Background: PerR is a metal-dependent H2O2 sensor in many Gram-positive bacteria.
Results: Staphylococcus aureus PerRSA, previously known as a Mn2+-specific repressor, uses Fe2+ to sense very low levels of H2O2.
Conclusion: The apparent lack of Fe2+-dependent repressor activity of PerRSA is due to the hypersensitivity of PerRSA under aerobic conditions.
Significance: Cells expressing hypersensitive PerRSA are less virulent than those expressing PerRBS.
Keywords: hydrogen peroxide, metal homeostasis, oxidative stress, redox regulation, Staphylococcus aureus (S. aureus), Fur family, histidine oxidation, PerR
Abstract
In many Gram-positive bacteria PerR is a major peroxide sensor whose repressor activity is dependent on a bound metal cofactor. The prototype for PerR sensors, the Bacillus subtilis PerRBS protein, represses target genes when bound to either Mn2+ or Fe2+ as corepressor, but only the Fe2+-bound form responds to H2O2. The orthologous protein in the human pathogen Staphylococcus aureus, PerRSA, plays important roles in H2O2 resistance and virulence. However, PerRSA is reported to only respond to Mn2+ as corepressor, which suggests that it might rely on a distinct, iron-independent mechanism for H2O2 sensing. Here we demonstrate that PerRSA uses either Fe2+ or Mn2+ as corepressor, and that, like PerRBS, the Fe2+-bound form of PerRSA senses physiological levels of H2O2 by iron-mediated histidine oxidation. Moreover, we show that PerRSA is poised to sense very low levels of endogenous H2O2, which normally cannot be sensed by B. subtilis PerRBS. This hypersensitivity of PerRSA accounts for the apparent lack of Fe2+-dependent repressor activity and consequent Mn2+-specific repressor activity under aerobic conditions. We also provide evidence that the activity of PerRSA is directly correlated with virulence, whereas it is inversely correlated with H2O2 resistance, suggesting that PerRSA may be an attractive target for the control of S. aureus pathogenesis.
Introduction
Reactive oxygen species, which are produced endogenously as a by-product of aerobic metabolism or exogenously by microbial competitors and eukaryotic hosts, can cause oxidative stress to bacteria by damaging cellular constituents (1, 2). To cope with reactive oxygen species, bacteria have evolved sophisticated oxidative stress response systems including transcription factors that efficiently sense specific reactive oxygen species and induce appropriate defense systems (2–4). For example, OxyR in the Gram-negative model bacterium Escherichia coli senses H2O2 using cysteine oxidation, and activates transcription of ∼20 genes, including genes involved in H2O2 detoxification (1). Whereas many Gram-negative bacteria use OxyR as the major H2O2 sensor, many Gram-positive bacteria use PerR as a functional equivalent of OxyR (5, 6).
Bacillus subtilis PerR (PerRBS)4 is a member of Fur family of metal-dependent regulators and is the prototype for a group of metal-dependent peroxide sensing repressors (5). PerRBS contains a structural Zn2+ coordinated by four cysteine residues (Cys4:Zn site, Site 1) and a second regulatory metal binding site (Site 2) composed of three N-donor ligands (His-37, His-91, and His-93) and two O-donor ligands (Asp-85 and Asp-104). Although the binding of either Fe2+ (PerRBS:Zn,Fe) or Mn2+ (PerRBS:Zn,Mn) at Site 2 activates PerRBS to bind DNA, only PerRBS:Zn,Fe can sense low levels of H2O2. Unlike cysteine thiol-based peroxide sensors such as OxyR and OhrR, PerRBS senses H2O2 by metal-catalyzed histidine oxidation. Reaction of Fe2+, bound to Site 2, with H2O2 leads to the rapid oxidation of either His-37 or, to a lesser degree, His-91 (two of the Site 2 ligands) with concomitant loss of iron binding (7). Structurally, this results in an opening of the DNA-binding competent caliper-like conformation, leading to a loss of DNA binding and thus allowing the induction of genes that are normally repressed by active PerRBS:Zn,Fe (8).
Staphylococcus aureus is a major human pathogen commonly causing nosocomial and community-acquired infectious diseases worldwide. S. aureus, which can be found as part of the normal skin flora and in anterior nares of the nasal passages, can cause a spectrum of illnesses from minor skin and soft tissue infections to more invasive and serious diseases such as pneumonia, meningitis, osteomyelitis, endocarditis, toxic shock syndrome, bacteremia, and sepsis (9). As a facultative anaerobic Gram-positive bacterium, S. aureus also uses PerR for the control of oxidative stress response (10, 11). The S. aureus PerR (PerRSA) regulon is similar to that described for PerRBS and includes genes encoding KatA, AhpCF, MrgA, Fur, and PerR, as well as others encoding thioredoxin reductase (TrxB), bacterioferritin comigratory protein (Bcp), and an iron storage protein ferritin (Ftn). Despite the similarity of the H2O2-dependent regulation of the PerRSA regulon, Fe2+ was reported to be completely ineffective as a corepressor for the PerRSA-regulated genes, which were only repressed by Mn2+. Indeed, expression of the PerRSA-regulated genes is induced rather than repressed by added Fe2+ (11–14). These observations led to the conclusion that PerRSA is a Mn2+-specific repressor and further suggest that PerRSA may use a fundamentally different and iron-independent mechanism to sense H2O2. Despite the importance of PerRSA in the regulation of virulence factors of S. aureus, the mechanism by which PerRSA senses H2O2 has not been elucidated.
Here we have analyzed the metal-dependent H2O2 sensing mechanisms of PerRSA in vitro and in vivo in comparison with those of PerRBS. PerRSA, like many other Fur family proteins, contains a structural Zn2+ site coordinated by four cysteine residues, which is resistant to oxidation by physiologically relevant H2O2 concentration. Contrary to the suggestion that PerRSA is a Mn2+-specific repressor, the regulatory metal binding site (composed of His-43, Asp-91, His-97, His-99, and Asp-110) can bind Fe2+ even with higher affinity than Mn2+ when measured under anaerobic conditions. Moreover, the Fe2+-bound PerRSA, but not the Mn2+-bound form, can sense H2O2 by Fe2+-dependent oxidation of His-43 and His-97. In cells grown under aerobic conditions most of PerRSA is detected in the fully oxidized state, whereas cells grown under oxygen-limited conditions exhibit Fe2+-dependent repression of the PerRSA regulon. The exquisite sensitivity of PerRSA to inactivation likely explains the previous observation of an apparent lack of Fe2+-dependent repressor activity. Finally, we provide evidence that the high H2O2 sensitivity of PerRSA (in comparison to PerRBS) is important for H2O2 resistance under aerobic conditions and that the low sensitivity of PerRBS (in comparison to PerRSA) increases virulence of S. aureus in host.
Experimental Procedures
Bacterial Strains, Media, and Growth Conditions
The bacterial strains used in this study are listed in Table 1. E. coli, B. subtilis, and S. aureus were grown in Luria-Bertani (LB) media at 37 °C with appropriate antibiotics unless otherwise indicated. As metal-limited minimal media (MLMM), MOPS-buffered minimal medium was used for B. subtilis (15), and phosphate-buffered minimal medium was used for S. aureus (10). Oxygen-limited cultures were grown in 15-ml rubber screw-top tubes with the addition of 0.2% potassium nitrate. To facilitate oxygen-limited growth of S. aureus in MLMM, 1% Chelex-treated tryptone was added.
TABLE 1.
