Background: Post-translational modification is an important approach to regulate NF-κB activity.
Results: Serine 316 (Ser-316) is a novel phosphorylation site on p65.
Conclusion: Phosphorylation of Ser-316 on p65 is essential for NF-κB activation and its related biological functions.
Significance: Our data shed light on how NF-κB transcriptional specificity is achieved through site-specific phosphorylation.
Keywords: mass spectrometry (MS), NF-κB (NF-κB), phosphorylation, post-translational modification (PTM), serine
Abstract
Nuclear factor κB (NF-κB) is a central coordinator in immune and inflammatory responses. Constitutive NF-κB is often found in some types of cancers, contributing to oncogenesis and tumor progression. Therefore, knowing how NF-κB is regulated is important for its therapeutic control. Post-translational modification of the p65 subunit of NF-κB is a well known approach for its regulation. Here, we reported that in response to interleukin 1β, the p65 subunit of NF-κB is phosphorylated on the novel serine 316. Overexpression of S316A (serine 316 → alanine) mutant exhibited significantly reduced ability to activate NF-κB and decreased cell growth as compared with wtp65 (wild type p65). Moreover, conditioned media from cells expressing the S316A-p65 mutant had a considerably lower ability to induce NF-κB than that of wtp65. Our data suggested that phosphorylation of p65 on Ser-316 controls the activity and function of NF-κB. Importantly, we found that phosphorylation at the novel Ser-316 site and other two known phosphorylation sites, Ser-529 and Ser-536, either individually or cooperatively, regulated distinct groups of NF-κB-dependent genes, suggesting the unique role of each individual phosphorylation site on NF-κB-dependent gene regulation. Our novel findings provide an important piece of evidence regarding differential regulation of NF-κB-dependent genes through phosphorylation of different p65 serine residues, thus shedding light on novel mechanisms for the pathway-specific control of NF-κB. This knowledge is key to develop strategies for prevention and treatment of constitutive NF-κB-driven inflammatory diseases and cancers.
Introduction
Transcription factor NF-κB plays central roles in immune and inflammatory responses and in oncogenesis and tumor progression. NF-κB family has five members that include: RelA (p65), RelB, C-Rel, NF-κB1 (p50), and NF-κB2 (p52). The most common NF-κB prototype is the heterodimer of p65 and p50, with p65 as the major subunit. All family members share an N-terminal DNA binding region named Rel homology domain (1). Under normal cellular conditions, NF-κB localizes in cytoplasm as an inactive complex by binding to the inhibitor of NF-κB (IκB).3 Upon treatment with different stimuli, NF-κB is activated through specific pathways. In general, activation of NF-κB can be classified into canonical and non-canonical pathways. The canonical NF-κB activation pathway is well established and includes the activation of an IκB kinase complex followed by the phosphorylation-induced degradation of IκB. This process enables the p65/p50 heterodimer to enter the nucleus and activate the expression of specific NF-κB target genes (1). NF-κB activity is known to be regulated by different types of post-translational modifications, including phosphorylation, ubiquitination, acetylation, sumoylation, nitrosylation, and methylation, etc. (2–8), especially on its p65 subunit.
Phosphorylation is a type of post-translational modification that universally exists in almost every process of the cell life cycle. Protein kinases catalyze phosphorylation on serine, threonine, or tyrosine residues by adding a covalently bound phosphate group (9). This process switches the protein states between either active or inactive forms, leading to a change in protein function or localization. Improper regulation of protein phosphorylation is often related to many kinds of diseases, including cancer. Thus, identification of novel phosphorylation sites of a given protein is critical for the understanding of protein regulation in normal conditions as well as in disease states (10, 11).
As the major subunit of NF-κB, p65 has drawn great attention with respect to the nature of its post-translational modifications. To date, 12 phosphorylation sites have been identified on p65 in response to different stimuli and/or in different cell systems. Among them, four sites (Ser-205, Thr-254, Ser-276, Ser-281) are located in the N-terminal RHD, one site (Ser-311) is in the linker region that is adjacent to the N-terminal RHD, and seven residues (Thr-435, Ser-468, Thr-505, Ser-529, Ser-535, Ser-536, and Ser-547) are contained in the C-terminal transactivation domain (3, 12, 13). Most of these phosphorylation events positively modulate NF-κB transcriptional activity. For example, the catalytic subunit of PKA (PKAc) stays inactive by binding to IκB to form an NF-κB-IκB-PKAc complex. Extracellular signals such as lipopolysaccharide, which degrade IκB, lead to activated PKAc to phosphorylate p65 on Ser-276 in 70Z/3 pre-B cells (14). This PKAc-mediated phosphorylated p65 interacts with CBP/p300 to positively regulate the transactivation potential of p65 (15). Interestingly, Ser-276 of p65 is also reported to be phosphorylated by mitogen- and stress-activated protein kinase-1 (MSK1) upon tumor necrosis factor α (TNFα)-mediated NF-κB activation in mouse L929sA cells (16). Haegeman and co-workers (16) found that Ser-276 is a crucial residue for the regulation of the nuclear transactivation capacity of p65. Therefore, Ser-276 phosphorylation is a prerequisite for the induction of NF-κB-dependent genes (16). These findings raised the possibility that multiple kinases could target the same serine residue in different experiment systems. Furthermore, Toriumi and co-workers (17) reported that IκB kinase phosphorylates the NF-κB p65 subunit on serine 536, leading to enhanced p65 transactivation in HeLa cells. Moreover, Baldwin and co-workers (18) found that TNFα could induce Ser-529 phosphorylation on p65, a process that is controlled by casein kinase II. It is worth noting that, although most of the phosphorylation events lead to NF-κB activation, there are few instances in which phosphorylation on the same site could either positively or negatively regulate NF-κB activity depending on the specific experimental systems. A relevant example would be the phosphorylation of Ser-468. Ser-468 is phosphorylated by IκB kinase ϵ leading to an enhanced p65 transcriptional activity in T-cells in response to T-cell co-stimulation (19). On the other hand, Ser-468 may also be phosphorylated by GSK-3β, exerting a negative regulatory effect on basal NF-κB activity in HeLa cells (20). This interesting phenomenon clearly exemplifies the extremely complex biological consequences caused by the phosphorylation of p65.
In this study, using mass spectrometry we identified the novel phosphorylation of Ser-316 on p65 upon interleukin 1β-induced NF-κB activation. We showed that phosphorylation of Ser-316 regulates a distinctive group of NF-κB-dependent genes as compared with those of Ser-529 or Ser-536. Thus, we suggest that novel phosphorylation of Ser-316 is critical for NF-κB-dependent differential gene regulation and leads to specific biological consequences. Our finding provides an important piece of evidence regarding how the complicated NF-κB regulation is finely tuned and achieved.
Experimental Procedures
Cell Lines and Materials
The 293C6 cell line has been described previously (21). Briefly, human 293C6 is a pool of 293IL1R cells (which carry transfected IL1 receptors and accessory protein). Due to the overexpression of the IL1 receptor, 293C6 shows robust NF-κB activation upon cytokine stimulation like IL-1β. Also 293C6 shows no constitutively active NF-κB and thus has normal levels of p65 (22).
The mouse embryo fibroblasts (MEF) cell line was a kind gift from Dr. Alexander Hoffman (University of California at San Diego) (23). Anti-phosphorylated Ser-529-p65 and phosphorylated Ser-536-p65 antibodies were from Cell Signaling, and the anti-p65 antibody was from Santa Cruz Biotechnology. Anti-β-actin was supplied by Sigma. Alexa Fluor 488 goat anti-mouse IgG was from Life Technologies. Casein kinase I (CKI) inhibitor D4476 was purchased from Sigma.
