Background: ATP-DnaA molecules oligomerize and form two subcomplexes on the replication origin.
Results: The Arg fingers of DnaA bound at the outer edges of the DnaA complexes are oriented inward within the origin.
Conclusion: The Arg fingers, but not bound ATP, of the outer edge DnaA protomers promote construction of active initiation complexes.
Significance: An important mechanical basis in the initiation complex is revealed.
Keywords: bacteria, DNA replication, DNA-protein interaction, protein assembly, protein chimera, protein complex, DnaA
Abstract
ATP-DnaA binds to multiple DnaA boxes in the Escherichia coli replication origin (oriC) and forms left-half and right-half subcomplexes that promote DNA unwinding and DnaB helicase loading. DnaA forms homo-oligomers in a head-to-tail manner via interactions between the bound ATP and Arg-285 of the adjacent protomer. DnaA boxes R1 and R4 reside at the outer edges of the DnaA-binding region and have opposite orientations. In this study, roles for the protomers bound at R1 and R4 were elucidated using chimeric DnaA molecules that had alternative DNA binding sequence specificity and chimeric oriC molecules bearing the alternative DnaA binding sequence at R1 or R4. In vitro, protomers at R1 and R4 promoted initiation regardless of whether the bound nucleotide was ADP or ATP. Arg-285 was shown to play an important role in the formation of subcomplexes that were active in oriC unwinding and DnaB loading. The results of in vivo analysis using the chimeric molecules were consistent with the in vitro data. Taken together, the data suggest a model in which DnaA subcomplexes form in symmetrically opposed orientations and in which the Arg-285 fingers face inward to mediate interactions with adjacent protomers. This mode is consistent with initiation regulation by ATP-DnaA and bidirectional loading of DnaB helicases.
Introduction
Higher order complexes are constructed at the replication origin region during initiation of chromosomal replication (1). The molecular architecture of these complexes is crucial for the regulation of the replication cycle under diverse cell growth conditions. In Escherichia coli, the initiator protein ATP-bound DnaA (ATP-DnaA) and IHF, a DNA-binding protein that causes a sharp bend in DNA, respectively, bind to multiple DnaA-binding sites and an IHF-specific site within the chromosomal replication origin, oriC (Fig. 1A). This binding produces a highly ordered complex (i.e. an initiation complex) that stimulates local duplex unwinding (2–7). DnaB helicase is loaded onto the resultant single-stranded DNA via interaction with DnaA and DnaC helicase loader (3). The loaded DnaB triggers the formation of the sister replisomes needed for bidirectional DNA synthesis (8). ATP-DnaA is converted to ADP-DnaA, which is inactive for initiation, during replication (2, 7, 9, 10).
FIGURE 1.
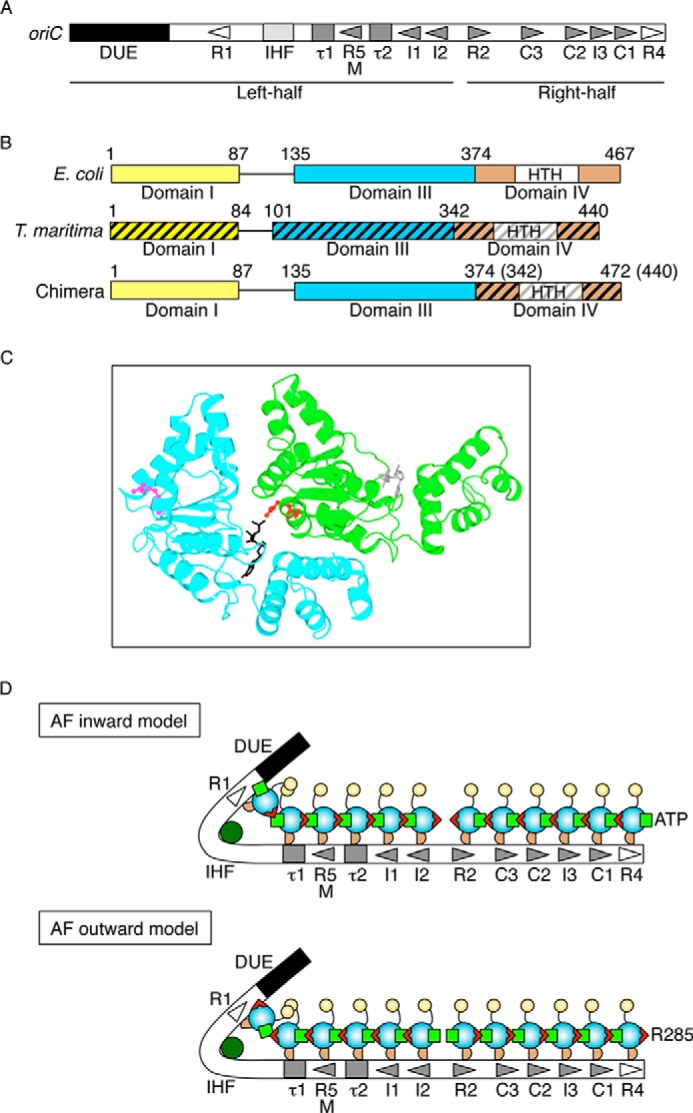
Structures of oriC and DnaA and models for DnaA complexes. A, overall structure of oriC. oriC (245 bp) includes DUE (black bar), IHF-binding site (IHF; light gray box), and multiple DnaA boxes (triangles and gray boxes). High affinity sites (R1 and R4) are indicated by an open symbol. Moderate (R2) and low affinity (R5M, τ1-2, I1-3, and C1-3) sites are indicated by filled gray symbols. The left-half and right-half regions of oriC are also indicated. B, DnaA domain structure of chiDnaA. Domains of EcoDnaA (E. coli) and TmaDnaA (T. maritima) are indicated by open and hatched boxes, respectively, corresponding to domains I, III, and IV. Lines indicate domain II. Boxes labeled HTH indicate the helix-turn-helix motif within domain IV. Domain boundary amino acid numbers are indicated. The chimeric protein constructed in this work (chiDnaA; 472 amino acid residues) contains EcoDnaA domains I–III (amino acids 1–374) and the TmaDnaA domain IV (amino acids 342–440). C, model of a dimeric structure of DnaA domain III. A homology model constructed previously using TmaDnaA sequence is shown (19). Each protomer is colored differently. The Arg finger motif of each protomer is shown using a ball-and-stick model and colored in red or pink. The bound ADP is shown using a stick model and colored in black or gray. D, models for DnaA assembly structure on oriC. DnaA domains I, II, III, and IV are indicated by a yellow circle, a curved line, a cyan circle, and a brown half circle, respectively. Domain III carries the Arg finger motif (red triangle) and bound ATP (light green square) at opposite sides of the tertiary structure. DnaA forms a homo-oligomer with a head-to-tail configuration that depends on the interaction between the Arg finger and ATP of flanking protomers. Each DnaA binds to one DnaA box. Generally, DnaA boxes in the left half of oriC and those in the right half of oriC have opposite orientations. IHF is shown in dark green. In the Arg finger-inward model (AF-inward; top), subcomplexes of DnaA formed on each oriC region have the Arg finger facing inward within oriC and the bound ATP facing outward from oriC. Conversely, in the Arg finger-outward model (AF-outward; bottom), subcomplexes of DnaA formed on each oriC region have the Arg finger facing outward from oriC and the bound ATP facing inward within oriC.
The E. coli oriC consists of an AT-rich duplex unwinding element (DUE)2 and a DnaA oligomerization region (DOR) that bears DnaA-binding sequences (EcoDnaA box) termed R1-2, R4, R5M, I1-3, τ1-2, and C1-3 (2, 11–13) in addition to an IHF-binding site (Fig. 1A). The R1 and R4 boxes reside at the outer edges of the DOR and have a typical 9-mer consensus sequence of TTATNCACA (2). R1 and R4 have the highest affinities of the EcoDnaA boxes for both ATP-DnaA and ADP-DnaA (2, 3, 12, 14, 15) and are suggested to act as assembly cores for DnaA oligomerization (13, 16). The other EcoDnaA boxes reside between the R1 and R4 boxes and have moderate (R2) or low affinities (R5M, I1-3, τ1-2, and C1-3) for DnaA. ATP-DnaA molecules form oligomers at these low affinity sites more readily than do ADP-DnaA molecules (11–14). IHF binds to a specific site between the R1 and τ1 boxes (Fig. 1A) and stimulates ATP-DnaA assembly on oriC and unwinding of the DUE (4, 11, 17).
The DnaA-binding consensus sequence is asymmetric, and bound DnaA proteins would therefore also be expected to exhibit directionality. Consistent with this suggestion, a previous study demonstrated that cell growth was inhibited when the direction of the R1 box was inverted (18). However, the directionality of DnaA proteins bound at the oriC sequences, including R1 and R4, has not yet been determined.
Previously, we suggested that oriC can be subdivided into two structurally and functionally distinct subregions: the left-half and right-half oriC (Fig. 1A) (4, 19). The left-half oriC plays a crucial role in DUE unwinding and DnaB helicase loading and contains the DUE, IHF-binding site, and DnaA-binding sites R1, R5M, τ1–2, and I1–2. The right-half oriC plays a stimulatory role in DnaB helicase loading and contains DnaA-binding sites R2, C1–3, I3, and R4. The R1 and R4 box sequence directions are opposed to one another. The right-half oriC sequences (R2, I3, and C1-3) are in the same direction as the R4 box sequence (Fig. 1A). The R5M and I1-2 sequences (left-half oriC) are in the same orientation as the R1 box sequence (Fig. 1A). The τ1 and τ2 sites (left-half oriC) exhibit degenerate sequence similarity with respect to the consensus, allowing the possibility of either direction (12, 19), but a recent study suggests that the direction is the same as that of the R1 box (13).
DnaA contains four functional domains (Fig. 1B) (3, 20). Domain I interacts with DnaB and DiaA, a protein that stimulates assembly of DnaA molecules on oriC, and has a low affinity site for homodimerization (21–23). Domain II is a flexible linker (23, 24). Domain III includes AAA+ family motifs (e.g. Walker A/B, Sensor 1/2, and Arg finger) in addition to B/H motifs and plays crucial roles in nucleotide binding, ATP hydrolysis, inter-DnaA interactions, and single-stranded DUE (ssDUE) binding (12, 19, 25–28). In particular, the Arg finger plays a predominant role in the recognition of ATP bound to DnaA (12). Domain IV contains a helix-turn-helix motif and is crucial for direct binding to DnaA boxes (Fig. 1B) (28–32).
ATP-DnaA molecules in initiation complexes form homo-oligomers that can adopt a spiral structure (12, 27, 31, 33). The Arg finger motif, Arg-285, of a DnaA protomer plays a crucial role in the cooperative binding of DnaA to the low affinity DnaA boxes of oriC (12). DnaA R285A retains the activities in DnaA box binding to the R1 and R4 boxes, nucleotide binding, and interaction with DnaB at levels similar to the wild-type DnaA; however, the ATP-DnaA R285A is similar to the ADP form of wild-type DnaA in that it is specifically impaired in binding to the low affinity DnaA boxes of oriC. Structurally, within a DnaA homo-oligomer, the Arg-285 of a DnaA protomer is thought to interact with ATP bound to the adjacent DnaA protomer, resulting in a head-to-tail interaction of DnaA protomers (Fig. 1C) (12, 27, 31).
As mentioned above, the DnaA box sequence is asymmetric, and DnaA protomers construct homo-oligomers using a head-to-tail interaction. The overall directions of the DnaA boxes and of the subregions within oriC suggest at least two possibilities for the orientation of oriC-bound DnaA protomers (Fig. 1D). The first possibility is that the adenine nucleotides of DnaA protomers bound to the R1 and R4 boxes face outward from oriC. In this case, the Arg fingers of the R1- and R4-bound protomers would be oriented inward within oriC and would be expected to mediate important interactions with adjacent protomers during assembly on each oriC subregion (AF-inward model in Fig. 1D). The second possibility is that the Arg fingers of DnaA protomers bound to the R1 and R4 boxes are oriented outward from oriC. In this case, the ATP of the R1- and R4-bound protomers would be oriented inward within oriC and would be expected to interact with adjacent protomers during assembly on each oriC subregion (AF-outward model in Fig. 1D).
To distinguish between these two possibilities, we used a DnaA ortholog (TmaDnaA) and the cognate DnaA box (TmaDnaA box) from Thermotoga maritima, one of the most ancient hyperthermophile eubacteria (34). The consensus sequence for the EcoDnaA box (TTATNCACA) is highly conserved in many bacterial species (35); however, the consensus for the TmaDnaA box, AAACCTACCACC, differs substantially (36). By contrast, TmaDnaA has 47% amino acid sequence similarity with E. coli (EcoDnaA), and the domain structures of TmaDnaA and the affinities of TmaDnaA for the TmaDnaA boxes are similar to those of EcoDnaA for the cognate consensus sequence (Fig. 1B). As with EcoDnaA, TmaDnaA has high affinity for ATP and ADP, and ATP-TmaDnaA, but not ADP-TmaDnaA, unwinds the cognate oriC in vitro (36). In this study, chimeric oriC sequences were constructed in which the R1 or R4 box of the E. coli oriC was substituted with the TmaDnaA box. Chimeric DnaA (chiDnaA) was also constructed, in which domain IV of EcoDnaA was substituted with that of TmaDnaA (Fig. 1B). These oriC and DnaA derivatives facilitated the analysis of DnaA protomer binding to the R1 and R4 boxes in vitro and in vivo. Experimental results were consistent with the AF-inward model (Fig. 1D).
