Background: Plasmid pLS20 conjugates only during growth of cells.
Results: DNA binding activity of repressor protein Rco is repressed through binding of Rap protein, which is counteracted by a Phr peptide.
Conclusion: Plasmid-encoded Rap and Phr directly restrict the timing of conjugation to exponential growth, leading to bistable expression of conjugation genes.
Significance: A Rap/Phr/repressor system mediates the unusual timing of plasmid conjugation.
Keywords: bacterial conjugation, gene regulation, plasmid, protein-nucleic acid interaction, protein-protein interaction, quorum sensing, Bacillus subtilis, Rap-Phr module
Abstract
Conjugation of plasmid pLS20 from Bacillus subtilis is limited to a time window between early and late exponential growth. Genetic evidence has suggested that pLS20-encoded protein RcoLS20 represses expression of a large conjugation operon, whereas Rap protein RapLS20 relieves repression. We show that RapLS20 is a true antirepressor protein that forms dimers in vivo and in vitro and that it directly binds to the repressor protein RcoLS20 in a 1:1 stoichiometry. We provide evidence that RapLS20 binds to the helix-turn-helix-containing domain of RcoLS20 in vivo, probably obstructing DNA binding of RcoLS20, as seen in competitive DNA binding experiments. The activity of RapLS20 in turn is counteracted by the addition of the cognate PhrLS20 peptide, which directly binds to the Rap protein and presumably induces a conformational change of the antirepressor. Thus, a Rap protein acts directly as an antirepressor protein during regulation of plasmid conjugation, turning on conjugation, and is counteracted by the PhrLS20 peptide, which, by analogy to known Rap/Phr systems, is secreted and taken back up into the cells, mediating cell density-driven regulation. Finally, we show that this switchlike process establishes a population heterogeneity, where up to 30% of the cells induce transcription of the conjugation operon.
Introduction
Horizontal gene transfer is a central driving force of bacterial genome evolution and plasticity. Beside transformation and transduction, conjugation is the major determinant in the spread of genetic information among bacteria, endowing recipient cells with new traits, such as catabolic pathways, antibiotic resistance, or even virulence (1, 2).
Conjugative elements are either carried on plasmids or maintained in genomes as integrative conjugative elements (ICEs)2 (2). Transfer of ssDNA occurs via type IV secretion systems (3) and needs to be tightly regulated to minimize the metabolic burden on the host caused by transcription and synthesis of conjugation genes/proteins and the conjugational transfer itself. Therefore, conjugation genes are either kept in a default OFF state and need to be induced by signaling molecules to be switched ON, or conjugation genes are constitutively expressed but limited in their expression in order to not increase the fitness costs of the host. Constitutive systems, such as those of the IncF plasmid family, integrate transcriptional cues of plasmid and host factors as well as environmental stimuli to control the expression of their transfer region. Inducible conjugation systems either rely on sensing of phenolic compounds like the Ti-plasmids from Agrobacterium tumefaciens or on quorum-sensing systems using homoserine lactones as signaling molecules in Gram-negative bacteria or signaling peptides in Gram-positive bacteria (4). Among the peptide-regulated systems, regulation of conjugation of the Enterococci plasmids pCF10 and pAD1 has been extensively studied. Conjugative transfer of these plasmids is controlled through the ratio of two peptides; a plasmid-encoded peptide (iCF10/iAD1) promotes the inhibitory function of the master regulator (TraA/PrgX) by keeping the conjugation operon in an OFF state, and a chromosomally encoded peptide (cCF10/iAD1) found in many Enterococci strains relieves transcriptional repression of the conjugation operon through competitive binding to the master regulator at high concentrations. Thus, in the presence of plasmid-free cells, the conjugation operon is turned ON, whereas at high donor cell densities, the operon is turned OFF. In Bacillus subtilis, the quorum-sensing RapI-PhrI module regulates the transfer of the mobile genetic element ICEBs1, which can also be activated by the RecA-dependent SOS response. In contrast to PrgX, RapI activates excision and conjugation of ICEBs1 through induction of cleavage of the master regulator ImmR, which is inhibited at high concentrations of PhrI (5).
Rap proteins have been best characterized for their role in the regulation of developmental processes, such as sporulation and competence in B. subtilis (6). Phr peptides are secreted and are generally modified; their gene is located downstream of the gene encoding for their cognate Rap protein, which in most cases is a phosphatase. Dephosphorylation of aspartate response regulators prevents the activation of the master regulator of sporulation, Spo0A. Spo0A receives a phosphoryl group through a relay including the response regulator Spo0F, which the Rap protein acts upon. Increasing levels of processed Phr peptides are taken back up into the cells as cell density rises, and during the transition to stationary phase, enough peptide has accumulated to efficiently bind to the Rap protein, displacing it from the response regulator, such that the phosphorelay can proceed toward initiation of sporulation. During competence development, ComP phosphorylates the response regulator ComA in response to high cell density. Phosphorylation of ComA triggers its DNA binding activity, which leads to the expression of several genes, ultimately resulting in the expression of late competence genes. Unlike sporulation-inhibiting Rap proteins, Rap proteins RapF and RapC act on ComA not via inhibition of its function through dephosphorylation, but through allosterically blocking its DNA-binding domain (7–9) and thereby delaying activation of competence genes until a high concentration of the corresponding Phr is reached.
Recently, it was found that a Rap/Phr-like module also plays a role in the regulation of transfer of the conjugative plasmid pLS20 from B. subtilis natto (10), which shows an unusual activity for conjugation; whereas ICEBs1 becomes active during the transition from exponential growth to stationary phase (11), pLS20 conjugates with some delay after cells have been resuspended into fresh medium and shuts down DNA transfer during late exponential phase, well before cells cease to grow (12). Interestingly, several components of the conjugation machinery of pLS20 assemble and disassemble at a single cell pole or a single site at the lateral membrane, in parallel with conjugation activity (13). How these kinetics are achieved has been hinted at through the identification of a repressor-like protein, RcoLS20, whose deletion results in higher and constitutive conjugation activity, whereas its overproduction leads to a reduction in conjugation efficiency (10). Induction of pLS20 conjugation is also achieved through overexpression of a plasmid-encoded Rap protein, RapLS20, which may be counteracted by the expression of its cognate peptide PhrLS20, encoded just downstream of the Rap protein-encoding gene. We show that RcoLS20-mediated repression of pLS20 transfer is relieved through direct interaction with RapLS20, which is blocked in the presence of PhrLS20. Thus, although the Rap-Phr-mediated regulation is similar to already known mechanisms involved in sporulation or competence development, we show that a Rap-Phr quorum-sensing system can also be applied for biotechnological applications needed during ongoing growth.
