
Sedimentation equilibrium analyses of hSMN·Gemin2 complexes. Linearized radial distributions are shown at four rotor speeds for hSMN·Gemin2 at 3.8 μm (A) and hSMNΔ5·Gemin2 at 3.0 μm (B). The slopes are proportional to Mw at a given value of r2. Single-species plots with calculated slopes for idealized SMN·Gemin2 dimer, tetramer, and octamer are shown for each rotor speed as dashed lines.
SMNΔ5·Gemin2 and hSMN·Gemin2 Complexes Have Similar Oligomeric Properties
To gain further insight into the nature of the oligomers formed by human SMN, we studied a hSMN·Gemin2 complex that lacks the segment coded by exon 5 of SMN. The hSMN Δexon5 (SMNΔ5) variant is a naturally occurring isoform that resembles SMN from zebrafish, fly, and worm, which lack much of this 32-residue proline-rich region (45). As we previously reported, the 58-kDa SMNΔ5·Gemin2 complex has improved solubility at physiological ionic strength and shows a reduced tendency to aggregate, making it more amenable to solution analyses than the full-length hSMN·Gemin2 complex (20).
SEC-MALS and SV analyses (Fig. 3, C and D) indicate that the oligomeric behavior of SMNΔ5·Gemin2 is similar to that observed for hSMN·Gemin2. The primary difference we observe is a shift toward lower mass averages, which would be consistent with a weakened stability of the octamer and could also be related to the reduced tendency of the SMNΔ5 complex to aggregate. To examine the concentration dependence of these masses, we measured Mw distributions at several SMNΔ5·Gemin2 complex concentrations (Table 1). The masses observed ranged from ∼200–450 kDa, indicating that the SMNΔ5·Gemin2 complex also forms oligomers spanning the dimer-octamer molecular weight range.
The SMNΔ5·Gemin2 complex shows the same temperature-dependent oligomerization that we observed for hSMN·Gemin2. The larger 11 S species observed at 25 °C dissociates to form smaller species at 4 °C (Fig. 3D). For SMNΔ5·Gemin2, two distinct species can be observed with sedimentation coefficients of 4.5 S and 5.9 S. As noted above for hSMN·Gemin2, a sedimentation coefficient of 5.9 S would be consistent with a tetramer, with f/f0 ∼1.8. The smaller 4.5 S species observed for SMNΔ5·Gemin2 could represent a dimer, with f/f0 = 1.5.
We previously reported SE results for the SMNΔ5·Gemin2 complex at 4 °C, where the centrifugation data could be fit by a dimer-tetramer-octamer equilibrium model, where Kd for dimer-tetramer is 0.5 μm and Kd for tetramer-octamer is 3 μm (Table 3). At the concentrations examined, dimers, tetramers, and octamers are all present according to this model, yielding an overall Mw close to that of a tetramer, as indicated by linearized plots of radial distributions (Fig. 4B).
To obtain additional evidence for a tetramer-octamer equilibrium, we carried out SAXS experiments for SMNΔ5·Gemin2, where the scattering intensities were extrapolated to zero scattering angle for a series of concentrations that were well above the tetramer-octamer Kd of 3 μm estimated from SE. The Mw values obtained were close to that of an octamer of SMNΔ5·Gemin2 heterodimers, in agreement with the highest masses observed in SEC-MALS experiments (Table 2). The SAXS experiments also provide an indication of the spatial size of the SMNΔ5·Gemin2 complexes in solution. Over the range of concentrations examined, the radius of gyration (Rg) is 90–100 Å, and the maximum dimension (Dmax) is ∼300 Å, explaining the large frictional coefficients inferred from SV experiments.
The SMN Δ7 Isoform Is Defective in Oligomerization
The hSMN Δexon 7 isoform (SMNΔ7) is deficient in self-interaction assays (18, 19), and the isolated YG box of this truncated variant is monomeric (20). To confirm that the SMNΔ7·Gemin2 complex is oligomerization-deficient, we co-expressed SMNΔ7 and hGemin2, purified the complex, and examined its behavior using SEC-MALS (Fig. 5). As anticipated, a heterodimer of SMNΔ7 bound to Gemin2 (∼60 kDa) is the largest species observed.
FIGURE 5.
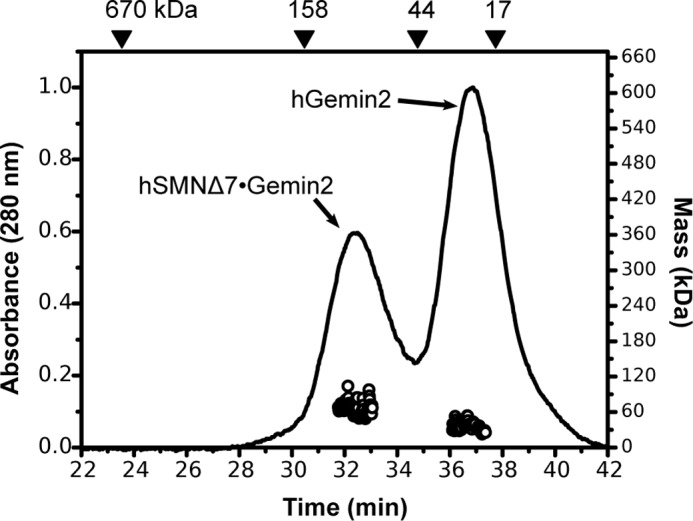
SEC-MALS analysis of SMNΔ7·Gemin2 at 20 °C. Baseline separation of SMNΔ7·Gemin2 complex from excess Gemin2 was not possible in the preparative sizing column during purification, resulting in a mixture on the analytical SEC column shown here. No species larger than the SMNΔ7·Gemin2 heterodimer is observed.
Schizosaccharomyces pombe SMN Forms Dimers and Tetramers
To determine which properties of SMN·Gemin2 are evolutionarily conserved, we studied the complex from fission yeast. ySMN is essential and oligomeric, and reduced expression adversely affects snRNP levels and RNA splicing (46–49). We co-expressed ySMN and yGemin2 in E. coli and purified the complex to homogeneity using a scheme similar to that described above for the human complex. A higher expression level of yGemin2 relative to ySMN also ensured saturation of the Gemin2-binding domains in the ySMN oligomers, resulting in a stoichiometric 1:1 complex upon purification (Fig. 6A).