Experimental Procedures
Strains
E. coli strains are listed in Table 1. A DNA fragment that included the tet gene was amplified from pBR322 using tet-oriC f and tet-oriC r primers (Table 2). This fragment was introduced into MG1655 using the λRed recombination system (38), resulting in strain SYM1 (MG1655 gidA::tet). For construction of strain NY20 (MG1655 asnA::kan), a DNA fragment including the frt-flanked kan and oriC regions was amplified from pRSoriC (see below) using primers pRS1 and pRS2 (Table 2). The resultant fragment was introduced into SYM1 cells using the λRed recombination system, and colonies that were resistant to kanamycin and sensitive to tetracycline were selected (39). A representative strain was named NY20. Strains NY21 (MG1655 oriCΔR4::TmaDnaA box asnA::kan), NY24 (MG1655 oriCΔR1::TmaDnaA-box asnA::kan), and NY25 (MG1655 oriCΔR4::R4-inverted asnA::kan) were similarly constructed using DNA fragments amplified from the pRSoriC derivatives pRSR1Tma, pRSR4Tma, and pRSR4inv (see below), respectively. The altered oriC structures in these strains were confirmed by nucleotide sequencing. Strain NY26 was constructed by transduction using a P1 phage lysate of KP7364.
TABLE 1.
Strain list
| Strain | Relevant genotype | Reference |
|---|---|---|
| MG1655 | Wild type | Laboratory stock |
| SYM1 | MG1655 gidA::tet | This work |
| NY20 | MG1655 asnA::kan | This work |
| NY21 | MG1655 asnA::kan oriCΔR1-box::TmaDnaA-box | This work |
| NY24 | MG1655 asnA::kan oriCΔR4-box::TmaDnaA-box | This work |
| NY25 | MG1655 asnA::kan oriCΔR4-box::R4-inverted | This work |
| NY26 | MG1655 ΔdnaA::spec rnhA::kan | This work |
| KH5402-1 | ilv thyA tyrA(Am) trpE9829(Am) metE deo supF6(Ts) | Ref. 37 |
| NA001 | KH5402–1 dnaAcos | Ref. 37 |
| KA451 | KH5402–1 dnaA::Tn10 rnhA::cat | Ref. 19 |
| KP7364 | ΔdnaA::spec rnhA::kan | Ref. 26 |
TABLE 2.
Primer list
| Primer | Sequence |
|---|---|
| coliDAf | GTTAGGTGGTCGTCCTACGCTACCG |
| coliDAr | CAGTTTTTCCTGCAATGCCAGCAAG |
| tmadnaa4f | CTTGACTGCAGATCCAATAGATGAACTCATAGAGATCG |
| tmadnaa4r | CATGTCTCGAGCAGACCGCTTCTGCGTTCTG |
| SUE260 | ATCCCATGGCCCGGGCCGTGGATTCTAC |
| SUE261 | CTTATGCATGAAGATCAACATTCTTGATCACG |
| DnaAI-f | CCTGTACGCGCCAAACGCGTTTGTCCTCGATTGGG |
| ori1 | ATCGCACTGCCCTGTGG |
| orir1tmafb | GGATCCGGCTTTTAAGATCAAC |
| R1tma12r | AACCTACCACCCAGTGCGATCCTAATAAG |
| R1tma12rb | TTAAACCTACCACCGCAGTGCGATCCTAATAAG |
| R1tma12rc | TAAACCTACCACCGGCAGTGCGATCCTAATAAG |
| R1tma12rd | AAACCTACCACCGGGCAGTGCGATCCTAATAAG |
| R4tmafb | GATCGCACGATCTGTATACTTATTTG |
| R4tma12rb | GGTGGTAGGTTTTGTCAGGAAGCTTG |
| ChDAB-M28-up | GATCTGTTCTATTGTGATCTCTTATTAGGATCGCACTGGGTGGTAGGTTTCAAGGA |
| ChDAB-M28-low | TCCTTGAAACCTACCACCCAGTGCGATC |
| tet-oriC f | GGGCCGTGGATTCTACTCAACTTTGTCGGCTTGAGAAAGACCTGGGATTCTCATGTTTGACAGCTTATCATCGATAAGCTTTAATGCGG |
| tet-oriC r | ATAGAACAGATCTCTAAATAAATAGATCTTCTTTTTAATACCCAGGATCAGGTCGAGGTGGCCCGGCTCCATGCACCGCG |
| pSA4 | GCTCTGCCTGATGCCCAGTTGTGTAGGCTGGAGCTGCTTC |
| pSA5 | GACGGGTGTGGTCGCCATGACATATGAATATCCTCCTTA |
| pSA6 | TCATGGCGACCACACCCGTCCTGTGGATCCTCTACGCCGG |
| pSA7 | ATCGATGATAGCTGTCAAACATGAGAATTCTTGAAGACGA |
| pSA8 | TTTGACAGCTTATCATCGATCCGAGTGCATCCACTTCTTT |
| pRS1 | TGTTCACCCATACGCGCCGCGGCCATCGCGGCCTCGGTGC |
| pRS2 | GGTCTGACGTTTCCACTTCGCCAGTGAATGAACCACTTCGCATATGAATATCCTCCTTA |
| R4inv-f | CTACCGGTTGATCCAAGCTTCCTGACAGAGTGTGGATAAGTAGATCGCACGATCTGTATACTTATTTG |
| R4inv-r | CAAATAAGTATACAGATCGTGCGATCTACTTATCCACACTCTGTCAGGAAGCTTGGATCAACCGGTAG |
Oligonucleotides and Plasmids
Oligonucleotides are listed in Table 2. Plasmids pKA234 and pTHMA-1 were used for overproduction of EcoDnaA and TmaDnaA, respectively, as described previously (19, 36). Plasmids pECTMA, pECTMAR285A, pECTMAD269A, and pECTMAdnaAcos were constructed and used for the expression of chiDnaA, chiDnaA R285A, chiDnaA D269A, and DnaAcos, respectively. To construct pECTMA, first a DNA fragment encoding TmaDnaA domain IV and a part of the vector region was amplified by reverse PCR using pTHMA-1 and primers of tmadnaA4f and tmadnaA4r (Table 2), digested with restriction enzyme PstI, and blunted using a Blunting High kit (TOYOBO). Next, a DNA fragment encoding EcoDnaA domains I–III and the vector region was amplified by PCR using pKA234 and primers of coliDAf and coliDAr (Table 2). Because the entire vector region contained a single XhoI site, each of the resultant fragments was digested with XhoI and ligated, resulting in pECTMA. pECTMAR285A and pECTMAD269A were constructed by site-directed mutagenesis using mutagenic primers as described previously (12, 26). To construct a plasmid of chiDnaA bearing the dnaAcos-specific mutations, a DNA fragment encoding DnaAcos domain III, which carries the mutations, was amplified by PCR using NA001 strain and primers DnaAR45f and coliDAr (Table 2), digested with AgeI and BsiWI, and then ligated with pECTMA fragment digested with the same restriction enzymes, resulting in pECTMAdnaAcos.
A 419-bp DNA fragment containing oriC was amplified by PCR using primers SUE260 and SUE261 and inserted in the NruI site of pBR322, resulting in pBRoriC. To construct a set of oriC mutants containing the TmaDnaA box at or near the site of the R1 box, DNA fragments were amplified by PCR using pBRoriC, a forward primer (orir1tmafb), and a set of reverse primers (R1tma12r, R1tma12rb, R1tma12rc, and R1tma12rd) and self-ligated, resulting in pR1Tma12, pR1Tma12-b, pR1Tma12-c, and pR1Tma12-d, respectively. To construct an oriC plasmid bearing the TmaDnaA box at the site of the R4 box, a fragment was similarly amplified using primers R4tma12rb and R4tma12fb and self-ligated, resulting in pR4Tma12.
pRSoriC was a pBR322-derivative oriC plasmid that contained a region including gidA′-oriC-mioC-asnC-asnA′::kan. To construct pRSoriC, first a DNA fragment containing oriC and its flanking regions was amplified by PCR using MG1655-derived genomic DNA and primers pSA3 and pSA8. In addition, an frt-kan fragment was amplified by PCR using pTH5 and primers pSA4 and pSA5 (40). In addition, a pBR322 fragment containing the origin and bla regions was amplified by reverse PCR using pBR322 and primers pSA6 and pSA7. These three fragments were ligated by a sequence- and ligation-independent cloning system using T4 DNA polymerase, resulting in pRSoriC (41). pRSoriC derivatives bearing the TmaDnaA box at the site of the R1 or R4 box were constructed by PCR as for pR1Tma12 and pR4Tma12, resulting in pRSR1Tma and pRSR4Tma. A pRSoriC derivative bearing the inverted R4 box was constructed by site-directed mutagenesis using pRSoriC and mutagenic primers R4inv-f and R4inv-r, resulting in pRSR4inv. The inverted R4 box sequence was the same as that described previously (18).
DORΔR1 was described previously as DARΔR1 (4, 19). A right-half oriC fragment R2R4 and its derivatives, R2R4-R4Tma and R2R4-R4inv were prepared by PCR using template DNA, pBRoriC, pR4Tma12, and pRSR4inv, respectively, as described previously (4, 19).
ssDUE-dsR1Tma was prepared by annealing radiolabeled ssDNA, ChDAB-M28-up, and ChDAB-M28-low (Table 2). ssDUE-dsR1 and ssDUE-dsNon were described previously (4).
Buffers
Buffer M contained 20 mm Tris-HCl (pH 7.5), 0.1 mg/ml bovine serum albumin, 8 mm dithiothreitol, 10 mm magnesium acetate, 125 mm potassium glutamate, and 2 mm ATP. Buffer G″ contained 20 mm Hepes-KOH (pH 7.6), 1 mm EDTA, 4 mm dithiothreitol, 5 mm magnesium acetate, 10% (v/v) glycerol, 1 mm ATP, 0.1% Triton X-100, 0.1 mg/ml bovine serum albumin, 4 μg/ml poly(dA-dT)-(dA-dT), and 4 μg/ml poly(dI-dC)-(dI-dC). Other buffers were described previously (19, 36).
Purification of chiDnaA Proteins
chiDnaA and its derivative proteins were overproduced using KA451 (ΔdnaA ΔrnhA) (Table 1) cells bearing pECTMA or pECTMAR285A. Proteins were purified using a previously described method for EcoDnaA purification (12, 19).
EMSA Using DNA with a Single DnaA Box
EMSA was performed with 18-bp DNA fragments containing either the TmaDnaA box consensus or a nonsense sequence and with 15-bp DNA fragments containing either the EcoDnaA R1 box or a nonsense sequence, as described previously (36, 42). Briefly, ATP-EcoDnaA or ATP-chiDnaA was incubated in buffer for 10 min at 30 °C with the appropriate DNA fragment and λDNA (50 ng) as a competitor. This incubation was followed by electrophoresis on a 5% polyacrylamide gel (PAGE) and GelStar staining (Lonza).
oriC Plasmid Replication Assay
The oriC replication assay was performed as described previously (19). Briefly, EcoDnaA was incubated on ice in buffer M containing DnaB helicase, DnaC helicase loader, DnaG primase, histone-like protein HU, gyrase, SSB (single-stranded binding protein), DNA polymerase III holoenzyme, rNTPs, dNTPs including [α-32P]dATP, and a supercoiled form of pBRoriC or its derivative. Subsequently, chiDnaA or EcoDnaA was added, and reactions were incubated for 20 or 30 min at 30 °C prior to filtration using a GF/C glass filter and liquid scintillation counting.
DUE Unwinding (P1 Nuclease) Assay
This assay was performed as described previously (4, 19). Briefly, EcoDnaA was incubated on ice in buffer containing IHF (55 nm) and a supercoiled form of the oriC plasmid. Next, chiDnaA or EcoDnaA was added, and the reaction was incubated for 3 min at 38 °C, followed by incubation for 200 s at 38 °C in the presence of P1 nuclease (4 units; Yamasa Co.). Reactions were terminated by the addition of 0.5% SDS, and DNA was purified using a WIZARD spin column (Promega). A portion of the eluate was digested using the restriction enzyme AlwNI, and the digested DNA was subjected to electrophoresis using a 1% agarose gel and ethidium bromide staining. The oriC plasmids (4.7 kb) used had only a single site for AlwNI digestion. If the unwound DUE and the AlwNI site are cut, two fragments (2.5 and 2.2 kb) are yielded.