Site Mutagenesis of p65
Different p65 mutants (S316A, S529A, S536A, S316A/S529A, S316A/S536A, S529A/S536A, S316A/S529A/S536A) were generated by using QuikChange II XL site-directed mutagenesis kit from Agilent Technologies (24). Primers were designed using Agilent Technologies QuikChange Primer Design online software.
Purification of Endogenous p65
293C6 cells were cultured to ∼95% confluence then were treated with 10 ng/ml IL-1β for 1 h. Cells were lysed, and endogenous p65 was immunoprecipitated the same way as described before (23). The purified p65 band was excised and further subjected to mass spectrometry analysis.
Digestion and Liquid Chromatography-Tandem Mass Spectrometric Analysis
The Coomassie-stained SDS-PAGE gel band containing p65 protein was subjected to in-gel tryptic digestion. First, p65 gel pieces were destained with 50% acetonitrile in 100 mm ammonium bicarbonate followed by 100% acetonitrile. Next, cysteine residues were reduced by incubating the sample with 20 mm DTT at room temperature for 60 min and alkylated with 55 mm iodoacetamide for 30 min in the dark. The solution was removed, and the gel pieces were washed with 100 mm ammonium bicarbonate and dehydrated in acetonitrile. Gel pieces were then dried in a SpeedVac centrifuge and rehydrated in 50 mm ammonium bicarbonate containing sequencing grade modified trypsin (Promega, WI) for overnight digestion at 37 °C. After proteolytic digestion, peptides were extracted from the gel with 50% acetonitrile in 5% formic acid, dried, and reconstituted in 0.1% formic acid for mass spectrometry analysis.
Analysis of proteolytic digests was performed by using an LTQ Orbitrap XL linear ion-trap mass spectrometer (Thermo Fisher Scientific) coupled with an Ultimate 3000 HPLC system (Dionex). The digests were injected onto a reverse-phase C18 column (0.075 × 150 mm, Dionex) equilibrated with 0.1% formic acid, 4% acetonitrile (v/v). A linear gradient of acetonitrile from 4 to 40% in water in the presence of 0.1% formic acid over a period of 45 min was used at a flow rate of 300 nl/min. The spectra were acquired by data-dependent methods, consisting of a full scan (m/z 400–2,000) and then tandems on the five most abundant precursor ions. The previously selected precursor ions were scanned once during 30 s and then were excluded for 30 s. The obtained data were analyzed by Mascot software (Matrix Science) against customized p65 protein database with the setting of 10 ppm for precursor ions and 0.8 Da for product ions. Carbamidomethylation of cysteine was set as fixed modification, whereas oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. The tandem mass spectra of candidate-modified peptides were further interpreted manually.
Transfection and Luciferase Assay
Constructs were transfected into cell lines by using the Lipofectamine and PLUS Reagents (Invitrogen). To establish stable pools, cells were co-transfected with a plasmid encoding a puromycin resistance gene, and selected in 1 μg/ml puromycin 48 h later. For NF-κB luciferase assays, the κB-luciferase construct p5XIP10 κB (23) was transfected transiently into the cells, and luciferase activity was quantified 48 h later. A β-galactosidase construct was co-transfected to normalize for transfection efficiency. Transfections and luciferase assays were carried out essentially as described by Lu et al. (23).
Western Analysis
Cells cultured to ∼90–95% confluence were treated with IL-1β at different time points. Samples were collected and lysed by radio immunoprecipitation assay buffer (150 mm NaCl, 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 50 mm Tris-HCl, pH 8.0, and protease inhibitors), and whole cell lysates were separated by SDS/PAGE gels and further assayed by Western blotting (23). Different antibodies were used to detect the target proteins.
Immunofluorescence
Coverslips were coated with 0.1% sterile gelatin for 2 h and dried for 30 min at room temperature. 1 × 105 cells were then seeded onto coverslips per well in a 24-well plate. After overnight culture, cells were treated with IL-1β for 1 h to continue with immunofluorescence experiments. Cells were fixed with 4% of formaldehyde for 30 min and then blocked with blocking buffer for 10 min at room temperature. Coverslips were further probed with anti-FLAG antibody and Alexa Fluor 488 goat anti-mouse IgG. Before sealing the coverslips, mounting media with DAPI was used to stain the nucleus. The slides were examined under Nikon Eclipse 80i.
Quantitative PCR Analyses
Cells cultured to 80–90% confluence were transfected with different p65 constructs. RNA samples were collected 48 h later. cDNA was generated using reverse PCR. FastStart Universal SYBR Green Master ROX (Roche Applied Science) was then used for the quantitative PCR reactions. Primers were designed by Primer Express 3.0 software (8). Primers were: TRIM73 forward (5′-TGAAGCAGGAGCAGAAGAAGGT-3′) and reverse (5′-TCCGACTCATTGACGATTCG-3′; TTLL2 forward (5′-TCTTGAAGCCGCTGGTTTTT-3′) and reverse (5′-CCAGGAGGACGCTTTGCA-3′); USP28 forward (5′-AGAGACCCCCACCTCTCACA-3′) and reverse (5′-TACCCTTTTGGGTGCTTCATTT-3′; NKG7 forward (5′-CTTTGAGCACCGATTTCTGGTT-3′) and reverse (5′-TGTCCCCATGCCCTGTTG-3′; SLC32A1 forward (5′-ATCTTCGCCGCCGTTGT-3′) and reverse (5′-CCGTCTTCATTCTCCTCGTACAG-3′).
Cell Growth Assay
Cells were plated at 2 × 104/well in triplicate per time point in 6-well plates. Cells were trypsinized, resuspended, and counted using a cell counting chamber on days 3, 5, 7, and 9 (23).
Illumina Microarray Analysis
250 ng of RNA was reverse-transcribed into cRNA and labeled with biotin-UTP using the Illumina TotalPrep RNA Amplification kit (Ambion/Applied Biosystems). cRNA was quantified using a nanodrop spectrophotometer, and the cRNA quality (size distribution) was further analyzed on a 1% agarose gel. cRNA was hybridized to the Illumina Human Ref-v3 v1Expression BeadChip and scanned into a BeadArray Reader using standard protocols (provided by Illumina). Illumina BeadStudio software was used for data analysis (23).
Conditioned Media Assay
293C6 cells and MEFp65−/− cells were seeded into 12-well plates and transfected with different plasmids: empty vector, wtp65, or S316A-p65. Twenty-four hours after transfection, media in the cell culture plates were replaced with fresh media, and cells were cultured for an additional 48 h. Conditioned media were then collected and used to treat stable 293-NF-κB reporter cells for 4 h before carrying out luciferase assay (23).
Human Cytokine ELISA Arrays
Human cytokine ELISA arrays were purchased from Signosis. Experiments were carried out according to the manufacturer's protocol. Briefly, conditioned media collected from 293C6 control, wtp65, or S316A-p65 stable cell lines were added to specific cytokine capture antibody-precoated wells for 2 h at room temperature. After incubation, the wells were washed to remove unbound labeled antibodies. The plate was further detected with HRP luminescent substrate. The level of expression for each specific cytokine was directly proportional to the luminescence that was emitted.
Statistical Analysis
The associations between relative luciferase activity and relative gene expression in different groups were analyzed by Student's t test. The data represent the mean ± S.D. from three independent experiments. A p value < 0.05 was considered to be statistically significant.