Strains used in this study
| Strains | Relevant genotype and feature | Source |
|---|---|---|
| S. aureus | ||
| Newman | Wild type, human clinical isolate | NCTC |
| RN451 | RN450 lysogenic for Φ11 | NARSA |
| RN4220 | Restriction-deficient transformation recipient | NARSA |
| LS0085 | Newman perR::cat | This study |
| LS0088 | LS0085 pLL29::perRSA-FLAG | This study |
| LS0093 | LS0085 pLL29 | This study |
| LS0106 | Newman pCN33::PmrgA-lacZ | This study |
| LS0107 | Newman pCN33::PkatA-lacZ | This study |
| LS0111 | LS0085 pLL29::perRSA-FLAG pCN33::PmrgA-lacZ | This study |
| LS0114 | LS0085 pLL29::perRBS-FLAG pCN33::PmrgA-lacZ | This study |
| LS0124 | LS0085 pLL29 pCN33::PmrgA-lacZ | This study |
| LS0134 | LS0085 pLL29:: perRBS-FLAG | This study |
| LS0166 | LS0085 pCN48::perRSA-FLAG | This study |
| LS0241 | LS0085 pLL29::perRSA-FLAG pCN33::PkatA-lacZ | This study |
| LS0242 | LS0085 pLL29::perRSA (H43A)-FLAG pCN33::PkatA-lacZ | This study |
| LS0243 | LS0085 pLL29::perRSA (H97A)-FLAG pCN33::PkatA-lacZ | This study |
| LS0244 | LS0085 pLL29::perRSA (C102S)-FLAG pCN33::PkatA-lacZ | This study |
| LS0245 | LS0085 pLL29 pCN33::PkatA-lacZ | This study |
| LS0304 | LS0085 pLL29::perRSA (D91A)-FLAG pCN33::PkatA-lacZ | This study |
| LS0305 | LS0085 pLL29::perRSA (H99A)-FLAG pCN33::PkatA-lacZ | This study |
| LS0306 | LS0085 pLL29::perRSA (C105S)-FLAG pCN33::PkatA-lacZ | This study |
| LS0307 | LS0085 pLL29::perRSA (D110A)-FLAG pCN33::PkatA-lacZ | This study |
| LS0308 | LS0085 pLL29::perRSA (C142S)-FLAG pCN33::PkatA-lacZ | This study |
| LS0309 | LS0085 pLL29::perRSA (C145S)-FLAG pCN33::PkatA-lacZ | This study |
| B. subtilis | ||
| HB9703 | perR::tet | Ref. 15 |
| HB9738 | HB9703 amyE::perRBS-FLAG SPβC2Δ2::Tn917::Φ (PmrgA-cat-lacZ) | Ref. 15 |
| LB1530 | HB9703 amyE::perRSA-FLAG SPβC2Δ2::Tn917::Φ (PmrgA-cat-lacZ) | This study |
| LB1532 | HB9703 amyE::spc SPβC2Δ2::Tn917::Φ (PmrgA-cat-lacZ) | This study |
| E. coli | ||
| HE9501 | BL21 (DE3) pLysS pET-16b::perRBS | Ref. 15 |
| LE0003 | BL21 (DE3) pLysS pET-16b::perRSA | This study |
| LE0031 | BL21 (DE3) pLysS pET-16b::perRSA (H43A) | This study |
| LE0032 | BL21 (DE3) pLysS pET-16b::perRSA (D91A) | This study |
| LE0033 | BL21 (DE3) pLysS pET-16b::perRSA (H97A) | This study |
| LE0034 | BL21 (DE3) pLysS pET-16b::perRSA (H99A) | This study |
| LE0035 | BL21 (DE3) pLysS pET-16b::perRSA (C102S) | This study |
| LE0036 | BL21 (DE3) pLysS pET-16b::perRSA (C105S) | This study |
| LE0037 | BL21 (DE3) pLysS pET-16b::perRSA (D110A) | This study |
| LE0038 | BL21 (DE3) pLysS pET-16b::perRSA (C142S) | This study |
| LE0039 | BL21 (DE3) pLysS pET-16b::perRSA (C145S) | This study |
| LE2302 | BL21 (DE3) pLysS pET-11a::oxyREC | This study |
Construction of a perR Deletion Mutant Strain of S. aureus Newman
The perRSA::cat cassette was constructed by joining PCR using two 1-kb DNA fragments (upstream and downstream of perRSA ORF) and a fragment with a chloramphenicol resistance marker (cat) from pDG1661. This cassette was cloned into the BamHI and EcoRI sites of pMAD (16) having a temperature-sensitive replication origin and a erythromycin-resistant marker (em) resulting in pJL954. Then, pJL954 was introduced into S. aureus Newman (NCTC8178) after passage through a restriction-deficient host S. aureus RN4220. S. aureus Newman strain having pJL954 integrated into the chromosome by a Campbell-type event was selected based on light blue colony color on LB agar plates containing chloramphenicol, erythromycin, and X-Gal after growing cells at 43 °C. After an overnight subculture two times in LB broth containing chloramphenicol at 30 °C, erythromycin-sensitive but chloramphenicol-resistant colonies were selected on LB plate at 43 °C. After confirmation of perRSA deletion by PCR, the resultant strain was named LS0085.
Construction of perR-FLAG Fusion and Reporter Fusion in B. subtilis and S. aureus
For the expression of perRSA-FLAG fusion in B. subtilis, the PCR fragment containing perRSA ORF and about 200 bp upstream region was cloned into BamHI and EagI sites of pJL070 (15) generating pJL361. The ScaI digest of pJL361 was introduced to the perR null mutant B. subtilis strain HB9703 (15) to generate a transformant containing perRSA-FLAG in the amyE locus. Then the PmrgA-cat-lacZ reporter fusion stain (LB1530) was constructed by transduction with SPβ phage from HB1122 (17). For the expression of perRSA-FLAG or perRBS-FLAG fusions in S. aureus, the DNA fragment of pJL070 containing perRBS-FLAG and that of pJL361 containing perRSA-FLAG, were each cloned into BamHI and EcoRI sites of pLL29 (18) producing pJL1434 and pJL1430, respectively. Mutant alleles of perRSA-FLAG were generated by the QuikChange method (Stratagene) using pJL1430 as templates. Each of these plasmids was integrated into the chromosome of S. aureus RN4220 with the help of int gene encoded in pLL2787, and then transferred into the perR null mutant S. aureus Newman strain (LS0085) by phage transduction using Φ11 (19). For the construction of reporter fusion plasmids, the lacZ gene from pDG1661 was PCR-amplified with the introduction of an NcoI site just after the KpnI site, and cloned into the KpnI and EcoRI sites of pCN33 resulting in pJL901. Then, DNA fragment containing the mrgA or katA promoter regions was introduced into the BamHI and NcoI sites of pJL901. The resulting reporter fusion plasmids were introduced into S. aureus Newman by electroporation after passage through S. aureus RN4220. To increase the copy number of perRSA-FLAG, perRSA-FLAG was cloned into the BamHI and EcoRI sites of the high copy number plasmid pCN48 (20) resulting in pJL643. This plasmid was introduced into the perR null mutant S. aureus Newman (LS0085) by electroporation after passage through S. aureus RN4220, generating a strain named LS0166. PerRBS-FLAG and PerRSA-FLAG proteins are fully functional as judged by reporter fusion assays, and were used for complementation of the perR null mutant strains and pulldown assays (15, 21).
Overexpression and Purification of Proteins
The PCR-amplified DNA fragments containing the perRSA ORF were digested with BspHI and BamHI, and cloned into the NcoI and BamHI sites of pET-16b (Novagen) producing a plasmid named pJL203. Mutant alleles of perRSA were generated by the QuikChange method (Stratagene) using pJL203 as template. E. coli oxyR was cloned into the NdeI and BamHI sites of pET-11a (Novagen) producing a plasmid named pJL1282. The encoded proteins were overexpressed using E. coli BL21(DE3) pLysS cells. Wild type (WT) PerRSA proteins were purified after overexpression in E. coli BL21(DE3) pLysS cells harboring pJL203 as previously described for PerRBS proteins (15). Briefly, the cell lysates were clarified by centrifugation and then PerRSA was purified by heparin-Sepharose and Mono-Q chromatography using buffer A (20 mm Tris-HCl, pH 8.0, 0.1 m NaCl, and 5% glycerol (v/v)) containing 10 mm EDTA with the application of a linear gradient of 0.1–1 m NaCl. Further purification was performed using a Superdex-200 (HiLoad 16/60) column equilibrated with Chelex-treated buffer A. The concentration of PerRSA was determined using a molar extinction coefficient of 10,430 m−1 cm−1 at 280 nm.