Form I* Assay
The form I* assay was performed as described previously (4, 22). Briefly, EcoDnaA was incubated on ice in buffer M containing SSB, IHF, DnaB, DnaC, gyrase, and the oriC plasmid. Next, chiDnaA or EcoDnaA was added, and the reaction was incubated at 30 °C for 15 min. Reactions were stopped by the addition of phenol and chloroform. DNA was precipitated in ethanol and resuspended in Tris-EDTA buffer, followed by 0.65% agarose gel electrophoresis and ethidium bromide staining. The relative amounts of form I* DNA were quantified using densitometry.
EMSA of DnaA Oligomer Formation
This analysis was performed as described previously (19). Briefly, oriC DNA fragments were incubated at 30 °C for 10 min in buffer containing ATP-EcoDnaA and λDNA (200 ng) as a competitor. Reactions were analyzed at room temperature using 2% agarose gel electrophoresis followed by GelStar staining and densitometric scanning. For chiDnaA analysis, EcoDnaA was added on ice in the initial stage, and chiDnaA was added subsequently.
EMSA of ssDUE Recruitment
This analysis was performed as described previously (4, 19). Briefly, 12.5 nm 32P-labeled DNA containing a 28-mer ssDUE was incubated with chiDnaA for 5 min on ice in buffer G″. EcoDnaA and a truncated oriC DNA, DORΔR1 (5 nm), were added to a portion of this mixture, and the reaction was incubated for 10 min at 30 °C in buffer G″. Reactions were analyzed using 4% PAGE at room temperature.
Results
Construction of Chimeric DnaA
To investigate the role of an individual DnaA molecule within the context of DnaA homo-oligomers at oriC, we constructed a chiDnaA consisting of EcoDnaA domains I–III and TmaDnaA domain IV (Fig. 1B). The consensus sequence of the EcoDnaA box (TTATNCACA) is widely conserved in eubacterial species, but the TmaDnaA box consensus differs substantially (AAACCTACCACC) (36). These distinct binding characteristics were used to develop the experimental approach. We reasoned that, when a specific DnaA box within oriC is substituted with a TmaDnaA box and both chiDnaA and EcoDnaA are co-incubated with the mutant oriC, chiDnaA could specifically bind to the substituted DnaA box, and, at the same time, EcoDnaA could bind to other intact DnaA boxes. This strategy appeared suitable to elucidate the role for an individual DnaA molecule within the DnaA homo-oligomers assembled on oriC.
EcoDnaA and TmaDnaA share a high degree of overall similarity in all protein regions except for domain II, which is a structurally flexible linker that varies in length and sequence between eubacterial species (Fig. 1B). The boundary between domains III and IV includes a short loop (i.e. a hinge) that connects the α helix of the domain III C terminus with that of the N terminus of domain IV (28, 31). The DNA-binding sites of domain IV reside in a helix-turn-helix motif in the middle region of this domain (29, 30). We therefore selected a site in the EcoDnaA domain IV N-terminal helix region for substitution with TmaDnaA domain IV (Fig. 1B). This resulted in the construction of chiDnaA, which consisted of amino acids 1–374 of EcoDnaA and amino acids 342–440 of TmaDnaA. Overproduction and purification of chiDnaA was performed as for EcoDnaA purification, and this resulted in a sample of >90% purity, as determined by SDS-PAGE.3
Nucleotide Binding Activity and DNA Binding Specificity of chiDnaA
To assess the basic activity of chiDnaA, the binding activities with adenine nucleotides and DnaA boxes were analyzed. A filter retention assay using EcoDnaA and chiDnaA revealed that the two purified proteins were similarly active in binding to both ATP and ADP (Table 3).
TABLE 3.
Nucleotide binding of chiDnaA
EcoDnaA (EcoWT), chiDnaA (ChiWT), and chiDnaA R285A (ChiRA) (1.9 pmol) were incubated at 0 °C for 15 min in the presence of various amounts (0–1 μm) of [α-32P]ATP or [3H]ADP, followed by filtration on a nitrocellulose membrane. Dissociation constant (Kd) and binding stoichiometry were determined using a Scatchard plot.
| DnaA |
Kd |
Stoichiometry |
||
|---|---|---|---|---|
| ATP | ADP | ATP | ADP | |
| nm | ||||
| EcoWT | 76 | 19 | 0.45 | 0.1 |
| ChiWT | 52 | 55 | 0.42 | 0.23 |
| ChiRA | 77 | 35 | 0.53 | 0.28 |
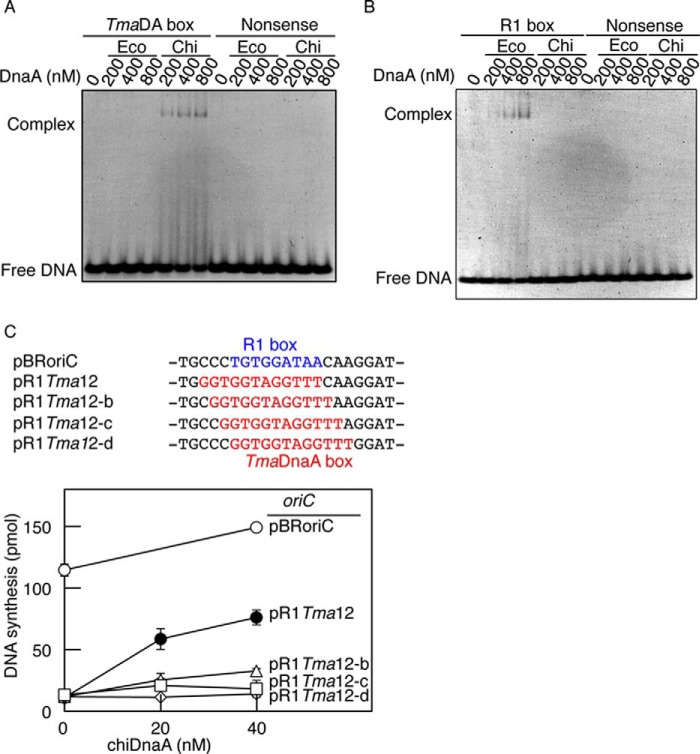
EMSA demonstrated that chiDnaA was able to bind specifically to DNA bearing the TmaDnaA box consensus sequence but was practically unable to bind to DNA bearing the EcoDnaA R1 box sequence (Fig. 2, A and B). In this assay, chiDnaA exhibited TmaDnaA box binding activity, even at 200 nm; this was consistent with previous data for TmaDnaA binding to the TmaDnaA box (36). Similarly, EcoDnaA bound to the R1 box DNA (Fig. 2B), but no substantial binding of EcoDnaA to the TmaDnaA box was detected (Fig. 2A). Neither EcoDnaA nor TmaDnaA exhibited detectable affinity for the nonsense control sequence used in this assay. These results showed that chiDnaA and EcoDnaA bound with high specificity to the TmaDnaA and R1 boxes, respectively, and demonstrated the utility of chiDnaA for the functional categorization of individual DnaA molecules within an initiation complex.
FIGURE 2.
DNA binding and replication activity of chiDnaA. A and B, affinity of chiDnaA for DnaA boxes. DNA fragments bearing a nonsense control sequence and a TmaDnaA box (TmaDA box) (A) or an EcoDnaA box R1 (R1 box) (B) (400 nm) were incubated at 30 °C for 10 min in buffer containing the indicated amounts of ATP-EcoDnaA (Eco) or ATP-chiDnaA (Chi). Complexes were analyzed by 5% PAGE and GelStar staining. Representative gel images are shown in a black-white inverted mode. C, replication activity of chiDnaA in a replication-reconstituted system. oriC plasmid pBRoriC (○) and its derivatives (pR1Tma12 (●), pR1Tma12-b (▵), pR1Tma12-c (♢), and pR1Tma12-d (□)), which bear TmaDnaA boxes at the R1 position, were assayed. The positions of the R1 and TmaDnaA boxes within the plasmid sequences are indicated in blue and red type, respectively. ATP-chiDnaA was incubated at 30 °C for 20 min in buffer containing the supercoiled form of the oriC plasmid (600 pmol), EcoDnaA (40 nm), and purified replicative proteins. DNA synthesis, as determined by nucleotide utilization, is indicated in the graph. Two independent experiments were done, and both data and mean values are shown.
Initiation Activity of chiDnaA
An oriC replication system reconstituted with purified proteins was used to evaluate the ability of chiDnaA to stimulate initiation in vitro. The in vitro system utilized a plasmid with oriC alongside DnaA, DnaB helicase, DnaG primase, and DNA polymerase III holoenzyme. In this experiment, we used chimeric oriC in which the DnaA box R1 was substituted with the TmaDnaA box. The R1 box is separated from the adjacent DUE by a 13-bp intervening region (Fig. 1A). The length of the intervening region is also important in initiation (4). Because the EcoDnaA box is a 9-mer and the TmaDnaA box is a 12-mer, we constructed four different chimeric oriC plasmids bearing the TmaDnaA box at positions shifted by 1 bp. The resultant plasmids were pR1Tma12 and its derivatives (Fig. 2C). To evaluate the initiation activity of chiDnaA and the chimeric oriC sequences, mixtures of EcoDnaA and various amounts of chiDnaA were included in the oriC replication-reconstituted systems (Fig. 2C). Plasmid pBRoriC (wild-type oriC) was replicated in a chiDnaA-independent manner. By contrast, pR1Tma12 (TmaDnaA box) exhibited substantial chiDnaA-dependent activity. A slight structural difference between EcoDnaA and chiDnaA might cause partial inhibition of DnaB helicase loading and replication when chiDnaA is used (see below), and this would explain the lower maximal DNA synthesis observed with pR1Tma12. Replication with pR1Tma12-derivative plasmids (i.e. pR1Tma12-b, -c, and -d) was severely inhibited even in the presence of chiDnaA (Fig. 2C). This confirmed the importance of the length of the intervening region between the DUE and the R1 box.
These results indicate that chiDnaA is fundamentally active in initiation when the TmaDnaA box is located at a proper position in oriC. The results also suggest that the right side of the EcoDnaA box corresponds to the right side of the TmaDnaA box in the construction of DnaA-DNA complexes (Fig. 2C).
Role of the DnaA Arg Finger of the R1 Box-DnaA in DUE Unwinding
To determine the orientation of DnaA protomers in an initiation complex, we used a chiDnaA R285A mutant protein to further investigate DnaA bound to the R1 box. The Arg-285 residue corresponds to the AAA+ Arg finger motif that is crucial for activation of an initiation complex (12). This residue is suggested to directly interact in a head-to-tail manner with ATP bound to the adjacent DnaA protomer (12, 27, 31). However, because the R1 DnaA box resides at the left edge of the DnaA assembly region of oriC, there are two functional possibilities for the Arg-285 residue of the DnaA protomer bound to the R1 box during initiation. First, if Arg-285 is oriented inward within oriC (AF-inward model in Fig. 1D) and interacts with ATP bound to the adjacent DnaA protomer, then this residue should be crucial for initiation. Second, if Arg-285 is oriented outward from oriC (AF-outward model in Fig. 1D), then it would not interact with the adjacent DnaA protomer and should therefore be irrelevant for initiation. In addition, if the AF-inward model is correct, the resultant initiation complexes should be active in initiation even when the R1 box-bound DnaA is in the ADP form. In contrast, initiation complexes should be inactive if the AF-outward model is correct, and the R1 box-bound DnaA is in the ADP form. We can thus infer using chiDnaA R285A and ADP-chiDnaA the orientation of the R1 box-bound DnaA protomer.
First, the filter retention assay confirmed that the purified chiDnaA R285A protein retained nucleotide binding activity at levels similar to the wild-type form of chiDnaA (Table 3). In addition, EMSA confirmed that TmaDnaA box-specific binding activity was equivalent between chiDnaA R285A and the wild-type form of chiDnaA.3 Next, unwinding activity was assessed using a P1 nuclease assay with pR1Tma12, EcoDnaA, and chiDnaA proteins with and without R285A substitution. If active initiation complexes are successfully constructed, then the DUE duplex is unwound, and the resultant ssDUE becomes susceptible to cleavage by P1 nuclease. In these experiments, ATP-EcoDnaA or ADP-EcoDnaA was first preincubated on ice with pR1Tma12. Varied amounts of chiDnaA or chiDnaA R285A (preincubated with ATP or ADP) were then added, and the mixtures were incubated at 38 °C.
Quantification of P1 nuclease products indicated that the addition of the ATP form of chiDnaA (ATP-chiDnaA) promoted DUE unwinding of pR1Tma12 to a level similar to that of pBRoriC in the presence of ATP-EcoDnaA alone (Fig. 3, A and B), consistent with the initiation activity of chiDnaA (Fig. 2C). By contrast, unwinding of pR1Tma12 did not occur when ATP-EcoDnaA was added in the second stage. These data indicate that the EcoDnaA included in the first stage constituted a basal level that did not cause binding to the R1 box or unwinding of pR1Tma12 and that unwinding was dependent upon the subsequent addition of ATP-chiDnaA. These results underline the specificity of the assay, whereby additional chiDnaA specifically binds to the R1-TmaDnaA box on pR1Tma12 and stimulates initiation.