FIGURE 6.
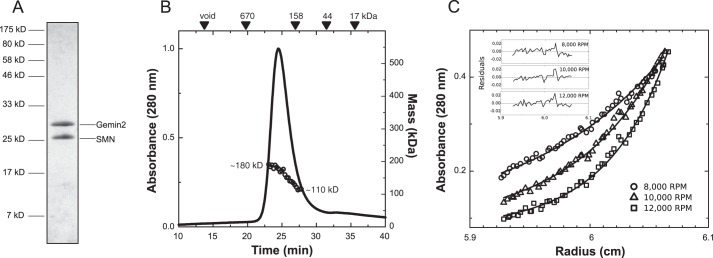
ySMN·Gemin2 forms dimers and tetramers. A, SDS-PAGE of purified ySMN·Gemin2 complex. B, SEC-MALS analysis indicates a distribution of Mw values from 110 to 180 kDa and a larger than expected Stokes radius based on elution volumes of globular standards. C, SE analysis indicates a dimer-tetramer equilibrium of SMN·Gemin2 heterodimers, with Kd = 1.0 ± 0.9 μm. The Kd was determined from a global fit of three concentrations and three rotor speeds (Table 3). One concentration is shown here.
When the ySMN·Gemin2 complex was analyzed using SEC-MALS, the Mw values observed across the peak eluting from an analytical Superdex-200 column spanned 110–180 kDa, consistent with the presence of dimers through tetramers of the 45 kDa complex (Fig. 6B). As observed for the human complex, the lack of a flat Mw plateau across the sizing peak indicates that two or more oligomeric forms are present. The SEC-MALS results also indicate a lack of higher order oligomeric species because masses higher than 180 kDa were not observed. These mass profiles are concentration-dependent, with a shift toward 180 kDa when larger amounts of complex were injected and a shift toward 100 kDa when lower amounts were injected.
In order to further investigate the nature of the ySMN·Gemin2 oligomeric distribution, we performed SE experiments (Fig. 6C and Table 3). The best global fit to the radial distributions measured at three concentrations and three rotor speeds was for a dimer-tetramer equilibrium, with Kd = 1.0 ± 0.9 μm. Alternative models, such as a single species tetramer or a monomer-trimer equilibrium of ySMN·Gemin2, led to poor fits. Thus, at concentrations near 1 μm, fission yeast SMN·Gemin2 exists as an equimolar mixture of dimers and tetramers.
Shape of ySMN·Gemin2 Oligomers
The SEC elution time of the ySMN·Gemin2 complex corresponds to a unusually large Stokes radius (Rs), based on comparison with globular protein standards (Fig. 6B). This indicates that, like the human complex, ySMN·Gemin2 does not adopt a compact, globular structure. To investigate this further, we analyzed ySMN·Gemin2 using sedimentation velocity ultracentrifugation. At a concentration of 8.7 μm, ySMN·Gemin2 sediments as a 6.4 S species at 20 °C, with an apparent molecular mass of 168 kDa and f/f0 value of 1.7 (Fig. 7A). Globular ySMN·Gemin2 tetramers (i.e. with f/f0 = 1.2) would be expected to have a sedimentation coefficient of ∼7.9. Similar results were obtained when the SV experiments were carried out at 4 °C.
FIGURE 7.
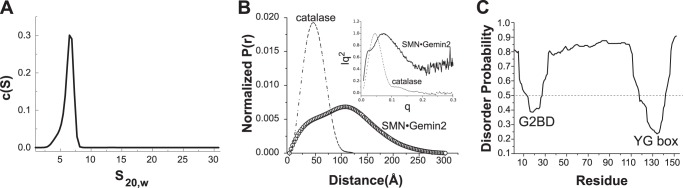
Shape of ySMN·Gemin2 oligomers. A, sedimentation velocity analysis of ySMN·Gemin2 complex at 8.7 μm. The best fit to 200 boundaries measured over 5 h gave s20,w = 6.4, Mf = 168 kDa, and f/f0 = 1.7. Similar results were obtained using only the initial 20 min of data. B, interatomic distance distribution for ySMN·Gemin2, calculated by transformation of SAXS I(q) data. P(r) for catalase, a globular, tetrameric enzyme with molecular mass of 232 kDa, is shown for comparison. The radii of gyration (Rg) and maximum dimensions (Dmax) derived from SAXS analyses for both are summarized in Table 2. Kratky plots for ySMN·Gemin2 and catalase are shown in the inset. C, prediction of natively unstructured regions for ySMN, based on the metadisorder2 algorithm (60). Values greater than 0.5 are predicted to be unstructured.
SAXS measurements for ySMN·Gemin2 at concentrations from 30 to 220 μm indicate Rg = 80–85 Å and Dmax = 275–300 Å (Table 2). The shapes of the interatomic distance distribution function and the Kratky plot for ySMN·Gemin2 both indicate a spatially extended complex that lacks overall compactness (Fig. 7B). For comparison, catalase is a tetramer with a similar molecular mass (232 kDa) but adopts a compact structure with Rg = 37 Å and Dmax = 124 Å (Fig. 7B). Together, the SEC, SV, and SAXS results indicate that the ySMN·Gemin2 complex adopts a highly extended shape.
The SAXS experiments also provide an independent estimate of Mw for the ySMN·Gemin2 complex at high concentrations, where tetramers are expected to predominate (Table 2). The average Mw derived from I(0) measurements over five independent experiments is 180 kDa, with a range of 140–210 kDa. This result agrees well with the values obtained from SEC-MALS and SE experiments and underscores the finding that fission yeast SMN·Gemin2 does not form stable oligomeric species larger than a tetramer.
Based on structures of hGemin2 bound to the Gemin2-binding domain of hSMN (23, 37, 38) and the structure of the hSMN YG box (20), the conserved N-terminal and C-terminal domains of ySMN are expected to form folded, well defined structures in the ySMN·Gemin2 complex. The central region of ySMN, however, is predicted with high probability to be natively unstructured (Fig. 7C). Thus, the large spatial extent of the ySMN·Gemin2 complex may be primarily due to an unstructured 80-amino acid segment linking the G2BD to the YG box (Fig. 1A).