Results
Identification of Novel Phosphorylation of Ser-316 on p65 by Mass Spectrometry Analysis
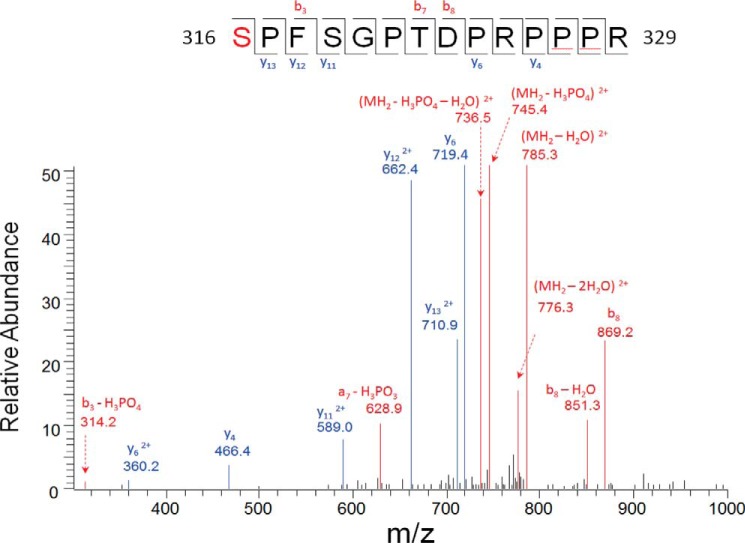
NF-κB is known to be phosphorylated on different sites in response to different stimuli. To date more than a dozen different phosphorylated sites have been reported. Under inflammation conditions or within certain tumor microenvironments, secreted cytokines such as IL-1β are important factors to keep NF-κB constitutively activated (25). To determine whether p65 can be phosphorylated on novel sites after IL-1β treatment, we treated 293C6 cells with IL-1β for 1 h and further isolated endogenous p65 protein by using anti-p65 antibody (8, 23). The purified p65 band was further subjected to mass spectrometry analysis. An 80-kDa mass shift was observed clearly on Ser-316 in the IL-1β-treated but not in the untreated samples, suggesting that p65 is phosphorylated on Ser-316 upon IL-1β treatment (Fig. 1). This exciting finding indicates that Ser-316 is a novel phosphorylation site during the process of IL-1β-triggered NF-κB activation.
FIGURE 1.
Mass spectrometry identified Ser-316 phosphorylation of p65 in response to IL-1β. Tandem mass spectrometry (ms2) of precursor ions in the phosphorylated p65 peptide (amino acids 316–329; sequence SPFSGPTDPRPPPR, where S (red) indicates phosphorylated serine). Black lines indicate peptide cleavage. Compared with unmodified 316–329 peptide, an 80-Da mass shift was observed on precursor ion and b series ions such as b3, b7, and b8, but not on y series ions from y4 to y13, indicating phosphorylation of Ser-316.
Effect of the S316A Mutation on NF-κB Activity
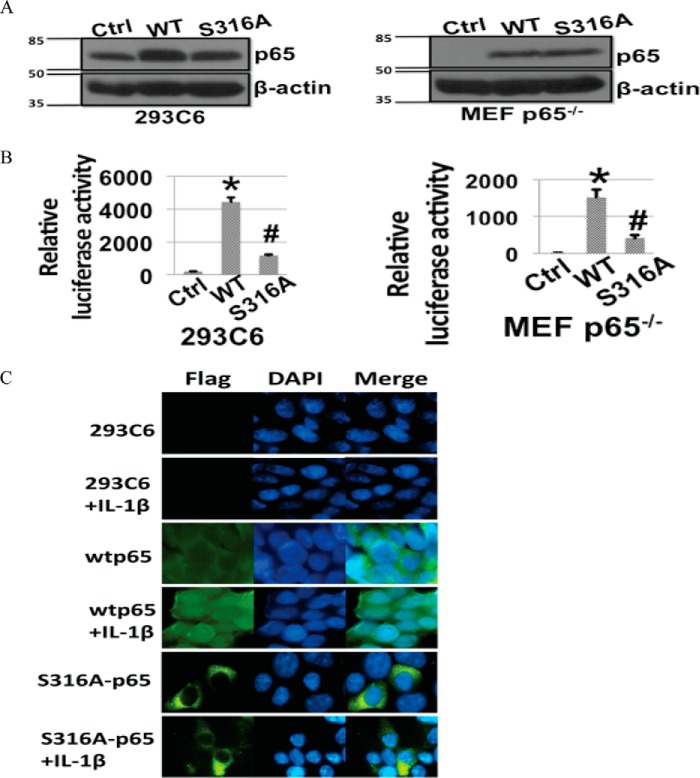
To test whether phosphorylation of Ser-316 is important for NF-κB activation, we generated S316A-p65 mutant and further expressed it at a similar level to that of wtp65 in either 293C6 (Fig. 2A, left panel) or MEFp65−/− cells (Fig. 2A, right panel). The effect of S316A-p65 mutation on NF-κB activity was further determined by carrying out the κB-specific luciferase assay. As shown in Fig. 2B, overexpression of wtp65 significantly activated NF-κB, and it was similar to what we observed previously (8). However, overexpression of the S316A-p65 mutant protein lost this ability in both 293C6 (Fig. 2B, left panel) and MEFp65−/− cells (Fig. 2B, right panel), confirming that phosphorylation of Ser-316 of p65 is critical for the activation of NF-κB. We wondered whether the decrease of NF-κB activity in S316A-p65 overexpression cells was related to the altered NF-κB nuclear translocation. We carried out immunofluorescence assays in 293C6, wtp65, or S316A-p65 cells (Fig. 2C). Because both wtp65 and S316A-p65 are tagged with FLAG, anti-FLAG antibody was used to detect cellular localization of the overexpressed wtp65 and S316A-p65 proteins (shown in green). DAPI was used for nuclear staining. 293C6 cells (without any FLAG tagged p65 protein) were used as negative control for the anti-FLAG antibody (black). Data suggested that before IL-1β treatment of the wtp65 overexpressing cell line, p65 was observed in both the cytoplasm and the nucleus but mainly in the cytoplasm, confirming the well known phenomenon that overexpression of wtp65 leads to constitutive activation of NF-κB. However, after IL-1β treatment, more p65 was further translocated into the nucleus, suggesting that IL-1β could further induce NF-κB activation. In contrast, S316A-p65 was largely distributed into the cytoplasm before or post IL-1β treatment, indicating that mutation at Ser-316 led to the defect of nuclear translocation of the p65 subunit. In short, our data support the notion that S316A-p65 mutant led to decreased NF-κB activity via compromised ability of NF-κB nuclear translocation.
FIGURE 2.
Effects of the S316A mutation on NF-κB activity. A, Western assays showing the overexpression of wtp65 and S316A-p65 in 293C6 cells (left panel) or in MEFp65−/− cells (right panel). The molecular marker is marked on the left. Units are in kDa. B, luciferase assay of NF-κB activity. Overexpression of wtp65 significantly activated NF-κB, whereas overexpression of the S316A-p65 mutant protein reduced this activity in both 293C6 cells (left panel) and MEFp65−/− cells (right panel). Results of triplicate luciferase assays are shown as the mean ± S.D. *, p < 0.01 versus control (Ctrl) group; #, p < 0.01 versus wt group. C, immunofluorescence, showing the localization of wtp65 and S316A-p65. Both FLAG-tagged wtp65 and S316A-p65 were probed with anti-FLAG antibody. DAPI was used for nuclear staining. In the wtp65 overexpression cell line, without IL-1β treatment the localization of p65 was observed in both cytoplasm and nucleus but mainly in the cytoplasm; however, after IL-1β treatment, p65 was largely translocated into the nucleus. In contrast, p65 translocation was substantially decreased in cell lines with S316A mutation in the presence or absence of IL-1β treatment, indicating that mutation at Ser-316 compromised the nuclear translocation of p65 subunit. Control 293C6 cell lines without overexpression of FLAG-tagged p65 did not show the expression of FLAG-tagged p65 and was used as a negative control.