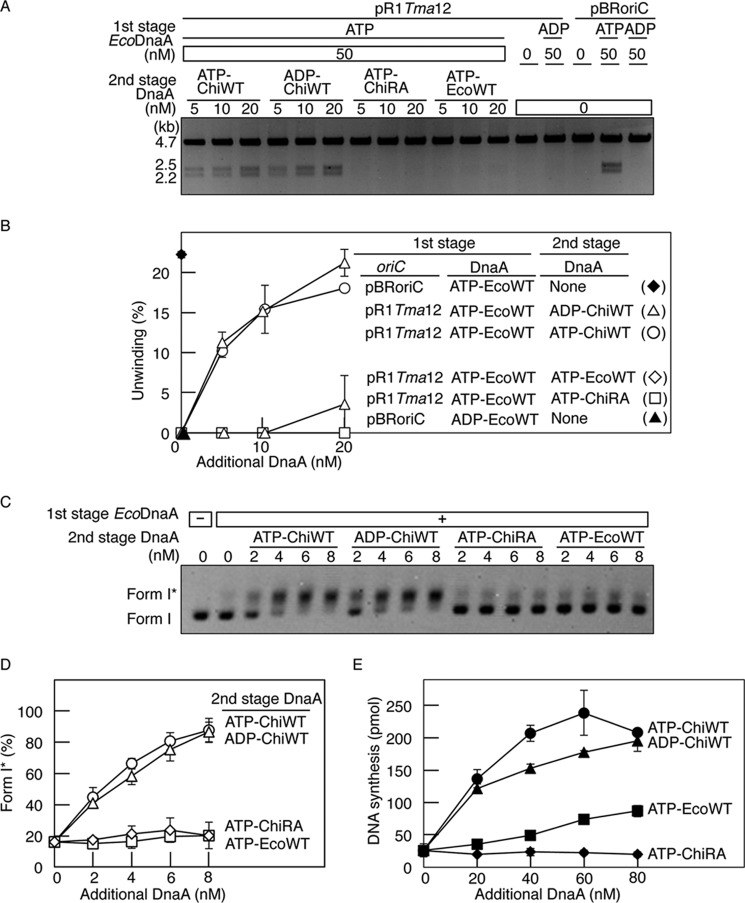
FIGURE 3.
Role for the Arg finger of R1 box-bound DnaA in initiation. A and B, DUE unwinding assay. In the first stage, pBRoriC (WT) or pR1Tma12 (4 nm as plasmid) was incubated on ice in buffer containing ATP-EcoDnaA (ATP-EcoWT) or ADP-EcoDnaA (ADP-EcoWT) (50 nm). In the second stage, the indicated amounts of ATP-chiDnaA (ATP-ChiWT), ADP-chiDnaA (ADP-ChiWT), ATP-chiDnaA R285A (ATP-ChiRA), or ATP-EcoDnaA were added to each mixture. Reactions were incubated at 38 °C for 3 min prior to digestion with P1 nuclease and AlwNI. Digestion products were analyzed by 1% agarose gel electrophoresis (A). The gel image is shown in a black-white inverted mode. The relative amounts of digested DNA to input DNA were quantified and used as a measure of DUE unwinding (%) (B). Two independent experiments were done, and both data and mean values are shown. C and D, form I* assay for DnaB helicase loading. The indicated amounts of DnaA proteins, as described above, were incubated at 30 °C for 20 min in buffer containing pR1Tma12 (1.6 nm as plasmid) and ATP-EcoDnaA (16 nm) in the presence of DnaB helicase, DnaC helicase-loader, SSB, and gyrase. Reactions were assessed using agarose gel electrophoresis analysis (C). The gel image is shown in a black-white inverted mode, and the migration positions of form I and form I* DNA are indicated. The amounts of form I* relative to total DNA were quantified and used as a measure of form I* (%) (D). Two independent experiments were done, and both data and mean values are shown. E, reconstituted replication assay. The indicated amounts of DnaA proteins, as described above, were incubated at 30 °C for 30 min in buffer containing pR1Tma12 (5.1 nm as plasmid; 600 pmol as nucleotides), ATP-EcoDnaA (60 nm), and replicative proteins. Two independent experiments were done, and both data and mean values are shown.
The addition of ADP-chiDnaA in the second stage of the assay also promoted DUE unwinding of pR1Tma12 at levels similar to that of ATP-chiDnaA (Fig. 3, A and B). By contrast, the ATP form of chiDnaA R285A (ATP-chiDnaA R285A) did not support unwinding (Fig. 3, A and B).
These results suggest that the DnaA Arg finger, but not the bound ATP, of the R1 box-bound DnaA protomer is required for DUE unwinding. Thus, these results support the AF-inward model (Fig. 1D), in which the nucleotide-binding site of the R1 box-bound DnaA is oriented outward from oriC, and the Arg finger of the same DnaA molecule is oriented inward within oriC and is required for specific interaction with the adjacent DnaA protomer.
The DnaA Arg Finger of the R1 Box-bound DnaA Is Required for Initiation
During initiation, DUE unwinding is followed by loading of DnaB helicase onto the resultant ssDNA region. Loading of DnaB is dependent on the temporal interaction with DnaC helicase loader and the DnaA complex on oriC. The activity of the R1 box-bound chiDnaA in DnaB loading was assessed using a form I* assay. This assay uses a supercoiled form (form I) of oriC plasmid with DnaA, DnaB, DnaC, and DNA gyrase. DnaB is loaded on the ssDNA and, in the presence of DNA gyrase, migrates along the ssDNA by unwinding the duplex DNA. This unwinding results in the production of a highly negative supercoiled form of DNA, form I* (4, 22). The presence of form I* DNA therefore acts as an indicator of DnaB loading. Form I and form I* are distinguished using agarose gel electrophoresis and fluorescent staining. In the present experiments, mixtures of pR1Tma12, DnaB, DnaC, DNA gyrase, and a low amount of EcoDnaA were prepared. Various amounts of EcoDnaA or chiDnaA were subsequently added, and reactions were incubated at 30 °C.
Quantification of the relative levels of form I* pR1Tma12 DNA demonstrated that ADP-chiDnaA and ATP-chiDnaA promoted DnaB loading at similar levels (Fig. 3, C and D). DnaB loading was minimal in the presence of ATP-chiDnaA R285A or ATP-EcoDnaA. The inactivity of ATP-EcoDnaA in this assay supports the specific requirement for DnaA binding to the R1-TmaDnaA box. These results are consistent with those of the DUE unwinding assay (Fig. 3, A and B) and support the idea that the Arg finger, but not the bound ATP, of the R1 box-bound DnaA is required for constructing DnaA complexes active in DnaB loading as well as DUE unwinding.
Replication activity was also assessed using an in vitro system reconstituted with purified protein. In the present experiments, pR1Tma12 was incubated with replicative proteins and DnaA. To exclude the possibility that chiDnaA could be non-specifically involved in DnaA complexes during the cooperative binding process, EcoDnaA was first added at a level insufficient to initiate replication. EcoDnaA and chiDnaA were added subsequently, and reactions were incubated at 30 °C (Fig. 3E). DNA synthesis was active with ATP-chiDnaA and ADP-chiDNA but not with ATP-chiDnaA R285A. Slight activity was detected with ATP-EcoDnaA. This activity was more pronounced at higher protein concentrations and might have occurred as a result of nonspecific binding to the R1-TmaDnaA box. Substantially similar results were exhibited even when EcoDnaA and chiDnaA were mixed at various ratios and these mixtures were added to replication reactions, including pR1Tma12.3 These results, which are consistent with those of the oriC unwinding and DnaB loading assays, indicate that the Arg finger, but not ATP, of the R1 box-bound DnaA is required for replication initiation in vitro.
Role of the DnaA Arg Finger of the R1 Box-bound DnaA in ssDUE Binding
To investigate the specific roles of the Arg finger of the R1 box-bound DnaA in initiation reactions, we analyzed the interaction of oriC-DnaA complexes with ssDUE. We previously proposed a model suggesting that after DUE unwinding in an initiation complex, the resultant upper strand of ssDUE binds to an ATP-DnaA homo-oligomer constructed on the left-half oriC (4, 19). This reaction is dependent on the DnaA complex constructed on the ATP-DnaA-preferential low affinity boxes (i.e. R5M, I1-2, and τ1-2) and is stimulated by DnaA bound to the R1 box (Fig. 4A). This stimulation is explained by a probable interaction between the R1 box-bound DnaA and the DnaA cluster bound to the low affinity boxes. This interaction is thought to assist recruitment of ssDUE to the DnaA cluster (4, 19) (Fig. 4A).
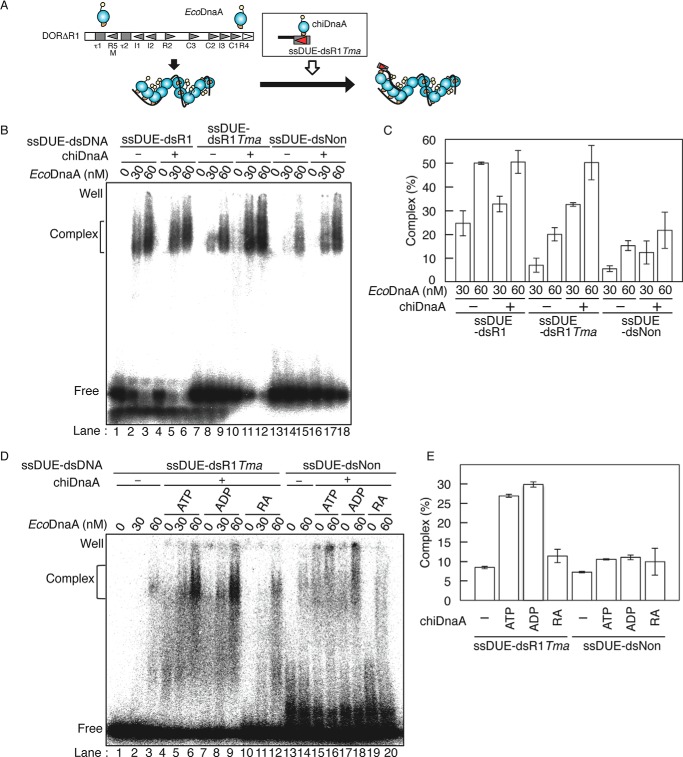
FIGURE 4.
Role of the Arg finger of the R1 box-bound DnaA in ssDUE binding. A, schematic of ssDUE recruitment assay. [32P]ssDUE ligated to dsDNA bearing the DnaA box is incubated with DnaA, followed by further incubation in the presence of ATP-EcoDnaA oligomers complexed with DORΔR1. Resultant complexes are analyzed by EMSA. DnaA domains and oriC sequences are illustrated as in Fig. 1. B and C, ssDUE recruitment assay using EcoDnaA and chiDnaA. In the first stage, ATP-chiDnaA (100 nm) was incubated on ice for 5 min with [32P]ssDUE ligated to dsDNA bearing the R1 box (ssDUE-dsR1), TmaDnaA box (ssDUE-dsR1Tma), or nonsense sequence (ssDUE-dsNon) (12.5 nm). In the second stage, one-fifth portions of these mixtures were incubated at 30 °C for 5 min in buffer containing DORΔR1 (5 nm) and the indicated amounts of ATP-EcoDnaA. EMSA was performed using 4% PAGE and BAS2500 image analysis (B). The amounts of ssDUE-dsDNA complexed with EcoDnaA-DORΔR1 were quantified, and the levels relative to the input ssDUE-dsDNA are shown as Complex (%) (C). Two independent experiments were done, and both data and mean values are shown. D and E, ssDUE recruitment assay using chiDnaA R285A. In the first stage, ATP-chiDnaA (ATP), ADP-chiDnaA (ADP), or ATP-chiDnaA R285A (RA) (100 nm) was incubated on ice for 5 min with ssDUE-dsR1Tma or ssDUE-dsNon (12.5 nm). In the second stage, one-fifth portions of these mixtures were incubated with DORΔR1 and ATP-EcoDnaA as described above, followed by EMSA using 4% PAGE and BAS2500 image analysis (D). The amounts of ssDUE-dsDNA complexed with EcoDnaA-DORΔR1 at 60 nm EcoDnaA were quantified, and the levels relative to the input ssDUE-dsDNA were shown as Complex (%) (E). Two independent experiments were done, and both data and mean values are shown.
To analyze the interaction of ssDUE with the DnaA complex, we performed EMSA using a truncated oriC fragment (DORΔR1) and [32P]ssDUE ligated to oligo-dsDNA (ssDUE-dsDNA) (Fig. 4A) using our previously described method (4, 19). DORΔR1 contained an oriC region but lacked a region containing DUE, the R1 box, and the IHF binding site (Fig. 4A). [32P]ssDUE-dsDNA carried the 28-mer ssDUE upper strand and dsDNA bearing the DnaA R1 box, TmaDnaA box, or nonsense sequence (ssDUE-dsR1, ssDUE-dsR1Tma, or ssDUE-dsNon, respectively) (Fig. 4, A and B). In these experiments, ssDUE-dsDNA was first incubated in the presence or absence of ATP-chiDnaA. Second, a portion of each initial reaction was added to buffer containing ATP-EcoDnaA complexes formed on the DORΔR1. ATP-EcoDnaA molecules assembled efficiently on the left-half oriC region with only slight inhibition even when the R1 box was deleted,3 in good agreement with previous observations (4, 19, 43).