Enzyme Assays
On-gel catalase activity was assayed using 1:1 mixture of 5% ferric chloride and 5% potassium ferricyanide after gel-soaking in 2 mm H2O2. β-Galactosidase assays were performed with or without 100 μm H2O2 treatment for 30 min as described previously (15), except that lysostaphin (10 μg/ml) was used for the lysis of S. aureus cells. Measurement of Zn2+ release by H2O2 was performed as described previously (15) using 2.5 μm dimeric PerRSA and 100 μm 4-(2-pyridylazo)resorcinol (PAR). The Zn2+ content of purified PerRSA by PAR assay was measured using a molar extinction coefficient of 85,000 m−1 cm−1 at 494 nm for Zn2+-PAR complex (15).
Western Blot Analysis
At A600 = 0.6, 10-ml cultures were harvested by centrifugation after the addition of 1.1 ml of trichloroacetic acid. Then, cells were resuspended in 500 μl of 10% trichloroacetic acid and sonicated. After recovering sonicated samples by centrifugation, the pellets were resuspended with 60 μl of alkylating buffer (100 mm iodoacetamide, 0.5 m Tris, pH 8.0, 5% glycerol, 100 mm NaCl, 2% SDS) and incubated for 1 h in the dark to alkylate-free thiols. Alkylated samples of 10 μl (75 μg of protein) were separated on 13.3% non-reducing SDS-PAGE gel using a Tris-Tricine buffer system and blotted to a polyvinylidene difluoride membrane. FLAG-tagged proteins were probed with mouse monoclonal anti-FLAG antibody and anti-mouse antibody conjugated with alkaline phosphatase (Sigma).
Fluorescence Anisotropy (FA) Experiments
FA experiments were performed using an LS55 luminescence spectrometer (PerkinElmer Life Sciences) installed in an anaerobic chamber (Coy). A 6-carboxyfluorescein (6-FAM)-labeled katA-PerR box DNA fragment was generated by annealing 5′-6-FAM-TTAAATTATAATTATTATAAATTGT-3′ (Integrated DNA Technology) and its unlabeled complement. FA measurements (λex = 492 nm, slit width = 15 nm; λem = 520 nm, slit width = 20 nm, integration time = 1 s) were performed in 3 ml of Chelex-treated anaerobic buffer A. The percentage activity and Kd for DNA of purified PerRSA were determined to be ∼20% and ∼1 nm, respectively, by titration of PerRSA into 3 ml of buffer A containing 10 nm DNA and 1 mm manganese as reported previously (7). For the metal binding and H2O2 sensitivity assays, buffer A containing 100 nm DNA and 100 nm active dimeric PerRSA were used, and FA was measured after each addition.
MALDI-TOF and LC-ESI MS/MS Mass Analyses
To analyze in vivo oxidation of PerRSA (Fig. 4), LS0166 cells expressing PerRSA-FLAG were grown in MLMM containing 50 μm FeSO4 or 50 μm MnCl2. At an A600 of ∼1, cells were treated with or without 100 μm H2O2 for 2 min and lysed with 50 μg of lysostaphin for 1 h in 0.5 ml of buffer A. PerRSA-FLAG protein recovered using anti-FLAG M2-agarose beads (Sigma) was incubated with 100 mm iodoacetamide in the presence of 50 mm EDTA and 1% SDS for 1 h in the dark, and separated on SDS-PAGE gel. The protein bands corresponding to PerRSA-FLAG were analyzed by MALDI-TOF MS using a 4700 Proteomics Analyzer instrument (Applied Biosystems) after in-gel tryptic digestion as described previously (15, 22). The MALDI-TOF MS analysis of in vitro oxidation of purified PerRSA (Fig. 3D) was performed as described previously (7, 15) using a Voyager-DE STR instrument (Applied Biosystems). The analysis of protein oxidation after overexpression in E. coli (Fig. 5) was performed as previously described (22), except that sample preparations for Fig. 5B were carried out in an anaerobic chamber. The sites of oxidation were identified by LC-MS/MS analyses using an Agilent nanoflow-1200 series HPLC system connected to a linear ion trap mass spectrometer (Thermo Scientific).
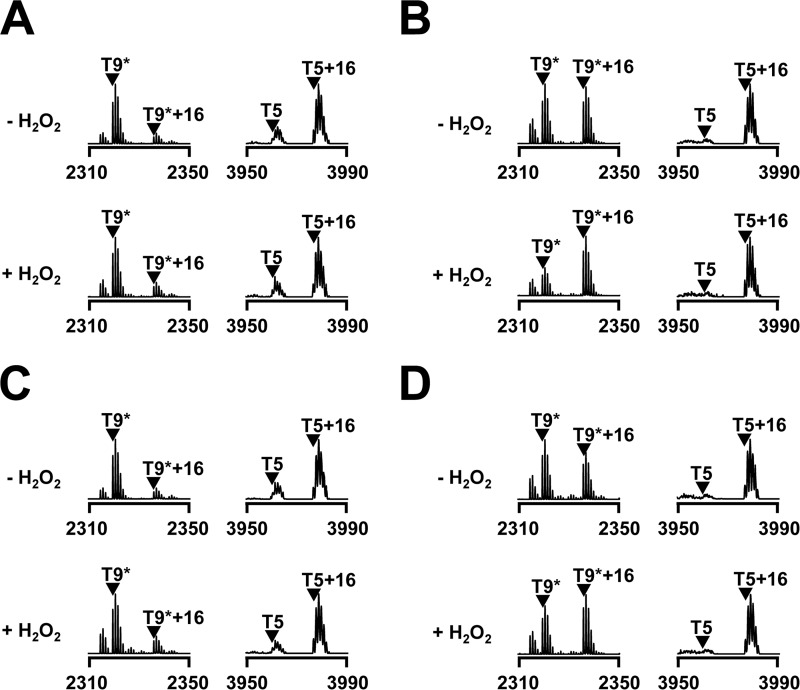
FIGURE 4.
In vivo oxidation of PerRSA. Oxidation of PerRSA from S. aureus cells (LS0166) grown aerobically in MLMM supplemented with no metal ion (A), 50 μm FeSO4 (B), 50 μm MnCl2 (C), or both 50 μm FeSO4 and 50 μm MnCl2 (D). PerRSA-FLAG proteins were recovered by immunoprecipitation from S. aureus cells treated with no H2O2 or 100 μm H2O2 for ∼2 min, and analyzed by MALDI-TOF MS after SDS-PAGE separation and in-gel tryptic digestion.