Phosphorylation of p65 on Ser-316 Is Critical for NF-κB-inducible Gene Expression
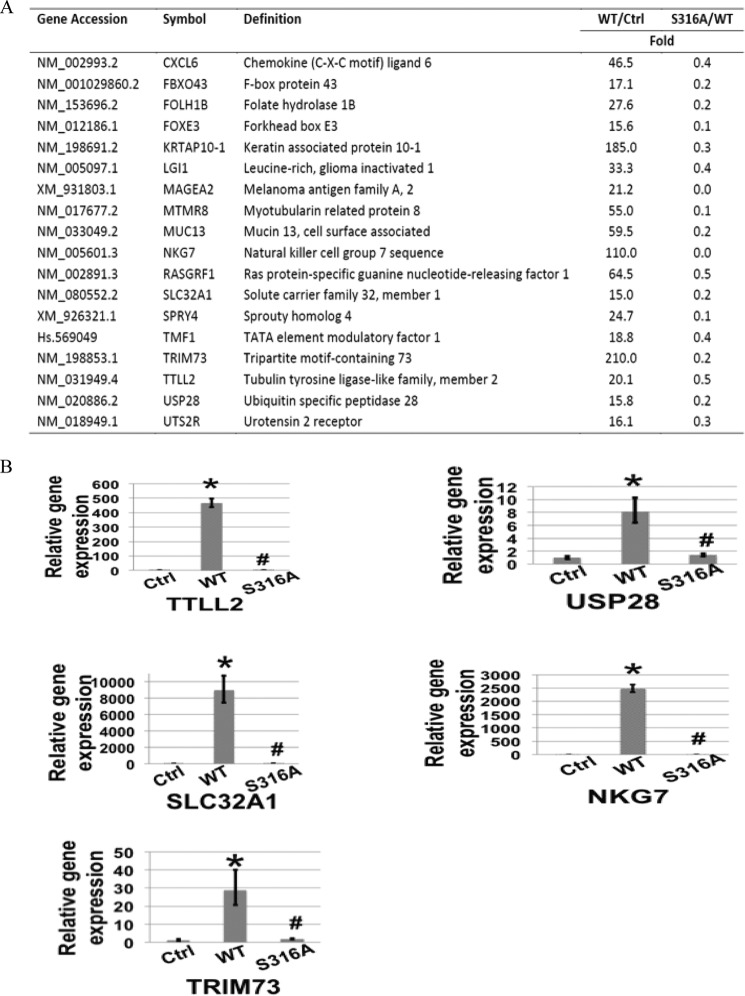
Often a specific post-translational modification site is associated with the regulation of some specific genes. To assess the role of Ser-316 in NF-κB-inducible gene expression, RNA samples from 293C6 cells with either the overexpression of wtp65 or the S316A-p65 mutant protein were used for the Illumina microarray assay. Our data suggested that ∼72% of wtp65-inducible genes were down-regulated by 2-fold or more by the S316A-p65 mutation, supporting the notion that Ser-316 phosphorylation is important for the activation of a majority of NF-κB-inducible genes. A short list of typical genes whose expression levels were affected by S316A mutation is shown in Fig. 3A. These genes encode proteins exhibiting a broad range of functions, such as chemokines, important transcription factors, tumor promoting factors, etc. We further carried out quantitative PCR to confirm the expression levels of several candidate genes. As shown in Fig. 3B, the mRNA levels of tubulin tyrosine ligase-like family, member 2 (TTLL2), ubiquitin specific peptidase 28 (USP28), solute carrier family 32A member 1 (SLC32A1), natural killer cell granule protein 7 (NKG7), and tripartite motif containing 73 (TRIM73) were strongly induced in cells with the overexpression of wtp65, confirming that they are NF-κB-inducible genes. In great contrast, the expression levels of these genes were much less induced by the overexpression of the S316A-p65 mutant (Fig. 3B). Collectively, our Illumina microarray data demonstrate the great importance of Ser-316 phosphorylation in the induction of the majority of NF-κB target genes.
FIGURE 3.
Regulation of NF-κB-dependent gene expression by Ser-316 phosphorylation. A, a short list of NF-κB-dependent genes that are down-regulated by S316A-p65 mutant. These genes encode proteins that have a broad range of functions. B, quantitative PCR analysis showing that TTLL2, USP28, SLC32A1, NKG7, and TRIM73 were induced by the overexpression of wtp65 as compared with control cells. However, this induction was greatly decreased after S316A-p65 mutation. The data represent the mean ± S.D. from three independent experiments. *, p < 0.01 versus control (Ctrl) group; #, p < 0.01 versus wt group.
Important Biological Effect of Ser-316 Phosphorylation
Activation of NF-κB is often associated with the induction and release of multiple cytokines, chemokines, and growth factors. We determined the effect of S316A mutation on the release of cytokines and growth factors in both 293C6 and MEFp65−/− cells. Conditioned media from cells with the overexpression of either wtp65 or S316A-p65 mutant were collected and used to treat stable 293-κB reporter cells (8) for the NF-κB-specific luciferase assay. As shown in Fig. 4A, a significantly higher NF-κB-inducing activity was observed from the media produced by wtp65 but not by S316A-p65 mutant-overexpressing cells, confirming that S316A-p65 mutation greatly compromised the expression and secretion of many NF-κB activating cytokines and growth factors.
FIGURE 4.
Important biological functions of Ser-316. A, assays of conditioned media showing that conditioned media from 293C6 cells (left panel) overexpressing wtp65 had much higher NF-κB-inducing activity than media from cells overexpressing the S316A-p65 mutant protein. Similar phenomenon was observed with MEFp65−/− cells (right panel). Stable 293-NF-κB reporter cells were used. The data were normalized to the total number of cells that generated the conditioned media and to the total amounts of protein. The results of triplicate luciferase assays are shown as the mean ± S.D. *, p < 0.05 versus control (Ctrl) group; #, p < 0.05 versus wt group. B, analysis of cytokine expression in the conditioned media from 293C6 control, wtp65, or S316A-p65 by using human cytokine ELISA plate array. Total 32 cytokines/chemokines were tested. C, cell growth rate assay. Top panel, Western assays showing overexpression of wtp65 and Ser-316-p65 in 293C6 stable cells. The molecular marker is marked on the left. Units, kDa. Bottom panels: cell growth rate, showing that wtp65-overexpressing cells grew much more rapidly than either control or S316A-p65-overexpressing cells. The results of triplicate luciferase assays are shown as the mean ± S.D. *, p < 0.05 versus control group.
To examine the secretory factor(s) regulated through phosphorylation of Ser-316 of p65, we carried out human cytokine ELISA arrays in 293C6 control and wtp65 or S316A-p65 stable cell lines (Fig. 4B). Data suggested that as compared with control group, 19 of 32 tested cytokines/chemokines have significant induction in wtp65-overexpressing cells, suggesting these cytokines/chemokines are NF-κB-inducible factors. Impressively, overexpression of S316A-p65 showed remarkably reduced expression of most of these factors in comparison to the media from wt-p65-overexpressing cells, supporting the notion that phosphorylation of Ser-316 of p65 specifically controls the secretion of a subgroup of cytokine/chemokines or growth factors. Among these factors, especially TNFα, interferon γ-induced protein 10 (IP-10), regulated on activation (normal T-cell expressed and secreted (Rantes)), vascular endothelial growth factor (VEGF), monocyte chemotactic protein 1 (MCP1), and IL-8, -10, -17a, etc. showed more dramatic effects than other factors. Interestingly, these factors are often shown to play important roles in inflammation, cell growth, and tumor microenvironment, confirming the important role of Ser-316 phosphorylation in the regulation of NF-κB activity.