Reaction mixtures were subjected to EMSA. ATP-EcoDnaA complexes formed on the oriC fragment were able to bind ssDUE-dsR1 at low (30–60 nm) concentrations of EcoDnaA, as we reported previously (Fig. 4, B, lanes 1–3, and C) (4, 19). The addition of chiDnaA in the first stage yielded similar results (Fig. 4, B (lanes 4–6) and C). Binding of ssDUE-dsR1 was slightly enhanced in the presence of chiDnaA, which may be due to minimal nonspecific binding of chiDnaA to ssDUE-dsR1 DNA.
Compared with the binding of ssDUE-dsR1, binding of ssDUE-dsR1Tma was substantially inhibited in the absence of chiDnaA (Fig. 4, B (lanes 7–9) and C). The minimal ssDUE-dsR1Tma binding observed in the absence of chiDnaA was probably due to the binding of the ssDUE region with EcoDnaA complexes bound to DORΔR1. This was supported by similar binding observed with ssDUE-dsNon (Fig. 4, B (lanes 13–15) and C; see below) and also corresponds with our previous results (4, 19). When chiDnaA was added, ssDUE-dsR1Tma binding was stimulated to a similar level as was observed for ssDUE-dsR1/EcoDnaA binding (Fig. 4, B (lanes 10–12) and C). This stimulation is plausibly explained by the interaction of chiDnaA (bound to the TmaDnaA box of ssDUE-dsR1Tma) with EcoDnaA (bound to DORΔR1). This interaction simulates the binding of ssDUE with the EcoDnaA complexes bound to DORΔR1.
In the presence of chiDnaA, binding of ssDUE-dsR1Tma was substantially higher than binding of ssDUE-dsNon (Fig. 4, B (lanes 11 and 12 and lanes 17 and 18) and C), supporting the specific binding of chiDnaA. The minimal binding of ssDUE-dsNon in the absence of chiDnaA is consistent with our previous results as explained above (Fig. 4B, lanes 13–15) (4, 19). Binding of ssDUE-dsNon was slightly higher when chiDnaA was present. This might also have been due to basal, nonspecific binding of chiDnaA (Fig. 4B, lanes 16–18).
Based on the results above, we next investigated the requirement for ATP and the Arg finger of chiDnaA in the stimulation of ssDUE-dsR1Tma binding (Fig. 4D). Binding of ssDUE-dsR1Tma was stimulated at similar levels with ADP-chiDnaA and ATP-chiDnaA; however, binding was not stimulated in the presence of ATP-chiDnaA R285A (Fig. 4, D (lanes 1–12) and E). Binding of ssDUE-dsNon was inefficient, as described above; however, ATP-chiDnaA and ADP-chiDnaA, but not chiDnaA R285A, could enhance binding slightly. This stimulation may have been due to the basal, nonspecific binding of chiDnaA to ssDUE-dsNon (Fig. 4, D (lanes 13–20) and E). These results are consistent with those shown in Fig. 3 and support the idea that the Arg finger, but not the bound ATP, of DnaA at the R1 box is crucial for the interactions with the DnaA complexes formed on DORΔR1 and hence the stimulation of ssDUE binding.
Role of the R4 Box-DnaA in DnaB Loading
Next, using similar methods as those used to assess the R1 box-bound DnaA (Fig. 3), we investigated the orientation of DnaA bound to R4, the high affinity binding site in the right-half oriC (Fig. 1A). DnaA binding at R4 is thought to trigger cooperative binding of DnaA molecules to the neighboring low affinity sites. DnaA boxes in the right-half oriC generally have the opposite orientation to those in the left-half oriC (Fig. 1A). Unlike the left-half oriC, the right-half oriC is dispensable for DUE unwinding (4). Loading of DnaB helicase onto the unwound region is stimulated by a DnaA complex constructed on the right-half oriC (4).
First, DUE unwinding was analyzed by P1 nuclease assay using pR4Tma12, which contained a TmaDnaA box in place of the R4 box (Fig. 5, A–C). A small amount of ATP-EcoDnaA or ADP-EcoDnaA was preincubated on ice with pBRoriC or pR4Tma12, and this was followed by incubation at 38 °C in the presence of additional EcoDnaA or chiDnaA (Fig. 5, B and C). Quantification of the P1 nuclease products showed that a moderate level of unwinding occurred in pBRoriC and pR4Tma12 without the second addition of DnaA. This is consistent with the dispensability of the right-half oriC for DUE unwinding. Additional ATP-EcoDnaA stimulated further unwinding of both pBRoriC and pR4Tma12. By contrast, the addition of chiDnaA did not stimulate further unwinding in either pBRoriC or pR4Tma12 (Fig. 5, B and C). These results are consistent with the above data and with the specificity of chiDnaA binding to the TmaDnaA box at the R4 position. When ADP-DnaA was used, unwinding did not take place, supporting the specificity of ATP-DnaA in this assay (Fig. 5, B and C).
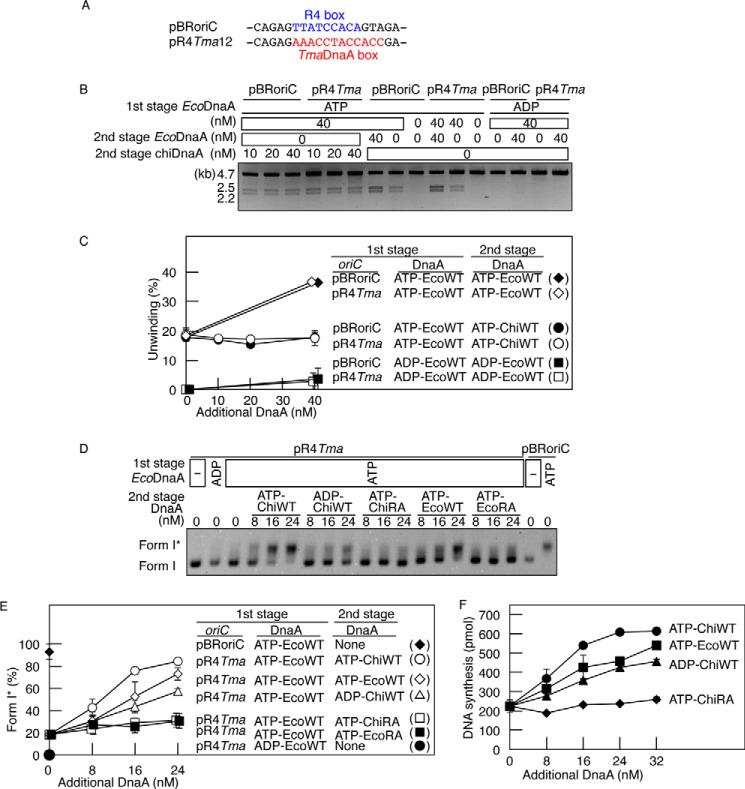
FIGURE 5.
Role for the Arg finger of the R4 box-bound DnaA in DnaB loading. A, substitution of the R4 box with TmaDnaA box. Sequences including the R4 box in oriC and in the relevant region of pR4Tma12 plasmid are shown. The R4 box and TmaDnaA box are indicated by blue and red type, respectively. B and C, DUE unwinding assay. In the first stage, pBRoriC (WT) or pR4Tma12 (pR4Tma), a pBRoriC derivative bearing the R4 box substituted with the TmaDnaA box (4 nm as plasmid) was incubated on ice in buffer containing ATP-EcoDnaA (ATP-EcoWT) or ADP-EcoDnaA (ADP-EcoWT) (50 nm). In the second stage, the indicated amounts of ATP-EcoDnaA or ATP-chiDnaA (ATP-ChiWT) were added. Reactions were incubated at 38 °C for 3 min before digestion with P1 nuclease and AlwNI. Digestion products were analyzed by 1% agarose gel electrophoresis (B). The gel image is shown in a black-white inverted mode. The relative amounts of digested DNA to input DNA were quantified and used as a measure of DUE unwinding (%) (C). Two independent experiments were done, and both data and mean values are shown. D and E, form I* assay for DnaB helicase loading. In the first stage, pBRoriC (WT) or pR4Tma12 (pR4Tma) (1.6 nm) was incubated on ice with ATP-EcoDnaA (ATP-EcoWT) or ADP-EcoDnaA (ADP-EcoWT) (16 nm) in the presence of DnaB helicase, DnaC helicase loader, SSB, and gyrase. In the second stage, the indicated amounts of ATP-chiDnaA (ATP-ChiWT), ADP-chiDnaA (ADP-ChiWT), ATP-chiDnaA R285A (ATP-ChiRA), ATP-EcoDnaA, or ATP-EcoDnaA R285A (ATP-EcoRA) were added. Reactions were incubated at 30 °C for 20 min and then assessed using agarose gel electrophoresis analysis (D). The gel image is shown in a black-white inverted mode, and the migration positions of form I and form I* DNA are indicated. The amounts of form I* relative to total DNA were quantified and used as a measure of form I* (%) (E). Two independent experiments were done, and both data and mean values are shown. F, reconstituted replication assay. ATP-EcoDnaA (32 nm) was added to buffer containing pR4Tma12 (5.1 nm as plasmid; 600 pmol as nucleotides) and replicative proteins. The indicated amounts of DnaA protein, as described above, were subsequently added before incubation at 30 °C for 30 min. Details are described under “Experimental Procedures.” Two independent experiments were done, and both data and mean values are shown.
Next, DnaB loading activity was assessed using the form I* assay, as performed for R1 box analysis (Fig. 3, C and D). Reaction mixtures containing pR4Tma12, a low level of ATP-EcoDnaA, DnaB, and other required proteins were incubated at 30 °C in the presence of additional EcoDnaA or chiDnaA. The addition of ATP-chiDnaA promoted DnaB loading onto pR4Tma12 at a level comparable with DnaB loading on pBRoriC stimulated by ATP-EcoDnaA (Fig. 5, D and E). ADP-chiDnaA was also able to stimulate DnaB loading onto pR4Tma12, albeit at a lower level than ATP-chiDnaA. These results indicate that both the ATP and ADP forms of R4 box-bound chiDnaA can activate DnaB loading. ATP-DnaA might interact more effectively with DnaB than ADP-DnaA (see below), and this would explain the reduced stimulation of DnaB loading with the ADP form. By contrast, ATP-chiDnaA R285A did not stimulate DnaB loading activity. ATP-EcoDnaA also stimulated DnaB loading on pR4Tma12 but at a lower level than ATP-chiDnaA/pR4Tma12 or ATP-EcoDnaA/pBRoriC. ATP-EcoDnaA would not bind to the TmaDnaA box at the R4 position; the DnaB loading observed with the ATP-EcoDnaA/pR4Tma12 combination might have occurred as a result of stimulation of unwinding and DnaB loading using the left-half subcomplex in addition to possible DnaA binding at the low affinity boxes in the right-half oriC. No substantial DnaB loading was observed when ATP-EcoDnaA R285A was used (Fig. 5, D and E).
Taken together, these results support a crucial role for the Arg finger of DnaA bound to the R4 box in stimulating DnaB loading. This is consistent with the idea that the DnaA Arg finger on the R4 box is oriented inward within oriC and is involved in interacting with DnaA bound to the C1 site in a head-to-tail manner (AF-inward model; Fig. 1D). The ATP of the R4-bound DnaA might stimulate interaction with DnaB; a specific site in domain III of DnaA is suggested to interact with DnaB during the process of DnaB loading (44, 45).
Using methods similar to those for R1 box analysis (Fig. 3E), replication initiation activity was assessed using an in vitro reconstituted system and pR4Tma12 (Fig. 5F). Basal ATP-EcoDnaA was added, followed by the addition of EcoDnaA or chiDnaA and incubation at 30 °C. Replication of the plasmid was stimulated by the addition of ATP-chiDnaA, ATP-EcoDnaA, or ADP-chiDnaA but not by the addition of ATP-chiDnaA R285A. As above, moderate stimulation occurred with ADP-chiDnaA and ATP-EcoDnaA. Substantially similar results were exhibited even when mixtures of EcoDnaA and chiDnaA in various ratios were added to reactions including pR4Tma12.3 These data are consistent with those of the form I* assay and support the AF-inward model.
