Background: How the repression activity of Tcf7l1 is regulated is not well understood.
Results: Jmjd6 binds to and derepresses Tcf71.
Conclusion: Jmjd6 is essential for derepression of Tcf7l1 and Xenopus body axis patterning.
Significance: Derepression of Tcf7l1 by Jmjd6 is a novel mechanism for the understanding of Wnt action in not only embryos but also in stem cells and cancers.
Keywords: development, embryo, transcription regulation, Wnt pathway, Xenopus, Jmjd6, Tcf7l1, body axis patterning, derepression
Abstract
Tcf7l1 (also known as Tcf3) is a bimodal transcription factor that plays essential roles in embryogenesis and embryonic and adult stem cells. On one hand, Tcf7l1 works as transcriptional repressor via the recruitment of Groucho-related transcriptional corepressors to repress the transcription of Wnt target genes, and, on the other hand, it activates Wnt target genes when Wnt-activated β-catenin interacts with it. However, how its activity is modulated is not well understood. Here we demonstrate that a JmjC-domain containing protein, Jmjd6, interacts with Tcf7l and derepresses Tcf7l. We show that Jmjd6 binds to a region of Tcf7l1 that is also responsible for Groucho interaction, therefore making it possible that Jmjd6 binding displaces the Groucho transcriptional corepressor from Tcf7l1. Moreover, we show that Jmjd6 antagonizes the repression effect of Tcf7l1 on target gene transcription and is able to enhance β-catenin-induced gene activation and that, vice versa, inhibition of Jmjd6 activity compromises gene activation in both cells and Xenopus early embryos. We also show that jmjd6 is both maternally and zygotically transcribed during Xenopus embryogenesis. Loss of Jmjd6 function causes defects in anterioposterior body axis formation and down-regulation of genes that are involved in anterioposterior axis patterning. The results elucidate a novel mechanism underlying the regulation of Tcf7l1 activity and the regulation of embryonic body axis formation.
Introduction
Tcf7l1 is a transcription factor of the Tcf/Lef family that contains a high mobility group DNA-binding domain (1). It plays distinct roles in transcriptional regulation depending on the cellular context. In the absence of Wnt signaling, it functions as a transcriptional repressor because of its recruitment of Groucho/transducin-like enhancer protein corepressors (2). Otherwise, upon Wnt activation, β-catenin competes with the corepressors for Tcf7l1 binding (3). The resulting β-catenin·Tcf7l1 complex interacts with the promoters of the target genes of Wnt signaling and activates the transcription of these genes. Wnt activation also leads to the phosphorylation of Tcf7l1 and subsequent derepression of target genes (4). Tcf7l1 plays essential roles not only in embryogenesis (5–7) but also in the maintenance of the stemness of both embryonic and somatic stem cells (8–10). At blastula stages during Xenopus embryogenesis, Tcf7l1 represses dorsal gene expression at the ventral side (11), where there is no Wnt activation, whereas, at the dorsal side, it transduces Wnt signaling to activate dorsal genes to pattern dorsoventral axis formation (5, 12, 13). Zebrafish homozygous for the tcf3 mutation develop without head formation (6), and the repression of Wnt/β-catenin by Tcf7l1 is a prerequisite for the normal development of the anterior structures (14). Tcf7l1 knockout in mice leads to disrupted axial structures and, ultimately, embryonic lethality (15). In embryonic stem cells, Tcf7l1 is an integral component of the core regulatory circuitry that maintains pluripotency (10). Tcf7l1 has also been demonstrated to maintain stem cell features in skin and neural stem cells by inhibition of the genes that promote differentiation (8, 16). However, how the activity of Tcf7l1 is regulated is not well understood.
Jmjd6 (previously known as the phosphatidylserine receptor, Psr) is one of the JmjC-domain-containing proteins, many of which exhibit lysine demethylase activity for histone substrates (17). Jmjd6 has been reported to have functions other than lysine demethylation, including protein arginine demethylation (18), lysyl hydroxylation (19–21), RNA splicing (19, 22), and transcriptional regulation (23). Knockdown or knockout phenotypes in zebrafish and mice demonstrate that Jmjd6 is required for embryonic development (24, 25). Moreover, Jmjd6 is overexpressed in and related to a poor prognosis of cancers (21, 26, 27). So far, our understanding of the underlying molecular events that regulate embryogenesis through Jmjd6 is very poor.
In this study, we found out that Jmjd6 is an interaction partner for Tcf7l1. In both cells and Xenopus embryos, we show that Jmjd6 binding alleviates the activity of transcriptional repression of Tcf7l1. We demonstrate that Jmjd6 is required for the normal expression of genes involved in body axis patterning during Xenopus embryonic development. Considering that Tcf7l1 and Wnt/β-Catenin signaling are master regulators for cell fate decision and that Jmjd6 is conserved in multicellular organisms, the mechanism of regulation of Tcf7l1 by Jmjd6 is also relevant to the research of stem cell and cancer biology.
Experimental Procedures
Plasmid Construction
The complete coding regions of Xenopus laevis jmjd6a (accession no. NM_001092479) and jmjd6b (accession no. NM_001087045) were amplified from the cDNA pool derived from stage 10 embryos and ligated to the pCS2+, pCS2+Hamcs, or pCS2+MTmcs vectors. Different Xenopus Jmjd6a and Tcf7l1 deletion mutants were constructed using a PCR-based method and subcloned to the pCS2+HAmcs, pCS2+MTmcs, or pCS2+NLS-MT vectors. To knock down the endogenous JMJD6 in HEK293T cells, a strategy of vector-based miRNA was used for RNA interference. Two miRNA sequences, miJMJD6-1 (5′-AGACTACAAGGTGCCAAAGTT-3′) and miJMJD6-2 (5′-ACGAAGCTATTACCTGGTTTA-3′), were designed using an online program and subcloned separately to the pcDNATM6.2-GW/EmGFP-miR vector (Invitrogen). A negative control miRNA RNAi (miCtrl) plasmid, pcDNATM6.2-GW/EmGFP-miR-neg, was purchased from Invitrogen. All constructs were confirmed by sequencing.
Cell Culture and Transfection
HEK293T cells were cultured at 37 °C under 5% CO2 in DMEM supplemented with 10% fetal bovine serum. Different plasmids were transfected into HEK293T cells with PEI for luciferase assays, immunoblotting, co-IP,2 or immunofluorescence. To perform luciferase assays, HEK293T cells at about 70% confluency in each well of 24-well plates were transfected with 100 ng of TopFlash or FopFlash reporter plasmid, 1 ng of pRL-TK Renilla luciferase reporter, and 100 ng of each expression plasmid or miRNA knockdown plasmid. For co-IP experiments, each 6 μg of plasmids for tagged proteins were transfected into HEK293T cells that were cultured in 10-cm culture dishes. An empty vector plasmid was used as a supplement to ensure that, in the same experiment, cells in different wells or dishes were transfected with the same quantity of plasmids.
Cell Extract Preparation
Whole cell lysates were prepared from untransfected or transfected cells. After being washed twice in ice-cold PBS, cells were lysed on ice for 30 min in lysis buffer containing 150 mm NaCl, 0.5% Nonidet P-40, 5 mm EDTA, 50 mm Tris (pH 8.0), and protease inhibitors (Roche). Cell lysates were cleared via centrifugation at 14,000 × g for 30 min, mixed with a ¼ volume of 5× loading buffer and boiled for 5 min. Xenopus embryonic total cell extracts were prepared exactly as described previously (28, 29). Briefly, when embryos reached the desired stages, they were homogenized in cold lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1% Nonidet P-40) supplemented with protease inhibitors (Roche, 10 μl of lysis buffer/embryo). After incubation on ice for 10 min, homogenates were centrifuged at 16,000 × g for 10 min. Cell debris was removed by centrifugation, and supernatants were boiled in 1× Laemmli buffer and centrifuged again at 16,000 × g for 5 min. Cell extracts were used for co-IP or/and immunoblotting.