Because cytokines, especially growth factors, could affect cell growth, we further determined the effect of S316A-p65 mutant on cell growth. Stable pools of 293C6 cells with either overexpression of vector control, wtp65, or S316A-p65 mutant were set up for this purpose (Fig. 4C, top panel). In comparison to control cells, overexpression of wtp65 had considerably higher cell numbers on the indicated days. Increased cell growth rate was observed as early as day 3. However, no statistically significant difference was observed for the cell growth rates between the control and the S316A-p65 mutant groups (Fig. 4C, bottom panel). Taken together, these interesting data strongly support the critical role that Ser-316 phosphorylation plays in NF-κB-dependent biological functions.
Phosphorylation of Multiple Serine Residues Triggered by IL-1β
Among the 12 phosphorylated amino acids that were reported, two (Ser-529 and Ser-536) are frequently observed and well known. In 1998, Ser-529 phosphorylation was first identified in HeLa cells under the treatment of TNFα (26). A year later, Ser-536 phosphorylation was also discovered in TNFα-treated HeLa cells (17), suggesting that the same cytokine could induce the phosphorylation of more than one amino acid residue. We had not previously tested the phosphorylation of either Ser-529 or Ser-536 in response to IL-1β in our 293C6 cell system. In the same p65 sample from which Ser-316 phosphorylation was identified, we observed the phosphorylation of Ser-529 and Ser-536. To further confirm the phosphorylation of Ser-529 and 536 of p65 and to analyze their induction kinetics in 293C6 cells upon IL-1β treatment, we treated 293C6 with IL-1β for indicated times and directly used p65 site-specific phosphorylation antibodies to examine the phosphorylation status of Ser-529 and Ser-536 by Western analysis. As shown in Fig. 5, both Ser-529 and Ser-536 are dramatically phosphorylated in a dynamic pattern after the treatment of IL-1β (Fig. 5, top two panels). Both Ser(P)-529-p65 and Ser(P)-536-p65 started to show up at ∼15 min or an earlier time point and reached the peak at ∼30 min then gradually started to decay at ∼2 h after the treatment of IL-1β. This interesting phenomenon proved that, in response to one of the most important cytokines in human system, i.e. IL-1β, p65 could be phosphorylated not only on Ser-316 but also on Ser-529 and Ser-536. Furthermore, our data clearly pointed out that IL-1β-induced IκBα degradation is an important step in the activation of NF-κB (Fig. 5, third panel), and phosphorylation of both Ser-529 and Ser-536 happens before IκB degradation even occurs.
FIGURE 5.
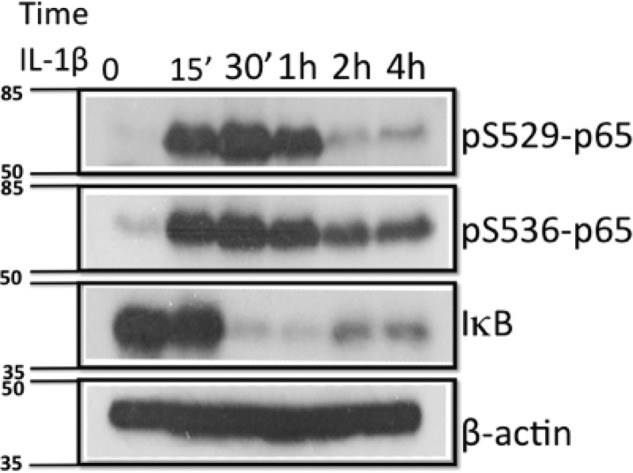
Dynamic phosphorylation of Ser-529 and Ser-536 of p65 in response to IL-1β in 293C6 cells. A Western assay shows that upon IL-1β treatment, both Ser(P)-529 and Ser(P)-536-p65 were induced within 15 min then started to decay after 2 h. Meanwhile, IκB was degraded, indicating the activation of NF-κB. The molecular marker is marked on the left. Units, kDa.
Role of Serine Phosphorylation in Differential Gene Regulation
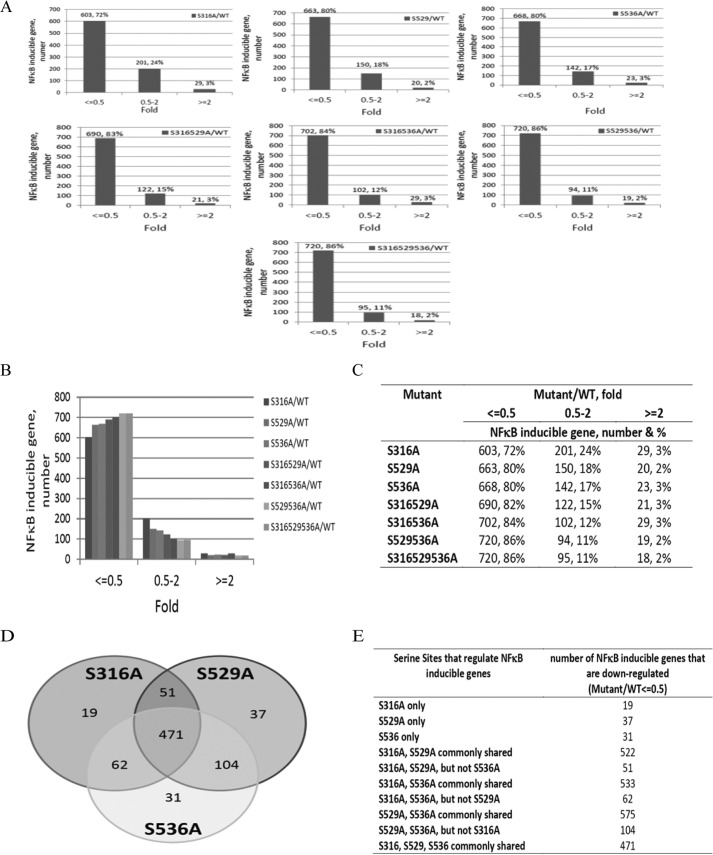
It is quite interesting to observe that p65 is phosphorylated on different serine residues in response to the same cytokine. We hypothesize that phosphorylation of each individual serine is responsible for the regulation of different subgroups of κB-inducible genes. To test this hypothesis, we generated Ser-316, Ser-529, and Ser-536 single, double, and triple alanine mutants. 293C6 cells overexpressed with wtp65 or different S-A-p65 mutants were used to carry out a thorough Illumina microarray assay to compare the gene induction profiles of these phosphorylation sites. As shown in Fig. 6A, in comparison to wtp65 cells, each single site mutant down-regulated (by 2-fold or more) ∼70∼80% of NF-κB target genes with S316A-p65 affected ∼72% (603 genes), S529A affected ∼80% (663 genes), and S536A affected ∼80% (668 genes) of NF-κB target genes, respectively. Surprisingly, double or triple mutants only moderately increased the number of genes that are down-regulated as compared with those of different single mutants. Overall, different double mutants reduced the expression of ∼85% of NF-κB target genes (by 2-fold or more). In comparison to wtp65 cells, cells that expressed S316A/S529A double mutant reduced ∼83% (by 2-fold or more) (690 genes), S316A/S536A reduced ∼84% (702 genes), and S529A/S536A reduced ∼86% (720 genes) of NF-κB target gene expression. Furthermore, S316A/S529A/S536A triple mutant down-regulated the expression of ∼86% (720 genes) of NF-κB target genes, which is not much different from those of double mutants, suggesting that there are rarely genes co-regulated by all three serine residues together. Detailed comparisons of genes that are affected by one, two, or three different serine residues are shown in Fig. 6, B–E. Particularly, as indicated in Fig. 6, D and E, phosphorylation of Ser-316, Ser-529, and Ser-536 can either share the regulation of some NF-κB-dependent genes or solely regulate a subgroup of genes. For example, Ser-316, Ser-529, and Ser-536 solely regulated 19, 37, or 31 genes, respectively. In contrast, Ser-316, Ser-529, and Ser-536 share the regulation of most of the genes at different regulation levels. For example, Ser-316 and Ser-529 share the regulation of 522 NF-κB target genes, which means these 522 genes can either be regulated by Ser-316 alone or by Ser-529 alone. Among these 522 genes, the third residue, Ser-536, cannot regulate 51 genes (Fig. 6, D and E). Likewise, Ser-316 and Ser-536 share the regulation of 533 common genes; among them, the third residue Ser-529 cannot regulate 62 genes. Ser-529 and Ser-536 share the regulation of 575 NF-κB target genes; 104 of them cannot be regulated by Ser-316. Finally, the majority of NF-κB target genes (∼471 genes) can be regulated by any of these three residues. This phenomenon supports a hierarchical regulation pattern for serine-dependent differential gene regulation.