Role of the Arg Finger of the R4 Box-bound DnaA in DnaA Assembly
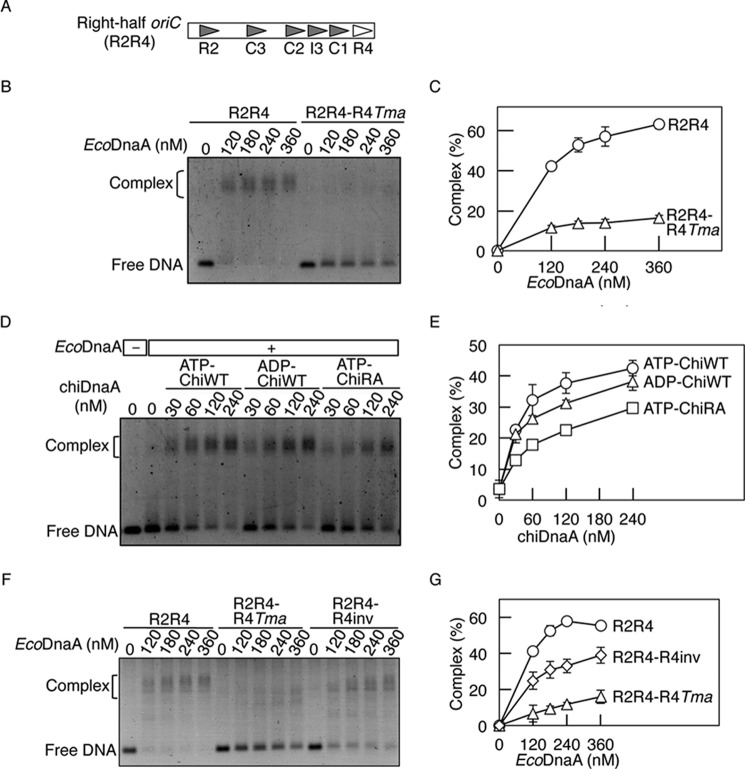
To analyze the role that R4-bound DnaA plays in DnaA assembly on oriC, EMSA was performed using right-half oriC DNA (R2R4), which encompasses the R2–R4 region. A derivative of R2R4 in which the R4 box was substituted with the TmaDnaA box (R2R4-R4Tma), was also used (Fig. 6A). DnaA complexes were formed without detectable levels of binding intermediates when ATP-EcoDnaA was co-incubated with R2R4 DNA (Fig. 6, B and C). This is consistent with previous research (19) and suggests that DnaA binds cooperatively in this region. In other previous papers that analyzed binding intermediates (12, 46, 47), competitor DNA was not used for stabilizing the intermediates.
FIGURE 6.
Role for the Arg finger of the R4 box-bound DnaA in DnaA assembly. A, structure of the right-half oriC. Symbols are as described in the legend to Fig. 1A. B and C, EMSA using EcoDnaA. The indicated amounts of ATP-EcoDnaA were incubated at 30 °C for 10 min with the right-half oriC (35 nm) bearing the wild-type sequence (R2R4) or the R4 box replaced with the TmaDnaA box sequence (R2R4-R4Tma). EMSA was performed using 2% agarose gel and GelStar staining. A representative gel image is shown in a black-white inverted mode (B). The amounts of DnaA-DNA complex relative to the total DNA were quantified and used as a measure of complex formation (%) (C). Two independent experiments were done, and both data and mean values are shown. D and E, EMSA using EcoDnaA and chiDnaA. ATP-EcoDnaA (80 nm) was incubated in buffer containing R2R4-R4Tma (35 nm). The indicated amounts of ATP-chiDnaA (ATP-ChiWT), ADP-chiDnaA (ADP-ChiWT), or ATP-chiDnaA R285A (ATP-ChiRA) were added, and reactions were incubated at 30 °C for 10 min. EMSA was performed as described above (D). The relative amounts of DnaA-DNA complexes were quantified. Error bars, S.D. from three independent experiments (E). F and G, EMSA using the right-half oriC fragments. The indicated amounts of ATP-EcoDnaA were incubated at 30 °C for 10 min with the right-half oriC (35 nm) bearing the wild-type sequence (R2R4), the R4 box replaced with the TmaDnaA box sequence (R2R4-R4Tma), or the inverted R4 box (R2R4-R4inv). EMSA was performed as described above (F). The relative amounts of DnaA-DNA complexes were quantified. Error bars, S.D. from three independent experiments (G).
DnaA assembly was severely inhibited when R2R4-R4Tma DNA was used (Fig. 6, B and C). This is consistent with previous research (43) and indicated that DnaA binding to the R4 box is crucial for DnaA assembly on the right-half oriC.
Next, a similar assay was performed using EcoDnaA, chiDnaA, and R2R4-R4Tma DNA (Fig. 6, D and E). ATP-chiDnaA and ADP-chiDnaA promoted DnaA assembly in the presence of ATP-EcoDnaA at similar levels, but DnaA assembly with ATP-chiDnaA R285A was impaired (Fig. 6, D and E). These results suggest that, when the TmaDnaA box sequence is present at the R4 position, ATP-chiDnaA and ADP-chiDnaA can promote cooperative binding of DnaA molecules to the low affinity DnaA boxes but that ATP-chiDnaA R285A is impaired in this ability. This is consistent with the idea that the DnaA Arg finger on the R4 box-bound DnaA is oriented toward the DnaA box C1 (AF-inward model). Moderate activity of ATP-chiDnaA R285A in DnaA assembly might be due to Arg finger-independent inter-DnaA interactions at other sites (48).
In addition, to investigate the importance of orientation of the R4 box-bound DnaA in DnaA assembly, EMSA was performed using a derivative of R2R4 in which the R4 box sequence was inverted (R2R4-R4inv). DnaA assembly was moderately impaired when R2R4-R4inv DNA was used (Fig. 6, F and G). This indicates that the orientation of the R4 box-bound DnaA is relevant to DnaA assembly on the right-half oriC, consistent with the above results and previous in vitro research (43). Residual activity of R2R4-R4inv in DnaA assembly was also observed, which might be due to Arg finger-independent inter-DnaA interactions, such as inter-domain I interactions (3).
In Vivo Examination of the Arg Finger of DnaA at the R1 and R4 Boxes
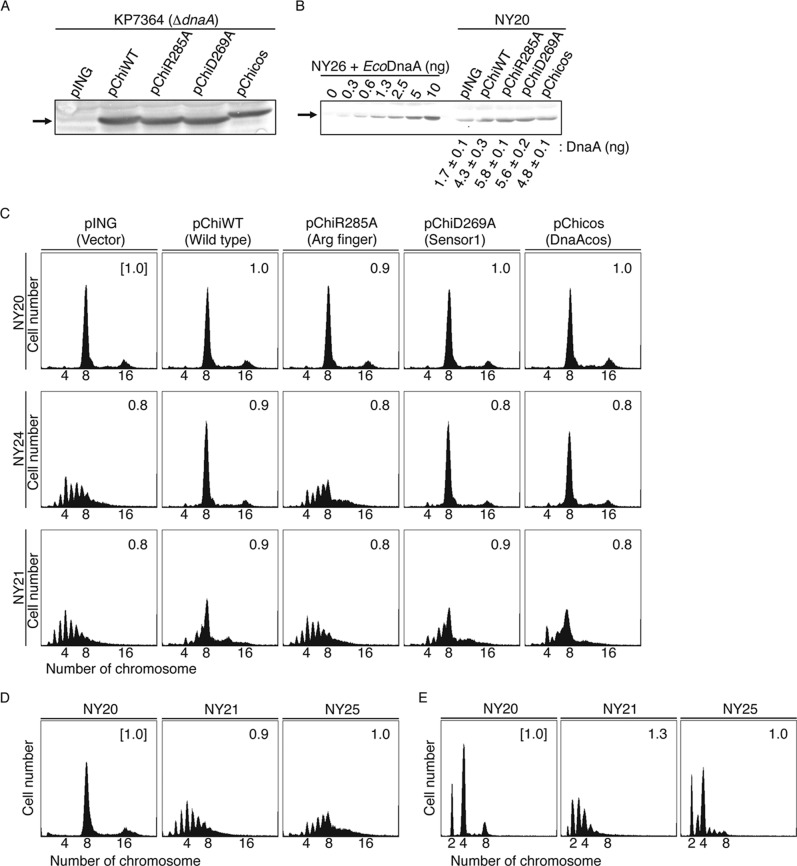
The orientation of DnaA bound to the R4 and R1 boxes was assessed in vivo using modified strains. MG1655-derivative mutant strains were constructed that had the TmaDnaA box sequence at the chromosomal R1 box (strain NY24) or R4 box (strain NY21) (Table 1). Strain construction details are provided under “Experimental Procedures.” NY24 and NY21 cells grew at the same rate (generation time of 20–23 min) as the parental strain containing wild-type oriC (NY20); however, regulation of initiation was impaired (see below). This is consistent with previous descriptions of strains with mutations in the R1 or R4 box (43).
NY20, NY24, and NY21 cells were transformed with pING1-derivative plasmids carrying genes encoding chiDnaA (pChiWT), chiDnaA R285A (pChiR285A), chiDnaA D269A (pChiD269A), or chiDnaAcos (pChiDnaAcos). The pING1 plasmid is derived from pBR322 and carries the arabinose promoter and the araC repressor gene. Plasmid dnaA expression occurred mainly as a result of leaky expression, which occurred at a similar level in all cells (Fig. 7, A and B). The total amounts of DnaA in NY20 cells bearing the dnaA expression plasmid were only 2.5–3.5-fold higher than the levels in cells bearing pING vector, suggesting that the amounts of DnaA expressed from plasmid were comparatively minimal relative to the EcoDnaA expressed from the chromosome (Fig. 7B). The DnaA D269A and DnaAcos proteins carry mutations in the AAA+ domain and are impaired in ATP and ADP binding. The DnaA Asp-269 residue is a crucial element within the AAA+ sensor I motif that supports the high affinity of DnaA for adenine nucleotide binding. Wild-type DnaA binds ATP and ADP with Kd of 10–100 nm, but DnaA D269A affinity for ATP and ADP is 100-fold lower (26). It is therefore conceivable that, in cells where ATP is abundant (millimolar levels) and concentrations are 10-fold higher than ADP levels, DnaA D269A accelerates the dissociation of ADP and binds primarily with ATP. Consistent with this, we previously demonstrated that overinitiation of replication occurred in cells bearing DnaA D269A (26). The DnaAcos protein, which bears mutations in domain III, maintains initiation activity at 30 °C but is severely impaired in ATP and ADP binding. This results in a severe overinitiation of replication that leads to inhibition of cell growth (50–52).
FIGURE 7.
Role for the Arg finger of DnaA bound to the R1 and R4 boxes in initiation in vivo. A and B, amounts of cellular chiDnaA were determined by Western blotting. Cells of strain KP7364 (ΔdnaA::spec rnhA::kan) (A) or NY20 (B) carrying the indicated plasmid were grown in LB medium at 37 °C to an absorbance (A660) of 0.1. A portion (500 μl) of each culture was analyzed by Western blotting using anti-EcoDnaA antibody as described previously (26). Arrows, bands corresponding to chiDnaA or to a mixture of chiDnaA and EcoDnaA. For a quantitative standard, the indicated amounts of purified EcoDnaA were mixed with whole cell extract of NY26 (ΔdnaA::spec rnhA::kan). The amounts of cellular DnaA were quantified in two independent experiments. Disruption of rnhA activates alternative origins independent of DnaA (49). chiDnaA has a molecular mass similar to EcoDnaA. C, in vivo analysis of R1 and R4 box-bound DnaA. Cells of strains NY20 (wild-type oriC; WT), NY24 (chromosomal R1 box substituted with TmaDnaA box; R1Tma), and NY21 (chromosomal R4 box substituted with TmaDnaA box; R4Tma) carrying pING (vector), pChiWT (chiDnaA), pChiR285A (chiDnaA R285A), or pChiD269A (chiDnaA R269A) were grown at 37 °C in LB medium containing ampicillin. When the absorbance A660 reached 0.1, culture portions were withdrawn and used for quantification of cell size (cell mass) using flow cytometry. Additional culture portions were further incubated for 4 h in the presence of rifampicin and cephalexin. DNA contents were quantified using flow cytometry. Equivalent chromosome numbers are shown. Cell mass relative to that of NY20 cells bearing pING1 (1.0) is indicated at the top right of each panel. Two independent experiments were performed, and indistinguishable results were exhibited. D, in vivo analysis of cells bearing the inverted R4 box. Cells of strains NY20, NY21, and NY25 (chromosomal R4 box inversion; R4inv) were grown and analyzed using flow cytometry as described above. Cell mass relative to that of NY20 cells (1.0) is indicated at the top right of each panel. For NY25, cells from three independent transductants were analyzed, and similar results were obtained for each; representative data are shown. E, analysis of NY20, NY21, and NY25 cells grown in a minimum medium. Flow cytometry analysis was performed as described above except that minimum medium described previously (18) was used. For NY25 cells, three independent transductants were analyzed, and similar results were obtained for each; representative data are shown.
Flow cytometry analysis was performed using the constructed cell lines (Fig. 7C). Cells were rapidly grown in LB medium at 37 °C. Cephalexin and rifampicin were then added to the culture to stop cell division and replication initiation while permitting progression of established replisomes. Incubation was continued to allow run-out replication of the chromosomes. The resulting cells therefore contained the same number of chromosomes as there were copies of oriC at the time of antibiotic addition (53). Flow cytometry of NY20 cells bearing pING1 revealed a major peak at eight chromosomes and a minor peak at 16 chromosomes. This indicated that a newborn cell contained eight sister oriC copies and that replication was synchronously initiated at these origins prior to cell division (Fig. 7C). Similar flow cytometry results were observed for NY20 cells bearing pChiWT or its derivatives (Fig. 7C). These results showed that introduction of the pChi plasmids did not significantly affect replication initiation in NY20 cells.