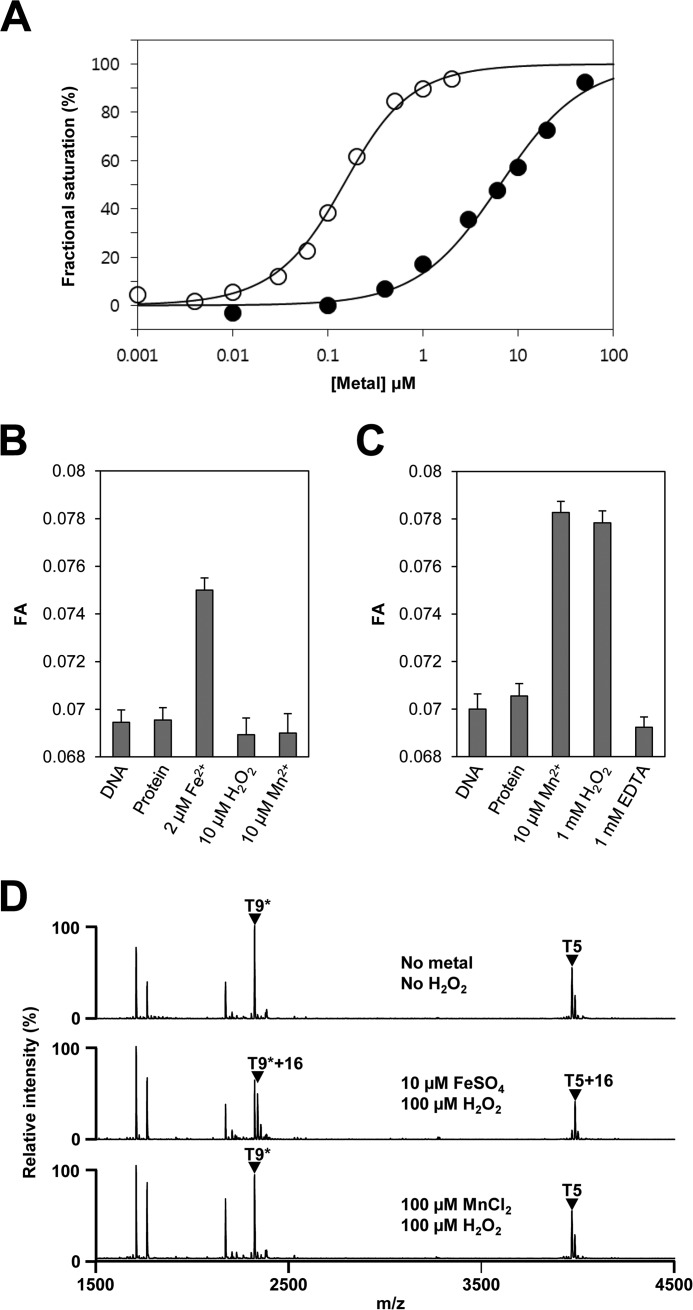
FIGURE 3.
Metal-dependent DNA binding and H2O2-mediated oxidation of PerRSAin vitro. A, metal-dependent DNA binding activity of PerRSA. Various concentrations of metal ions (open circle for Fe2+ and filled circle for Mn2+) are added to samples containing 100 nm DNA and 100 nm active PerRSA dimer, and metal-dependent DNA-binding of PerRSA was monitored by fluorescence anisotropy change. B, sensitivity of PerRSA to H2O2 in the presence of Fe2+. Protein (100 nm active PerRSA:Zn dimer), FeSO4, H2O2, and MnCl2 were sequentially added to buffer A containing 100 nm DNA with an interval of 2 min between each addition, and FA was measured after each addition. 10 μm H2O2 rapidly (<20 s) inactivated PerRSA in the presence of 2 μm Fe2+ and the loss of PerRSA activity was not recovered by the addition of Mn2+. C, sensitivity of PerRSA to H2O2 in the presence of Mn2+. Protein (100 nm active PerRSA:Zn dimer), MnCl2, H2O2, and EDTA were sequentially added to buffer A containing 100 nm DNA with an interval of 2 min between each addition except for 10 min between H2O2 and EDTA, and FA was measured after each addition. PerRSA was insensitive to 1 mm H2O2 for 10 min but lost DNA-binding activity rapidly by addition of 1 mm EDTA. D, metal-dependent oxidation of PerRSA. Oxidation of PerRSA by H2O2 in the presence of 10 μm Fe2+ or 100 μm Mn2+ was monitored by MALDI-TOF MS after tryptic digestion. Note that no cysteine oxidation was observed as judged by fully alkylated T9 peptide (T9*) containing Cys-102 and Cys-105.
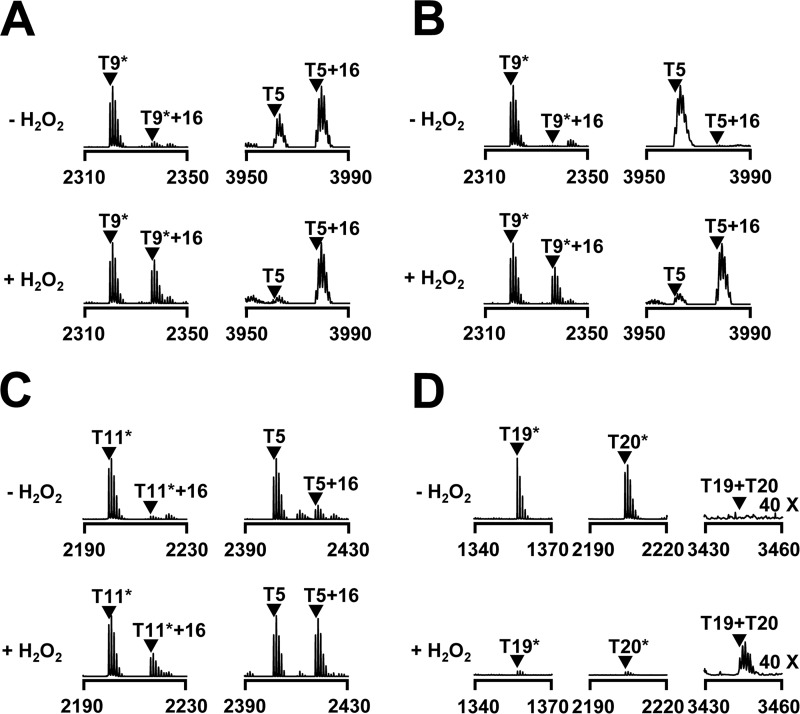
FIGURE 5.
Comparison of oxidation sensitivity among PerRBS, E. coli OxyR and PerRSA. Oxidation of PerRSA (A), PerRBS (C), or E. coli OxyR (D) in E. coli cells (LE0003, PerRSA; HE9501, PerRBS; LE2302, E. coli OxyR) grown under aerobic conditions, or PerRSA (B) in E. coli cells (LE0003) grown under oxygen-limited conditions. E. coli cells were grown in LB media under aerobic conditions or oxygen-limited conditions, and treated with or without 100 μm H2O2 for 1 min. Oxidation status of proteins was analyzed by MALDI-TOF MS after SDS-PAGE fractionation and in-gel tryptic digestion as reported previously (22). Asterisks represent peptides containing carboxyamidomethylated cysteine residue(s).
Caenorhabditis elegans Killing Assay
NGM agar plates (5.5 cm diameter) spread with 30 μl of A600 = 1 culture of E. coli OP50 (laboratory nematode food), S. aureus expressing no PerR (LS0093), S. aureus expressing PerRSA (LS0088), or S. aureus expressing PerRBS (LS0134) were used. For each assay 90 L4 stage C. elegans were used in triplicate of 30 worms/plate. The plates were incubated at 25 °C, and scored for live and dead worms at least every 24 h as described previously (23). For each assay, the survival of worms was calculated by the Kaplan-Meier method, and survival differences were tested by using OASIS (24).