FIGURE 6.
Differential regulation of NF-κB-dependent genes by phosphorylation of Ser-316, -529, and -536. A–C, microarray data show the different gene regulation effects by different p65 serine to alanine mutants, including S316A, S529A and S536A single mutants and S316/529, S316/536, S529/536 double mutants as well as the S316A/S529A/S536A triple mutant. In this experiment, the data showing the ratio of gene expression levels from mutant(s) p65 versus wtp65, the levels of gene expression from wtp65 sample were considered as 1. Overall, each single mutant was able to decrease 70∼80% that of NF-κB target gene expression. Double and triple mutants had slightly more dramatic effect than each single site mutant. D and E, phosphorylation of Ser-316, Ser-529, and Ser-536 can either commonly (all three sites) regulate some of NF-κB target genes or solely regulate their specific pools of genes. Moreover, two different sites, i.e. Ser-316/529, or Ser-316/536 or Ser-529/536 can commonly regulate some genes, which are not regulated by the third S site, suggesting a hieratical regulation pattern.
CKI Phosphorylates p65 at Ser-316
To determine which kinase(s) phosphorylates p65 at Ser-316, we carried out a mass spectrometry protein identification experiment to identify the kinases that can bind to p65 in response to IL-1β treatment. Casein kinase I was identified as a top candidate. We further used the Human Protein Reference Database to predict potential kinases that related Ser-316 phosphorylation (27). Once again, CKI was identified as the top candidate. We then used the CKI-specific inhibitor D4476 to determine whether it could block IL-1β-induced p65 phosphorylation. D4476 is a cell-permeable triaryl substituted imidazolo compound that acts as a selective inhibitor of CKI. 293C6, wtp65, and S316A-p65 cells were treated with 50 μm D4476 for 4 h, and samples were then collected for the luciferase assay. As shown in Fig. 7, D4476 dramatically decreased NF-κB activity in both IL-1β-induced 293C6 control cells and wtp65 overexpressing cells. In great contrast, D4475 barely had any effect on the NF-κB activity in S316A-p65-overexpressing cells. These data strongly suggested that CKI phosphorylated Ser-316 of p65.
FIGURE 7.
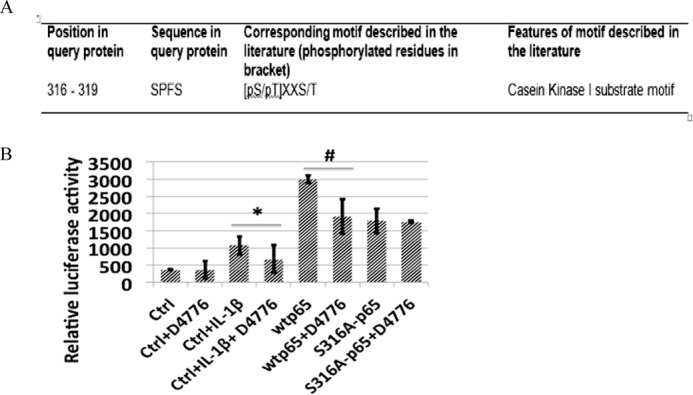
CKI Phosphorylates p65 at Ser-316. A, HPRD website predicted CKI motif in position 316–319 of p65. B, NF-κB activity was determined by luciferase assay, showing that CKI inhibitor D4476 significantly decreased the NF-κB-inducing activity in both 293C6 cells treated with IL-1β and wtp65 overexpressing cells but had no notable effect on the S316A-p65 overexpressing cells, suggesting that D4476 functioned through Ser-316 phosphorylation. The data represent the mean ± S.D. from three independent experiments. *, p < 0.01 versus control +IL-1β (Ctrl+IL-1β) group; #, p < 0.01 versus wtp65 group.
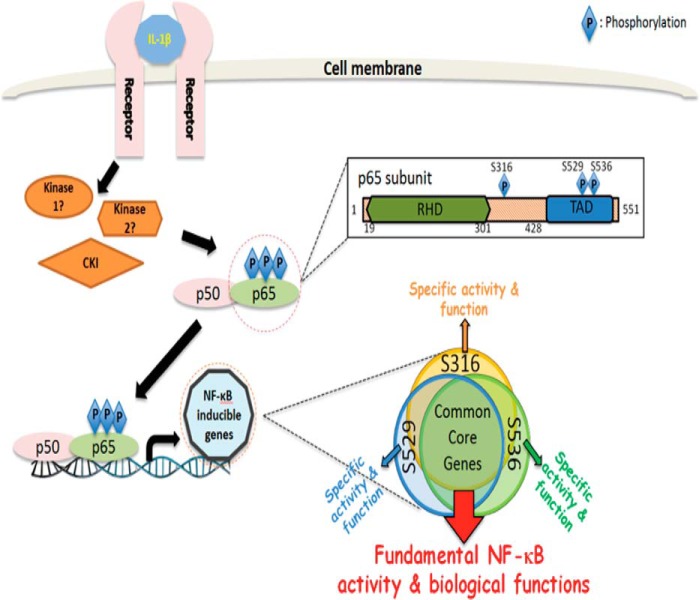
Hypothetical Model
Based on our current study, we propose the following. 1) Ser-316 is a novel phosphorylation site after the treatment of IL-1β. 2) Multiple serine residues can be phosphorylated by the same activating stimulus, such as IL-1β. 3) S316A mutation affects p65 nuclear translocation. 4) Phosphorylation of different serine residues leads to the activation of different subgroups of NF-κB target genes. 5) The majority of NF-κB target genes can be regulated by any of the three serine residues we studied; therefore, if one residue loses its function, it will affect the expression of the majority of NF-κB target genes. 6) The purpose of multiple serine regulation is to fine-tune the regulation of NF-κB genes, adding more derivative functions to the original NF-κB core function. 7) CKI is the kinase that phosphorylated p65 at Ser-316 site (Fig. 8).
FIGURE 8.
Hypothetical model. We hypothesize that upon treatment with IL-1β, Ser-316, Ser-529, or Ser-536 of p65 are phosphorylated by the same or different kinases, including CKI (inset: magnified image of the precise phosphorylation sites and their location in the p65 structure). Each phosphorylated site regulates the common core (∼80% of genes in our case) of NF-κB dependent genes. Additionally, each site further regulates ∼20% unique subgroups of genes (inset: illustrating common core genes and different specific subgroups of genes that are activated by each of the three phosphorylated sites individually or cooperatively). This regulation pattern not only will allow NF-κB to exhibit its major fundamental biological functions but also further provide NF-κB with more specificity and flexibility, thus leading to diversified NF-κB-driven biological consequences in specific cell systems under specific microenvironments and in response to a variety of different specific stimuli. TAD, transactivation domain.
Discussion
Although phosphorylation is the very original type of modification identified on p65, its role in NF-κB regulation is still not fully understood. Why do cells need to phosphorylate multiple serine residues on p65? How do different sites of serine phosphorylation contribute to NF-κB gene regulation? These interesting phenomena motivated us to further explore whether there are any unidentified serine phosphorylations on p65 beyond the existing knowledge.