The SMN YG Box Dictates Oligomeric State
To determine whether the YG box can independently mediate formation of oligomers similar to those observed for human and fission yeast SMN·Gemin2, we replaced hSMN residues 1–228 and ySMN residues 1–38 with the maltose-binding protein (MBP) and tested the oligomeric properties of the resulting fusion proteins.
SEC-MALS analyses indicate that both MBP-SMN fusions are oligomeric, with molecular mass profiles similar to those observed for the wild-type SMN·Gemin2 complexes. For MBP-hSMN(229–294), the Mw values span ∼200–400 kDa, corresponding to tetramers through octamers of the 50-kDa fusion protein (Fig. 8A). For MBP-ySMN(39–152), the fusion protein has a molecular mass close to that expected for a tetramer (Fig. 8B).
FIGURE 8.
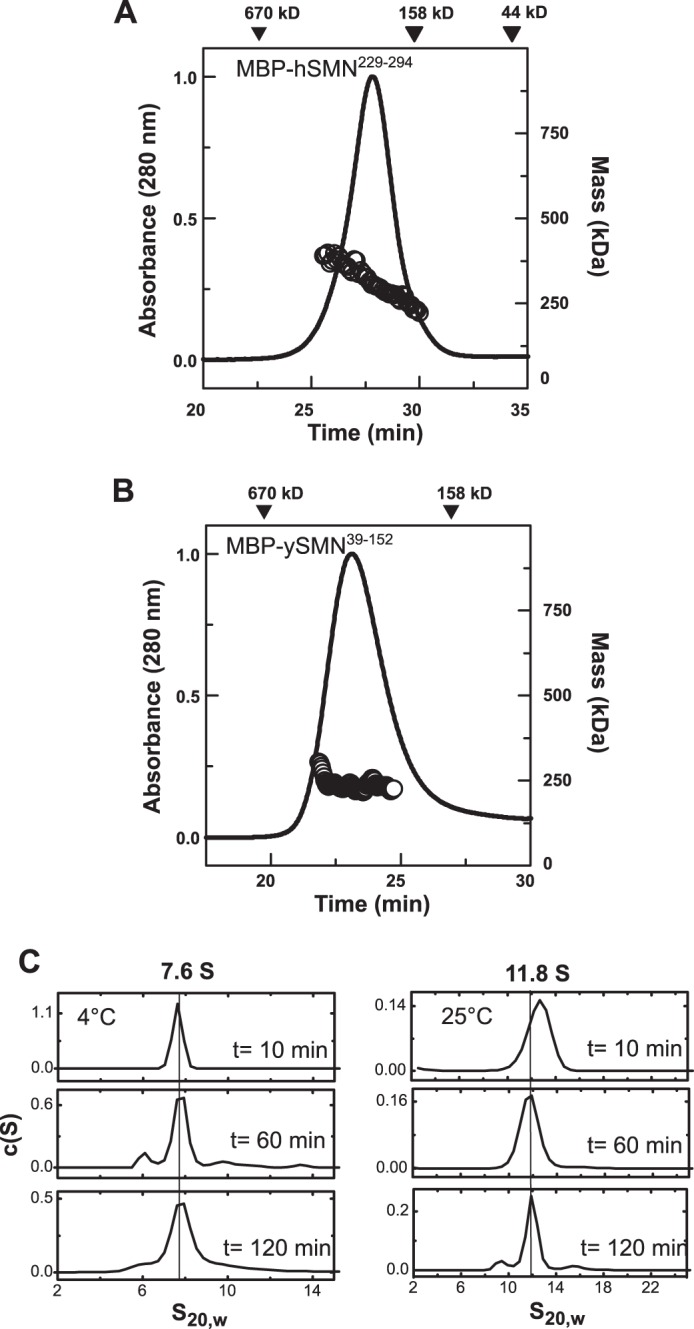
Oligomeric properties of MBP-SMN fusions. A, SEC-MALS analysis of MBP-hSMN(229–294) at 20 °C. The MBP-SMN fusion (48 kDa) forms oligomers with Mw = 200–400 kDa, spanning the tetramer to octamer range. B, SEC-MALS analysis of MBP-ySMN(39–152) at 20 °C. The 54-kDa fusion protein has an average Mw of 189 kDa across the eluted peak. C, SV analyses of MBP-hSMN(229–294) at 4 °C and 25 °C. Like hSMN·Gemin2, the MBP-hSMN fusion displays a temperature-dependent oligomerization. The larger 11–12 S oligomers observed at 25 °C are destabilized at 4 °C, where a 7.6 S species is observed.
Interestingly, the MBP-hSMN fusion shows a temperature dependence of oligomerization that is similar to that observed for hSMN·Gemin2. MBP-hSMN(229–294) sediments at 11 S at 25 °C, but at 7.6 S at 4 °C (Fig. 8C). SE analysis of MBP-hSMN(229–294) at 4 °C resulted in radial distributions that could be adequately fit by a tetramer-octamer model (Table 3). Given the strong similarities between the SMN·Gemin2 complex and the MBP-YG box fusion for both the human and fission yeast systems, we conclude that the YG box alone confers the oligomeric properties of the SMN·Gemin2 complex, and the oligomeric states neither require nor are strongly influenced by Gemin2 or other regions of SMN.
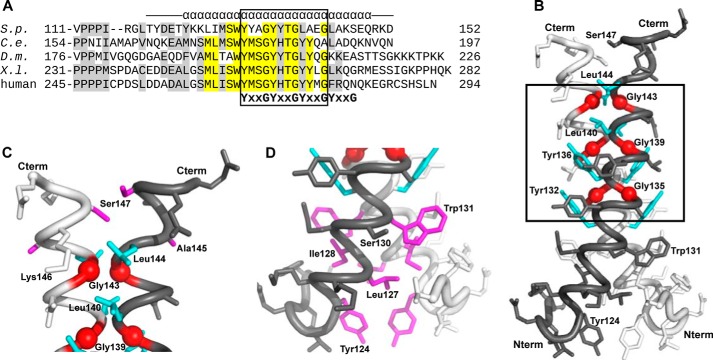
Structure of the ySMN YG Box Dimer
Although the YG box region of SMN is conserved from S. pombe to humans, there are some subtle differences. For example, the third YXXG repeat in ySMN is LXXG. The same variation is found in fly SMN (Fig. 9A). The ySMN YG box sequence is also less hydrophobic on the C-terminal end when compared with metazoan orthologs.
FIGURE 9.