Materials and Methods
Bacterial Strains, Plasmids, and Strain Construction
Bacterial strains, plasmid, and primers used in this study are listed in Tables 1 and 2. Escherichia coli strain DB3.1 was used to propagate ccdB-containing plasmids, Mach1 cells were used for construction of plasmids, and Rosetta (DE3) 2 or BL21 (DE3) 2 cells were used for expression of proteins in E. coli. E. coli and B. subtilis cells were grown in LB medium at 30 or 37 °C, supplemented with the appropriate antibiotics at the following final concentrations: 100 μg ml−1 ampicillin, 5 or 15 μg ml−1 chloramphenicol, (B. subtilis or E. coli), 10 or 50 μg ml−1 kanamycin (B. subtilis or E. coli), 25 μg ml−1 lincomycin, 1 μg ml−1 erythromycin, 100 μg ml−1 spectinomycin, and 5 μg ml−1 phleomycin.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source/Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DB 3.1 | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (strR) endA1 nupG fhuA::IS2 | Invitrogen |
| Rosetta (DE3) 2 | F− ompT hsdSB(RB− mB−) gal dcm λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) pLysSRARE (cm) | Novagen |
| Mach1 | ΔrecA1398 endA1 tonA Φ80ΔlacM15 ΔlacX74 hsdR (rK− mK+) | Invitrogen |
| BL21 (DE3) | F− ompT hsdSB (RB− mB−) gal dcm | Invitrogen |
| B. subtilis | ||
| PY79 pLS20neo | Ref. 13 | |
| 1012sigW | sigW::bleo | Gift of T. Wiegert |
| 1012rsiW | rsiW::spec | Ref. 32 |
| 1012clpX | clpX::cm amyE::PIPTG-rsiW mls | Ref. 33 |
| TCR16 | PY79 pLS20neo sigW::bleo | This study |
| TCR17 | PY79 pLS20neo rsiW::spec | This study |
| TCR18 | PY79 pLS20neo amyE::PIPTG-rsiW mls | This study |
| TCR19 | PY79 pLS20neo amyE::Pxyl-rco spec | This study |
| TCR20 | PY79 thrC::Pc-mcherry mls | This study |
| TCR21 | PY79 pLS20neo thrC::Pc-mcherry mls | This study |
| PG300 | PY79 Pctc::gfp cm | Laboratory collection |
| Plasmids | ||
| pENTR-D-TOPO | attL1 ccdB attL2 kan | Invitrogen |
| pHGWA | PT7-his6-rfa* cm amp | Ref. 34 |
| pNDIV | PBAD-divIVA-dest14 amp cm | Ref. 15 |
| pNGFP | PT7-gfp-dest17 cm | Ref. 15 |
| pCGFP | PT7-dest42-gfp cm | Ref. 15 |
| pDESTBs1 | Pxyl-dest42-yfp spec amp 5′amyE 3′amyE | This work |
| pTCR16 | Pxyl-rco yfp spec amp 5′amyE 3′amyE | This work |
| pTCR17 | pENTR-D-TOPO rco kan | This work |
| pTCR18 | pENTR-D-TOPO rap kan | This work |
| pTCR19 | pENTR-D-TOPO ΔC-rco kan | This work |
| pTCR20 | pENTR-D-TOPO ΔN-rco kan | This work |
| pTCR21 | pHGWA PT7-his6-rco amp | This work |
| pTCR22 | pHGWA PT7-his6-rap amp | This work |
| pTCR23 | pNGFP PT7-gfp-rap cm | This work |
| pTCR24 | pNGFP PT7-gfp-ΔC-rco cm | This work |
| pTCR25 | PBAD-divIVA-rap amp | This work |
| pTCR26 | PBAD-divIVA-rco amp | This work |
| pTCR27 | PBAD-divIVA-ΔC-rco amp | This work |
| pTCR28 | PBAD-divIVA-ΔN-rco amp | This work |
| pTCR29 | pDG1664 Pc-mcherry mls | This work |
a amp, ampicillin-resistant; cm, chloramphenicol-resistant; bleo, phleomycin-resistant; spec, spectinomycin-resistant; mls, lincomycin-erythromycin-resistant; kan/neo, kanamycin/neomycin-resistant. rfa*, reading frame cassette A (Invitrogen).
TABLE 2.
Oligonucleotides used in this study
Restriction sites are underlined.
| Oligonucleotide sequence (5′–3′) | |
|---|---|
| RT-PCR analysis | |
| 1_FW | AAGAGCTTCCATACTCTAAAACAT |
| 1_RV | TAAGGATCCTTTCTAACAAGTATTTAGAGAGATCAA |
| 2_FW | TCCGTTGCATTGTCTAACACAC |
| 2_RV | CACCATGACTGAAAAAGATGTTTTAGAGTAT |
| 3_FW | TAAGCATGCCTACATTAGAAGCAGAACACTGC |
| 3_RV | AGCAGCAATAAATCATCAAGCA |
| 4_FW | TAAGCTAGCAAGGAGATTCCTAGGATGGATGAAAAAGCTATTTTAAGTGATGA |
| 4_ RV | TAAGCATGCCTATACCTTTAATTCACCATTCTTCT |
| 5_FW | TAAGGGCCCTGGATTTGAATAAAGGTATTGAAG |
| 5_RV | TTTGATGTAAGCTGATTCATG |
| 6_FW | TAAGGGCCCTCGATTTTAAAGTACCTGAAGAG |
| 6_RV | TAAATCGATCATTTTCTCACCTCGATTTTCT |
| 7_FW | ATCGAATTCTTGGTAAGTGCCAGCTTTGG |
| 7_RV | TAAGGGCCCCAGCCGATAGACGATTTATATACA |
| 8_FW | ACTGGGCCCACACAAAAGATATTCATAGCTG |
| 8_RV | TAATTTTACCTCCATTAAATTGTCG |
| 9_FW | CACCATGAAGAAAAAGGTATATTCGCTAG |
| 9_RV | TCGAATCAACGATAAAGCAG |
| 10_FW | CACCATGCTTGATGGAGCAGTAATG |
| 10_RV | GTTGTGTTGCTCTGAAGTTGC |
| 11_FW | CACCATGGCTGCTACAAAAGCC |
| 11_RV | TTAGATACCCCCACTTTCATTTAG |
| 12_FW | CACCATGCCAGATAATATTGTAGATATGCT |
| 12_RV | AAGCTTTTTGTAAGAGGCTACTAG |
| 13_FW | ATGGAATTCAACTTTACATCAACGAGATACA |
| 13_RV | CAAGCTCATTTTGTTTGCCA |
| 14_FW | CCTATCGATCGAACTAAAAGGACTTGAAAG |
| 14_RV | ATGGAATTCAACTTTACATCAACGAGATACA |
| 15_FW | ACTGGGCCCGTTTTGATAATAAGATTGCTC |
| 15_RV | ATCGTCAGCTTGGCCAATGTAATCTGT |
| 16_FW | ATATATGATTCAAAGTCAGTAAGA |
| 16_RV | AATAAAGTTATTCATACCATGTAAGATCTT |
| 17_FW | ATGGAATTCTTGATCGAATCAACTGTGTATAT |
| 17_RV | ATCTTCTGTGACACTCATCAAGTATAA |
| 18_FW | GAATTCAAATATTTAATGCGCTCAG |
| 18_RV | CTGAAGCAAAGTTTGCAACAC |
| 19_FW | ACTGGGCCCGGATGTATGGTGATCAGC |
| 19_RV | GAGGAATATCTTGTAGAGAATCTGTGATC |
| 20_FW | TCCAATGAAAGCAAAATTTCTTC |
| 20_RV | TCCTTCATTTACTCTGATGAATTC |
| 21_FW | CACCATGCCGGATCTCAACATC |
| 21_RV | CTACCGGCTCTGTTCCATTTC |
| 22_FW | TAACAATGCTTTGAATAATATTCG |
| 22_RV | TTAATGCTGTTCACCAACATAAAA |
| 23_FW | CACCATGATTTTTAAACTAGATCACTATATCAATG |
| 23_RV | GAAAGCTGTTTTTCTCTTTATCAAT |
| 24_FW | ATGATGTGCTGAAGGTCC |
| 24_RV | TTATTGAGTGTTGCAACAAGA |
| 25_FW | TATAGTTCAAAATCCGACTCTACAGTT |
| 25_RV | TCTGATTTCGTTGTTTCTGATACTC |
| 26_FW | TGAGATCAATTATGGAGAAAAATCC |
| 26_RV | AACGAATTCGTTAAAAAGGAAGGTCATCATCACTAAC |
| 27_FW | ATGTTGAATAGAGTAGTTCTAGTAGGA |
| 27_RV | TCCTTATAAAGTTTAATCGGTATTCTT |
| 28_FW | ATGTTGAATAGAGTAGTTCTAGTAGGA |
| 28_RV | GGTAATGGTCAAGGTTCCC |
| Amplification of genes | |
| Rco_FW | CACCGTGGGCAATAGAGAGCAAT |
| Rco +STOP_RV | CTAGTCCTTTTTTAATTTCATGTATTC |
| ΔC-Rco_FW | CACCGTGGGCAATAGAGAGCAATT |
| ΔC-Rco_RV | TTATTCTATATTAGGAATGGAGAGCG |
| ΔN-Rco_FW | CACCATGCTTCAAATAATGGAGTATATAGC |
| Rap_FW | CACCATGTTGTCCAAAGTAAAAAAAGTACC |
| Rap_RV | TCATCCTAACGCCTCCGT |
| DEST42_KpnI | TAAGGTACCACAAGTTTGTACAAAAAAGCTGAA |
| DEST42_XhoI | TAACTCGAGCACCACTTTGTACAAGAAAGCT |
| Pc_FW | TAAGGATCCTTATACCACCTCGCAAAATAAA |
| Pc_RV | TAAGCTAGCCATTTCCTCTCCTCCTAAATTTTCAATCAGTGTAAAGA |
| mcherry_FW | TAAGCTAGCATGGTGAGCAAGGGC |
| mcherry_RV | TAAGAATTCTTACTTGTACAGCTCGTCCA |
| PCR fragments used for electrophoretic mobility shift assays | |
| PUTR_FW | CACCATGTATGAATACAATAAATGGGG |
| PUTR _RV | TCAGTTAATAATAAGTTTAGTAAAAACAGG |
| Pcds57_FW | TTTTAGAAATTGTAAGGGAGGC |
| Pcds57 _RV | GCCTCCCTTGTTTCCAGTA |
| Pcds80_FW | ACCCAAAACAGCTGCTATT |
| Pcds80_RV | AAAATTTCACCAGTGAGAAAAC |
| Pcds82_FW | ATCATCTTCTCCCCCCAA |
| Pcds82_RV | GAAAAAAACCTCCTTTAATATGGTAA |
| qRT-PCR analysis | |
| Rco_qRT-PCR_FW | GGGTTAAGCCAAACACAAGTAGC |
| Rco_qRT-PCR_RV | CTGCCTGTGTCGGTCTTTTTC |
| VirB11_qRT-PCR_FW | TGAAGATACGCGGGAAGGAC |
| VirB11_qRT-PCR_RV | TACCCCAGGAGAAGTAAGCC |
| RpsJ_qRT-PCR_FW | GCGGTGCACAAATACAAAG |
| RpsJ_qRT-PCR_RV | TCGCATAAGAGCATCAACAG |
| SigA_qRT-PCR_FW | GCAACTTCACCTTCTGACCAC |
| SigA_qRT-PCR_RV | CCGAATCGAAGACGCAATAC |
To transfer the deletion of sigW, rsiW, and the overexpression construct of rsiW in the background of the PY79 pLS20neo strain, PY79 pLS20neo cells were transformed with 0.1–1 μg of chromosomal DNA from the cognate strains listed in Table 1.