In this study we discovered that Ser-316 was dramatically phosphorylated upon the treatment of IL-1β. We showed that phosphorylation of Ser-316 enhanced p65 transcription activity. κB-specific luciferase assay proved that overexpressing wtp65 but not the S316A-p65 mutant significantly activated NF-κB in both 293C6 cells and MEFp65−/−cells. S316A-p65 mutant compromised p65 nuclear translocation, leading to the loss of NF-κB activation. Because many of NF-κB target genes are cytokines, chemokines, and growth factors, we further tested the effect of Ser-316 mutation on the secretion of these factors. Conditioned media from 293C6 cells overexpressing either wtp65 or the S316A-p65 mutant were used to treat NF-κB reporter cells. Media from cells with the S316A mutant protein decreased NF-κB-inducing activity significantly more than that from wtp65 cells. Similar results were obtained in p65-null MEFs with the expression of either wt or the S316A-p65 mutant p65 (Fig. 4A), supporting that Ser-316 is important for the secretion of many NF-κB stimulating factors. By using human cytokine ELISA arrays we identified Ser-316-specific cytokines/chemokines such as TNFα, IP-10, Rantes, VEGF, MCP-1, IL8, IL-10, and IL17a, etc. that are critical for the process of inflammation, cell growth, angiogenesis, and tumor microenvironment. For instance, VEGF promotes endothelial cell proliferation and migration in vitro (28). Inflammatory mediators have a significant effect on the process of angiogenesis and tumor growth, mainly through up-regulation of VEGF. Therefore, VEGF has been a therapeutic target for multiple types of cancers (29, 30). Moreover, we further determined the effect of Ser-316 on cell growth. Cell proliferation assays showed that overexpression of wtp65 increased cell growth rate, whereas no significant difference between control and S316A mutant was observed. Taken together, these data further support the notion that phosphorylation of S316A is crucial for the regulation of NF-κB related biological functions.
In addition to Ser-316 phosphorylation, it is interesting that we found both Ser-529 and Ser-536 could also be phosphorylated under the same experimental conditions. The co-existence of three different serine phosphorylations on p65 raises a very interesting question, i.e. are these phosphorylation sites functionally unique or redundant? By generating a group of single, double, or triple mutants of Ser-316, -529, and -536 sites and examining their function in gene regulation, we show that phosphorylation of each individual serine not only can control the majority of NF-κB targeted genes, which are commonly shared with other two phosphorylation sites, but also can control its own subgroup of unique signature genes. Based on our data, it is fair to say that the existence of each unique phosphorylation site has its own function. Each phosphorylation site can regulate the common core (∼80%, in our study) of NF-κB target genes. Therefore, logically, if any phosphorylation site fails to function properly, the entire commonly shared core gene population will be affected. This is possibly a smart system that cells have adopted for the regulation of NF-κB. By doing so each phosphorylation can not only support the basic regulation of NF-κB target genes expression but can also further fine-tune NF-κB-dependent gene expression to achieve both the flexibility and specificity in response to a given cellular stimulus or under a specific cell system and cellular environment. Importantly, we identified CKI as the kinase to phosphorylate Ser-316 on p65. CKI belongs to the CK serine/threonine kinase family. It is ubiquitously expressed in eukaryotic organisms and highly conserved from yeast to human. The CKI kinase has been shown to play an important role in cancer development and is considered an important cancer therapeutic target (31). CKI is involved in different but rather important cellular functions such as regulation of membrane transport, cell division, DNA repair, circadian rhythms, and nuclear localization by phosphorylating its target proteins (32). Therefore, identification of CKI as the kinase to phosphorylate Ser-316 of p65 added another significant and interesting function to CKI.
In the future there is a great deal of work that needs to be done. For example, although both Ser-529 and Ser-536 were reported to be phosphorylated, the number and nature of the kinases that catalyze these phosphorylation events are very complex. There are multiple reports regarding the kinases that phosphorylate Ser-529 or Ser-536, including IκB kinase (α, β, and ϵ), TRAF (TNF receptor-associated factor) family member-associated (TANK)-binding kinase 1 (TBK1)), PI3 kinase, p38 mitogen-activated protein kinase, and CKII (18) (33, 34). The existing information certainly suggests that a number of kinases may have the ability to phosphorylate the same serine residue under specific conditions. Adding the CKI we identified in this study, the combination of kinase and the residues that are phosphorylated possibly allows for a different group of genes to be activated/repressed and is surely a fascinating research field to further pursue.
Furthermore, it is worth noting an interesting phenomenon regarding the distribution of phosphorylation on p65. Among the 12 known phosphorylation sites, 4 are located in RHD domain, 7 are located on the transactivation domain, and only one site, the Ser-311, is located in the linker region of p65 (35). Ser-311 is phosphorylated by the atypical protein kinase Cζ (PKCζ). Ser-311 phosphorylation is required for CBP-enhanced NF-κB transcriptional activity in embryo fibroblasts in response to TNFα stimulation. Our current study identified a novel Ser-316 phosphorylation site in the p65 linker region. Thus, it would be interesting to investigate whether location of the phosphorylated site has any functional effect on NF-κB and correlates with induction of specific gene families.
In this paper we have discussed the phosphorylation sites in domains of the p65 subunit, which is specifically involved in activation of the canonical arm of the NF-κB pathway. However, as mentioned before, few phosphorylation sites are also located in the RHD domain, which is conserved in all the family members of the NF-κB superfamily. In this regard another aspect to investigate would be to see if phosphorylation of sites in the conserved RHD domain affects the activity of the NF-κB members involved in the non-canonical arm of the NFκB signaling pathway. The non-canonical pathway has distinct factors involved in comparison to the canonical arm, and hence it would be interesting to see if there is cross-regulation between these pathways linked by phosphorylation of the conserved sites of RHD domain and how that may affect cell function.
In summary, our study identified Ser-316 as a novel IL-1β-inducible phosphorylation site on p65. We showed that phosphorylation of Ser-316 is critical for NF-κB nuclear translocation, NF-κB-dependent gene regulation, cytokines/chemokines secretion, and biological functions. Furthermore, we provided evidence of the co-existence of Ser-529 and Ser-536 phosphorylation on p65. Our microarray data strongly support our hypothetical model that phosphorylation of different serine residues regulate the same common core (in our study, ∼80%) of NF-κB target genes. On top of this basic function, each individual phosphorylated serine residue exhibits its own uniqueness for gene regulation, adding multiple facets and layers of complexity to NF-κB regulation. This strategy for serine regulation may well exist in other types of post-translational modifications of NF-κB, such as ubiquitination, acetylation, and methylation, etc. Our study and hypothesis further justify the necessity and strategy of targeting specific protein kinases or other types of post-translation modification-mediating enzymes as specific and effective ways to control excessive NF-κB activity in NF-κB-driven inflammatory diseases and cancers. This pathway and post-translational modification site-specific inhibition of NF-κB is impossible to achieve by simply targeting NF-κB transcription factor itself.
Author Contributions
B. W., H. W., L. P., W. Z., and T. L. carried out the experimental work. T. L. designed the experiments. T. L., H. W., B. W., L. P., M. M., and A. H. contributed analysis of the data and writing the manuscript.
Acknowledgment
We thank Lisa King at the Department of Pharmacology and Toxicology at Indiana University School of Medicine for professional help with revising this manuscript.
The authors declare that they have no conflicts of interest with the contents of this article.
- IκB
- inhibitor of nuclear factor κB
- CKI
- casein kinase I
- IP-10
- interferon γ-induced protein 10
- NF-κB
- nuclear factor κB
- NKG7
- natural killer cell granule protein 7
- PKAc
- catalytic subunit of protein kinase A
- RHD
- Rel homology domain
- SLC32A1
- solute carrier family 32A member 1
- threonine
- T
- TRIM73
- tripartite motif containing 73
- TTLL2
- tubulin tyrosine ligase-like family member 2
- USP28
- ubiquitin specific peptidase 28
- MEF
- mouse embryo fibroblast.