Results
Structural Zn2+ and Regulatory Metal Binding Sites of PerRSA
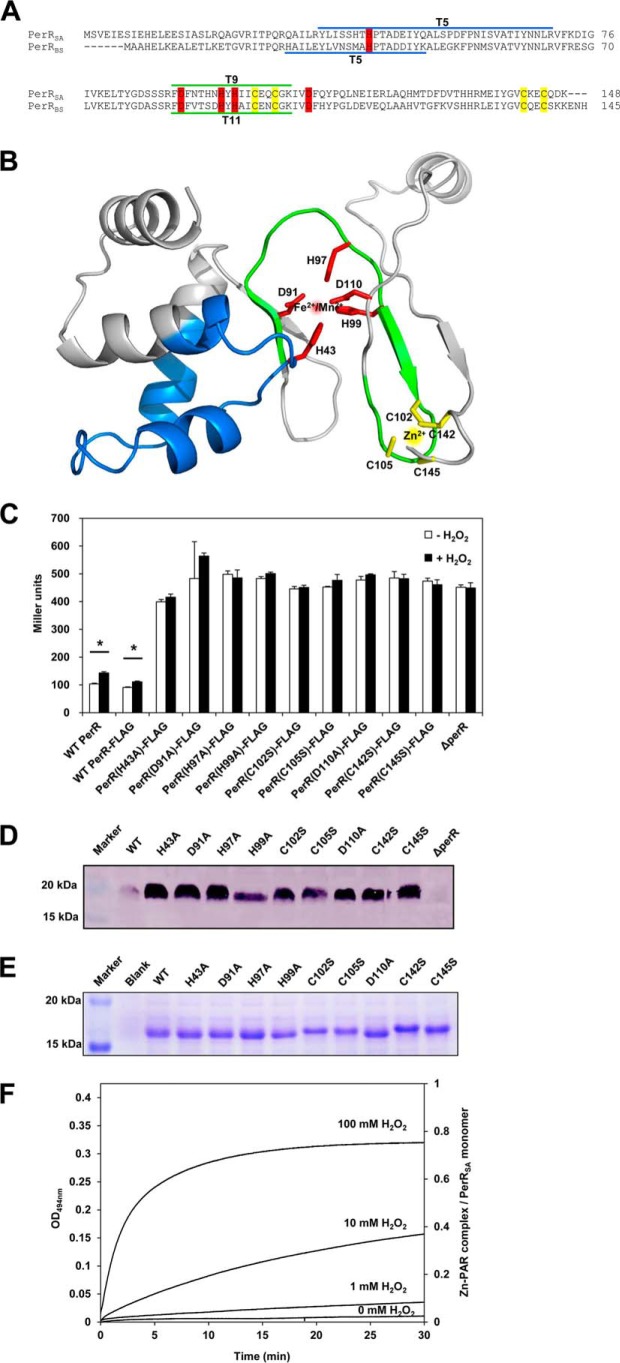
PerRSA is highly similar (67% sequence identity) to PerRBS, but previous results have highlighted some striking differences in their response to added metal ions (11–14). To provide a structural context for our investigation of PerRSA reactivity we generated a homology model of PerRSA based on the known structure of PerRBS (25). As shown in Fig. 1, A and B, PerRSA retains four highly conserved cysteine residues (Cys-102, Cys-105, Cys-142, and Cys-145) corresponding to those involved in high affinity structural Zn2+ binding (Site 1) in most Fur family proteins as well as in PerRBS (5, 15, 26). PerRSA also has five other residues (His-43, Asp-91, His-97, His-99, and Asp-110), which correspond to the N/O donor ligands for the regulatory metal binding (Site 2) in PerRBS (7, 8). To investigate the role of these predicted metal-binding residues in protein function, we generated PerRSA mutants and examined the in vivo repressor activities of WT and mutants using a PerR-regulated katA promoter-lacZ fusion (PkatA-lacZ) (Fig. 1C). As expected, the PkatA-lacZ reporter fusion was repressed in cells expressing WT PerRSA-FLAG but derepressed in the perR null mutant cells. Note that the repression levels of PkatA-lacZ reporter fusion by WT PerRSA-FLAG were similar to those observed with the WT S. aureus strain, indicating that the perR null mutant strain complemented with WT PerRSA-FLAG behaves like WT strain. All nine mutants exhibited no repressor activity for the PkatA-lacZ reporter fusion, and furthermore, these mutant proteins were present at levels greater than WT protein (Fig. 1D) indicative of a loss of repression of the autoregulated perR promoter. These results indicate that these amino acid residues proposed to be metal ligands are essential for in vivo repressor function.
FIGURE 1.
Metal binding sites of PerRSA. A, sequence alignment of PerRSA with PerRBS. The candidate ligands for Zn2+ (yellow) and Fe2+/Mn2+ (red) are conserved in PerRSA. The two tryptic peptides of PerRSA, T5 (blue) containing His-43 and T9 (green) containing His-97, are the sites of oxidation. The tryptic peptides of PerRBS, T5 containing His-37 and T11 containing His-91, are also shown. B, predicted structure of PerRSA monomer based on PerRBS structure (Protein Data Bank code 3F8N) (25) using Swiss Model (40). C, mutational analyses of PerRSA. Wild type S. aureus cells (WT PerR, LS0107), S. aureus cells expressing no PerRSA (ΔperR, LS0245), or S. aureus cells expressing PerRSA-FLAG variants as indicated, were grown in LB medium and treated with 100 μm H2O2 (+H2O2) for 30 min or not (−H2O2). Repressor activities of PerRSA variants were measured using PkatA-lacZ reporter fusion and were expressed as Miller units of β-galactosidase activity (values represent the mean ± S.D. from three separate experiments). Statistical analysis was performed with a Student's t test (*, p < 0.05). D, analysis of expression levels for PerRSA variants. S. aureus cells, expressing no PerRSA (ΔperR) or expressing PerRSA-FLAG variants as indicated, were grown in LB medium. The expression levels of FLAG-tagged PerR variants were analyzed by Western blot using anti-FLAG antibody. E, structural Zn2+-binding assay of PerRSA variants on SDS-PAGE gel. The crude extracts of E. coli cells, overexpressing no PerRSA (Blank) or overexpressing PerRSA variants as indicated, were preincubated with 10 mm DTT and separated by SDS-PAGE using Tris glycine buffer system. Protein bands were detected by Coomassie Brilliant Blue R staining. F, oxidation of Cys4:Zn site by H2O2. Release of Zn2+ from PerRSA:Zn (5 μm purified PerRSA) was measured by monitoring Zn2+-PAR complex at 494 nm after treatment of 0, 1, 10, and 100 mm H2O2 as described previously (15). The Zn2+ content was calculated using a molar extinction coefficient of 85,000 m−1 cm−1 at 494 nm for Zn2+-PAR complex (15).
Previously we have demonstrated that the structural Zn2+ binding status of PerRBS can be monitored by mobility difference on non-reducing SDS-PAGE: monomeric PerRBS containing bound Zn2+ migrates faster than PerRBS lacking bound Zn2+ (15). To investigate the Zn2+ binding status of PerRSA, WT and mutant proteins were separated on SDS-PAGE after overexpression in E. coli. WT and Site 2 mutants migrated with the mobility characteristic of the Zn2+-bound form, whereas all four Site 1 mutants migrated with the mobility characteristic of the Zn2+-lacking form (Fig. 1E). This result indicates that PerRSA contains a tightly bound Zn2+ coordinated by four cysteine residues, and that mutations at the proposed regulatory metal binding site do not affect the Zn2+ binding.
PerRSA was shown previously to be a Mn2+-specific repressor, which suggested that this protein might use an Fe2+-independent H2O2 sensing mechanism (11, 14). We therefore wondered whether the cysteine residues coordinating Zn2+ might serve a role in peroxide sensing. To test this, we measured the rate of Zn2+ release from purified PerRSA upon H2O2 treatment (Fig. 1F) by monitoring the formation of a Zn2+-PAR complex as reported previously (15). The second-order rate constant of Zn2+ release and the Zn2+ content of PerRSA were determined to be ∼0.05 m−1 s−1 and ∼0.8 Zn2+/monomer, respectively, which are comparable with those of PerRBS (15). The slow rate of H2O2-mediated Zn2+ release, along with the retention of Zn2+ despite the use of 10 mm EDTA during the purification procedures, further supports the idea that the Zn2+ site of PerRSA plays a structural rather than a peroxide sensing role. All these data together indicate that PerRSA has a structural Zn2+ site coordinated by four cysteine residues and a regulatory metal binding site composed of His-43, Asp-91, His-97, His-99, and Asp-110.