To generate a Gateway-accessible vector for B. subtilis, the dest42 cassette containing a ccdB gene, a chloramphenicol resistance gene, and the required attachment (att) sites were introduced into plasmid pSG1193 (14). The dest42 cassette was amplified by PCR using primer pair DEST42_KpnI/DEST42_XhoI and plasmid pCGFP (15) as template. The PCR product was digested with KpnI and XhoI and cloned into correspondingly digested pSG1193, creating plasmid pDESTBs1.
All expression plasmids were generated using the Gateway cloning system (Invitrogen). To generate entry clones of rco, rap, ΔC-rco, and ΔN-rco, the genes were amplified by PCR using pLS20neo as DNA template and primer pairs Rco_FW/Rco+STOP_RV, Rap_FW/Rap_RV, ΔC-Rco_FW/ΔC-Rco_RV, and ΔN-Rco_FW/Rco+STOP_RV. All PCR products were cloned in pENTR-D-TOPO according to the protocol of the manufacturer. To generate the expression plasmids of the individual genes, we used the Gateway LR Clonase II enzyme mix (Invitrogen). All plasmids were sequenced to verify that the genes were integrated in the right orientation.
To generate a transcriptional fusion between the promoter of the conjugation operon (Pc) and a fluorescent reporter gene, the promoter region and the mcherry gene were PCR-amplified using primer pairs Pc_FW/Pc_RV and mcherry_FW/mcherry_RV and the templates pLS20 and pSG1164-mcherry. PCR products were digested using BamHI/NheI (Pc) and NheI/EcoRI (mcherry) and ligated into the similarly digested pDG1664 plasmid.
Conjugation Assays
Mating experiments were performed as described previously (16) using strain PG300 as a recipient. For induction of rco gene expression, xylose was added to the medium at a final concentration of 0.5%. Accordingly, the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside to the growth medium induced expression of rsiW.
Qualitative and Quantitative RT-PCR
Isolation of total RNA, DNase I treatment, and reverse transcription were performed as described previously (16). For the qualitative RT-PCR analysis of the conjugation operon, 1.5 μl of the cDNA reactions, 1.5 μl of the cDNA reactions omitting reverse transcriptase, and 1.5 μl of pLS20neo DNA were used in a 30-μl PCR performed with Q5 High Fidelity DNA polymerase and the primers indicated in Fig. 1A and listed in Table 2. Quantitative RT-PCRs of rco and virB11 in the corresponding background strains harvested during exponential growth were conducted as indicated previously (16).
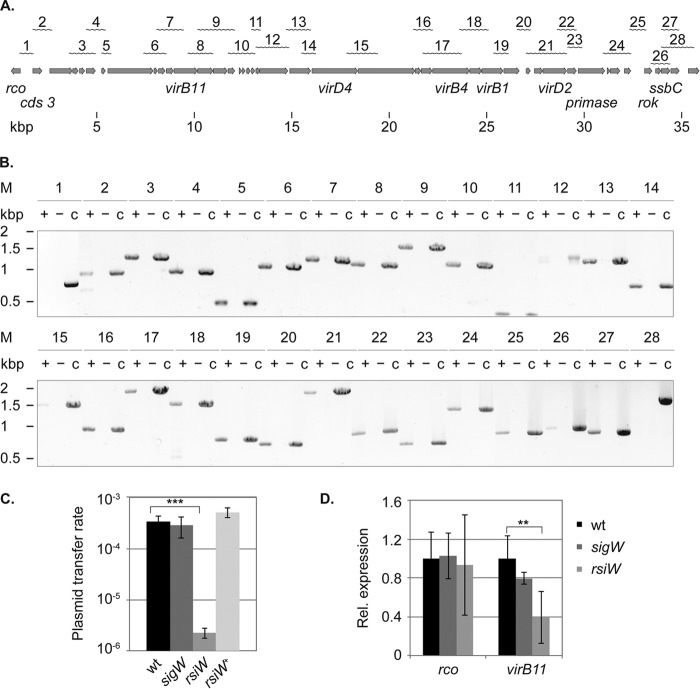
FIGURE 1.
Mapping of the conjugation operon and identification of the promoter driving expression of the rco gene. A, physical organization of the operon on the plasmid. Gray arrows, individual genes. The wavy lines at the top indicate the products of the RT-PCR analysis, and the numbers correspond to the products shown in B. B, agarose gel electrophoresis of the RT-PCR analysis. Total RNA was extracted from exponentially growing cultures of PY79/pLS20neo and DNase I-digested, and PCRs were performed following cDNA synthesis (+). For every RT-PCR analysis, the product of a cDNA synthesis reaction omitting reverse transcriptase (−) and a PCR with pLS20neo DNA as template were included (c). C, transfer of plasmid pLS20 in strains modulated in the activity of SigW. The plasmid transfer rate is expressed as transconjugants/ml/donor cell and was determined in three independent experiments. Error bars, S.D.; statistical significance was calculated using a two-sided independent sample t test. ***, p < 0.001 compared with the wild type. D, quantitative RT-PCR analysis showing transcript levels of rco and virB11 in the background of sigW and rsiW mutants in relation to the wild type. The sigA and rpsJ genes served as references for internal normalization. Total RNA was extracted from two independently growing cultures, and qRT-PCRs were conducted in duplicate (n = 4). Error bars, S.D.; statistical significance was calculated using the Relative Expression Software Tool version 2.0.13 (**, p < 0.01).
Protein Expression and Purification
Rco and RapLS20 were overexpressed as N-terminal His6 tag fusion proteins. Single colonies of freshly transformed Rosetta (DE3) 2 cells were inoculated in LB medium containing ampicillin and grown overnight at 30 °C. The next day, cells were diluted 1:100 in LB medium containing ampicillin and grown to an optical density of 0.4 at 30 °C. Overexpression of Rco and RapLS20 was induced by the addition of 1 and 0.25 mm isopropyl 1-thio-β-d-galactopyranoside, and incubation of cells continued for 3–4 h before cells were harvested by centrifugation (4 °C, 4000 rpm, 20 min). Cells were washed with LEW buffer (50 mm NaH2PO4, 300 mm NaCl; 4 °C, 4000 rpm, 20 min) and frozen at −20 °C.