Coimmunoprecipitation and Immunoblotting
Protein co-IP was carried out using conventional methods. In short, protein complexes were precipitated from whole cell lysates with anti-c-Myc-agarose affinity gel (Sigma-Aldrich, catalog no. A7470). Agarose beads were pelleted by centrifugation and washed three times with TBST buffer (25 mm Tris-Cl, 150 mm NaCl, and 0.05% Tween 20 (pH 7.2)). Immunocomplexes were eluted by boiling the beads in loading buffer and subsequent centrifugation. Cell lysates or immunoprecipitates were subjected to SDS-PAGE and Western blotting using conventional methods. The antibodies and dilutions of each antibody were as follows: FLAG epitope (Santa Cruz Biotechnology, catalog no. sc-807, 1:2000), c-Myc epitope (Santa Cruz Biotechnology, catalog no. sc-789, 1:4000), HA epitope (Santa Cruz Biotechnology, catalog no. sc-805, 1:4000), β-Actin (CWBIO, catalog no. CW0097, 1:4000), JMJD6 (Abcam, catalog no. ab65770, 1:5000), and TCF7L1/TCF3 (CST, catalog no. 2883, 1:1000). Secondary antibodies were horseradish peroxidase-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology, catalog no. sc-2005) and goat anti-rabbit IgG (Santa Cruz Biotechnology, catalog no. sc-2030), which were used at 1:10,000.
Immunofluorescence
Immunofluorescence was performed as described previously (30). HEK293T cells were cultured on coverslips in 6-well plates. Cells grown at about 70% confluency were transfected with expression plasmids. Twenty-four hours after transfection, cells were washed with PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in PBS. Cells were then blocked with 0.2% fish skin gelatin (Sigma-Aldrich) to avoid unspecific staining. An antibody against HA epitope (CST, catalog no. 2367, 1:1000) was used for the detection of HA tags, and antibodies against the c-Myc epitope (1:1000) (CST #2278 was used for Fig. 2C, and Santa Cruz sc-40 was used for Fig. 3D) were used for detecting c-Myc tagged proteins. Secondary antibodies were FITC-conjugated anti-mouse IgG (Sigma-Aldrich, catalog no. F5897, 1:500) and Cy3-conjugated anti-rabbit IgG (Sigma-Aldrich, catalog no. C2306, 1:500). DAPI staining was performed to view cell nuclei. Then cells were rinsed, and coverslips were mounted with antifade mounting medium. Cells were then detected using a fluorescence microscope (FluoView FV1000, Olympus).
FIGURE 2.
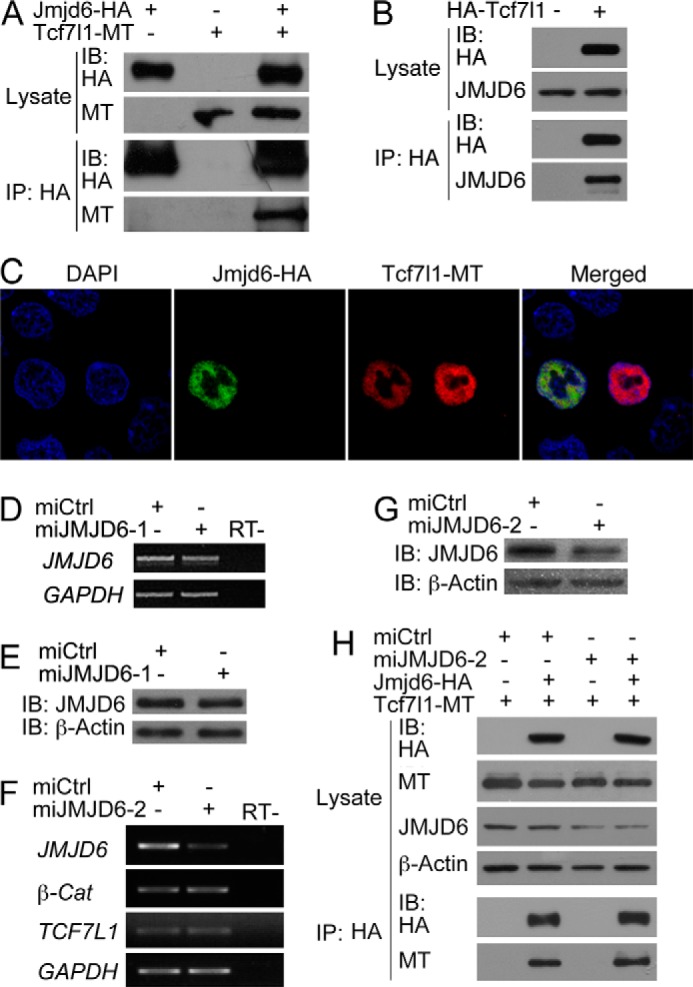
Jmjd6 interacts with Tcf7l1. A, co-IP detection of the interaction between overexpressed Jmjd6 and Tcf7l1 in HEK293T cells. IB, immunoblot. MT, myc tag. B, overexpressed Tcf7l1 precipitated endogenous JMJD6 in HEK293T cells. C, nuclear colocalization of Jmjd6 and Tcf7l1 in HEK293T cells as detected by immunofluorescence staining. DAPI staining reveals nuclei. D and E, test of the knockdown efficiency of miJMJD6-1. Transfection of the plasmid for miJMJD6-1 did not cause a significant reduction in both the transcript (D) and protein (E) levels of JMJD6 in HEK293T cells. RT−, transcription without reverse transcriptase. F and G, test of the knockdown efficiency of miJMJD6-2 and the effect of JMJD6 knockdown on the transcription of TCF7l1 and β-Catenin (β-Cat). Transfection of the plasmid for miJMJD6-2 resulted in a significant decrease in both the transcript (F) and protein (G) levels of JMJD6. Meanwhile, the transcription of TCF7L1 and β-Catenin was not affected in response to efficient JMJD6 knockdown. GAPDH was used as a loading control for RT-PCR detection of the JMJD6 transcript in D and F, and β-Actin was used as a loading control for the detection of JMJD6 protein with immunoblotting in E and G. H, detection of the effect of endogenous JMJD6 knockdown on the interaction between exogenous Jmjd6 and Tcf7l1.
FIGURE 3.
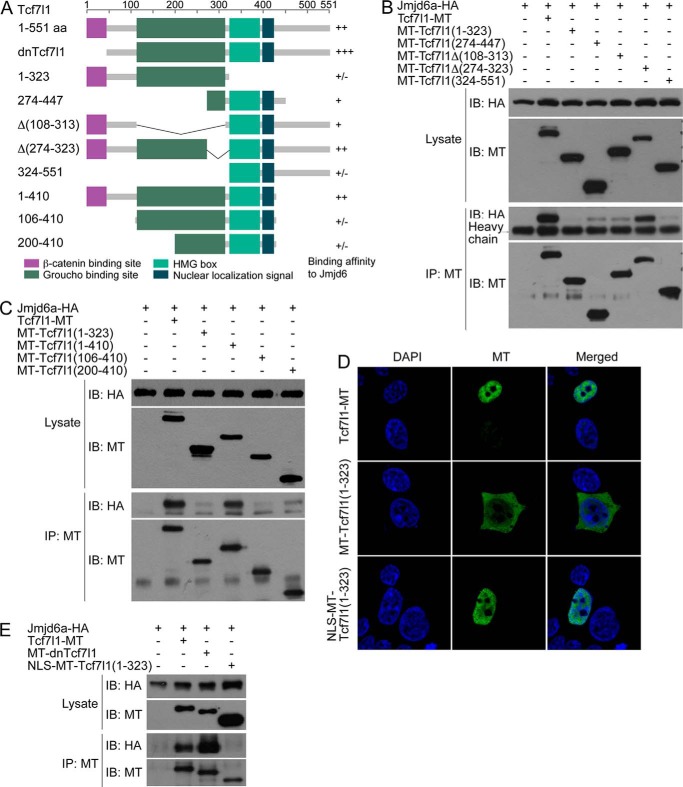
Mapping of the region in the Tcf7l1 protein responsible for Jmjd6 interaction. A, domain structure, the construction of the deletion mutants of Tcf7l1 used in the study, and their binding affinities to Jmjd6 after co-IP assays. +++, very strong binding; ++, strong binding; +, weak binding; ±, trace or no binding. B and C, Tcf7l1 deletion mutants exhibited different binding affinities to Jmjd6 as shown by co-IP assays. IB, immunoblot. MT, myc tag. D, immunofluorescence showed that the aa 1–323 region of Tcf7l1 primarily distributed in the cytosol, whereas addition of an NLS to the region relocalized it into the nucleus. E, addition of an NLS to the 1–323 region of Tcf7l1 did not increase its binding affinity to Jmjd6.