In Vivo Repressor Activity of PerRSA in Comparison with PerRBS
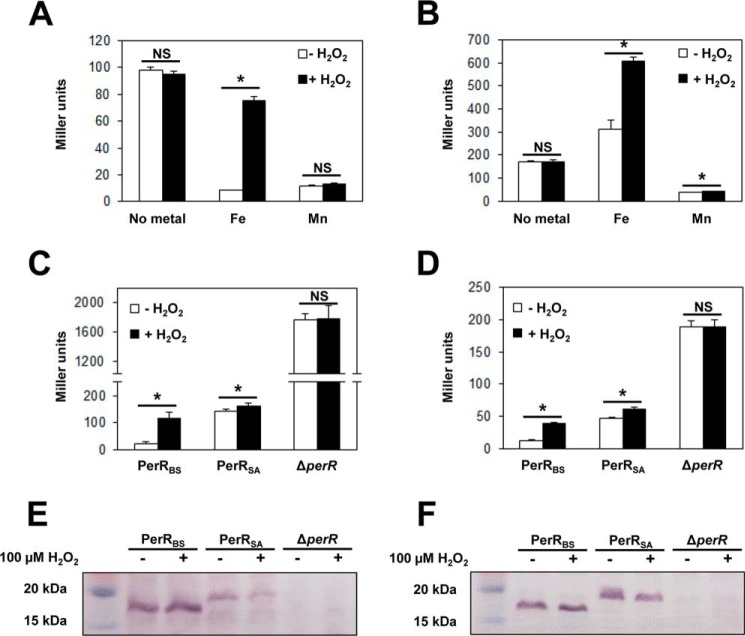
To investigate the difference in metal- and H2O2-sensing ability of PerRBS and PerRSA, the repressor activities of PerR proteins were examined in MLMM using an mrgA promoter lacZ-fusion (PmrgA-lacZ). As reported previously, PerRBS repressed the PmrgA-lacZ reporter fusion in the presence of either Fe2+ or Mn2+, and Fe2+-dependent repression was relieved upon H2O2 treatment (Fig. 2A) (7). PerRSA repressed the PmrgA-lacZ reporter fusion in the presence of Mn2+ but not in the presence of Fe2+, consistent with the previous finding that PerRSA is a Mn2+-dependent repressor (Fig. 2B) (11). Interestingly, however, β-galactosidase expression was increased by about 2-fold in the presence of Fe2+ as reported previously (12) and further increased upon H2O2 treatment. The previous observation that this Fe2+-dependent induction of the mrgA gene is not observed with the perR null mutant S. aureus (12) suggests that the Fe2+- and H2O2-dependent increase of β-galactosidase expression is mediated by PerRSA, although it is not clear whether Fe2+ directly interacts with PerRSA or not.
FIGURE 2.
Comparison of activities between PerRSA and PerRBSin vivo. A and B, metal-dependent repressor activities of PerRBS (A) and PerRSA (B). B. subtilis cells expressing PerRBS-FLAG (HB9738) and S. aureus cells expressing PerRSA-FLAG (LS0111) were grown aerobically in MLMM supplemented with 10 μm FeSO4 or 10 μm MnCl2, and treated with 100 μm H2O2 (+H2O2) for 30 min or not (−H2O2). Repressor activities of PerRBS and PerRSA were measured using B. subtilis PmrgA-lacZ and S. aureus PmrgA-lacZ reporter fusions, respectively. Statistical analysis was performed with a Student's t test (*, p < 0.05; NS, not significant). C and D, repressor activities of PerRBS and PerRSA in B. subtilis (C) and in S. aureus (D). B. subtilis cells expressing no PerR (LB1532), PerRBS-FLAG (HB9738), or PerRSA-FLAG (LB1530), and S. aureus cells expressing no PerR (LS0124), PerRBS-FLAG (LS0114), or PerRSA-FLAG (LS0111) were used. Cells were grown aerobically in LB medium and treated with 100 μm H2O2 (+H2O2) for 30 min or not (−H2O2). Repressor activities of PerR proteins in B. subtilis were measured using B. subtilis PmrgA-lacZ and those in S. aureus were measured using S. aureus PmrgA-lacZ reporter fusions. A–D, repressor activities were expressed as Miller units of β-galactosidase activity (values represent the mean ± S.D. from three separate experiments). Statistical analysis was performed with a Student's t test (*, p < 0.05; NS, not significant). E and F, Western blot analysis of PerRBS and PerRSA in B. subtilis (E) and in S. aureus (F). B. subtilis cells expressing no PerR (LB1532), PerRBS-FLAG (HB9738), or PerRSA-FLAG (LB1530), and S. aureus cells expressing no PerR (LS0124), PerRBS-FLAG (LS0114), or PerRSA-FLAG (LS0111) were used. Cells were grown aerobically in LB medium and treated with 100 μm H2O2 (+H2O2) for 30 min or not (−H2O2). The FLAG-tagged PerR proteins were probed by anti-FLAG antibody.
To test whether differences in cellular milieu between B. subtilis and S. aureus affect the repressor activity of PerR proteins, PerRBS and PerRSA were expressed both in B. subtilis (Fig. 2, C and E) and S. aureus (Fig. 2, D and F). PerRBS expressed in S. aureus repressed the S. aureus PmrgA-lacZ reporter fusion with even higher repressor activity than PerRSA, and responded normally to H2O2 as in B. subtilis (Fig. 2D). PerRSA expressed in B. subtilis or in S. aureus repressed the B. subtilis PmrgA-lacZ fusion or S. aureus PmrgA-lacZ fusion, respectively, although not quite as efficiently as PerRBS (Fig. 2, C and D). Interestingly, the repression levels of both the B. subtilis and S. aureus PmrgA-lacZ reporter fusions by PerRSA were similar to those by PerRBS treated with H2O2 for 30 min. Furthermore, PerRSA responded poorly to H2O2 (∼1.2-fold induction) both in B. subtilis and S. aureus when compared with the responsiveness of PerRBS to H2O2 (more than 3-fold induction). Thus it is likely that the difference in responsiveness to metal and H2O2 between PerRSA and PerRBS is due to differences between the PerR proteins rather than the cellular environments. In summary the in vivo data indicate that Fe2+ addition appears to lead to apparent activation or derepression, rather than repression, of PerRSA-regulated genes under our experimental conditions.
In Vitro PerRSA Senses H2O2 by Iron-mediated Histidine Oxidation
To test whether PerRSA is activated to bind DNA by both Mn2+ and Fe2+, we measured the apparent affinity of PerRSA for Fe2+ and Mn2+ using a fluorescence anisotropy-based DNA-binding assay (Fig. 3A). Because PerR is immediately oxidized in the presence of Fe2+ under aerobic conditions as reported previously (7), all the FA experiments were performed under anaerobic conditions. Consistent with the observed Mn2+-dependent repressor activity of PerRSA in vivo, the DNA-binding affinity of PerRSA was increased by the addition of Mn2+. The apparent Kd for the Mn2+-dependent activation of PerRSA was determined to be 9 μm, which is slightly weaker than that of PerRBS (∼3 μm). Interestingly, despite the apparent lack of Fe2+-dependent repressor activity of PerRSA in vivo, Fe2+ could also increase the DNA binding of PerRSA in a concentration-dependent manner. The apparent Kd for the Fe2+-dependent activation of PerRSA (0.1 μm) appeared to be the same as that of PerRBS. These results therefore suggest that the apparent lack of Fe2+-dependent repressor activity and poor responsiveness to H2O2 of PerRSA in vivo is not due to a decreased Fe2+ binding affinity of PerRSA per se.
Because both Fe2+ and Mn2+ increase the DNA binding affinity of PerRSA, we next investigated the effect of H2O2 on the DNA-binding activity of different metal-bound forms of PerRSA (PerRSA:Zn,Fe and PerRSA:Zn,Mn) under anaerobic conditions (Fig. 3, B and C). Upon addition of 10 μm H2O2, PerRSA:Zn,Fe completely lost DNA-binding activity in 20 s, whereas PerRSA:Zn,Mn retained DNA-binding activity for 10 min even in the presence of 1 mm H2O2, indicating that PerRSA utilizes Fe2+, but not Mn2+, for the sensing of low levels of H2O2 in vitro. Furthermore, loss of the DNA-binding ability of PerRSA:Zn,Fe by H2O2 treatment could not be restored by the addition of Mn2+, indicating that H2O2 sensing by PerRSA:Zn,Fe likely accompanies protein modification rather than simply the loss of bound Fe2+.