Frozen cells were resuspended in LEW buffer containing Complete Protease Inhibitor Mixture (Roche Applied Science), 1 μg of DNase, and 0.1 mg/ml lysozyme. Cells were homogenized by three passages through a French press cell and centrifuged at 15,000 rpm for 30 min at 4 °C. The lysates were cleared through a 0.45-μm syringe filter (Filtropur S, Sarstedt) and applied to a Ni-TED 2000 column (Macherey-Nagel). The column was washed with 20 column volumes of LEW, and the proteins were eluted in increasing steps of 25 mm imidazole. Fractions containing His6-Rco were pooled and concentrated by ultrafiltration (Vivaspin 20, 5,000 molecular weight cut-off; Sartorius Stedim). The concentrate was further polished by size exclusion chromatography equilibrated in LEW buffer. Fractions containing Rco were concentrated by ultrafiltration. Glycerol was added to a final concentration of 50%, and aliquots were stored at −20 °C.
His6-Rap was identically purified by affinity chromatography as His6-Rco. Fractions containing His6-Rap were concentrated by ultrafiltration (Vivaspin 20, 10,000 molecular weight cut-off; Sartorius Stedim) and further polished by size exclusion chromatography. For EMSA experiments, the size exclusion chromatography column was equilibrated in Buffer A (12 mm HEPES, 4 mm Tris-HCl, 60 mm KCl, 1 mm EDTA, 1 mm EGTA, pH 8.0), and for all other experiments, the column was equilibrated in LEW100 buffer (50 mm NaH2PO4, 100 mm NaCl).
Peptide Synthesis
The synthetic pentapeptide PhrLS20 (NH2-QKGMY-COOH) was purchased from GenScript. According to the recommendations of the manufacturer, the peptide was resuspended in H2O and stored at −20 °C.
Chemical Cross-linking of Rco
Aliquots of 10 μg of Rco were incubated in Buffer B (100 mm Bicine (pH 8), 300 mm NaCl, 1 mm DTT) with increasing concentrations of glutardialdehyde. After 15 min at room temperature, the reactions were stopped through the addition of glycine (pH 8) to a final concentration of 140 mm and incubated for 5 min at room temperature. Aliquots were precipitated by acetone and resuspended in 1× SDS loading buffer. Samples were applied to a Tris-glycine PAGE (Nusep) and visualized by silver staining.
EMSA
Fragments Pc and PUTR as well as the upstream regions of cds57, cds80, and cds82 were PCR-amplified using Q5 high-fidelity DNA polymerase, pLS20neo as template, and the primers indicated in Table 2. For DNA binding reactions, Rco was diluted in Binding Buffer A (12 mm HEPES (pH 8), 4 mm Tris-HCl (pH 8), 60 mm KCl, 1 mm EDTA, 1 mm EGTA, 2.5 mm DTT, and 100 μg/ml BSA) and incubated in Binding Buffer A for 20 min at 37 °C with a 50 nm concentration of the PCR fragments. For competitive DNA binding reactions, the repressor was incubated with the antirepressor RapLS20 and the PhrLS20 peptide for 20 min at room temperature prior to the addition of a 50 nm concentrations of fragments X. All reactions were stopped through the addition of DNA loading buffer (TBE, 3% (v/v) glycerol and bromphenol blue) and loaded on 1% agarose gels prepared with 0.5× TBE. Gels were run in the refrigerator for 45 min at 100 V using 0.5× TBE as running buffer and stained with Midori Green Advance (NIPPON Genetics).
Gradient Sedimentation Analysis
Aliquots of 10, 20, and 30 μm Rco were incubated for 30 min on ice in LEW buffer, and 50 μl were layered on top of a 12-ml sucrose gradient (5–20%) prepared in LEW buffer and centrifuged in a Beckmann SW41 rotor for 16 h at 35,000 rpm and 4 °C. 1-ml fractions were sequentially taken up, precipitated by TCA, washed twice with acetone, resuspended in 1× SDS loading buffer, and analyzed by SDS-PAGE.
Size Exclusion Chromatography
20 μm RapLS20, 40 μm Rco, and 40 μm PhrLS20 were incubated or co-incubated for 30 min on ice in LEW200 buffer (50 mm NaH2PO4, 200 mm NaCl), centrifuged at 10,000 × g for 10 min at 4 °C, and applied to a Superdex 200 10/300 GL column (GE Healthcare). Fractions of 0.5 ml were collected and analyzed by 15% SDS-PAGE.
Fluorescence Microscopy
For the GFP translocation assay in E. coli cells, freshly transformed colonies were incubated overnight at 30 °C in LB medium containing 1% glucose and the appropriate antibiotics. The next day, cells were diluted 1:100 in LB medium containing the appropriate antibiotics and grown at 30 °C and 200 rpm. After 2 h of growth, the culture was split, and expression of the DivIVA fusion proteins were induced by adding 0.2% arabinose to one of the growing cultures. Localization of the GFP fusion proteins was examined 60–90 min after induction. For single-cell gene expression analysis of the reporter gene fusion Pc-mcherry, cells were grown overnight in selective LB medium, diluted to an OD of 0.05, and grown at 37 °C and 200 rpm in LB medium. The GFP translocation assays shown in Fig. 5 and the time course experiment shown in Fig. 7A were performed using a Zeiss Axio Imager AX10 microscope equipped with a 100× objective (Zeiss Plan Fluor 100×, numerical aperture = 1.45) and a CCD camera (Coolsnap HQ2, Photometrics). Pictures shown in Fig. 6A were acquired using a Nikon Eclipse Ti equipped with a 100× objective (CFI Plan Apo Lambda DM 100× oil, numerical aperture = 1.45, Ph3, Nikon) and a CCD camera (DS-Qi1, Nikon). For time lapse microscopy, cells were first grown to OD 0.4 in LB medium, diluted to an OD of 0.01 in minimal medium, and seeded on an agarose pad made with S750 medium containing 0.1% (w/v) glucose, which was placed into a small imaging dish (μDish 35 mm, Ibidi). Growth of cells was followed at 37 °C using a Nikon Eclipse Ti equipped with a motorized stage, a 100× objective (CFI Apo TIRF 100× oil, numerical aperture = 1.49), a thermostage (Tokai Hit), and an EM-CCD camera (ImagEM X2, Hamamatsu). Fluorescence microscopy pictures were processed using ImageJ version 2.0. Single-cell gene expression was measured either by using MicrobeTracker (17), where phase-contrast images were available, or by manually extracting fluorescence values from each cell in ImageJ version 2.0.
FIGURE 5.
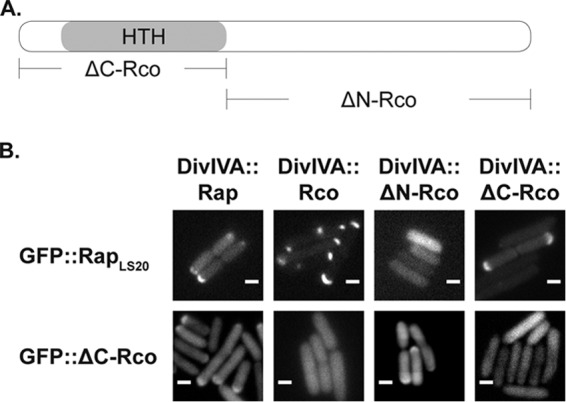
RapLS20 interacts with the N terminus of the repressor containing the DNA binding motif. A, graphical representation of the repressor protein and its derivatives used for the translocation assay. B, top, heterologous co-expression of GFP::RapLS20 with DivIVA fusions of RapLS20, Rco, and C- or N-terminally truncated versions of Rco. Bottom, heterologous co-expression of GFP::ΔC-Rco with DivIVA fusions of RapLS20, Rco, and C- or N-terminally truncated versions of Rco. Scale bar, 2 μm.
FIGURE 7.