Structure of the S. pombe SMN YG box. A, alignment of diverse SMN sequences, with only the C-terminal regions of the proteins shown. S.p., S. pombe; C.e., Caenorhabditis elegans; D.m., Drosophila melanogaster; X.l., Xenopus laevis. Helical secondary structure is indicated for S. pombe. The three conserved YXXG repeats found in SMN orthologs are given below the sequences. A fourth, weaker repeat is also shown. This motif defines the glycine zipper region of SMN (boxed). B, structure of the ySMN YG box dimer. Conserved glycine residues are represented by red spheres at Cα, and conserved tyrosine/leucine side chains are colored cyan. The glycine zipper region is boxed. C, close-up of the C-terminal end of the YG box, illustrating packing of Leu140, Leu144, and Lys146 side chains around Gly143. The residues used in cross-linking experiments, Ser147 and Ala145, are shown in magenta. D, close-up of the N-terminal end of the YG box. Residues involved in hydrophobic packing in this region are colored magenta.
To examine structural differences that may result from changes in the YG box sequence relative to hSMN, we crystallized the ySMN YG box dimer, determined its structure using molecular replacement, and refined the resulting model to 1.7 Å resolution. Because the YG box is poorly soluble in isolation, we used an MBP fusion to improve solubility and facilitate crystallization, as described for the hSMN YG box (20). A summary of crystallographic data and results is given in Table 4.
TABLE 4.
Crystallographic data
Rmerge = Σ|Ih − 〈Ih〉|/ΣIh, where 〈Ih〉 is the average intensity over symmetry-equivalent measurements. Rwork = Σ|Fo − Fc|/ΣFo, where summation is data used in refinement. The summation for Rfree was calculated with data (5%) not used in refinement. Numbers in parentheses represent values in the highest resolution shell.
| Parameter | Value |
|---|---|
| Data collection | |
| Resolution (Å) | 1.7 |
| Space group | P212121 |
| Cell constants (Å) | a = 96.4, b = 105.0, c = 107.7 |
| Mosaicity (degrees) | 0.33–0.48 |
| Wavelength (Å) | 1.0750 |
| Completeness (%) | 99.6 (99.6) |
| Rmerge | 0.067 (0.734) |
| Total reflections | 119,867 |
| Unique reflections | 27,872 |
| Average I/σ | 11.3 (2.89) |
| Redundancy | 7.0 (6.9) |
| Refinement | |
| Rwork | 0.169 |
| Rfree | 0.191 |
| No. of atoms | |
| Protein | 5,578 |
| Solvent | 1,077 |
| Ligand | 48 |
| Average B factors (Å2) | |
| Protein | 20.7 |
| Solvent | 26.1 |
| Ligand | 18.4/ 20.9 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.007 |
| Bond angles (degrees) | 1.322 |
| Ramachandran (%) | |
| Most favored | 96.4 |
| Allowable | 3.3 |
| Outlier | 0.4 |
As anticipated, the S. pombe YG box forms a glycine zipper helical dimer, with a right-handed crossing of helices (Fig. 9B). The first two conserved tyrosines in the YXXG repeats (Tyr132 and Tyr136) pack against the conserved glycines (Gly135 and Gly139), as observed for hSMN. However, Leu140 in the third (LXXG) repeat forms only minimal contact with Gly143. Instead, Leu144 packs against the Gly143 main chain in the opposing helical subunit (Fig. 9C).
A superposition of the human and S. pombe YG box structures indicates that they are very similar, with a root mean square deviation of 0.47 Å for Cα atoms (Met263–Phe280 of hSMN). However, the construct used for crystallization of the S. pombe YG box is longer by several residues at the N-terminal end than that present in the human YG box structure, and more of the YG box is ordered at the C-terminal end. Thus, the yeast YG box structure extends the current structural model of this region of SMN to include an additional turn leading into the glycine zipper region (Fig. 9D) and an extended helical region following the conserved YXXG repeats (Fig. 9C).
Although the ySMN YG box structure includes residues starting from Tyr119, the N-terminal end of the helix (Tyr119–Thr123) is distorted (Fig. 9, B and D). This appears to be the result of the connection to the C-terminal helix of MBP and is therefore unlikely to represent the native SMN structure. Given the pattern of conserved hydrophobic residues present in this region of SMN (Fig. 9A), it seems likely that the YG box helices are extended an additional 1–2 turns in the N-terminal direction before yeast Tyr124 (human Leu160).
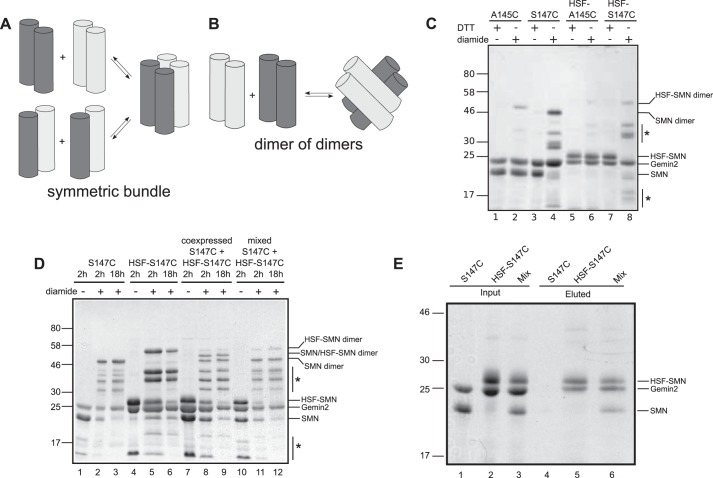
SMN Forms Dimers of Dimers
ySMN·Gemin2 exists as an equilibrium mixture of dimers and tetramers at concentrations near 1 μm. The structure shown in Fig. 9 represents the oligomeric core of dimeric SMN. To investigate how the YG boxes are organized within tetrameric SMN, we considered two different models.
The first model is that of a 4-fold symmetric helical bundle (Fig. 10A) resembling the oligomers found in glycine zipper-containing transmembrane proteins, such as the KcsA potassium channel (21). In a symmetric bundle, all four subunits are equivalent, and similar residues are involved in forming the intersubunit interfaces in the dimeric and tetrameric forms. The alternative model is a dimer of YG box dimers (Fig. 10B). This model presumes that a tightly associated dimer is the fundamental structural unit of SMN, and these dimers have the capacity to self-associate to form a tetramer. In this case, the interface used to form the dimer is distinct from that involved in forming the tetramer.