To test the hypothesis that H2O2 leads to a metal-dependent modification of PerRSA, we analyzed the effect of H2O2 on different metallated forms of PerRSA using MALDI-TOF MS (Fig. 3D). PerRSA:Zn,Mn displayed no detectable changes in tryptic peptide peaks after 100 μm H2O2 treatment. However, H2O2-treated PerRSA:Zn,Fe exhibited a significant decrease in the intensity of the T5 peptide (Tyr-36 to Arg-70) and T9* peptide (Phe-90 to Lys-107) with a concomitant increase in the intensity of two tryptic peptides corresponding to T5 + 16 and T9* + 16. The sites of oxidation responsible for this 16-Da mass increase were mapped to His-43 (corresponding to His-37 in PerRBS) in the T5 peptide and His-97 (corresponding to His-91 in PerRBS) in the T9* peptide using LC-ESI MS analysis (Fig. 1A and data not shown). Note that the T9* peptide also contains Cys-102 and Cys-105, which were detected in their fully alkylated form, indicative of no oxidation at cysteine residues after 100 μm H2O2 treatment, consistent with the structural role for these zinc-coordinating cysteine residues. These data indicate that PerRSA, like PerRBS, senses low levels of H2O2 by Fe2+-mediated oxidation of either of two histidine residues, His-43 and/or His-97, which are used as regulatory metal binding ligands.
Apparent Lack of Fe2+-dependent Repressor Activity of PerRSA in Vivo Is Due to the Hypersensitivity of PerRSA to Iron-mediated Oxidation under Aerobic Conditions
Since we observed the Fe2+-dependent DNA-binding activity and Fe2+-mediated histidine oxidation of PerRSA in vitro, we wondered whether H2O2 sensing by histidine oxidation would also occur in vivo. To monitor the oxidation of PerRSA in vivo, we analyzed the oxidation status of PerRSA recovered by immunoprecipitation from S. aureus cells grown in MLMM supplemented with Fe2+ or Mn2+ using MALDI-TOF MS (Fig. 4). Interestingly, even without H2O2 treatment, almost all of the T5 peptide was detected as oxidized form (T5 + 16) and a significant amount of the T9* peptide was also detected as oxidized form (T9* + 16) for PerRSA from cells grown in MLMM supplemented with Fe2+ or both Fe2+ and Mn2+. However, less oxidation was observed at both T5 and T9* peptides for PerRSA from cells grown in MLMM supplemented with Mn2+ or no metal ion, and no significant further oxidation of these peptides was detected upon H2O2 treatment. These observations indicate that under aerobic growth conditions the majority of PerRSA:Zn,Fe exists in an oxidized form even without external addition of H2O2.
To compare the sensitivity of PerRSA with those of well known H2O2 sensors, E. coli OxyR and PerRBS, we analyzed protein oxidation in E. coli using MALDI-TOF MS as described previously (22). As noted for PerRSA recovered from S. aureus cells (Fig. 4), PerRSA from aerobically grown E. coli cells exhibited a significant oxidation at the T5 peptide even in the absence of H2O2 treatment (Fig. 5A). However, PerRSA from E. coli cells grown under oxygen-limited conditions exhibited no detectable oxidation at both T5 and T9* peptides, and oxidation of these peptides was observed upon external H2O2 treatment (Fig. 5B). In contrast, under aerobic conditions, PerRBS exhibited less oxidation at both T5 and T11* peptides when compared with PerRSA (Fig. 5C), and E. coli OxyR exhibited no detectable oxidation of both T19 and T20 peptides, which contain the peroxidatic cysteine (Cys-199) and resolving cysteine (Cys-208), respectively (Fig. 5D). These results indicate that PerRSA is more sensitive than PerRBS or E. coli OxyR to oxidation by low levels of H2O2, which are normally encountered during the aerobic growth of E. coli.
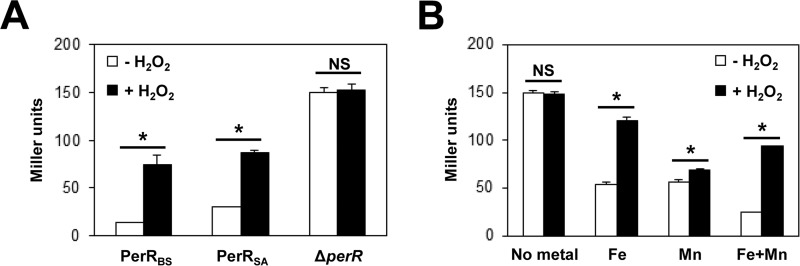
The above observations suggest that the poor H2O2 responsiveness and the apparent lack of Fe2+-dependent repressor activity of PerRSA can be overcome under oxygen-limited growth conditions where limited amounts of H2O2 are generated. Consistent with this hypothesis, PerRSA exhibited an increased repressor activity under oxygen-limited growth conditions (Fig. 6A) when compared with that under aerobic growth conditions (Fig. 2D). Furthermore, under these conditions PerRSA responded to H2O2 (∼3-fold induction) much like PerRBS, as judged by an increased β-galactosidase activity upon H2O2 treatment. We also investigated the metal-dependent H2O2-sensing ability of PerRSA by growing cells in MLMM under oxygen-limited growth conditions. As shown in Fig. 6B, addition of either Fe2+ or Mn2+ enabled PerRSA to repress the PmrgA-lacZ reporter fusion, and β-galactosidase activity was increased by more than 2-fold upon H2O2 treatment in the presence of Fe2+ or in the presence of both Fe2+ and Mn2+, but only slightly (∼1.2-fold) in the presence of only Mn2+. These data demonstrate that PerRSA functions as a Fe2+-dependent repressor and senses H2O2 in an Fe2+-dependent manner in vivo under oxygen-limited growth conditions (Fig. 6), as observed in vitro under anaerobic conditions (Fig. 3). We also note that the efficient sensing of H2O2 in the presence of both Fe2+ and Mn2+ is consistent with the higher affinity of PerRSA for Fe2+ than Mn2+ as observed in vitro (Fig. 3A). In general, these results indicate that PerRSA behaves in oxygen-limited cells much like PerRBS does in B. subtilis, and that the apparent lack of Fe2+-dependent repressor activity (and thus poor H2O2 responsiveness) of PerRSA under aerobic conditions is due to an efficient oxidation of PerRSA by low levels of endogenous H2O2.
FIGURE 6.
In vivo repressor activities of PerRSA under oxygen-limited conditions. A, repressor activities and H2O2 sensing abilities of PerRBS and PerRSA under oxygen-limited conditions. S. aureus cells expressing no PerR (LS0124), PerRBS-FLAG (LS0114), or PerRSA-FLAG (LS0111) were grown in LB medium under oxygen-limited conditions, and treated with 100 μm H2O2 (+H2O2) for 30 min or not (−H2O2). Repressor activities of PerR proteins were measured using S. aureus PmrgA-lacZ reporter fusions. B, metal-dependent repressor activities of PerRSA under oxygen-limited conditions. S. aureus cells expressing PerRSA-FLAG (LS0111) were grown in MLMM supplemented with no metal ion, 10 μm FeSO4, 10 μm MnCl2, or both 10 μm FeSO4 and 10 μm MnCl2 under oxygen-limited conditions, and treated with 100 μm H2O2 (+H2O2) for 30 min or not (−H2O2). Repressor activities were measured using S. aureus PmrgA-lacZ reporter fusion. A and B, repressor activities were expressed as Miller units of β-galactosidase activity (values represent the mean ± S.D. from three separate experiments). Statistical analysis was performed with a Student's t test (*, p < 0.05; NS, not significant).