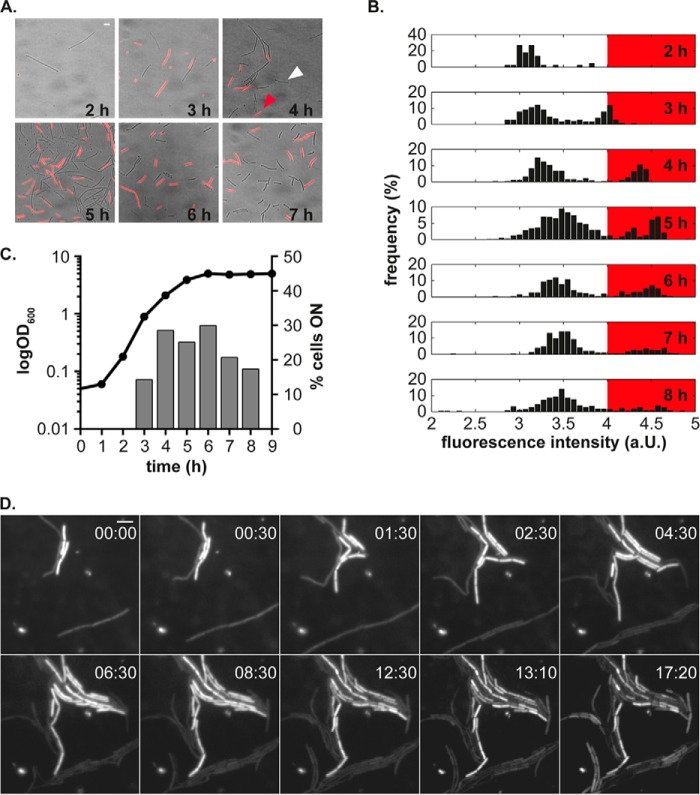
Phenotypic heterogeneity of the conjugation operon during growth of B. subtilis. A, the promoter of conjugation shows a heterogonous expression pattern in the presence of regulatory elements encoded by plasmid pLS20. Fluorescence microscopy pictures were overlaid on bright field microscopy pictures to indicate the coexistence of two states in the clonal population of pLS20-containing cells carrying a Pc-mcherry fusion. Scale bar, 4 μm. B, fluorescence distribution of the mCherry reporter protein under control of the promoter of conjugation (Pc) at different time points during growth. Histograms derived from the time course experiment show a bimodal distribution. The red box highlights the signal intensities above the threshold used to calculate the number of cells switching the conjugation operon to the ON state. Fluorescence intensities were log-transformed and plotted linearly. C, percentage of cells exceeding the fluorescence threshold during growth. Left y axis, growth of cells; right y axis, number of cells expressing the fluorescent reporter protein above the threshold. D, snapshots from a representative time lapse experiment illustrate the stable expression of the fluorescent reporter protein mCherry from the Pc promoter during microcolony development. The time scale is represented in hours, and the scale bar corresponds to 4 μm. a.U., arbitrary units.
FIGURE 6.
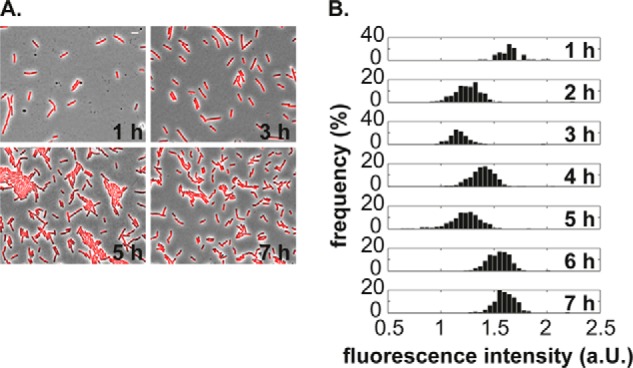
Cells devoid of the plasmid show a homogeneous expression of the promoter of conjugation when ectopically integrated into the chromosome of B. subtilis. A, fluorescence pictures of PY79 cells carrying Pc-mcherry. Shown are overlays of fluorescent and phase-contrast images taken at the indicated time points. Scale bar, 4 μm. B, cells devoid of the plasmid exhibit a unimodal distribution of the fluorescent signal. Shown are log-transformed signal intensities plotted on a linear scale. a.U., arbitrary units.
Results
Mapping of the Conjugation Operon Encoded by pLS20
To define the transcriptional organization of the conjugation operon present on pLS20, we performed RT-PCR analysis of total RNA from exponentially growing cultures containing pLS20neo, using primer pairs to link adjacent genes (Table 2). For every RT-PCR, we included a cDNA synthesis reaction omitting reverse transcriptase to confirm the absence of DNA contamination and a DNA control reaction to verify the specificity of the reactions. In accordance with our previous study (16), we confirmed co-transcription of genes cds12 (designated as pLS20_014 in the NCBI database) to cds35 (pLS20_039) and extended the analysis to the regions located down- and upstream. Fig. 1B shows that the operon starts at open reading frame cds3 (pLS20_005), which corresponds to the second RT-PCR product of the analysis (Fig. 1A), and ends at open reading frame cds43 (pLS20_049), which corresponds to the second to last RT-PCR product (Fig. 1A). Thus, the conjugation operon spans in total more than 33 kbp and also includes genes presumably not involved in the conjugative transfer itself, like roK (pLS20_046), the repressor of competence (18), or a putative DNA primase encoded by cds36 (pLS20_040), which may be required for the establishment of pLS20 in the recipient cell. Our analysis reveals all genes that are up-regulated after cells have been diluted into fresh medium and suggests that their expression is driven by a single promoter region.
Conjugation Is Affected by Enhanced SigW Activity
Rco has been shown to negatively regulate conjugation in B. subtilis strain 168 (10). We have found that this is also the case for strain PY79 (data not shown), confirming that the protein acts as a repressor independent of strain background. Ramachandran et al. (19) identified two convergent and overlapping promotors that drive the expression of the repressor gene and the conjugation operon. In silico analysis of the intergenic region between the first gene of the conjugation operon and the repressor gene using the DBTBS database (20) revealed a sigW promoter-like sequence 27 bp upstream of the rco gene (5′-cGAAAa-N16-CGTATA). SigW is an extracytoplasmic σ factor, which controls expression of several membrane proteins and which is fully activated upon cell envelope stress (21). It has been shown that the transition state regulator AbrB regulates expression of sigW and that transcription of sigW-dependent genes increases in a growth phase-dependent manner, with highest expression during stationary phase (21). To test whether the regulatory region of the rco gene is a target of SigW, we performed conjugation assays with a sigW deletion strain and strains modulating the activity of SigW. Fig. 1C shows that the deletion of sigW or overexpression of rsiW, its cognate anti-σ factor, resulted in slightly higher transfer rates of pLS20. Interestingly, the transfer rate seen in the rsiW deletion strain was 2 orders of magnitude lower than that of the wild type strain. Quantitative RT-PCR showed that the relative transcript levels of rco and virB11 as a marker for the conjugation operon were not affected in the background of the sigW strain but that deletion of rsiW resulted in a lower abundance of virB11 transcripts (Fig. 1D). These experiments reveal a link between SigW activity and transcription of the conjugation operon, which does not occur via the transcriptional level of the repressor gene.
Rco and Rap Are Homodimers in Solution, and Rco Binds to Several Sites on the Plasmid
To determine the native weight of the transcriptional regulator and its putative modulator protein, RapLS20, we performed size exclusion chromatography of affinity-purified His6-Rco and His6-RapLS20. Rco eluted at a column volume of 52.6 ml (Fig. 2A, solid line), which corresponds to a hexameric state of the protein, whereas RapLS20 eluted after 55 ml (Fig. 2A, dashed line), which corresponds to a dimeric state. Due to the large Stokes radius of Rco (RS = 4.6) we subsequently performed a sucrose gradient sedimentation analysis of Rco in varying concentrations to further evaluate its oligomeric state. SDS-PAGE analysis and application of the Monte-Siegel analysis combining the Stokes radius and the sedimentation coefficient showed that the repressor mainly exists as a dimer in solution (Fig. 2B). At higher concentrations, the repressor was also found to be in trimeric and tetrameric forms. Calculation of the Smax/S ratio shows that the monomeric conformation probably represents the most elongated shape, whereas the oligomeric conformations have moderately elongated shapes compared with proteins of known structure. Finally, glutardialdehyde cross-linking experiments confirmed the observation that Rco mainly exists in a dimeric conformation (Fig. 2C). Dimer formation was even present without the addition of the cross-linking agent. In its presence, the protein was also seen as trimer, tetramer, and hexamer.
FIGURE 2.