FIGURE 10.
ySMN forms dimers of dimers. A, symmetric tetramer model of SMN. Dissociation of the tetramer results in formation of all possible pairings of helical dimers. Subunits are shaded to distinguish, for example, tagged from untagged subunits. B, dimer of dimers model of SMN. Subunits within dimers do not exchange when tetramers associate and dissociate. This model assumes that dimers are highly stable and do not exchange subunits on the time scale of minutes to hours. C, disulfide cross-linking of tagged and untagged SMN mutants. Ser147 is within the dimer interface, and the S147C mutant forms disulfide cross-links that can be identified as dimers on non-reducing SDS-PAGE. Ala145 is on the surface of the YG box dimer, and A145C is not efficiently cross-linked. HSF is a 28-residue His7-Strep-FLAG tag. All four cysteine mutants were prone to degradation during expression in E. coli, resulting in a weak background of truncated SMN subunits following purification (indicated by asterisks), which are also cross-linked to form SMN dimers for S147C constructs. D, untagged and tagged S147C ySMN·Gemin2 complex were incubated separately in the presence of oxidizing agent, resulting in formation of cross-linked SMN dimers (lanes 2 and 3) and HSF-SMN dimers (lanes 5 and 6) on non-reducing SDS-PAGE. When the two are co-expressed, a cross-linked HSF-SMN·SMN heterodimer is readily formed (lanes 8 and 9). When the two complexes are incubated together, no cross-linked heterodimer is observed (lanes 11 and 12). E, when tagged and untagged S147C SMN·Gemin2 complexes are incubated together and purified on Ni-NTA beads, untagged SMN is co-purified (lane 6), indicating the formation of mixed tetramers. The untagged complex alone does not bind efficiently to the nickel beads (lane 4).
To distinguish between these models, we designed a disulfide cross-linking experiment that exploits our finding that Ser147 lies at the dimer interface (Fig. 9C). We first purified a ySMN S147C mutant as a complex with yGemin2 and found that it can be readily cross-linked by mild oxidation, resulting in ySMN dimers on non-reducing SDS-PAGE (Fig. 10C). As a control, we purified a complex containing the A145C substitution. Ala145 lies on the surface of the YG box dimer, where direct intersubunit cross-linking within a dimer should not be possible (Fig. 9C). As expected, the A145C complex is not efficiently cross-linked (Fig. 10C).
For each cysteine mutant, we also generated an N-terminal HSF-tagged variant that would allow us to identify when subunit exchange has occurred upon mixing of tagged and untagged ySMN·Gemin2 complexes. If symmetric tetramers are formed, the subunits are expected to undergo exchange to form heterodimers of tagged and untagged ySMN (Fig. 10A). If tetramers are instead formed by association of stable dimers, then only homo-dimeric cross-links should be observed within a mixed population (Fig. 10B).
As shown in Fig. 10D, we do not observe disulfide cross-linked heterodimers containing both HSF-ySMN and ySMN subunits upon mixing the respective Gemin2 complexes, even after prolonged incubations. Mixed dimers can, however, be formed by co-expression of HSF-ySMN and ySMN with Gemin2 (Fig. 10D). To confirm that mixed tetramers containing tagged and untagged ySMN homodimers are formed under our experimental conditions, we incubated a mixture of previously purified HSF-ySMN·Gemin2 and ySMN·Gemin2 with Ni-NTA beads, washed the beads, and eluted bound proteins with imidazole. ySMN is co-purified from the mixture but is not retained on the beads in the absence of HSF-ySMN (Fig. 10E). Together, these data support a dimer of dimers model but are inconsistent with a symmetric tetramer model for the ySMN·Gemin2 complex.
Discussion
Despite the well documented functional importance of SMN oligomerization, the nature of the complexes formed by SMN has been elusive. Here, we have shown that hSMN·Gemin2 exists as an equilibrium mixture of species that spans dimers through octamers. Thus, hSMN·Gemin2 forms discrete oligomers covering a much smaller molecular weight range than is implied by SEC experiments or sedimentation coefficients alone.
The fission yeast SMN complex has provided important insights into the nature of SMN·Gemin2 oligomerization. In this system, dimers and tetramers are the only species observed. The primary difference between the two systems at the level of oligomerization is that hSMN forms larger complexes that appear to involve self-association of tetramers. The strong temperature dependence of these larger species suggests a hydrophobic driving force for their formation. The more hydrophilic nature of the C-terminal end of the yeast YG box (e.g. Ala140 and Glu142; Fig. 9A) could limit the ability of ySMN·Gemin2 to form higher order oligomers and could account for the difference in oligomeric properties between the two systems. We are currently testing this hypothesis.
The simplest model for SMN oligomerization that is consistent with our experimental data is an evolutionarily conserved dimer-tetramer equilibrium, where SMN dimers are the core building blocks used to assemble larger complexes. For human SMN and, presumably, other metazoan SMN orthologs, further self-association to form larger species is favored at elevated concentrations. This type of extended oligomeric structure could be particularly relevant in the nucleus, where SMN has been found to be a component of nuclear bodies (50). The multivalent interaction scaffold resulting from SMN oligomerization could play important structural or organizational roles in these condensed states (51).
Our experiments do not exclude the possibility that hexamers of hSMN·Gemin2 could also be formed as intermediate oligomeric forms (i.e. from three dimers or from one dimer and one tetramer). A detailed understanding of how YG box dimers are arranged in the tetrameric and octameric forms of SMN would provide useful insights into whether hexamers are likely to be stable oligomeric species. Our SE data can readily accommodate such a model, but the fits cannot be clearly distinguished from those of the dimer-tetramer-octamer model.
The finding that ySMN·Gemin2 forms stable dimers that self-associate with a Kd of ∼1 μm raises a number of intriguing questions about the oligomeric form of SMN that might be required for snRNP assembly. A transcriptome and proteome study of S. pombe has revealed that ySMN is present at ∼2,500 copies/cell during logarithmic growth (52). With an average cell volume of 128 μm3 (52), this implies a concentration of 30 nm ySMN or 15 nm dimeric ySMN. Even if these SMN levels are underestimated by a factor of 10, it still appears that the concentration of ySMN·Gemin2 in fission yeast cells is well below the Kd of tetramer formation. Immunofluorescence experiments have indicated that ySMN is found in both cytoplasmic and nuclear compartments, with no indication of highly enriched localization (46). Thus, ySMN dimers are likely to be abundant species in fission yeast and could represent a functionally active species.