Luciferase Assays
Luciferase assays with cultured cells were carried out essentially as described previously (29). Data are presented as mean ± S.E. of Topflash/Fopflash ratios that were obtained from at least four independent experiments. Significance was calculated using Student's t test.
Xenopus Embryos, in Vitro Transcription, Antisense Morpholino Oligonucleotides (MOs), and Microinjection
X. laevis embryos were obtained with in vitro fertilization and cultured in 0.1× MBSH (1× MBSH: 88 mm NaCl, 2.4 mm NaHCO3, 1 mm KCl, 0.82 mm MgSO4, 0.41 mm CaCl2, 0.33 mm Ca(NO3)2, and 10 mm HEPES (pH 7.4)). Antisense RNA probes for whole-mount in situ hybridization and mRNAs for microinjection were prepared as described previously (29). To prepare antisense RNA probes for whole-mount in situ hybridization, plasmids for X. laevis jmjd6a, jmjd6b, sia, xnr3, cer1, dkk1, gsc, chrd, and xag2 were linearized and transcribed with T7 RNA polymerase. To prepare mRNAs for microinjection, plasmids pCS2+Tcf7l1, pCS2+dnTcf7l1, pCS2+Tcf7l1-MT, pCS2+Jmjd6a, and pCS2+Jmjd6a-HA were linearized and transcribed with Sp6 mMessage mMachine kits (Ambion). All probes and mRNAs were cleaned up with RNeasy kit (Qiagen). An antisense MO (Jmjd6MO, TGCGCTTCTTACTCTTGTGGTTCAT) that targets the first 25 bases of the ORF of both X. laevis jmjd6a and jmjd6b transcripts was designed to knock down endogenous Jmjd6a and Jmjd6b in embryos. An MO for the knockdown of endogenous Tcf7l1 in Xenopus embryos, Tcf7l1MO, has been reported previously (31). The standard control MO (ctrlMO, CCTCTTACCTCAGTTACAATTTATA) was used as a control. All MOs were purchased from GeneTools. The injected doses of mRNAs or MOs are described in the text.
Gene Expression Microarray Analysis
To examine the possible cooperation of Tcf7l1 and Jmjd6 on gene regulation during germ layer formation of Xenopus embryos, we injected the embryos with ctrlMO (30 ng) or Tcf7l1 mRNA (300 pg) alone or with Tcf7l1 mRNA (300 pg) and Jmjd6MO (30 ng) together. When control sibling embryos reached stage 10 gastrula, each 20 injected embryos were collected, and total RNAs were extracted with QIAzol (Qiagen), followed by cleanup with the RNeasy kit (Qiagen) according to the protocols provided by the manufacturer. mRNAs from each sample were amplified, labeled, and purified using the GeneChip 3′IVT express kit (Affymetrix, Santa Clara, CA) to obtain biotin-labeled cRNA probes. After hybridization to the Affymetrix X. laevis 2.0 microarray using the protocols provided by the manufacturer, slides were scanned by a GeneChip® Scanner 3000 (Affymetrix, Santa Clara) and Command Console software 3.1 (Affymetrix) with default settings. Raw data were normalized by MAS 5.0 algorithm using Gene Spring software 11.0 (Agilent Technologies). Microarray hybridization and analysis were carried out by Shanghai Biotechnology Cooperation (Shanghai, China).
RT-PCR
To analyze gene expression using RT-PCR, cells were harvested 72 h post-transfection, or Xenopus embryos were collected at the desired stages. Total RNAs were extracted from cells or embryos with QIAzol lysis reagent (Qiagen) and cleaned up with the RNeasy kit (Qiagen). cDNAs were transcribed from the purified total RNAs using the RevertAid first strand cDNA synthesis kit (Fermentas) and the provided work manual. Semiquantitative PCR was performed on a conventional thermocycler. GAPDH was used as a loading control for gene expression in cells, whereas odc was used as a loading control in Xenopus embryos. Primers were as follows: β-Catenin, 5′-GATGTAGAAACAGCTCGTTGTACCG-3′ (forward) and 5′-GCAGCATCTGAAAGATTCCTGAGA-3′ (reverse); JMJD6, 5′-TCCCAGGGAACTCATCAAAG-3′ (forward) and 5′-TACCGTCTTGTGCCATACCA-3′ (reverse); TCF7L1, 5′-CCCAGAGATCGATCCAAAGA-3′ (forward) and 5′-AGAGAACCGACTGGAGACCA-3′ (reverse); GAPDH, 5′-GTCAGTGGTGGACCTGACCT-3′ (forward) and 5′-CCCTGTTGCTGTAGCCAAAT-3′ (reverse); jmjd6a, 5′-GGGATAACGTTGAGCGTGCG-3′ (forward) and 5′-CCATAGCTGCTATCAAAGATGTACAAA-3′ (reverse); jmjd6b, 5′-GTGCTGGATAATGTTGAGCGTGTT-3′ (forward) and 5′-ATAGCTGCTATCAAAGATGTACAGG-3′ (reverse); and odc, 5′-GGGCAAAGGAGCTTAATGTG-3′ (forward) and 5′-CATTGGCAGCATCTTCTTCA-3′ (reverse).
Whole-mount in Situ Hybridization
Control or injected embryos were collected at the desired stages, and whole-mount in situ hybridization was carried out essentially as described previously (32).
Results
Jmjd6 Interacts with Tcf7l1
In a screening of the interaction partners for Tcf7l1, we identified that Jmjd6 bound to Tcf7l1. Jmjd6 is a highly conserved protein during evolution. Sequence analysis showed that Jmjd6, in vertebrates such as human, Xenopus, and zebrafish, shares more than 80% similarity (Fig. 1A). The Jmjd6 in Drosophila is more than 60% similar to the protein sequence in vertebrates (Fig. 1A). Even in the basal species of multicellular animal Trichoplax adhaerens, there exists a putative Jmjd6 homologous protein that is about 60% similar to the proteins in higher animals (Fig. 1A). Phylogenetic analysis demonstrates that Jmjd6 proteins in Trichoplax and Drosophila are evolutionarily close to each other and that the proteins in vertebrates are closer (Fig. 1B). The conservation implies that Jmjd6 might have fundamental functions that are necessary for the survival of an organism. In HEK293T cells, we could confirm by using protein co-IP that overexpressed Xenopus Jmjd6a and Tcf7l1 interacted with each other (Fig. 2A). Moreover, overexpressed Xenopus Tcf7l1 was able to precipitate the endogenous JMJD6 in HEK293T cells (Fig. 2B). In immunofluorescence assays, both transfected Jmjd6 and Tcf7l1 colocalized in the cell nuclei (Fig. 2C). We then tested whether the endogenous JMJD6 protein would interrupt the interaction between Jmjd6 and Tcf7l1 using a vector-based miRNA knockdown approach. Two miRNAs against JMJD6, miJMJD6-1, and miJMJD6-2, were designed, and a standard negative miRNA was used as a control (miCtrl). Transfection of the plasmid for miJMJD6-1 did not cause a significant decrease in the levels of JMJD6 transcript (Fig. 2D) and protein (Fig. 2E), showing an ineffective knockdown. By contrast, transfection of the plasmid for miJMJD6-2 resulted in a significant decrease in the JMJD6 transcript (Fig. 2F) and, accordingly, a reduction in JMJD6 protein (Fig. 2G), indicating a better efficiency to block endogenous JMJD6 in HEK293T cells. In this case, we observed that there was no significant change in the level of β-Catenin and TCF7L1 transcripts (Fig. 2F). We then tested the interaction of transfected Jmjd6 and Tcf7l1 in response to JMJD6 knockdown. Co-IP demonstrated that a slightly stronger Jmjd6-Tcf7l1 complex was formed when JMJD6 was knocked down (Fig. 2H), suggesting that the reduction in endogenous JMJD6 resulted in a weaker competition with Jmjd6 for Tcf7l1 binding. These lines of evidence demonstrate that Jmjd6 is an interaction partner for Tcf7l1. In the Wnt/β-catenin signaling pathway, β-catenin forms a complex with Tcf7l1 to activate target gene transcription. In Co-IP assays, we did not observe the interaction between Jmjd6 and β-catenin (data not shown).
FIGURE 1.