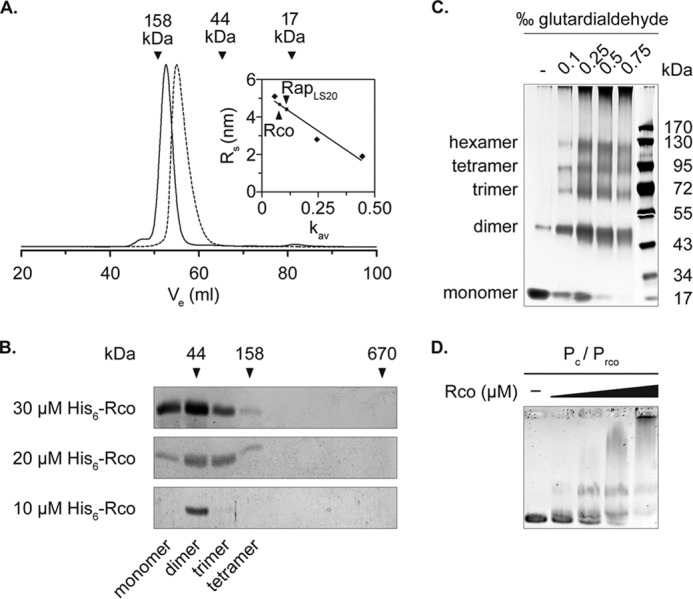
Rco and RapLS20 form dimers in solution. A, size exclusion chromatography of Rco and RapLS20. Vertical arrows, elution volumes of the standard proteins used to determine the molecular weight of Rco and RapLS20 (solid and dashed lines). The inset shows the standard curve used for calculating the Stokes radius of Rco (RS = 4.64) and RapLS20 (RS = 4.38). Size exclusion chromatography was performed on a HiLoad 16/600 Superdex 75 prep grade column (GE Healthcare). B, SDS-PAGE analysis of the 5–20% sucrose gradient sedimentation performed with different concentrations of Rco. Shown are fractions 1–9; arrows indicate the sedimentation behavior of the standards run in parallel. The molecular conformations of Rco below the SDS-PAGE analysis were calculated using the Monte-Siegel formula. C, chemical cross-linking of Rco with increasing concentrations of glutardialdehyde. Samples were run on 10% Tris-glycine PAGE (Nusep). D, binding of His6-Rco to the intergenic region of the repressor gene and the first gene of the conjugation operon containing the promoter of the conjugation operon (Pc) and the repressor gene (Prco). A 50 nm concentration of the DNA fragment was mixed with 0.2, 0.4, 0.8, and 1.6 μm Rco.
Previous work has shown that Rco regulates transcription of the conjugation operon through binding to two operator sites within the intergenic region of rco and the first gene of the conjugation operon, possibly by forming a DNA loop of 75 bp (19). Although we did not observe a preference of the repressor to form tetramers as stated before, we found a DNA binding behavior comparable with that described in the literature (Fig. 2D). Specifically, Rco binds with high affinity to sequences containing 5′-CAGTGAAA-3′, which we confirmed using EMSAs and a 125-bp fragment containing an identical sequence, which is located 175 bp upstream of the first gene of the conjugation operon (named PUTR; Fig. 3, first panel). However, binding affinity was lower in this region compared with binding to the previously described operator binding sites (19) (Fig. 2D), suggesting that the newly identified binding region may act as a secondary repressor binding site.
FIGURE 3.

Identification of additional binding sites of the Rco repressor. Rco additionally binds to DNA fragments immediately upstream of the first gene of the conjugation operon (fragment PUTR) and upstream of gene cds80 (first and second panels) but not to upstream regions of cds57 and cds82 (third and fourth panels). Increasing concentrations of isolated Rco were incubated with 50 nm DNA.
We wondered if Rco might regulate the expression of other pLS20-encoded genes, and we performed EMSA experiments with the regulatory regions of genes, which were shown to be differently regulated in the background of Rco- or RapLS20-overexpressing strains, as indicated by RNA sequencing analysis of Meijer and co-workers (10). We found that Rco bound to the promoter region of cds80 (pLS20_102; Fig. 3, second panel), a protein of unknown function with a predicted signal peptide sequence, but not to regions preceding gene cds57 (pLS20_101) and the putative operon of cds81 and cds82 (pLS20_105 and pLS20_106). Similar to fragment PUTR, the region upstream of cds80 contains a conserved consensus sequence, which differs only in 1 base (5′-CAGTGAgA-3′) from the sequence shown to be the consensus binding site of Rco. Nevertheless, it seems that additional purine residues at the 3′-end of the consensus sequence increase the binding affinity of the repressor. These experiments show that Rco binds to several sites in the conjugation promoter region and to at least one additional site on the plasmid.
RapLS20 Binds Directly to Rco to Inhibit Its DNA Binding Activity, whereas the Addition of PhrLS20 Relieves the Inhibitory Function of RapLS20 on Rco
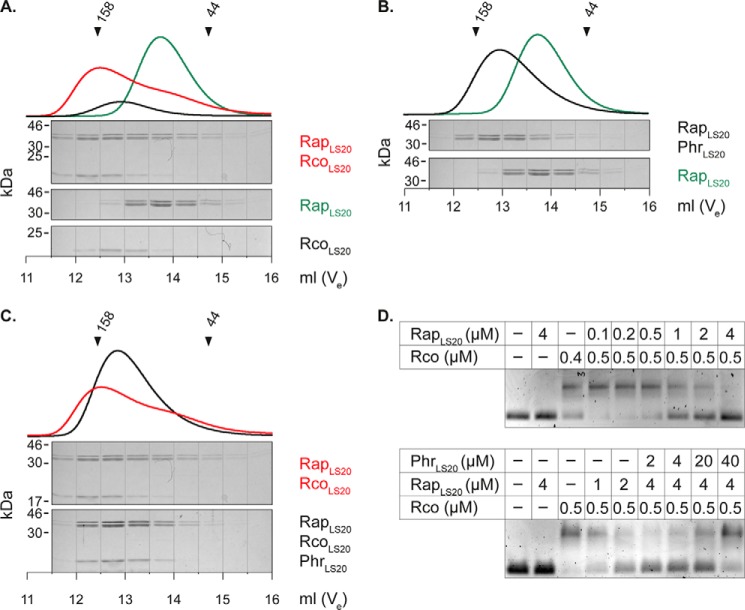
Genetic evidence has shown that an endogenous Rap-Phr module on pLS20 (RapLS20 and PhrLS20) affects the activity of the repressor of conjugation (10). To determine whether RapLS20 directly regulates the activity of the repressor protein through protein-protein interactions, we performed analytical size exclusion chromatography of RapLS20, Rco, and a mixture of both proteins. Fig. 4A shows that RapLS20 eluted much earlier from the column upon the addition of Rco than alone, providing evidence for complex formation of these proteins. The complex migrated at a molecular mass of 154 kDa, suggesting the formation of a heterotetrameric complex ((2 × 46.6 kDa) + (2 × 21.8 kDa)) with a 1:1 stoichiometry, as determined by densitometric measurements of the corresponding bands.
FIGURE 4.
RapLS20 binds to Rco and inhibits the DNA binding activity of Rco, whereas PhrLS20 relieves the inhibitory effect of RapLS20. A, in vitro interaction of RapLS20 and Rco. Size exclusion chromatography of RapLS20 (20 μm), Rco (40 μm), and RapLS20-Rco in complex (20 and 40 μm) was performed on a Superdex 200 10/300 GL column. Top, chromatograms; bottom, SDS-PAGE analysis of the eluted fractions. B, PhrLS20 binds to RapLS20. The addition of 40 μm PhrLS20 to 20 μm RapLS20 changes the elution profile of RapLS20. C, the addition of 40 μm PhrLS20 disrupts complex formation of RapLS20 and Rco (20 and 40 μm). D, top, RapLS20 inhibits DNA binding activity of Rco; bottom, PhrLS20 relieves the inhibitory effect of RapLS20 on DNA binding activity of Rco. Rco was mixed with RapLS20 and PhrLS20 at the indicated concentrations prior to the addition of 50 nm PUTR.
It is known that the function of Rap proteins is controlled by their cognate Phr peptides through protein-peptide interactions (6, 22). Meijer and co-workers (10) showed that the 5 C-terminal residues of the PhrLS20 prepeptide (corresponding to QKGMY) efficiently repressed conjugation of plasmid pLS20 and that repression could be relieved by deletion of the oligopeptide permease opp.
To verify that PhrLS20 acts on RapLS20, thereby possibly interfering with complex formation of RapLS20-Rco, we added the synthetic pentapeptide to RapLS20 and to RapLS20-Rco complexes. Indeed, synthetic PhrLS20 bound to RapLS20, causing RapLS20 to elute earlier from the column (Fig. 4B) than when in complex with Rco. Additionally, co-incubation of PhrLS20 with RapLS20 and Rco prevented complex formation between the two proteins (Fig. 4C), at the expense of the RapLS20-PhrLS20 complex.