References
- 1. Gilmore T. D. (2006) Introduction to NF-κB: players, pathways, perspectives. Oncogene 25, 6680–6684 [DOI] [PubMed] [Google Scholar]
- 2. Perkins N. D. (2006) Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene 25, 6717–6730 [DOI] [PubMed] [Google Scholar]
- 3. Viatour P., Merville M. P., Bours V., Chariot A. (2005) Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 30, 43–52 [DOI] [PubMed] [Google Scholar]
- 4. Chen Z. J. (2005) Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen L. F., Greene W. C. (2003) Regulation of distinct biological activities of the NF-κB transcription factor complex by acetylation. J. Mol. Med. 81, 549–557 [DOI] [PubMed] [Google Scholar]
- 6. Nasr R., Chiari E., El-Sabban M., Mahieux R., Kfoury Y., Abdulhay M., Yazbeck V., Hermine O., de Thé H., Pique C., Bazarbachi A. (2006) Tax ubiquitylation and sumoylation control critical cytoplasmic and nuclear steps of NF-κB activation. Blood 107, 4021–4029 [DOI] [PubMed] [Google Scholar]
- 7. Marshall H. E., Stamler J. S. (2001) Inhibition of NF-κB by S-nitrosylation. Biochemistry 40, 1688–1693 [DOI] [PubMed] [Google Scholar]
- 8. Wei H., Wang B., Miyagi M., She Y., Gopalan B., Huang D. B., Ghosh G., Stark G. R., Lu T. (2013) PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc. Natl. Acad. Sci. U.S.A. 110, 13516–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson L. N. (2009) The regulation of protein phosphorylation. Biochem. Soc. Trans. 37, 627–641 [DOI] [PubMed] [Google Scholar]
- 10. Foster J. S., Wimalasena J. (1996) Estrogen regulates activity of cyclin-dependent kinases and retinoblastoma protein phosphorylation in breast cancer cells. Mol. Endocrinol. 10, 488–498 [DOI] [PubMed] [Google Scholar]
- 11. Cohen P. (2001) The role of protein phosphorylation in human health and disease: The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 268, 5001–5010 [DOI] [PubMed] [Google Scholar]
- 12. Sabatel H., Di Valentin E., Gloire G., Dequiedt F., Piette J., Habraken Y. (2012) Phosphorylation of p65(RelA) on Ser-547 by ATM represses NF-κB-dependent transcription of specific genes after genotoxic stress. PLoS ONE 7, e38246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hochrainer K., Racchumi G., Anrather J. (2013) Site-specific phosphorylation of the p65 protein subunit mediates selective gene expression by differential NF-κB and RNA polymerase II promoter recruitment. J. Biol. Chem. 288, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhong H., SuYang H., Erdjument-Bromage H., Tempst P., Ghosh S. (1997) The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89, 413–424 [DOI] [PubMed] [Google Scholar]
- 15. Zhong H., Voll R. E., Ghosh S. (1998) Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1, 661–671 [DOI] [PubMed] [Google Scholar]
- 16. Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., Haegeman G. (2003) Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakurai H., Chiba H., Miyoshi H., Sugita T., Toriumi W. (1999) IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 274, 30353–30356 [DOI] [PubMed] [Google Scholar]
- 18. Wang D., Westerheide S. D., Hanson J. L., Baldwin A. S., Jr. (2000) Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser-529 is controlled by casein kinase II. J. Biol. Chem. 275, 32592–32597 [DOI] [PubMed] [Google Scholar]
- 19. Mattioli I., Geng H., Sebald A., Hodel M., Bucher C., Kracht M., Schmitz M. L. (2006) Inducible phosphorylation of NF-κB p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J. Biol. Chem. 281, 6175–6183 [DOI] [PubMed] [Google Scholar]
- 20. Buss H., Dörrie A., Schmitz M. L., Frank R., Livingstone M., Resch K., Kracht M. (2004) Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J. Biol. Chem. 279, 49571–49574 [DOI] [PubMed] [Google Scholar]
- 21. Lu T., Jackson M. W., Singhi A. D., Kandel E. S., Yang M., Zhang Y., Gudkov A. V., Stark G. R. (2009) Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NF-κB. Proc. Natl. Acad. Sci. U.S.A. 106, 16339–16344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu T., Burdelya L. G., Swiatkowski S. M., Boiko A. D., Howe P. H., Stark G. R., Gudkov A. V. (2004) Secreted transforming growth factor β2 activates NF-κB, blocks apoptosis and is essential for the survival of some tumor cells. Proc. Natl. Acad. Sci. U.S.A. 101, 7112–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu T., Jackson M. W., Wang B., Yang M., Chance M. R., Miyagi M., Gudkov A. V., Stark G. R. (2010) Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. U.S.A. 107, 46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu T., Yang M., Huang D. B., Wei H., Ozer G. H., Ghosh G., Stark G. R. (2013) Role of lysine methylation of NF-κB in differential gene regulation. Proc. Natl. Acad. Sci. U.S.A. 110, 13510–13515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoesel B., Schmid J. A. (2013) The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang D., Baldwin A. S., Jr. (1998) Activation of nuclear factor-κB-dependent transcription by tumor necrosis factor-α is mediated through phosphorylation of RelA/p65 on serine 529. J. Biol. Chem. 273, 29411–29416 [DOI] [PubMed] [Google Scholar]
- 27. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 28. Bernatchez P. N., Soker S., Sirois M. G. (1999) Vascular endothelial growth factor effect on endothelial cell proliferation, migration, and platelet-activating factor synthesis is Flk-1-dependent. J. Biol. Chem. 274, 31047–31054 [DOI] [PubMed] [Google Scholar]
- 29. Angelo L. S., Kurzrock R. (2007) Vascular endothelial growth factor and its relationship to inflammatory mediators. Clin. Cancer Res. 13, 2825–2830 [DOI] [PubMed] [Google Scholar]
- 30. Casey S. C., Amedei A., Aquilano K., Azmi A. S., Benencia F., Bhakta D., Bilsland A. E., Boosani C. S., Chen S., Ciriolo M. R., Crawford S., Fujii H., Georgakilas A. G., Guha G., Halicka D., Helferich W. G., Heneberg P., Honoki K., Keith W. N., Kerkar S. P., Mohammed S. I., Niccolai E., Nowsheen S., Vasantha Rupasinghe H. P., Samadi A., Singh N., Talib W. H., Venkateswaran V., Whelan R. L., Yang X., Felsher D. W. (2015) Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin Cancer Biol. 10.1016/j.semcancer.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knippschild U., Krüger M., Richter J., Xu P., García-Reyes B., Peifer C., Halekotte J., Bakulev V., Bischof J. (2014) The CK1 family: contribution to cellular stress response and its role in carcinogenesis. Front. Oncol. 4, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Price M. A. (2006) CKI, there's more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 20, 399–410 [DOI] [PubMed] [Google Scholar]
- 33. Buss H., Dörrie A., Schmitz M. L., Hoffmann E., Resch K., Kracht M. (2004) Constitutive and interleukin-1-inducible phosphorylation of p65 NF-κB at serine 536 is mediated by multiple protein kinases including IκB kinase (IKK)-α, IKKβ, IKKϵ, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279, 55633–55643 [DOI] [PubMed] [Google Scholar]
- 34. Madrid L. V., Wang C. Y., Guttridge D. C., Schottelius A. J., Baldwin A. S., Jr., Mayo M. W. (2000) Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol. Cell Biol. 20, 1626–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duran A., Diaz-Meco M. T., Moscat J. (2003) Essential role of RelA Ser311 phosphorylation by ζPKC in NF-κB transcriptional activation. EMBO J. 22, 3910–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]