The situation in human cells is more complicated because the distribution of SMN between cytoplasmic and nuclear compartments often involves a high degree of localization in Cajal bodies and related nuclear structures (53). The specifics of this localization appear to vary between cell types and developmental stages of a given cell type (54). One possibility is that dimeric SMN·Gemin2 could function in the early, cytoplasmic steps of core snRNP assembly and that this could be the form that is imported into the nucleus (55). This would be consistent with the more diffuse cytoplasmic localization (and therefore lower local concentration) of SMN and with the presence of these smaller oligomers even at the concentrations used in in vitro experiments (e.g. see Fig. 3). This idea would also help to explain how SMN·snRNP core complexes are escorted through the nuclear pore complex, given that the hydrodynamic sizes of the larger SMN·Gemin2 oligomers alone are near the limits of what has been observed for nuclear import (56).
It is important to note that our investigations of hSMN·Gemin2 oligomerization have been performed in the absence of Gemin8 and Gemin3. Both proteins require the SMN YG box for binding (8, 57), and one or both proteins could interact directly with the conserved YG box dimer surface (8). Indeed, Gemin3 binding is abolished by specific mutations in the fly YG box, leading to the suggestion that Gemin3 interacts with the dimeric form of SMN (58). Thus, Gemin3 or Gemin8 could modulate oligomerization by interacting with the same residues that are used to form higher order SMN complexes. Ongoing studies in our laboratory seek to address this question.
We also note that some of the results described here conflict with the interpretations of data from previous studies. For example, Ogawa et al. (39) reported that hGemin2 is dimeric based on gel filtration and GST pull-down assays. In contrast, we have shown that hGemin2 is monomeric in isolation and when bound to truncated SMN fragments, even at extremely high concentrations. The apparent dimer inferred from gel filtration can be readily explained by the extended N terminus, and therefore non-globular overall shape, of Gemin2. It is not clear why hGemin2 is retained by GST-hGemin2 (which is expected to be dimeric) on glutathione-agarose beads (39). We have also argued here that regions of SMN outside of the YG box do not strongly influence the oligomeric state of the SMN·Gemin2 complex. However, weak interactions involving Gemin2 and/or exons 2b, 3, and 4 could still play organizational roles within oligomeric SMN complexes, particularly within SMN dimers.
A second difference from previous studies concerns the oligomeric behavior of SMNΔ5·Gemin2 and SMNΔ7·Gemin2 relative to hSMN·Gemin2. Lorson et al. (18) used a GST pull-down assay to measure the ability of these SMN isoforms to self-interact. They found that in vitro translated SMNΔ5 binds less efficiently to GST-SMNΔ5 compared with hSMN binding to GST-hSMN, and the SMNΔ7 isoform was slightly impaired relative to SMNΔ5. We have shown that SMNΔ5·Gemin2 has similar oligomeric properties compared with hSMN·Gemin2, but octamer formation appears weaker, and the complex has less of a tendency to aggregate. We have also shown that SMNΔ7·Gemin2 does not form stable oligomers, in agreement with the properties of isolated SMNΔ7 (19).
One explanation for the discrepancies between these results could lie in the different properties of SMN versus SMN·Gemin2 complexes. A second explanation could be that interpreting the pull-down experiments could be difficult, given what we now know about SMN oligomerization and the likelihood that GST-SMN will be dimeric regardless of the isoform or mutant examined. Indeed, we have found that the hSMN G279V SMA mutant is even more oligomeric than wild-type hSMN,3 yet GST pull-down assays indicated a deficiency in self-association (18). The corresponding G210V mutant in fly SMN has also been shown to be functional at self-association in Drosophila S2 cells yet has a strong (pupal lethal) in vivo phenotype (58).
The ySMN YG box dimer structure shown in Fig. 9 reinforces the view that the YG box is evolutionarily conserved at the structural level. In addition to the residues directly involved in the dimerization interface, several surface residues are also conserved and are obvious candidates for involvement in tetramer formation and/or participation in RNP assembly steps that are shared between eukaryotes that use SMN.
Recent studies of the mechanism of SMN-mediated snRNP assembly have focused on the role of Gemin2 in organizing five Sm proteins into a partial ring in preparation for snRNA binding (37, 38, 59). A key step in this process is the transfer of the Sm pentamer from pICln, where the Sm proteins are prevented from interacting directly with RNA, to the SMN complex. An assembly intermediate mimic composed of the Sm pentamer bound to Gemin2 (Fig. 1A) can be formed from free Sm proteins, Gemin2, and truncated SMN variants, but transfer of the Sm pentamer from pICln requires full-length, oligomeric SMN (38, 59). It is currently unclear how the SMN YG box might be involved in this process.
Important goals related to SMN oligomerization are therefore to understand how missense mutations found in SMA patients affect the oligomers that can be formed in SMN-containing complexes, to understand what roles the YG box plays in SMN biology beyond simply serving as an oligomerization scaffold, and to understand how those roles are affected by the mutations associated with SMA. The experimental approaches described here are well suited for directly assessing changes in size and shape of the complexes formed by SMN variants. Together with crystallographic and electron microscopy data revealing the role of Gemin2 in organizing Sm proteins (37, 38, 59), structural models of the human (20) and yeast YG box dimers should facilitate structure-function studies directed toward improving our understanding of SMN.
Author Contributions
K. G. carried out analytical ultracentrifugation, SV, SE, SAXS, and cross-linking experiments, analyzed data, and wrote the paper. R. M. crystallized the yeast YG box and determined the structure. R. S. expressed and purified most protein complexes used in the study. K. L. S. performed SEC-MALS, analytical ultracentrifugation, and SAXS experiments on the yeast SMN complex and the human Gemin2·peptide complex. N. S. N. carried out SEC-MALS and analytical ultracentrifugation experiments on human Gemin2 alone and the SMNΔ5·Gemin2 complex. G. D. V. designed the study and wrote the paper.