To test whether an interaction between RapLS20 and Rco results in an inhibition of the DNA binding activity of the repressor protein, we conducted EMSA experiments in the presence of increasing amounts of RapLS20. As indicated in Fig. 4D, RapLS20 showed no DNA binding activity by itself but diminished the binding activity of Rco when the molar ratio reached a 1:1 stoichiometry and completely repressed DNA binding at an 8-fold molar excess (Fig. 4D, top). To see whether PhrLS20 relieves the inhibitory function of RapLS20 on Rco, we added increasing amounts of PhrLS20 to the (8-fold) molar ratio of RapLS20 and Rco at which no more DNA binding activity occurred. Interestingly, the addition of the peptide restored the DNA binding activity of Rco, although not to the same extent as without the addition of RapLS20 (Fig. 4D). These results show that the Rco/RapLS20/PhrLS20 module on pLS20 operates via direct protein-protein and protein-peptide interactions to regulate the conjugative activity of the plasmid.
RapLS20 Interferes with Rco Function through Interaction with the N Terminus of Rco
Previous studies revealed that RapF prevents binding of ComA to its target promoters by blocking the DNA-binding domain of ComA through a direct interaction (7, 8). We wondered whether this might be a general scheme of Rap proteins that regulate the activity of transcriptional regulators. To shed light on the interaction interface of RapLS20 and Rco, we applied an in vivo GFP translocation assay based on the intrinsic property of DivIVA to localize at negative membrane curvatures (23, 24). Therefore, we fused RapLS20 to GFP and co-expressed it pairwise with DivIVA fusions to full-length Rco or with DivIVA fusions to the C- or the N-terminal ends of Rco (ΔC-Rco and ΔN-Rco; Fig. 5A) in E. coli BL21 (DE3) cells. In all cases, we observed strong GFP signals even in the absence of the inducing agent (isopropyl 1-thio-β-d-galactopyranoside); thus, we split the cultures before induction of the DivIVA fusion proteins to clearly distinguish DivIVA-like localization from polar localization caused by overexpression of the GFP fusion proteins.
We found that GFP-RapLS20 localized in a half-moon-like manner to the cell pole when co-expressed with a DivIVA fusion of RapLS20, of full-length Rco, or of ΔC-Rco, but not when it was co-expressed with a DivIVA fusion of ΔN-Rco (Fig. 5B, top). To test whether these proteins indeed interact via the N terminus of Rco, we also assayed the localization of GFP-ΔC-Rco only containing the helix-turn-helix motif upon co-expression with DivIVA-RapLS20. As shown in Fig. 5B (bottom), DivIVA-RapLS20 targeted GFP-ΔC-Rco to the cell pole and to a certain extent also DivIVA-ΔN-Rco, but no translocation was observed when GFP-ΔC-Rco was co-expressed with DivIVA-Rco or with DivIVA-ΔC-Rco. Taken together, we provide evidence that RapLS20 recognizes the N-terminal part of Rco containing the helix-turn-helix motif.
Expression of the Conjugation Operon is Heterogeneous
To test whether, during growth, all cells or just a subset induce the conjugation operon, as is often characteristic of developmental pathways in bacteria, we monitored the activity of the promoter driving expression of the large conjugation operon at the single cell level. Therefore, we generated a transcriptional fusion of the promoter (Pc) with the fluorescent mCherry protein and integrated the reporter gene fusion into the thrC locus in the chromosome of B. subtilis PY79. First, we analyzed its expression during growth in cells devoid of the plasmid and its regulatory elements. Signal intensities of the fluorescent reporter were homogeneously distributed during all growth stages in cells lacking the plasmid (Fig. 6, A and B), although the mean fluorescence intensity varied between the time points (Fig. 6B). Contrary to plasmid-free cells, cells containing the plasmid exhibited a phenotypic heterogeneity in the expression pattern of the fluorescent reporter (Fig. 7A, red and white arrows). To quantitatively analyze expression of the promoter of conjugation, we measured the mCherry fluorescence signal of individual cells at each time point during growth and plotted the log-transformed fluorescence intensities in histograms. Fig. 7B shows that the overall signal intensity increases with ongoing growth and that the fluorescence intensity shifts from a rather unimodal distribution to a bimodal distribution upon entry into mid-exponential growth (first and second panel from the top). Similarly, after 8 h of growth, the fluorescence intensity shifted back to a unimodal distribution. To determine the number of cells switching on the conjugation operon, we set a threshold that separated the two populations from each other and calculated the number of cells exceeding the intensity threshold (Fig. 7B). Fig. 7C shows that under our conditions, a maximum of about 30% of the cells switched the operon to the ON state during the transition from the lag phase to exponential phase of growth. To further analyze the phenotypic heterogeneity of the promoter of conjugation at the single cell level, we performed time lapse microscopy. In accordance with the previous time course experiment, we observed two distinct populations. Additionally, we found that once induced, expression of the conjugation promoter was stably inherited in dividing cells and slowly disappeared when cells ceased to divide (Fig. 7D and supplemental Movie S1). These experiments reveal that the Rap/Phr/repressor system on pLS20 operates in a bistable manner, similar to the ComS/MecA/ComK system in competence and to the phosphorelay in sporulation. Our findings extend the previous observation that only a subset of cells assemble the conserved type IV secretion proteins at the membrane (13) and provide evidence that the assembly of the type IV secretion is already regulated at the transcriptional level.
Discussion
Rap-Phr modules play a pivotal role in determining the fate of individual cells in bacterial communities and are best characterized in B. subtilis (22, 25). So far, several modules in the genome of B. subtilis have been discovered that affect sporulation, competence development, secretion of extracellular proteases, or the horizontal transfer of the mobile genetic element ICEBs1. In this work, we provide in vitro and in vivo evidence that the Rap/Phr module on conjugative plasmid pLS20 directly regulates conjugation via binding to the Rco master repressor and that induction of conjugation occurs in a bistable manner, similar to the developmental processes of competence development and sporulation, leading to heterogeneous expression of the type IV secretion machinery encoded on pLS20. We show that RapLS20 acts as a direct antirepressor of the Rco protein and that its activity is regulated by the cognate PhrLS20 peptide. We show that the repressor and its antirepressor preferentially form dimers in solution and that complex formation occurs in a 1:1 stoichiometry that disrupts DNA binding of the repressor protein through obscuring the DNA-binding domain of the repressor. Unlike the plasmid-encoded Rap60, which presumably inactivates ComA activity by occluding its interaction with the RNA polymerase (26), inactivation of the Rco repressor by RapLS20 resembles that of RapC, -F, and -H, which prevent DNA binding of ComA, which, however, is an activator, rather than a repressor, like Rco. Structural analysis of the RapF-ComA interaction showed that the N-terminal 3-helix bundle of RapF captures the helix-turn-helix-containing domain of ComA and thereby prevents binding of ComA to its target promoters (8). Concurrent with its different binding partner, RapLS20 shows substitutions in all amino acid residues shown to be essential for the interaction of RapF and ComA (8).
Recent work showed that high levels of Phr peptide inhibit horizontal transfer of the plasmid, whereas at lower levels, the Rap protein becomes active and switches on the expression of the conjugation operon (10). Therefore, the known in vivo data nicely corroborate our in vitro findings.
Interestingly, several plasmids, such as pBS32 of the undomesticated B. subtilis strain NCIB3610, pX01 of Bacillus anthracis, and pTA1060 of B. subtilis were found to carry Rap/Phr modules known to affect the developmental fate of their host (27–29), showing that the use of Rap/Phr modules is widespread.
The interaction of RapF-ComA is antagonized through binding of PhrF to the C-terminal tetratricopeptide repeat domain of RapF. In the PhrF-bound state, RapF constricts and exerts a conformational change that leads to dissociation of the RapF-ComA complex (30, 31). PhrLS20 seems to induce a conformational change in RapLS20 as well, but our size exclusion chromatography data indicate that RapLS20 rather elongates or expands upon binding to the peptide, because the protein-peptide complex is much larger than RapLS20 alone. In fact, although Rap proteins share a high degree of overall sequence homology, it was not possible to model the structure of RapLS20 based on the previously solved structures. Thus, it would be interesting to reveal how the structures of RapLS20 and RapLS20-PhrLS20 differ from already known structures.