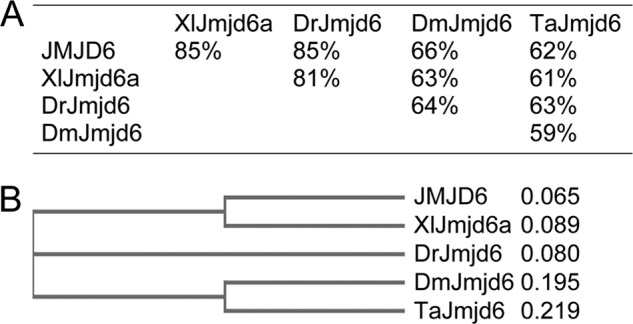
Conservation of Jmjd6 in multicellular organisms. A, sequence similarity analysis for Jmjd6 proteins in humans (JMJD6, accession no. NP_055982.2), X. laevis (XlJmjd6a, accession no. NP_001085948.1), zebrafish (DrJmjd6, accession no. NM_170761.2), fruit fly (DmJmjd6, accession no. NP_651026.1), and T. adhaerens (TaJmjd6, accession no. XM_002107775.1). B, phylogenetic tree showing the evolutionary distances between the Jmjd6 proteins in different species. The sequence comparison was made using an online program with default settings.
To figure out the region in Tcf7l1 that is responsible for Jmjd6 interaction, we made a series of deletion mutants of Tcf7l1 (Fig. 3A) and tested the interaction between Jmjd6 and the mutants. As a positive control, wild-type Tcf7l1 interacted with Jmjd6, whereas the mutants showed a different binding affinity to Jmjd6. Both the N-terminal region, aa 1–323, and the C-terminal region, aa 324–551, exhibited barely detectable binding to Jmjd6 (Fig. 3B), implying that the interaction region might be in the middle region of Tcf7l1. However, the isolated middle region, aa 274–447, also showed a rather weak interaction with Jmjd6 (Fig. 3B). When the aa 108–313 region, which is the Groucho binding domain (2), was deleted, the resulting mutant, Tcf7l1Δ(108–313), bound weakly to Jmjd6. In contrast, a strong interaction was observed between Jmjd6 and the mutant with truncation of a narrower region, aa 274–323 (Fig. 3B).
In another experiment, when the N-terminal region, aa 1–323, was extended to aa 1–410, a strong interaction with Jmjd6 was regained, comparable with the interaction between the wild-type protein (Fig. 3C). The result suggests that the 323–410 aa region significantly enhances the binding affinity between Jmjd6 and Tcf7l1. We further deleted the N-terminal sequences aa 1–105 and aa 1–199 from the aa 1–400 region. However, both mutants, Tcf7l1(106–410) and Tcf7l1(200–410), lost binding to Jmjd6. Therefore, the aa 1–105 region is also required for the interaction between Jmjd6 and Tcf7l1. There is a nuclear localization signal (NLS), aa 399–405 (KKKKRKR), in Tcf7l1. This raised the question of whether the inability of the mutant Tcf7l1(1–323) to bind to Jmjd6 was due to the inappropriate subcellular distribution of the Tcf7l1 mutant. Immunofluorescence showed that Tcf7l1(1–323) was indeed distributed primarily in the cytosol, with a minor part in the nucleus. Addition of an NLS enabled the mutant to distribute only in the nucleus, resembling the wild-type Tcf7l1 (Fig. 3D). We observed that nuclear distribution of Tcf7l1(1–323) did not enhance any interaction (Fig. 3E). In addition, the mutant dnTcf7l1, in which the β-catenin binding region, aa 1–32, was deleted, displayed an even stronger interaction with Jmjd6 (Fig. 3E).
Taking these results together, we concluded that the aa 33–410 region of Tcf7l1 is the binding domain between Tcf7l1 and Jmjd6 and that disruption of this region will compromise Tcf7l1-Jmjd6 complex formation. Because the Groucho-related corepressor also binds to this region (2), Jmjd6 should compete with Groucho for Tcf7l1 interaction. The binding affinity of different Tcf7l1 mutants to Jmjd6 is also summarized in Fig. 3A.
Jmjd6 Up-regulates Wnt Signaling
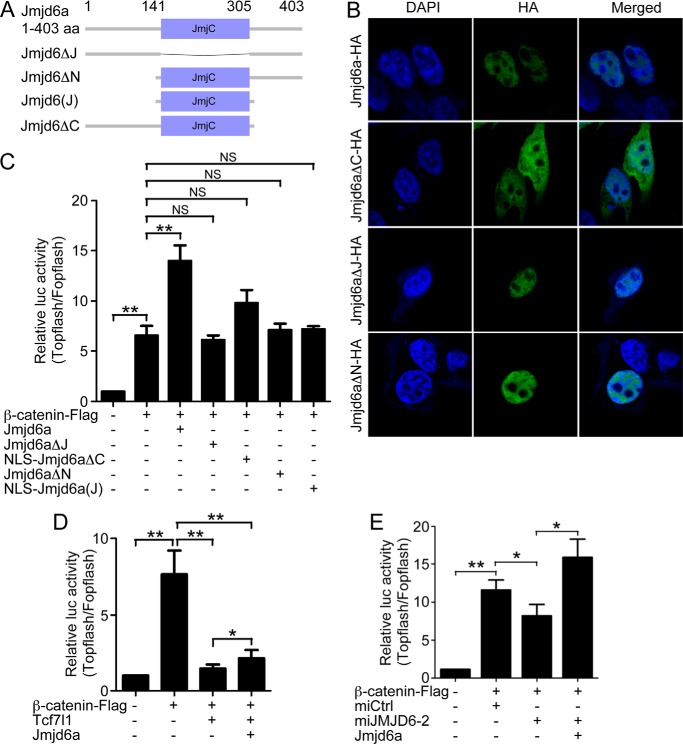
We next explored the functional significance of the interaction between Jmjd6 and Tcf7l1. Because Tcf7l1 transduces Wnt signaling upon Wnt activation, we tested the effect of Jmjd6 on Wnt transduction using the Wnt-responsive luciferase reporter Topflash, whereas Fopflash was used as a negative control. The Jmjd6 protein sequence is composed of a JmjC domain, which is responsible for the catalytic activity of the protein, in the middle and the flanking regions at both termini (Fig. 4A). We also constructed deletion mutants that did not contain the JmjC domain (Jmjd6ΔJ), the N-terminal region (Jmjd6ΔN), the C-terminal region (Jmjd6ΔC), or a mutant that contained only the JmjC domain (Jmjd6(J)) (Fig. 4A). Jmjd6 contains multiple putative NLSs. We observed in immunofluorescence assays that deletion of the JmjC domain or the N-terminal region did not affect nuclear localization, whereas loss of the C-terminal domain abolished nuclear distribution (Fig. 4B). This demonstrates that the C-terminal region is required for nuclear localization of Jmjd6. To avoid the possible effect of inappropriate subcellular distribution, we added an NLS to the mutants Jmjd6ΔC and Jmjd6(J). We then compared the effect of Jmjd6 and each mutant on reporter activity. In HEK293T cells, Topflash reporter activity was stimulated strongly in response to transfection of the β-catenin plasmid. When the plasmid for wild-type Jmjd6a was cotransfected, a much stronger stimulation was observed (Fig. 4C), suggesting that Jmjd6 enhances Wnt signaling. In contrast to the activity of wild-type Jmjd6, the mutants Jmjd6ΔJ, Jmjd6ΔN, and NLS-Jmjd6(J) showed no enhancing effect on β-catenin-stimulated Topflash activity (Fig. 4C). NLS-Jmjd6ΔC displayed a weak stimulating effect compared with the wild type and other mutant proteins (Fig. 4C). The result indicates that not only the catalytic domain but also the flanking sequences are required for Jmjd6 activity to enhance reporter activity.
FIGURE 4.