Acknowledgments
We are grateful to Richard Gillilan (Cornell University High Energy Synchrotron Source (CHESS)), Gregory Hura, Jane Tanamachi, Kevin Dyer, and Michal Hammel (Advanced Light Source (ALS)), and Lin Yang and Marc Allaire (National Synchrotron Light Source (NSLS)) for technical expertise and support and G. V. laboratory members for helpful discussions. CHESS is supported by National Science Foundation Grant DMR 0225180, and the MacCHESS facility is supported by National Institutes of Health Grant RR-01646. Financial support for the NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and from the National Center for Research Resources of the National Institutes of Health. X-ray scattering and diffraction technologies and their applications to the determination of macromolecular shapes and conformations at the SIBYLS beamline at ALS, Lawrence Berkeley National Laboratory, are supported in part by the Department of Energy programs Integrated Diffraction Analysis Technologies (IDAT) and Molecular Assemblies Genes and Genomics Integrated Efficiently (MAGGIE) under Contract DE-AC02-05CH11231 with the United States Department of Energy.
The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 4RG5) have been deposited in the Protein Data Bank (http://wwpdb.org/).
K. Gupta, N. Ninan, and G. D. Van Duyne, unpublished observations.
- SMN
- survival motor neuron
- ySMN and hSMN
- yeast and human SMN, respectively
- SMA
- spinal muscular atrophy
- SAXS
- small angle x-ray scattering
- SE
- sedimentation equilibrium
- SV
- sedimentation velocity
- SEC
- size exclusion chromatography
- MALS
- multiangle light scattering
- MBP
- maltose-binding protein
- Ni-NTA
- nickel-nitrilotriacetic acid
- yGemin2 and hGemin2
- yeast and human Gemin2, respectively.
References
- 1. Chari A., Paknia E., Fischer U. (2009) The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol. 21, 387–393 [DOI] [PubMed] [Google Scholar]
- 2. Li D. K., Tisdale S., Lotti F., Pellizzoni L. (2014) SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol. 32, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderton R. S., Meloni B. P., Mastaglia F. L., Boulos S. (2013) Spinal muscular atrophy and the antiapoptotic role of survival of motor neuron (SMN) protein. Mol. Neurobiol. 47, 821–832 [DOI] [PubMed] [Google Scholar]
- 4. Fallini C., Bassell G. J., Rossoll W. (2012) Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 1462, 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanchez G., Dury A. Y., Murray L. M., Biondi O., Tadesse H., El Fatimy R., Kothary R., Charbonnier F., Khandjian E. W., Côté J. (2013) A novel function for the survival motoneuron protein as a translational regulator. Hum. Mol. Genet. 22, 668–684 [DOI] [PubMed] [Google Scholar]
- 6. Paushkin S., Gubitz A. K., Massenet S., Dreyfuss G. (2002) The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14, 305–312 [DOI] [PubMed] [Google Scholar]
- 7. Carissimi C., Saieva L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L. (2006) Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 281, 8126–8134 [DOI] [PubMed] [Google Scholar]
- 8. Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., Fischer U. (2007) A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 282, 5825–5833 [DOI] [PubMed] [Google Scholar]
- 9. Burghes A. H., Beattie C. E. (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 10, 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Le Paslier D., Frezal J., Cohen D., Weissenbach J., Munnich A., Melki J. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165 [DOI] [PubMed] [Google Scholar]
- 11. Crawford T. O., Pardo C. A. (1996) The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 3, 97–110 [DOI] [PubMed] [Google Scholar]
- 12. Monani U. R., Lorson C. L., Parsons D. W., Prior T. W., Androphy E. J., Burghes A. H., McPherson J. D. (1999) A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 8, 1177–1183 [DOI] [PubMed] [Google Scholar]
- 13. Lorson C. L., Hahnen E., Androphy E. J., Wirth B. (1999) A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 96, 6307–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tiziano F. D., Melki J., Simard L. R. (2013) Solving the puzzle of spinal muscular atrophy: what are the missing pieces? Am. J. Med. Genet. A 161A, 2836–2845 [DOI] [PubMed] [Google Scholar]
- 15. Nurputra D. K., Lai P. S., Harahap N. I. F., Morikawa S., Yamamoto T., Nishimura N., Kubo Y., Takeuchi A., Saito T., Takeshima Y., Tohyama Y., Tay S. K. H., Low P. S., Saito K., Nishio H. (2013) Spinal muscular atrophy: from gene discovery to clinical trials. Ann. Hum. Genet. 77, 435–463 [DOI] [PubMed] [Google Scholar]
- 16. Hamilton G., Gillingwater T. H. (2013) Spinal muscular atrophy: going beyond the motor neuron. Trends Mol. Med. 19, 40–50 [DOI] [PubMed] [Google Scholar]
- 17. Battle D. J., Kasim M., Wang J., Dreyfuss G. (2007) SMN-independent subunits of the SMN complex: identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 282, 27953–27959 [DOI] [PubMed] [Google Scholar]
- 18. Lorson C. L., Strasswimmer J., Yao J. M., Baleja J. D., Hahnen E., Wirth B., Le T., Burghes A. H., Androphy E. J. (1998) SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 19, 63–66 [DOI] [PubMed] [Google Scholar]
- 19. Pellizzoni L., Charroux B., Dreyfuss G. (1999) SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 11167–11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin R., Gupta K., Ninan N. S., Perry K., Van Duyne G. D. (2012) The survival motor neuron protein forms soluble glycine zipper oligomers. Structure 20, 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S., Jeon T.-J., Oberai A., Yang D., Schmidt J. J., Bowie J. U. (2005) Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 14278–14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Green M. R., Sambrook J. (2012) Molecular Cloning: A Laboratory Manual, 4th Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23. Sarachan K. L., Valentine K. G., Gupta K., Moorman V. R., Gledhill J. M., Jr., Bernens M., Tommos C., Wand A. J., Van Duyne G. D. (2012) Solution structure of the core SMN-Gemin2 complex. Biochem. J. 445, 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta K., Diamond T., Hwang Y., Bushman F., Van Duyne G. D. (2010) Structural properties of HIV integrase: lens epithelium-derived growth factor oligomers. J. Biol. Chem. 285, 20303–20315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R. A., Rouault T. A., Schuck P. (2004) Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 326, 234–256 [DOI] [PubMed] [Google Scholar]