In contrast, flow cytometry of NY24 cells bearing pING1 revealed a major peak at four chromosomes and additional peaks at 3 and 5–9 chromosomes (Fig. 7C). This indicated that initiation was inhibited and that asynchronous initiations occurred. This is consistent with substantial in vivo inhibition of EcoDnaA binding to the TmaDnaA box at position R1. A NY20-like peak pattern was observed in NY24 cells bearing pChiWT. This demonstrated that chiDnaA bound effectively to the TmaDnaA box sequence at position R1 and promoted initiation in vivo (Fig. 7C). In contrast, the peak pattern of NY24 cells bearing pChiR285A was similar to the pattern of NY24 cells bearing pING1 (Fig. 7C), indicating the importance of the Arg finger in initiation. The data indicating that NY24 cells bearing pChiD269A or pChiDnaAcos showed a peak pattern similar to that of NY24 cells bearing pChiWT are also consistent with the idea that the nucleotide bound to the R1 box-DnaA is not a crucial determinant for initiation regulation. Indistinguishable results were obtained in the presence of 0.05% arabinose.3 These in vivo results were consistent with the in vitro data (Figs. 3 and 4). Taken together, the data indicate that the DnaA Arg finger of the DnaA bound at the R1 box is important for initiation in vivo, and this is consistent with the AF-inward model (Fig. 1D).
Similar analysis was extended to NY21 cells (Fig. 7C). As with NY24, several peaks were observed in NY21 cells bearing pING1; this showed that initiations were asynchronous and severely inhibited. These results were consistent with significant in vivo inhibition of EcoDnaA binding to the TmaDnaA box sequence at position R4. Initiation was partially rescued by the introduction of pChiWT. Cells with eight chromosomes were predominant, but a moderate level of asynchronous initiation was demonstrated by the presence of 5–7 chromosome peaks. The peak pattern of NY21 cells bearing pChiR285A was similar to the pattern of cells bearing pING1. NY21 cells bearing pChiD269A or pChiDnaAcos exhibited peak patterns that were similar to those of NY21 cells bearing pChiWT, with only minimal inhibitions observed (Fig. 7C). Indistinguishable results were obtained in the presence of 0.05% arabinose.3 These in vivo results are consistent with the in vitro data (Figs. 5 and 6) and support the suggestion that the Arg finger of the DnaA bound to the R4 box is important for initiation in vivo and is oriented inward within oriC.
In addition, we used flow cytometry to analyze cells of the NY25 strain, which bears the inverted R4 box in the chromosomal oriC. Replication initiation in NY25 cells was significantly inhibited and asynchronous (Fig. 7D), which was consistent with the above results. A moderate level of asynchronous initiations was observed even in minimum medium (Fig. 7E). Data from a previous research indicated that cells with the inverted R4 box had a slightly elevated level of asynchronous initiations in the same minimum medium (18) (see “Discussion”). Replication initiation in NY21 cells was asynchronous in the minimum medium (Fig. 7E), which was consistent with the above results and with the previous reports (18, 43).
Discussion
Discovering the functional structure of DnaA homo-oligomers complexed with oriC is crucial for understanding the regulation and mechanism of replication initiation. However, attribution of specific roles to individual DnaA protomers in the complexes has remained elusive. In this study, we performed detailed analysis using chimeric DnaA molecules and chimeric oriC sequences. Chimeric DnaA molecules bound specifically to a typical TmaDnaA box rather than to a typical EcoDnaA box. Chimeric oriC sequences harbored the TmaDnaA box at the R1 or R4 box positions. Experimental data indicated that the Arg finger of DnaA bound at the R1 or R4 box is crucial for accurate replication initiation both in vitro and in vivo. Further results showed that replication initiation was active even when ADP-DnaA rather than ATP-DnaA was bound at the R1 or R4 box. For the R1 box-bound DnaA, the Arg finger-dependent interaction would be crucial for the construction of a specific DnaA complex conformation with DUE unwinding activity (Figs. 3 and 4). For the R4 box-bound DnaA, the Arg finger-dependent interaction would be crucial for the construction of a specific DnaA complex with DnaB helicase loading activity (Fig. 5) and would also be important for cooperative DnaA binding at low affinity sites (Fig. 6). Flow cytometry data from cells with chimeric DnaA molecules and chimeric oriC sequences were consistent with the in vitro data (Fig. 7C). Taken together, we propose that the Arg fingers, but not the bound nucleotide, of the R1- or R4-bound DnaA are oriented inward within oriC and are used for functional interactions with DnaA bound to the flanking low affinity site (AF-inward model in Figs. 1D and 8A). The concept of the Arg fingers of DnaA bound to the R1 and R4 facing inward is a two-dimensional description of what is a three-dimensional situation.
FIGURE 8.
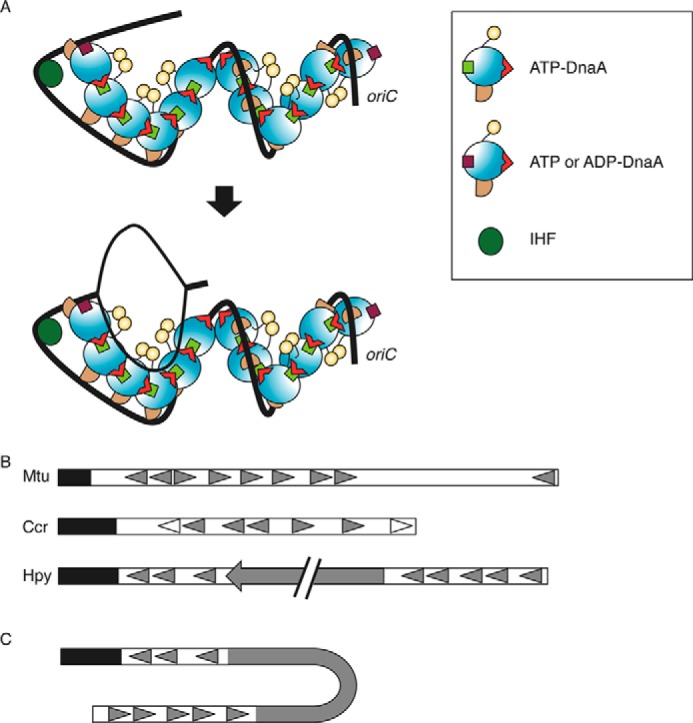
Model of initiation complex structure. A, DnaA domains I–IV are distinguished using different colors as in Fig. 1D. DnaA, IHF (dark green circle), and oriC (black line) are illustrated according to the AF-inward model (Fig. 1D). The position of the Arg finger in domain III is indicated with a red triangle. DnaA bound to the R1 and R4 binds ATP or ADP (brown square), whereas other DnaA molecules bound ATP (light green square). IHF bent oriC and promoted interaction between DnaA bound at the R1 box and the adjacent molecule (top). This led to unwinding of DUE and recruitment of the resultant ssDUE to a DnaA subcomplex formed on a low affinity box cluster of the left-half oriC (bottom). The two DnaA subcomplexes are oriented in opposite directions, and each would be expected to bind a single DnaB helicase. B, schematic presentation of the basic structure of oriC. Mtu, M. tuberculosis; Ccr, C. crescentus; Hpy, H. pylori. The DUE region is indicated by a black bar. DnaA boxes are indicated by filled or open triangles. In C. crescentus, the DnaA boxes indicated by open triangles have higher affinity for DnaA than others indicated by filled triangles. A filled arrow in H. pylori oriC indicates the dnaA gene. C, a model proposed for an initiation complex in H. pylori. When H. pylori DnaA is complexed with the cognate oriC, the oriC1-DnaA subcomplex might interact with the oriC2-DnaA subcomplex, depending on DNA bending in the dnaA gene region (62). DnaA protein is omitted in this figure for simplicity.
These conclusions are consistent with a previous in vivo study indicating the specific roles of the R1 and R4 boxes (18, 43) and with in vivo footprint experiments of DnaA binding showing that binding to the R1, R2, and R4 boxes was maintained stably during the cell cycle (54). ADP-DnaA is much more abundant than ATP-DnaA during the cell cycle, except at the time of replication initiation (10, 55). According to our results (Figs. 3 and 4), replication initiation is not inhibited even if ADP-DnaA binds to the R1 box. Rather, stable DnaA binding to the R1 box would allow efficient activation of initiation complexes. Supporting this is the observation that the addition of ADP-DnaA stimulates initiation of oriC replication when ATP-DnaA is present only at basal levels (56).
Consistent with the previous results mentioned above, our results also indicate that initiation remains active even when ADP-DnaA binds to the R4 box (Fig. 5, D–F). A moderate inhibition of DnaB loading was observed with ADP-chiDnaA and oriC with the TmaDnaA box at the R4 position (Fig. 5, D and E). This inhibition could be as a result of slight structural differences in domain IV between EcoDnaA and TmaDnaA. Otherwise, because domain III of DnaA bears a possible DnaB-binding site (44, 45), it is possible that ATP binding promotes a conformational change at domain III that stimulates interaction with DnaB at this site. Although DnaB loading activity was reduced when ADP-DnaA was bound to the R4 box, the remaining activity as assessed in vitro would be sufficient to support timely initiation under the in vivo conditions.
The Arg finger of the R1 box-bound DnaA was crucial for DUE unwinding and ssDUE binding (Figs. 3 and 4). These results are consistent with the idea that the Arg finger of the R1 box-bound DnaA interacts with the ATP bound to DnaA residing on the neighboring low affinity region (AF-inward model in Fig. 1D). This interaction would promote the recruitment of ssDUE to DnaA complexes formed on the low affinity region of the left-half oriC via IHF binding-dependent DNA bending (Fig. 8A) (4, 19). Our results indicated that the ATP on the R1 box-bound DnaA was not important for initiation (Figs. 3 and 4). This is consistent with the idea that the R1 box-bound DnaA does not bind another DnaA on the DUE side. In addition, previous results of footprint experiments did not detect binding of DnaA molecules in the region between the DUE and the R1 box (14, 19, 27, 46). These observations also support ssDUE recruitment as described above rather than the model of a continuous ATP-DnaA filament at the DUE region formed from ATP-DnaA bound to the R1 box, as suggested previously (5, 15).
In the right-half oriC region, the presence of an intact R2 box was insufficient for DnaA complex formation at the right-half oriC in the absence of the R4 box (Fig. 6B). Moderate DnaA complex formation activity was maintained even when chiDnaA R285A was bound to the TmaDnaA at position R4 or when the R4 box was inverted (Fig. 6, D–G). This is consistent with the possibility that other domain III residues or domain I of the R4 box-bound DnaA might assist in inter-DnaA interactions in the right-half oriC. We therefore suggest that the Arg finger of the R4 box-bound DnaA is a stimulator, but not a prerequisite, in the formation of DnaA homo-oligomers on the right-half oriC.
Our data are consistent with the idea that the Arg finger of the R4 box-bound DnaA plays a crucial role in the loading of DnaB helicase (Fig. 5, D and E). Previous studies suggested that formation of stable DnaA complexes on oriC was required for loading of DnaB on unwound DNA (48). It is plausible that the Arg finger of the R4 box-bound DnaA supports the formation of stable DnaA complexes, although DnaA complexes can be formed with moderate efficiency in the absence of this finger, as shown in the EMSA data (Fig. 6, D and E) and discussed above. The in vivo analysis confirmed the importance of the Arg finger of the R4 box-bound DnaA in replication initiation (Fig. 7C). Consistently, when the R4 box was inverted, DnaA assembly on the right-half oriC and replication initiation were impaired (Figs. 6 (F and G) and 7D). In a previous study, cells with an oriC construct similar in the inversion of the R4 box were grown in a minimal medium and analyzed using flow cytometry (18). The asynchrony index indicating levels of asynchronous initiations was 8.1 for those cells. This value was intermediate between the index value (5.7) of a wild-type synchronous control and the index value (11.0) of moderately asynchronous cells with the R2 box mutation (18). Here, the R4 box-inverted cells showed moderate asynchrony even in minimal medium (Fig. 7E). The subtle difference in the level of asynchrony between the present and previous data might have been due to differences in genetic elements of the strains, growth conditions (e.g. aeration or the timing of harvest), or flow cytometry equipment. The right-half oriC was reported to be more important for stimulating initiation in cells growing in rich medium than in cells in poor medium (57), consistent with the present results (Fig. 7, D and E) and with the specific role for the right-half subcomplex in stimulation of DnaB loading (4).
Previous results from pull-down experiments with various oriC fragments suggest that a pair of DnaB helicases bind to an initiation complex (4). The data suggest that the first DnaB-binding site is located in DnaA domain I, which contains the Glu-21 and Phe-46 residues, and that the second binding site is found at the N terminus of domain III (22, 23, 44, 45). In this study, we propose a model in which the DnaA subcomplexes constructed on each half of the oriC region are oriented in opposite directions (AF-inward model; Figs. 1D and 8A). This symmetric direction of DnaA subcomplexes could be significant for the probable symmetric binding of a pair of DnaB helicases to the two DnaA subcomplexes. It would be reasonable to infer that the interaction between DnaA domain III and DnaB can contribute to DnaB orientation and that this binding mode of DnaB-DnaA subcomplexes can be a stimulating factor in efficient bidirectional and synchronous loading of a pair of DnaB helicases onto the single-stranded oriC region. Examination of DnaB helicase orientation when bound to an initiation complex is important for future studies.