Jmjd6 enhances Wnt signaling. A, domain structure and construction of the deletion mutants of X. laevis Jmjd6a used in this study. B, subcellular localizations of the Jmjd6 mutants revealed by immunofluorescence. Jmjd6 without the C-terminal region lost nuclear localization and distributed ubiquitously throughout the whole cell. C, Jmjd6 strongly enhanced β-catenin-stimulated reporter activity, whereas the deletion mutants of Jmjd6 did not show such an enhancing effect. Error bars represent mean ± S.E. of four replicates. **, p < 0.01; NS, not significant. D, Tcf7l1 overexpression showed a strong repressive effect on β-catenin-stimulated reporter activity, whereas cotransfection of the plasmid for Jmjd6 led to a significant alleviation of the repressive effect. Error bars represent mean ± S.E. of four replicates. *, p < 0.05; **, p < 0.01. E, Jmjd6 is required for β-catenin-stimulated transcription. In HEK293T cells, β-catenin strongly stimulated the reporter activity. Simultaneous knockdown of endogenous JMJD6 compromised the stimulation of the reporter activity, which was then rescued by the cotransfection of the plasmid for X. laevis Jmjd6a. Error bars represent mean ± S.E. of four replicates. *, p < 0.05; **, p < 0.01.
Next we tested the effect of Jmjd6 on the repressive activity of Tcf7l1. Transfection of the plasmid for Tcf7l1 strongly inhibited β-catenin-stimulated Topflash. When Jmjd6 was cotransfected, Tcf7l1 repression was relieved (Fig. 4D). In contrast to Jmjd6 overexpression, knockdown of endogenous JMJD6 significantly weakened β-catenin-stimulated Topflash activity (Fig. 4E). When the plasmid for Xenopus Jmjd6a was cotransfected, reporter activity rose up again (Fig. 4E), showing a significant rescue effect. In summary, by both overexpression and knockdown approaches, we demonstrate that Jmjd6 is an interaction partner for Tcf7l1 to alleviate the transcriptional repression to maintain the transcription of β-catenin target genes.
Jmjd6 Is Expressed during Early Embryogenesis of X. laevis
Wnt/β-catenin signaling plays critical roles for embryonic development and is conserved in all multicellular organisms. Jmjd6 is also a highly conserved protein throughout metazoans. We subsequently examined the tempospatial expression patterns of jmjd6 during early embryogenesis, which would provide some clues regarding the potential roles of Jmjd6 during early embryogenesis. There are two pseudoalleles in Xenopus, jmjd6a (accession no. NM_001092479) and jmjd6b (accession no. NM_001087045), which encode two proteins that share 94% identity (data not shown). During Xenopus embryogenesis, jmjd6a and jmjd6b were both maternally transcribed because they were detected in four-cell stage embryos by both RT-PCR and whole-mount in situ hybridization (Fig. 5, A–C). Zygotic transcription of jmjd6a and jmjd6b decreases during gastrulation and rises up again at neurula stage (st. 15, Fig. 5A). Spatially, both jmjd6a and jmjd6b transcripts were primarily detected at the animal pole of cleavage (four-cell and st. 8) and gastrula (st. 10.5) embryos (Fig. 5, B and C). In neurula embryos at st. 17, transcripts were enriched in the neural plates (Fig. 5, B and C). Afterward, at the late neurula and tailbud stages (st. 18, st. 19, st. 25, and st. 30), the transcripts were localized to the nervous system, including the forming brains, neural tube, eyes, otic vesicle, and branchial arches that are neural crest derivatives (Fig. 5, B and C). This pattern of expression was also observed for the jmjd6 homologous gene in zebrafish, psr (25). Interestingly, the spatial expression patterns of jmjd6a and jmjd6b are strikingly similar to that of tcf7l1 (33), suggesting that Jmjd6a/b and Tcf7l1 colocalize in embryos and function cooperatively during embryonic development.
FIGURE 5.
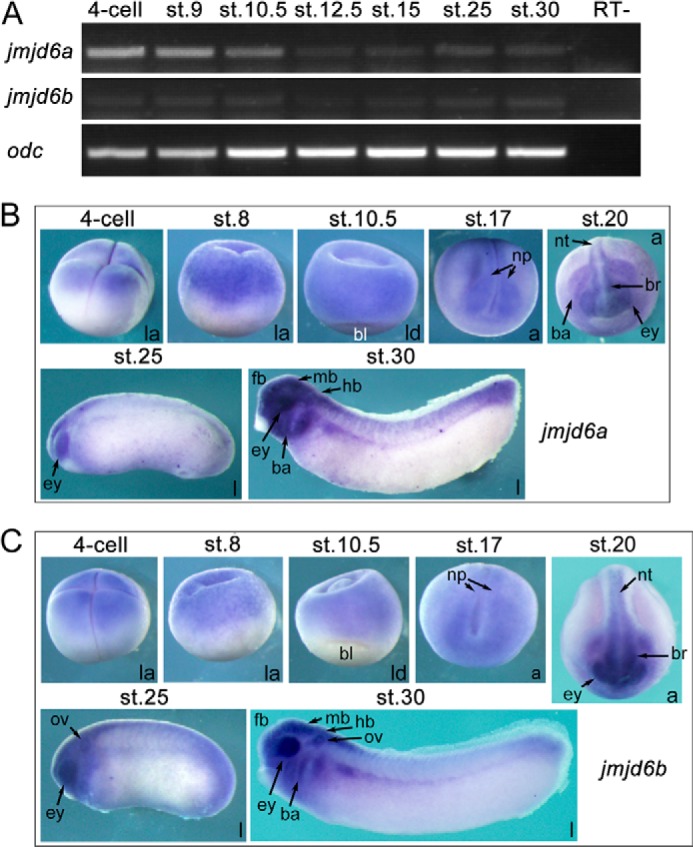
The spatiotemporal expression patterns of jmjd6a and jmjd6b during the embryogenesis of X. laevis. A, temporal expression of jmjd6a (accession no. NM_001092479) and jmjd6b (accession no. NM_001087045) in different stages (st.) of embryos as detected with RT-PCR. Expression of odc was used as a loading control. RT−, transcription without reverse transcriptase. B and C, spatial expression patterns of jmjd6a (B) and jmjd6b (C) as detected with whole-mount in situ hybridization. Embryo stages are indicated at the top of each panel. a, anterior view with the dorsal at the top of the panel; ba, branchial arch; bl, blastopore lip; br, brain; ey, eye; fb, forebrain; hb, hindbrain; l, lateral view with the anterior to the left; la, lateral view with animal pole at the top; ld, lateral-dorsal view with the animal pole at the top; mb, midbrain; np, neural plate; nt, neural tube; ov, otic vesicle.
Jmjd6 Is Required for Body Axis Formation in Xenopus Embryos
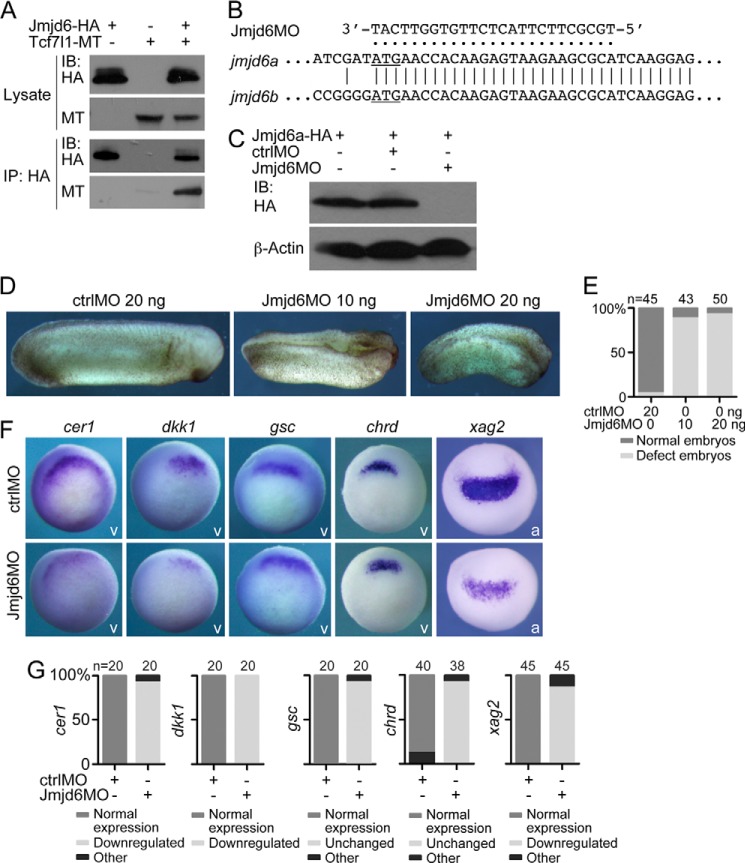
Before examination of the potential function of Jmjd6 during Xenopus embryogenesis, we investigated whether the two proteins bound together in Xenopus embryos as in HEK293T cells. Because the antibodies against Xenopus Jmjd6 and Tcf7l1 were not available, we overexpressed tagged Xenopus Jmjd6 and Tcf7l1 in embryos via microinjection of their mRNAs. Co-IP showed that the two proteins indeed interacted with each other in embryos (Fig. 6A). To examine the function of Jmjd6a/b during early embryonic development, we designed an antisense morpholino oligonucleotide, Jmjd6MO, that perfectly matches the transcripts of both jmjd6a and jmjd6b from the translational start site so that the function of both proteins could be blocked (Fig. 6B). Immunoblotting showed that Jmjd6MO could efficiently inhibit the translation of overexpressed Jmjd6a in Xenopus embryos via mRNA injection, whereas a standard control morpholino (ctrlMO) could not (Fig. 6C). In Xenopus embryos, injection of the ctrlMO did not disturb embryonic development. In contrast, injection of a low dose of Jmjd6MO at 10 ng resulted in shortening of the anterioposterior body axis and failure of neural tube closure and formation of anterior structures (Fig. 6, D and E). A higher injected dose at 20 ng caused a stronger phenotype, showing a dose-dependent effect (Fig. 6, D and E). In agreement, the expression of cer1 and dkk1, which are required for the formation of anterior structures (34, 35), and the expression of xag2, a marker gene for the most anterior structure, were reduced significantly in the Jmjd6 morphant, whereas chrd and gsc expression was not changed (Fig. 6, F and G). These results suggest that Jmjd6 is involved in the anterioposterior body axis formation of Xenopus embryonic development.