Furthermore, we show that Rco also binds to a second promoter region on the plasmid, suggesting that it regulates at least two transcriptional units. Interestingly, we found that increased activity of SigW through deletion of its antagonist RsiW resulted in a diminished plasmid transfer rate, whereas expression of the rco gene was not significantly affected in the background of these strains. Nevertheless, we found that expression of virB11 was slightly but significantly changed in the rsiW strain. Due to the only moderate effect, we think that the reduced plasmid transfer rate seen in the rsiW strain is probably due to secondary effects caused by enhanced SigW activity and not by direct transcriptional regulation of the repressor gene or the conjugation operon. In any event, it is intriguing to note that changes in the activity of a host-encoded σ factor affect conjugation activity of a plasmid, which may be important for the plasmid's decision to prevent conjugation in case of existing cell wall stress.
Last, we provide evidence that expression of the conjugation operon occurs in a heterogeneous but stable manner. Whereas in the absence of the plasmid, expression of the ectopic conjugation promoter showed a unimodal distribution, the presence of the plasmid elicited a bimodal distribution and thus a mutually exclusive expression pattern of the conjugation promoter in the population. Interestingly, time lapse microscopy revealed that once the ON state is established, expression of the conjugation promoter is propagated to the next generation of cells and vanishes with additional cell divisions. Interestingly, the Meijer group (19) recently showed that repressor protein RcoLS20 autoregulates itself and keeps the conjugation operon in the OFF state through binding to two operator sites, presumably forming a DNA loop. Loop formation may contribute to the relatively tight on/off regulation seen in our expression studies.
Our work shows that pLS20 harbors a classical Rap/Phr system, which modulates the activity of a repressor protein, but its output in vivo differs from known Rap/Phr systems, in that the partner switch shows an offset in vivo relative to known Rap/Phr systems as it occurs during a time window, when cells actively divide rather than arrest their cell cycle. Thus, the Rco/Rap/Phr regulatory circuit encoded by plasmid pLS20 could be employed for the rational design of expression systems that are limited to the cell's growth phase. It will be interesting to further analyze which factor(s) governs the heterogeneous expression of the conjugation operon.
Author Contributions
T. C. R. and P. L. G. conceived of and coordinated the study and wrote the paper. T. C. R. designed, performed, and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We thank Thomas Wiegert (University of Zittau, Germany) for the generous gift of the sigW mutant strain.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School), and by the LOEWE Centre for Synthetic Microbiology, SYNMIKRO, in Marburg. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Movie S1.
- ICE
- integrative conjugative element
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Chen I., Christie P. J., Dubnau D. (2005) The ins and outs of DNA transfer in bacteria. Science 310, 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thomas C. M., Nielsen K. M. (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 [DOI] [PubMed] [Google Scholar]
- 3. Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grohmann E., Muth G., Espinosa M. (2003) Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bose B., Grossman A. D. (2011) Regulation of horizontal gene transfer in Bacillus subtilis by activation of a conserved site-specific protease. J. Bacteriol. 193, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perego M., Brannigan J. A. (2001) Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22, 1541–1547 [DOI] [PubMed] [Google Scholar]
- 7. Bongiorni C., Ishikawa S., Stephenson S., Ogasawara N., Perego M. (2005) Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J. Bacteriol. 187, 4353–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baker M. D., Neiditch M. B. (2011) Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS Biol. 9, e1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Core L., Perego M. (2003) TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 49, 1509–1522 [DOI] [PubMed] [Google Scholar]
- 10. Singh P. K., Ramachandran G., Ramos-Ruiz R., Peiró-Pastor R., Abia D., Wu L. J., Meijer W. J. (2013) Mobility of the native Bacillus subtilis conjugative plasmid pLS20 is regulated by intercellular signaling. PLoS Genet. 9, e1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Auchtung J. M., Lee C. A., Monson R. E., Lehman A. P., Grossman A. D. (2005) Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 102, 12554–12559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Itaya M., Sakaya N., Matsunaga S., Fujita K., Kaneko S. (2006) Conjugational transfer kinetics of pLS20 between Bacillus subtilis in liquid medium. Biosci. Biotechnol. Biochem. 70, 740–742 [DOI] [PubMed] [Google Scholar]
- 13. Bauer T., Rösch T., Itaya M., Graumann P. L. (2011) Localization pattern of conjugation machinery in a Gram-positive bacterium. J. Bacteriol. 193, 6244–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feucht A., Lewis P. J. (2001) Improved plasmid vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene 264, 289–297 [DOI] [PubMed] [Google Scholar]
- 15. Edwards A. N., Fowlkes J. D., Owens E. T., Standaert R. F., Pelletier D. A., Hurst G. B., Doktycz M. J., Morrell-Falvey J. L. (2009) An in vivo imaging-based assay for detecting protein interactions over a wide range of binding affinities. Anal. Biochem. 395, 166–177 [DOI] [PubMed] [Google Scholar]
- 16. Rösch T. C., Golman W., Hucklesby L., Gonzalez-Pastor J. E., Graumann P. L. (2014) The presence of conjugative plasmid pLS20 affects global transcription of its Bacillus subtilis host and confers beneficial stress resistance to cells. Appl. Environ. Microbiol. 80, 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sliusarenko O., Heinritz J., Emonet T., Jacobs-Wagner C. (2011) High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80, 612–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singh P. K., Ramachandran G., Durán-Alcalde L., Alonso C., Wu L. J., Meijer W. J. (2012) Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ. Microbiol. 14, 2812–2825 [DOI] [PubMed] [Google Scholar]
- 19. Ramachandran G., Singh P. K., Luque-Ortega J. R., Yuste L., Alfonso C., Rojo F., Wu L. J., Meijer W. J. (2014) A complex genetic switch involving overlapping divergent promoters and DNA looping regulates expression of conjugation genes of a Gram-positive plasmid. PLoS Genet. 10, e1004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sierro N., Makita Y., de Hoon M., Nakai K. (2008) DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36, D93–D96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao M., Wang T., Ye R., Helmann J. D. (2002) Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σ(W) and σ(M) regulons. Mol. Microbiol. 45, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 22. Perego M. (2013) Forty years in the making: understanding the molecular mechanism of peptide regulation in bacterial development. PLoS Biol. 11, e1001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenarcic R., Halbedel S., Visser L., Shaw M., Wu L. J., Errington J., Marenduzzo D., Hamoen L. W. (2009) Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 28, 2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramamurthi K. S., Losick R. (2009) Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc. Natl. Acad. Sci. U.S.A. 106, 13541–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schultz D., Wolynes P. G., Ben Jacob E., Onuchic J. N. (2009) Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 106, 21027–21034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boguslawski K. M., Hill P. A., Griffith K. L. (2015) Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis. Mol. Microbiol. 96, 325–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bongiorni C., Stoessel R., Shoemaker D., Perego M. (2006) Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J. Bacteriol. 188, 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parashar V., Konkol M. A., Kearns D. B., Neiditch M. B. (2013) A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J. Bacteriol. 195, 2437–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koetje E. J., Hajdo-Milasinovic A., Kiewiet R., Bron S., Tjalsma H. (2003) A plasmid-borne Rap-Phr system of Bacillus subtilis can mediate cell-density controlled production of extracellular proteases. Microbiology 149, 19–28 [DOI] [PubMed] [Google Scholar]
- 30. Gallego del Sol F., Marina A. (2013) Structural basis of Rap phosphatase inhibition by Phr peptides. PLoS Biol. 11, e1001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parashar V., Jeffrey P. D., Neiditch M. B. (2013) Conformational change-induced repeat domain expansion regulates Rap phosphatase quorum-sensing signal receptors. PLoS Biol. 11, e1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schöbel S., Zellmeier S., Schumann W., Wiegert T. (2004) The Bacillus subtilis σW anti-σ factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 52, 1091–1105 [DOI] [PubMed] [Google Scholar]
- 33. Zellmeier S., Schumann W., Wiegert T. (2006) Involvement of Clp protease activity in modulating the Bacillus subtilis σ w stress response. Mol. Microbiol. 61, 1569–1582 [DOI] [PubMed] [Google Scholar]
- 34. Busso D., Delagoutte-Busso B., Moras D. (2005) Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343, 313–321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.