Sequence similarity analysis suggests that multiple DnaA boxes are found within the predicted oriC in many bacterial species; however, sequence analysis did not successfully predict the E. coli oriC low affinity sites (58). Experimental analysis of DnaA boxes has been performed only in a few species. In Caulobacter crescentus and Mycobacterium tuberculosis, DnaA boxes, including some with low affinity, were identified experimentally (59, 60) (Fig. 8B). The orientation of the oriC-DnaA boxes in these species resembled the arrangement in E. coli, whereby the direction of DnaA boxes in the left half of the DnaA box cluster region opposed the direction in the right-half region (Fig. 8B). Two oriC subregions, oriC1 and oriC2, were recently identified flanking the dnaA gene in Helicobacter pylori. The DnaA box sequences of oriC1 and oriC2 are oriented in the same direction, and it is suggested that the DnaA-oriC1 subcomplex interacts with the DnaA-oriC2 subcomplex via DNA looping of the dnaA gene region (61, 62). In the resultant complex, the directions of the DnaA complexes formed at oriC1 and oriC2 would then be opposed (Fig. 8C). The structural feature of two subregions bearing DnaA box clusters in opposite orientations might therefore be a common feature in eubacteria and could be important for bidirectional loading of replicative helicases.
Formation of specific DnaA complexes is required also at important sites other than oriC. The DnaA-reactivating sequences and datA, which each contain at least three DnaA boxes, require construction of specific DnaA homo-oligomers for the promotion of reactivation and inactivation of DnaA, respectively (10, 63–65). The datA locus promotes the hydrolysis of ATP bound to DnaA for repression of untimely initiations (10). Exchange of ADP to ATP is stimulated when ADP-DnaA interacts with DnaA-reactivating sequences, resulting in reactivation of DnaA (64, 65). These complexes have indispensable roles in the regulation of the replication cycle (2, 7, 9). The chimeric DnaA resources that were developed in this study will be invaluable in investigating the roles of these additional complexes.
Author Contributions
Y. N. and T. K. designed the study, Y. N. performed most of the experiments, Y. S. and H. K. partly performed in vivo experiments, and all authors analyzed and discussed the data. Y. N. and T. K. wrote the manuscript. All authors reviewed the manuscript and approved the final version.
This work was supported by grants-in-aid for scientific research (KAKENHI Grants 22370064 and 26291004) from the Japan Society of Promotion of Sciences and the Ministry of Education, Culture, Sports, Science, and Technology, Japan. The authors declare that they have no conflicts of interest with the contents of this article.
Y. Noguchi and T. Katayama, unpublished observation.
- DUE
- duplex unwinding element
- DOR
- DnaA oligomerization region
- ssDUE
- single-stranded DUE
- EcoDnaA
- E. coli DnaA
- TmaDnaA
- T. maritima DnaA
- chiDnaA
- chimeric DnaA.
References
- 1. O'Donnell M., Langston L., Stillman B. (2013) Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leonard A. C., Grimwade J. E. (2011) Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65, 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaguni J. M. (2011) Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15, 606–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ozaki S., Katayama T. (2012) Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duderstadt K. E., Berger J. M. (2013) A structural framework for replication origin opening by AAA+ initiation factors. Curr. Opin. Struct. Biol. 23, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dillon S. C., Dorman C. J. (2010) Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195 [DOI] [PubMed] [Google Scholar]
- 7. Saxena R., Fingland N., Patil D., Sharma A. K., Crooke E. (2013) Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int. J. Mol. Sci. 14, 8517–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Langston L. D., Indiani C., O'Donnell M. (2009) Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8, 2686–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katayama T., Ozaki S., Keyamura K., Fujimitsu K. (2010) Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170 [DOI] [PubMed] [Google Scholar]
- 10. Kasho K., Katayama T. (2013) DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 110, 936–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGarry K. C., Ryan V. T., Grimwade J. E., Leonard A. C. (2004) Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawakami H., Keyamura K., Katayama T. (2005) Formation of an ATP-DnaA-specific initiation complex requires DnaA arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430 [DOI] [PubMed] [Google Scholar]
- 13. Rozgaja T. A., Grimwade J. E., Iqbal M., Czerwonka C., Vora M., Leonard A. C. (2011) Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su'etsugu M., Fujimitsu K., Kagawa W., Yokoyama S., Kurumizaka H., Katayama T. (2007) The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 21, 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Speck C., Messer W. (2001) Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20, 1469–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller D. T., Grimwade J. E., Betteridge T., Rozgaja T., Torgue J. J., Leonard A. C. (2009) Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. U.S.A. 106, 18479–18484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ryan V. T., Grimwade J. E., Nievera C. J., Leonard A. C. (2002) IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 46, 113–124 [DOI] [PubMed] [Google Scholar]
- 18. Weigel C., Messer W., Preiss S., Welzeck M., Morigen, Boye E. (2001) The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40, 498–507 [DOI] [PubMed] [Google Scholar]
- 19. Ozaki S., Noguchi Y., Hayashi Y., Miyazaki E., Katayama T. (2012) Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozaki S., Katayama T. (2009) DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 62, 71–82 [DOI] [PubMed] [Google Scholar]
- 21. Felczak M. M., Simmons L. A., Kaguni J. M. (2005) An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 280, 24627–24633 [DOI] [PubMed] [Google Scholar]
- 22. Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T. (2009) DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T. (2007) Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827 [DOI] [PubMed] [Google Scholar]
- 24. Nozaki S., Ogawa T. (2008) Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154, 3379–3384 [DOI] [PubMed] [Google Scholar]
- 25. Nishida S., Fujimitsu K., Sekimizu K., Ohmura T., Ueda T., Katayama T. (2002) A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 277, 14986–14995 [DOI] [PubMed] [Google Scholar]
- 26. Kawakami H., Ozaki S., Suzuki S., Nakamura K., Senriuchi T., Su'etsugu M., Fujimitsu K., Katayama T. (2006) The exceptionally tight affinity of DnaA for ATP/ADP requires a unique aspartic acid residue in the AAA+ sensor 1 motif. Mol. Microbiol. 62, 1310–1324 [DOI] [PubMed] [Google Scholar]
- 27. Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S. Y., Yokoyama S., Kurumizaka H., Katayama T. (2008) A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362 [DOI] [PubMed] [Google Scholar]
- 28. Erzberger J. P., Pirruccello M. M., Berger J. M. (2002) The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21, 4763–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Yokoyama S. (2003) Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asklund M., Atlung T. (2005) New non-detrimental DNA-binding mutants of the Escherichia coli initiator protein DnaA. J. Mol. Biol. 345, 717–730 [DOI] [PubMed] [Google Scholar]
- 31. Erzberger J. P., Mott M. L., Berger J. M. (2006) Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683 [DOI] [PubMed] [Google Scholar]
- 32. Roth A., Messer W. (1995) The DNA binding domain of the initiator protein DnaA. EMBO J. 14, 2106–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zorman S., Seitz H., Sclavi B., Strick T. R. (2012) Topological characterization of the DnaA-oriC complex using single-molecule nanomanipuation. Nucleic Acids Res. 40, 7375–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bocchetta M., Gribaldo S., Sanangelantoni A., Cammarano P. (2000) Phylogenetic depth of the bacterial genera Aquifex and Thermotoga inferred from analysis of ribosomal protein, elongation factor, and RNA polymerase subunit sequences. J. Mol. Evol. 50, 366–380 [DOI] [PubMed] [Google Scholar]
- 35. Leonard A. C., Méchali M. (2013) DNA replication origins. Cold Spring Harb. Perspect. Biol. 5, a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ozaki S., Fujimitsu K., Kurumizaka H., Katayama T. (2006) The DnaA homolog of the hyperthermophilic eubacterium Thermotoga maritima forms an open complex with a minimal 149-bp origin region in an ATP-dependent manner. Genes Cells 11, 425–438 [DOI] [PubMed] [Google Scholar]
- 37. Fujimitsu K., Su'etsugu M., Yamaguchi Y., Mazda K., Fu N., Kawakami H., Katayama T. (2008) Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 190, 5368–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bates D. B., Asai T., Cao Y., Chambers M. W., Cadwell G. W., Boye E., Kogoma T. (1995) The DnaA box R4 in the minimal oriC is dispensable for initiation of Escherichia coli chromosome replication. Nucleic Acids Res. 23, 3119–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hatano T., Yamaichi Y., Niki H. (2007) Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol. Microbiol. 64, 1198–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li M. Z., Elledge S. J. (2007) Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 [DOI] [PubMed] [Google Scholar]
- 42. Obita T., Iwura T., Su'etsugu M., Yoshida Y., Tanaka Y., Katayama T., Ueda T., Imoto T. (2002) Determination of the secondary structure in solution of the Escherichia coli DnaA DNA-binding domain. Biochem. Biophys. Res. Commun. 299, 42–48 [DOI] [PubMed] [Google Scholar]
- 43. Kaur G., Vora M. P., Czerwonka C. A., Rozgaja T. A., Grimwade J. E., Leonard A. C. (2014) Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 91, 1148–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sutton M. D., Carr K. M., Vicente M., Kaguni J. M. (1998) Escherichia coli DnaA protein. the N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273, 34255–34262 [DOI] [PubMed] [Google Scholar]
- 45. Seitz H., Weigel C., Messer W. (2000) The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 37, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 46. Margulies C., Kaguni J. M. (1996) Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J. Biol. Chem. 271, 17035–17040 [DOI] [PubMed] [Google Scholar]
- 47. Weigel C., Schmidt A., Rückert B., Lurz R., Messer W. (1997) DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 16, 6574–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Felczak M. M., Kaguni J. M. (2004) The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 279, 51156–51162 [DOI] [PubMed] [Google Scholar]
- 49. Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Katayama T., Kornberg A. (1994) Hyperactive initiation of chromosomal replication in vivo and in vitro by a mutant initiator protein, DnaAcos, of Escherichia coli. J. Biol. Chem. 269, 12698–12703 [PubMed] [Google Scholar]
- 51. Katayama T. (1994) The mutant DnaAcos protein which overinitiates replication of the Escherichia coli chromosome is inert to negative regulation for initiation. J. Biol. Chem. 269, 22075–22079 [PubMed] [Google Scholar]
- 52. Chodavarapu S., Felczak M. M., Simmons L. A., Murillo A., Kaguni J. M. (2013) Mutant DnaAs of Escherichia coli that are refractory to negative control. Nucleic Acids Res. 41, 10254–10267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Skarstad K., Bernander R., Boye E. (1995) Analysis of DNA replication in vivo by flow cytometry. Methods Enzymol. 262, 604–613 [DOI] [PubMed] [Google Scholar]
- 54. Nievera C., Torgue J. J., Grimwade J. E., Leonard A. C. (2006) SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell 24, 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T. (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18, 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yung B. Y., Crooke E., Kornberg A. (1990) Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J. Biol. Chem. 265, 1282–1285 [PubMed] [Google Scholar]
- 57. Stepankiw N., Kaidow A., Boye E., Bates D. (2009) The right half of the Escherichia coli replication origin is not essential for viability, but facilitates multi-forked replication. Mol. Microbiol. 74, 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gao F., Luo H., Zhang C. T. (2013) DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 41, D90–D93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taylor J. A., Ouimet M. C., Wargachuk R., Marczynski G. T. (2011) The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Mol. Microbiol. 82, 312–326 [DOI] [PubMed] [Google Scholar]
- 60. Madiraju M. V., Moomey M., Neuenschwander P. F., Muniruzzaman S., Yamamoto K., Grimwade J. E., Rajagopalan M. (2006) The intrinsic ATPase activity of Mycobacterium tuberculosis DnaA promotes rapid oligomerization of DnaA on oriC. Mol. Microbiol. 59, 1876–1890 [DOI] [PubMed] [Google Scholar]
- 61. Donczew R., Weigel C., Lurz R., Zakrzewska-Czerwinska J., Zawilak-Pawlik A. (2012) Helicobacter pylori oriC: the first bipartite origin of chromosome replication in Gram-negative bacteria. Nucleic Acids Res. 40, 9647–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Donczew R., Mielke T., Jaworski P., Zakrzewska-Czerwińska J., Zawilak-Pawlik A. (2014) Assembly of Helicobacter pylori initiation complex is determined by sequence-specific and topology-sensitive DnaA-oriC interactions. J. Mol. Biol. 426, 2769–2782 [DOI] [PubMed] [Google Scholar]
- 63. Kitagawa R., Ozaki T., Moriya S., Ogawa T. (1998) Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12, 3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fujimitsu K., Senriuchi T., Katayama T. (2009) Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23, 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kasho K., Fujimitsu K., Matoba T., Oshima T., Katayama T. (2014) Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42, 13134–13149 [DOI] [PMC free article] [PubMed] [Google Scholar]