FIGURE 6.
Jmjd6 is required for Xenopus embryonic development. A, co-IP detection of the interaction between Jmjd6 and Tcf7l1 in Xenopus embryos. Each 300 pg of mRNA for tagged Jmjd6 or/and Tcf7l1 was injected into two-cell stage embryos. At gastrula stage, embryos were collected and subjected to co-IP assays. IB, immunoblot. MT, myc tag. B, design of an antisense morpholino oligonucleotide, Jmjd6MO, against both the transcripts of jmjd6a and jmjd6b. The translational start site of each transcript is underlined. C, immunoblot detection of the knockdown efficiency of the Jmjd6MO compared with ctrlMO. One nanogram of mRNA for HA-tagged Jmjd6a that contains the Jmjd6MO binding site was injected alone or coinjected with 20 ng of either ctrlMO or Jmjd6MO into Xenopus embryos. Embryos were collected at gastrula stage and subjected to immunoblotting. D, typical Xenopus embryos after injection of ctrlMO or different doses of Jmjd6MO showing a severe developmental defect in response to Jmjd6 knockdown. The defect became stronger in response to a higher dose of injected Jmjd6MO. Embryos are shown in lateral view, with the anterior at the right. E, numbers and percentages of total and defective embryos in the experiments shown in D. F, different changes in the expression of genes that are involved in anterioposterior body axis patterning in gastrula (for cer1, dkk1, gsc, and chrd) and neurula (for xag2) embryos after injection of 20 ng of ctrlMO or Jmjd6MO. v, vegetal view with the dorsal being orientated to the top; a, anterior view with the dorsal being placed at the top. G, numbers and percentages of embryos showing different changes in gene expression after Jmjd6 knockdown.
Jmjd6 Alleviates the Repression Effect of Tcf7l1 in Xenopus Embryos
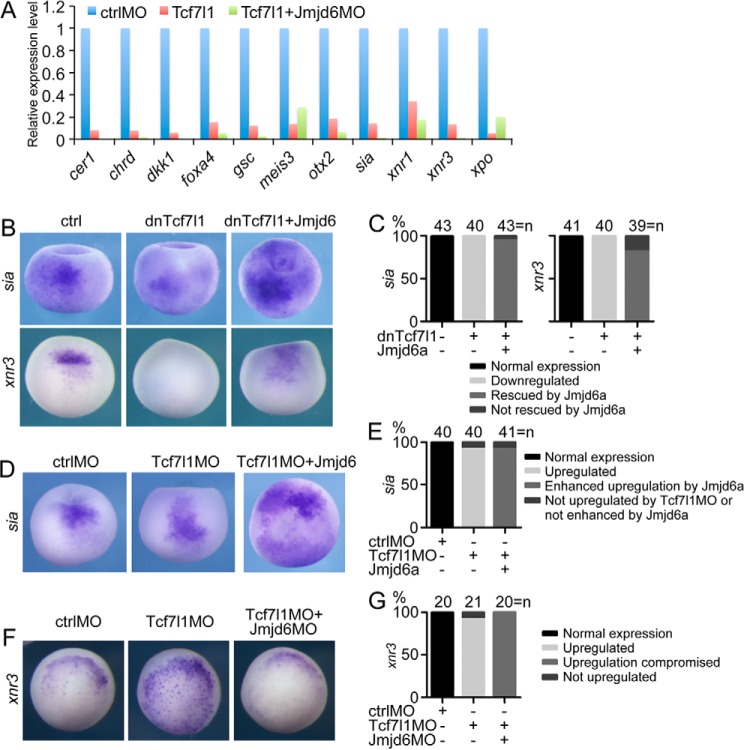
In the signaling cascade that triggers germ layer induction and patterning, the maternal β-catenin·Tcf7l1 complex stimulates the transcription of the gene for Sia, which subsequently activates dorsal genes such as cer1, nog, and so forth, in the Spemann organizer (36–38). The complex also activates xnr3, which is involved in the neurulization of ectoderm, and the Nodal related genes, including xnr1, at the dorsal side to establish a Nodal gradient (36–38). Because Jmjd6 interacts with Tcf7l1, we first detected the effect of Jmjd6 knockdown on Tcf7l1-mediated gene expression using microarray analysis. As reported previously (11), Tcf7l1 overexpression in Xenopus embryos led to the down-regulation of the β-catenin/Tcf7l1 direct target genes sia, xnr1, and xnr3 and other dorsal or anterior genes such as cer1, chrd, dkk1, foxa4, gsc, and otx2. Simultaneous knockdown of Jmjd6 via Jmjd6MO injection led to a more dramatic inhibition of dorsal genes (Fig. 7A). Tcf7l1 overexpression also repressed the ventroposterior genes meis3 and xpo. Instead, Jmjd6 knockdown did not enhance the repression of the genes (Fig. 7A). This implies that Jmjd6 knockdown tends to increase the repression effect mediated by Tcf7l1.
FIGURE 7.
Jmjd6 mediates Tcf7l1-regulated gene transcription. A, microarray result showing that Jmjd6 knockdown in X. laevis embryos enhanced the repressive effect of Tcf7l1 on the transcription of Wnt/β-catenin target genes involved in germ layer induction and patterning but did not enhance the effect on other genes. B, in Xenopus late blastula embryos, injection of mRNA (200 pg) for dominant-negative Tcf7l1 that lacks the β-Catenin binding site led to the repression of Wnt target genes. Coinjection of Jmjd6a mRNA (1 ng) reversed the repression effect. All embryos are shown in dorsal view with the animal pole being orientated to the top. ctrl, control. C, numbers and percentages of embryos showing different changes in gene expression in response to injection of dnTcf7l1 and Jmjd6 mRNA, as shown in the experiments in B. D, knockdown of endogenous Tcf7l1 in Xenopus late blastula embryos via injection of Tcf7l1MO (40 ng) up-regulated the Wnt target gene, whereas simultaneous injection of Jmjd6a mRNA (1 ng) enhanced the up-regulation of gene expression and even caused ectopic transcription. In the left and center panels, embryos are shown in dorsal view with the animal pole being placed at the top. In the right panel, the embryo is shown in animal view. sia was even ectopically expressed in the animal pole. E, numbers and percentages of embryos showing different changes in gene expression in response to injection of Tcf7l1MO and Jmjd6 mRNA, as shown in the experiments in D. F, injection of Tcf7l1MO (40 ng) caused an increase in expression of the Wnt target gene in Xenopus blastula embryos. However, the increase was weakened severely when Jmjd6MO (20 ng) was injected at the same time. G, numbers and percentages of embryos showing different changes in gene expression in response to injection of Tcf7l1MO and Jmjd6MO, as shown in the experiments in F.