- 26. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laue T. M., Shah B., Ridgeway T. M., Pelletier S. L., Harding S. E., Rowe A. J., Horton J. C. (1992) in Biochemistry and Polymer Science, pp. 90–125, Royal Society of Chemistry, London, UK [Google Scholar]
- 28. Nielsen S. S., Møller M., Gillilan R. E. (2012) High-throughput biological small-angle x-ray scattering with a robotically loaded capillary cell. J. Appl. Crystallogr. 45, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allaire M., Yang L. (2011) Biomolecular solution x-ray scattering at the National Synchrotron Light Source. J. Synchrotron Radiat. 18, 41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hura G. L., Menon A. L., Hammel M., Rambo R. P., Poole F. L., 2nd, Tsutakawa S. E., Jenney F. E., Jr., Classen S., Frankel K. A., Hopkins R. C., Yang S. J., Scott J. W., Dillard B. D., Adams M. W. W., Tainer J. A. (2009) Robust, high-throughput solution structural analyses by small angle x-ray scattering (SAXS). Nat. Methods 6, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta K., Brady T., Dyer B. M., Malani N., Hwang Y., Male F., Nolte R. T., Wang L., Velthuisen E., Jeffrey J., Van Duyne G. D., Bushman F. D. (2014) Allosteric inhibition of human immunodeficiency virus integrase: late block during viral replication and abnormal multimerization involving specific protein domains. J. Biol. Chem. 289, 20477–20488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Semenyuk A., Svergun D. (1991) GNOM: a program package for small-angle scattering data processing. J. Appl. Cryst. 24, 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 37. Zhang R., So B. R., Li P., Yong J., Glisovic T., Wan L., Dreyfuss G. (2011) Structure of a Key Intermediate of the SMN Complex Reveals Gemin2's Crucial Function in snRNP Assembly. Cell 146, 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grimm C., Chari A., Pelz J.-P., Kuper J., Kisker C., Diederichs K., Stark H., Schindelin H., Fischer U. (2013) Structural basis of assembly chaperone-mediated snRNP formation. Mol. Cell 49, 692–703 [DOI] [PubMed] [Google Scholar]
- 39. Ogawa C., Usui K., Aoki M., Ito F., Itoh M., Kai C., Kanamori-Katayama M., Hayashizaki Y., Suzuki H. (2007) Gemin2 plays an important role in stabilizing the survival of motor neuron complex. J. Biol. Chem. 282, 11122–11134 [DOI] [PubMed] [Google Scholar]
- 40. Young P. J., Man N. T., Lorson C. L., Le T. T., Androphy E. J., Burghes A. H., Morris G. E. (2000) The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding.[erratum appears in Hum. Mol. Genet. 2001 Jan 1;10(1):88]. Hum. Mol. Genet. 9, 2869–2877 [DOI] [PubMed] [Google Scholar]
- 41. Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M. (2001) SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8, 27–31 [DOI] [PubMed] [Google Scholar]
- 42. Scott D. J., Harding S. E., Rowe A. J. (2005) Analytical Ultracentrifugation: Techniques and Methods, pp. 1–23, Royal Society of Chemistry, London, UK [Google Scholar]
- 43. MacRaild C. A., Hatters D. M., Lawrence L. J., Howlett G. J. (2003) Sedimentation velocity analysis of flexible macromolecules: self-association and tangling of amyloid fibrils. Biophys. J. 84, 2562–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kegeles G., Rhodes L., Bethune J. L. (1967) Sedimentation behavior of chemically reacting systems. Proc. Natl. Acad. Sci. U.S.A. 58, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gennarelli M., Lucarelli M., Capon F., Pizzuti A., Merlini L., Angelini C., Novelli G., Dallapiccola B. (1995) Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem. Biophys. Res. Commun. 213, 342–348 [DOI] [PubMed] [Google Scholar]
- 46. Paushkin S., Charroux B., Abel L., Perkinson R. A., Pellizzoni L., Dreyfuss G. (2000) The survival motor neuron protein of Schizosacharomyces pombe: conservation of survival motor neuron interaction domains in divergent organisms. J. Biol. Chem. 275, 23841–23846 [DOI] [PubMed] [Google Scholar]
- 47. Hannus S., Bühler D., Romano M., Seraphin B., Fischer U. (2000) The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet. 9, 663–674 [DOI] [PubMed] [Google Scholar]
- 48. Owen N., Doe C. L., Mellor J., Davies K. E. (2000) Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein.[erratum appears in Hum. Mol. Genet. 2000 Apr 12;9(7):1142]. Hum. Mol. Genet. 9, 675–684 [DOI] [PubMed] [Google Scholar]
- 49. Campion Y., Neel H., Gostan T., Soret J., Bordonné R. (2010) Specific splicing defects in S. pombe carrying a degron allele of the survival of motor neuron gene. EMBO J. 29, 1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Q., Dreyfuss G. (1996) A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15, 3555–3565 [PMC free article] [PubMed] [Google Scholar]
- 51. Hebert M. D., Matera A. G. (2000) Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11, 4159–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bähler J. (2012) Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carvalho T., Almeida F., Calapez A., Lafarga M., Berciano M. T., Carmo-Fonseca M. (1999) The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 147, 715–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young P., Le T. T., thi Man N., Burghes A. H., Morris G. E. (2000) The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 256, 365–374 [DOI] [PubMed] [Google Scholar]
- 55. Narayanan U., Achsel T., Lührmann R., Matera A. G. (2004) Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell 16, 223–234 [DOI] [PubMed] [Google Scholar]
- 56. Panté N., Kann M. (2002) Nuclear pore complex is able to transport macromolecules with diameters of ∼39 nm. Mol. Biol. Cell 13, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Charroux B., Pellizzoni L., Perkinson R. A., Shevchenko A., Mann M., Dreyfuss G. (1999) Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 147, 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Praveen K., Wen Y., Gray K. M., Noto J. J., Patlolla A. R., Van Duyne G. D., Matera A. G. (2014) SMA-causing missense mutations in survival motor neuron (SMN) display a wide range of phenotypes when modeled in Drosophila. PLoS Genet. 10, e1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chari A., Golas M. M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. (2008) An assembly chaperone collaborates with the SMN complex to generate spliceosomal snRNPs. Cell 135, 497–509 [DOI] [PubMed] [Google Scholar]
- 60. Kozlowski L. P., Bujnicki J. M. (2012) MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 13, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]