We next examined the cooperative effect between Jmjd6 and Tcf7l1 in the regulation of gene expression by using different combinations of overexpression and knockdown of Jmjd6 and Tcf7l1. First we tested the effect of overexpression of both Tcf7l1 and Jmjd6. Injection of the mRNA for dnTcf7l1 resulted in a dramatic inhibition of sia and xnr3 expression in late blastula or early gastrula embryos, as reported previously (12, 13). In contrast, simultaneous injection of Jmjd6a mRNA strongly reversed the inhibitory effect of Tcf7l1, and the coinjected embryos showed elevated expression of sia and xnr3 compared with control embryos (Fig. 7, B and C). Second, we analyzed the effect of Tcf7l1 knockdown and overexpression of Jmjd6. When endogenous Tcf7l1 was blocked with a previously characterized morpholino (31), an increase in the expression of sia was observed, as reported previously (11). Coinjection of Jmjd6 mRNA synergistically augmented the increase in sia expression (Fig. 7, D and E). Finally, we tested the effect of knockdown of both Tcf7l1 and Jmjd6. In the Tcf7l1 morphant, the expression of xnr3 was also up-regulated. However, the up-regulation was weakened significantly in response to the simultaneous knockdown of Jmjd6 (Fig. 7, F and G). These results are in agreement with those in Topflash reporter assays in cells. In summary, from the results obtained with both cells and embryos, we conclude that Jmjd6 alleviates the repressive effect of Tcf7l1 on gene transcription.
On the basis of this study, we propose a novel model for the modulation of Tcf7l1 activity (Fig. 8). In the absence of Wnt activation or/and Jmjd6, Tcf7l1 is bound by Groucho-related transcriptional corepressors and functions as a transcription repressor. In the presence of either Wnt activation or Jmjd6, β-catenin or Jmjd6 binds to Tcf7l1, displaces Groucho, and turns the transcription repressor into an activator. When β-catenin and Jmjd6 interact simultaneously with Tcf7l1, the transcription activation by Tcf7l1 will be synergistically enhanced. This manner of regulation is important for the balance between gene repression and activation, which is critical for normal cellular functions.
FIGURE 8.
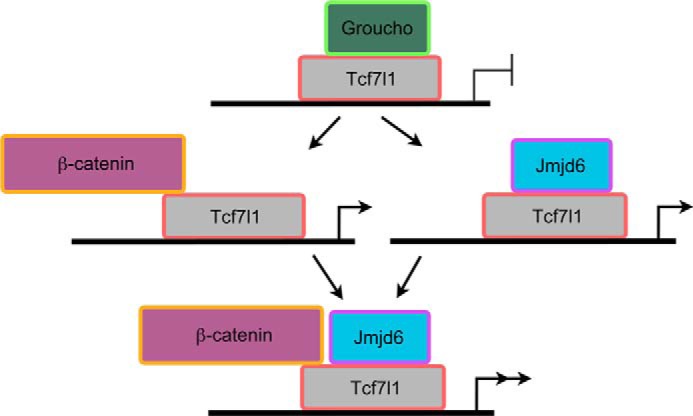
A model demonstrating the function of Jmjd6 in regulating the activity of Tcf7l1. In the absence of Wnt activation, Tcf7l1 recruits Groucho-related proteins and functions as a transcriptional repressor to inhibit the transcription of Wnt target genes. Otherwise, Wnt activation leads to β-catenin displacement of Groucho from Tcf7l1 and turns Tcf7l1 into an activator. Jmjd6 interacts with Tcf7l1 in the Groucho-binding domain, leading to alleviation of the repression activity of Tcf7l1 even in the absence of Wnt activation. This interaction enhances the transcriptional activation that is stimulated by Wnt signaling. See text for details.
Discussion
In this study, we identified a mechanism by which Jmjd6 antagonizes the repression activity of Tcf7l1 by direct interaction. Tcf7l1 plays two opposite roles in regulating Wnt pathway, which is switched by the accessibility to Wnt-activated β-catenin. This raises the question of how to control the repressive effect of Tcf7l1 to allow the transcription of Wnt target genes. Our results demonstrate that Jmjd6 should play a role in the regulation of Tcf7l1 activity. This regulation by Jmjd6 does not occur at the transcriptional level of tcf7l1 because Jmjd6 knockdown or overexpression did not result in the change of the tcf7l1 transcript in Xenopus embryos (data not shown) or the TCF7L1 transcript in cells. Instead, the regulation is manifested by the direct interaction between Jmjd6 and Tcf7l1 and by the analyses of reporter activity in cells and Wnt/β-catenin target gene expression in Xenopus embryos. Gain of Jmjd6 function enhances the transcription of Wnt/β-catenin target genes and alleviates Tcf7l1 repression, whereas loss of function leads to compromised target gene expression and more severe repression by Tcf7l1. These data suggest that Jmjd6 plays a role to derepress Tcf7l1 and maintains Wnt target gene transcription. Their functional relevance is also supported by the colocalization of their transcripts during Xenopus embryonic development.
Jmjd6 binds to Tcf7l1 in a region that includes the Groucho binding site. Jmjd6 binding displaces Groucho from Tcf7l1 and, consequently, relieves the repression activity of Tcf7l1. Therefore, Jmjd6 is similar to β-catenin in its ability to antagonize Tcf7l1 activity. β-catenin is able to recruit transcriptional coactivators like p300 to activate gene transcription (39). Nevertheless, it is not clear whether Jmjd6 can also recruit certain coactivators. Former screening studies have shown that the major part of Jmjd6-binding proteins is RNA-binding, and coactivators or corepressors were not identified in these studies that interact with Jmjd6 (19, 40). Arginine demethylation or lysyl hydroxylation modification by Jmjd6 regulates the functions of the interaction partners (19, 21, 41). Therefore, it is highly possible that Jmjd6 modifies Tcf7l1 and, consequently, alters the transcription activity of Tcf7l1. Modification-regulated Tcf7l1 activity has been reported. Wnt activation leads to the phosphorylation of Tcf7l1 and, subsequently, the derepression of target genes (4). Whether Jmjd6 modifies Tcf7l1 and the function of the modification remain intriguing topics to investigate.
Loss of Jmjd6 function in Xenopus embryos leads to severe developmental defects, including a decreased anterior structure and failure of neural tube closure. Abnormal neural development was also observed in the mouse Jmjd6 knockout phenotype (24). In Xenopus early embryos, Wnt/β-catenin signaling is responsible for pattern formation of the anterioposterior body axis and neuralization of the ectoderm. We show that Jmjd6 functional knockdown causes a deficiency in Wnt signaling and, accordingly, the embryonic defects that are similar to the phenotype after inhibition of β-catenin in Xenopus embryos (42). In agreement, the expression of genes that are required for body axis patterning is also down-regulated. These results provide further insights into the mechanisms that control body axis formation in Xenopus embryos.
It is well known that aberrant activation of Wnt/β-catenin signaling is involved in the progression of many cancers. Emerging evidence indicates that the elevated expression of Jmjd6 in some cancers predicts poor prognosis (21, 26, 27). A mechanistic study has demonstrated that Jmjd6 promotes colon carcinogenesis via negative regulation of the tumor repressor p53 (21). Our results suggest that enhancement of Wnt signaling by Jmjd6 via negative regulation of Tcf7l1 might account for the functions of Jmjd6 in cancer progression.
In summary, this study elucidates a novel mechanism for the regulation of canonical Wnt signaling via Jmjd6-mediated Tcf7l1 derepression. Considering the omnipresent roles of Wnt signaling and the high conservation of Jmjd6 in different species, this mechanism is worth further studies not only in embryogenesis but also in fields such as stem cell biology and cancer biology.
Author Contributions
Y. C. and X. Z. designed the research. X. Z., S. G., Z. Z., and L. X. performed the biochemical experiments. X. Z., Y. G., and L. L. carried out the experiments with Xenopus embryos. X. Z., S. G., and A. L. constructed the plasmids. Y. C. wrote the paper with consent from all authors.
This work was supported by Ministry of Science and Technology Grants 2011CB943804 and 2014CB964701, National Science Foundation of China Grants 31271544 and 31261160492, and Key Laboratory of Regenerative Biology of the Chinese Academy of Sciences Grant KLRB201306 (to Y. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- IP
- immunoprecipitation
- aa
- amino acid(s)
- NLS
- nuclear localization signal.
References
- 1. Clevers H., van de Wetering M. (1997) TCF/LEF factor earn their wings. Trends Genet. 13, 485–489 [DOI] [PubMed] [Google Scholar]
- 2. Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O., Clevers H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395, 608–612 [DOI] [PubMed] [Google Scholar]
- 3. Daniels D. L., Weis W. I. (2005) β-Catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12, 364–371 [DOI] [PubMed] [Google Scholar]
- 4. Hikasa H., Ezan J., Itoh K., Li X., Klymkowsky M. W., Sokol S. Y. (2010) Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev. Cell 19, 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Molenaar M., van de Wetering M., Oosterwegel M., Peterson-Maduro J., Godsave S., Korinek V., Roose J., Destrée O., Clevers H. (1996) XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell 86, 391–399 [DOI] [PubMed] [Google Scholar]
- 6. Kim C. H., Oda T., Itoh M., Jiang D., Artinger K. B., Chandrasekharappa S. C., Driever W., Chitnis A. B. (2000) Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 407, 913–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu C. I., Hoffman J. A., Shy B. R., Ford E. M., Fuchs E., Nguyen H., Merrill B. J. (2012) Function of Wnt/β-Catenin in counteracting Tcf3 repression through the Tcf3-β-Catenin interaction. Development 139, 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nguyen H., Rendl M., Fuchs E. (2006) Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127, 171–183 [DOI] [PubMed] [Google Scholar]
- 9. Tam W. L., Lim C. Y., Han J., Zhang J., Ang Y. S., Ng H. H., Yang H., Lim B. (2008) T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells 26, 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cole M. F., Johnstone S. E., Newman J.J., Kagey M. H., Young R. A. (2008) Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 22, 746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Houston D. W., Kofron M., Resnik E., Langland R., Destree O., Wylie C., Heasman J. (2002) Repression of organizer genes in dorsal and ventral Xenopus cells mediated by maternal XTcf3. Development 129, 4015–4025 [DOI] [PubMed] [Google Scholar]
- 12. Brannon M., Gomperts M., Sumoy L., Moon R. T., Kimelman D. (1997) A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 11, 2359–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McKendry R., Hsu S. C., Harland R. M., Grosschedl R. (1997) LEF-1/TCF proteins mediate Wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev. Biol. 192, 420–431 [DOI] [PubMed] [Google Scholar]
- 14. Andoniadou C.L., Signore M., Young R.M., Gaston-Massuet C., Wilson S.W., Fuchs E., Martinez-Barbera J.P. (2011) HESX1- and TCF3-mediated repression of Wnt/β-Catenin targets is required for normal development of the anterior forebrain. Development 138, 4931–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Merrill B. J., Pasolli H. A., Polak L., Rendl M., García-García M. J., Anderson K. V., Fuchs E. (2004) Tcf3: a transcriptional regulator of axis induction in the early embryo. Development 131, 263–274 [DOI] [PubMed] [Google Scholar]
- 16. Kuwahara A., Sakai H., Xu Y., Itoh Y., Hirabayashi Y., Gotoh Y. (2014) Tcf3 represses Wnt-β-Catenin signaling and maintains neural stem cell population during neocortical development. PLoS ONE 9, e94408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klose R.J., Kallin E.M., Zhang Y. (2006) JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 18. Chang B., Chen Y., Zhao Y., Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 19. Webby C. J., Wolf A., Gromak N., Dreger M., Kramer H., Kessler B., Nielsen M. L., Schmitz C., Butler D. S., Yates J. R., 3rd, Delahunty C. M., Hahn P., Lengeling A., Mann M., Proudfoot N. J., Schofield C. J., Böttger A. (2009) Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 [DOI] [PubMed] [Google Scholar]
- 20. Unoki M., Masuda A., Dohmae N., Arita K., Yoshimatsu M., Iwai Y., Fukui Y., Ueda K., Hamamoto R., Shirakawa M., Sasaki H., Nakamura Y. (2013) Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem. 288, 6053–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang F., He L., Huangyang P., Liang J., Si W., Yan R., Han X., Liu S., Gui B., Li W., Miao D., Jing C., Liu Z., Pei F., Sun L., Shang Y. (2014) JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 12, e1001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boeckel J.N., Guarani V., Koyanagi M., Roexe T., Lengeling A., Schermuly R.T., Gellert P., Braun T., Zeiher A., Dimmeler S. (2011) Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl. Acad. Sci. U.S.A. 108, 3276–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A. K., Rosenfeld M. G. (2013) Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M. O., Sarkisian M. R., Mehal W. Z., Rakic P., Flavell R. A. (2003) Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 302, 1560–1563 [DOI] [PubMed] [Google Scholar]
- 25. Hong J. R., Lin G. H., Lin C. J., Wang W. P., Lee C. C., Lin T. L., Wu J. L. (2004) Phosphatidylserine receptor is required for the engulfment of dead apoptotic cells and for normal embryonic development in zebrafish. Development 131, 5417–5427 [DOI] [PubMed] [Google Scholar]
- 26. Lee Y. F., Miller L. D., Chan X. B., Black M. A., Pang B., Ong C. W., Salto-Tellez M., Liu E. T., Desai K.V. (2012) JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 14, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J., Ni S.S., Zhao W.L., Dong X.C., Wang J.L. (2013) High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumour Biol. 34, 2397–2401 [DOI] [PubMed] [Google Scholar]
- 28. Cao Y, Siegel D, Oswald F, Knöchel W. (2008) Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. J. Biol. Chem. 283, 34168–34177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cao Q., Zhang X., Lu L., Yang L., Gao J., Gao Y., Ma H., Cao Y. (2012) Klf4 is required for germ-layer differentiation and body axis patterning during Xenopus embryogenesis. Development 139, 3950–3961 [DOI] [PubMed] [Google Scholar]
- 30. Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knöchel W., Borggrefe T. (2005) RBP-Jκ/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 25, 10379–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu F., van den Broek O., Destrée O., Hoppler S. (2005) Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/β-Catenin signalling in mesoderm development. Development 132, 5375–5385 [DOI] [PubMed] [Google Scholar]
- 32. Harland R. M. (1991) In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods. Cell Biol. 36, 685–695 [DOI] [PubMed] [Google Scholar]
- 33. Molenaar M., Roose J., Peterson J., Venanzi S., Clevers H., Destrée O. (1998) Differential expression of the HMG box transcription factors XTcf-3 and XLef-1 during early Xenopus development. Mech. Dev. 75, 151–154 [DOI] [PubMed] [Google Scholar]
- 34. Bouwmeester T., Kim S., Sasai Y., Lu B., De Robertis E.M. (1996) Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature 382, 595–601 [DOI] [PubMed] [Google Scholar]
- 35. Glinka A., Wu W., Delius H., Monaghan A.P., Blumenstock C., Niehrs C. (1998) Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391, 357–362 [DOI] [PubMed] [Google Scholar]
- 36. Moon R.T., Kimelman D. (1998) From cortical rotation to organizer gene expression: toward a molecular explanation of axis specification in Xenopus. BioEssays 20, 536–545 [DOI] [PubMed] [Google Scholar]
- 37. Wodarz A., Nusse R. (1998) Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14, 59–88 [DOI] [PubMed] [Google Scholar]
- 38. De Robertis E.M., Larraín J., Oelgeschläger M., Wessely O. (2000) The establishment of Spemann's organizer and patterning of the vertebrate embryo. Nat. Rev. Genet. 1, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hecht A., Vleminckx K., Stemmler M. P., van Roy F., Kemler R. (2000) The p300/CBP acetyltransferases function as transcriptional coactivators of β-Catenin in vertebrates. EMBO J. 19, 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weimann M., Grossmann A., Woodsmith J., Özkan Z., Birth P., Meierhofer D., Benlasfer N., Valovka T., Timmermann B., Wanker E.E., Sauer S., Stelzl U. (2013) A Y2H-seq approach defines the human protein methyltransferase interactome. Nat. Methods 10, 339–342 [DOI] [PubMed] [Google Scholar]
- 41. Poulard C., Rambaud J., Hussein N., Corbo L., Le Romancer M. (2014) JMJD6 regulates ERα methylation on arginine. PLoS ONE 9, e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heasman J., Kofron M., Wylie C. (2000) β-Catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev. Biol. 222, 124–134 [DOI] [PubMed] [Google Scholar]