Fe2+-mediated H2O2 Susceptibility of PerRSA Is Important for H2O2 Resistance and Pathogenicity of S. aureus
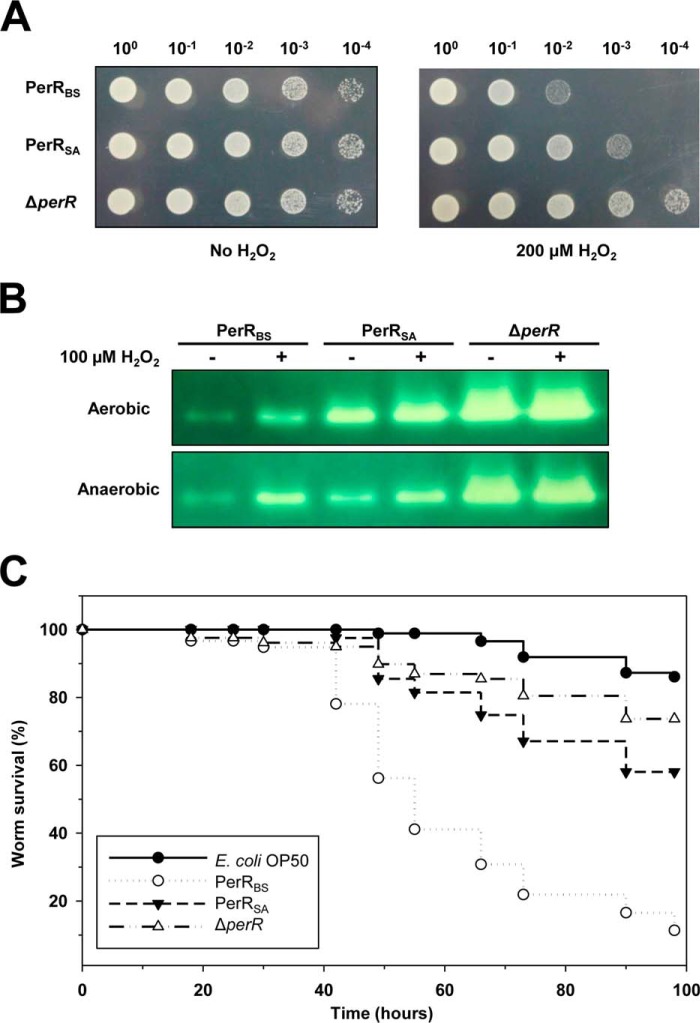
It has been suggested that resistance to oxidative stress is an important factor for the survival and persistence of S. aureus (14). To investigate whether the difference in sensitivity of PerR proteins to oxidation affects the H2O2 resistance of S. aureus, we measured the growth of cells in the presence and absence of H2O2 (Fig. 7A). As expected the perR null mutant S. aureus cells exhibited an increased H2O2 resistance compared with those expressing PerRSA. Also, compared with cells expressing PerRSA, S. aureus cells expressing PerRBS (which is less sensitive to Fe-mediated oxidation than PerRSA) exhibited an increased H2O2 sensitivity. These data indicate that the ability of PerRSA to respond to very low levels of H2O2 encountered during aerobic growth is important for the H2O2 resistance of S. aureus. Consistent with this, high levels of KatA activity were detected in the perR null mutant S. aureus cells, but low levels of KatA activity were detected from cells expressing PerRBS, when compared with those expressing PerRSA (Fig. 7B).
FIGURE 7.
Effects of PerR activity on H2O2 resistance and virulence of S. aureus. A, effects of PerR activity on the survival of S. aureus in the absence or presence of H2O2. S. aureus cells (3 μl of A600 = 1 culture, with indicated dilutions) expressing no PerR (LS0093), PerRBS-FLAG (LS0134), or PerRSA-FLAG (LS0088) were spotted on LB-agar plate containing no H2O2 or 200 μm H2O2, and grown under aerobic conditions at 37 °C. Note that fresh LB agar plate shielded from light was used to prevent the generation of H2O2 by photochemical reactions (41). B, effects of PerR activity on the expression of KatA. S. aureus cells expressing no PerR (LS0093), PerRBS-FLAG (LS0134), or PerRSA-FLAG (LS0088) were grown in LB medium under either aerobic or oxygen-limited conditions, and treated with 100 μm H2O2 or not. Catalase activity staining was performed after separation of cell extract (10 μg protein) on native PAGE gel. C, effects of PerR activity on the pathogenicity of S. aureus using C. elegans as a model host. C. elegans killing was presented as Kaplan-Meier survival plots of worms fed with E. coli OP50 (negative control) (n = 86), S. aureus strain expressing no PerR (LS0093) (n = 86), PerRBS-FLAG (LS0134) (n = 77), or PerRSA-FLAG (LS0088) (n = 84).
Although the H2O2 defense enzymes, such as KatA and AhpC, which are under the control of PerRSA play important roles in the nasal colonization and infection by S. aureus, it has been reported that they are not important for virulence (11, 14). However, interestingly, PerR is known to be required for virulence in other models of infection including murine skin abscess (11, 14), fruit fly (27), and zebrafish (28). We used a C. elegans model, which has been widely used as an invertebrate animal model for S. aureus pathogenesis (23), to investigate whether the difference in sensitivity of PerR proteins affects the virulence of S. aureus (Fig. 7C). As noted for other models of infection, the perR null mutant S. aureus was attenuated in the C. elegans model. Unexpectedly, S. aureus cells expressing PerRBS killed C. elegans more rapidly than did those expressing PerRSA. These results suggest that the virulence of S. aureus is somewhat directly correlated with the activity of PerR, given that heterologous PerRBS, which is less sensitive to oxidation than PerRSA, increases the virulence of S. aureus.
Discussion
PerR and PerR-like regulators have been described in a wide variety of organisms since the first characterization in B. subtilis (4, 5, 17, 29, 30). However, to date, the H2O2-sensing mechanism of PerR proteins has only been extensively studied for PerRBS (7, 15, 21). Here we demonstrate that PerRSA, previously regarded as a Mn2+-specific repressor, senses H2O2 using the same Fe2+-dependent histidine oxidation mechanism previously described for PerRBS. Moreover we show that the apparent lack of Fe2+-dependent repressor activity, and the consequent Mn2+-specific repressor activity of PerRSA in vivo, is due to the hypersensitivity of PerRSA to H2O2 under aerobic conditions, rather than due to a decreased Fe2+ binding affinity of PerRSA per se.
Several lines of evidence indicate that PerRSA is a more sensitive H2O2 sensor than either PerRBS or E. coli OxyR. The majority of PerRSA in aerobically grown S. aureus is detected in an oxidized form (Fig. 4), whereas only partial oxidation is observed with PerRBS from aerobically grown B. subtilis (7). However, the interpretation of this result can be complicated by potential differences in the levels of endogenous H2O2 between these two species. Therefore, we directly compared the levels of oxidized PerR proteins when both were expressed in either S. aureus or B. subtilis. Indeed the direct measurement of KatA activity (Fig. 7B) and reporter fusion assays (Fig. 2, C and D) indicate that the expression levels of PerRSA-regulated genes are higher in cells expressing PerRSA than in those expressing PerRBS. This indicates that under otherwise identical conditions, oxidation levels of PerRSA are higher than those of PerRBS in both B. subtilis and S. aureus, consistent with the hypothesis that PerRSA is intrinsically more susceptible to oxidation than PerRBS. Furthermore, we also used E. coli, where H2O2 detoxification systems are under the control of OxyR, as neutral host to directly compare the sensitivity of PerR proteins and OxyR protein. The rate of H2O2 generation in aerobically growing E. coli is ∼10 μm per s (31), however, the steady-state concentration of H2O2 is kept ∼50 nm by the action of scavenging enzymes such as AhpC (1). Normally, under these routine aerobic growth conditions, OxyR is inactive: OxyR is activated when the intracellular H2O2 concentration reaches ∼200 nm (1, 32, 33). PerRBS has a second-order rate constant of ∼105 m−1 s−1 for inactivation by H2O2, which is comparable with that of E. coli OxyR (7). Consistent with this, PerRBS and E. coli OxyR exhibit no significant oxidation in aerobically grown E. coli (Fig. 5, C and D). In contrast, a significant oxidation of PerRSA is observed in aerobically grown E. coli without external H2O2 treatment, indicating that endogenously produced H2O2 is sufficient to oxidize PerRSA (Fig. 5A). Collectively these indicate that PerRSA senses very low levels of H2O2 (as little as ∼50 nm) as generated during normal aerobiosis in E. coli, levels that do not significantly oxidize PerRBS or E. coli OxyR. Corroborating with this, it has recently been reported that OxyR2 from Vibrio vulnificus is activated under normal aerobic growth conditions, whereas OxyR1, an E. coli OxyR homologue, is only activated by exogenous H2O2 (34).
The efficient sensing of H2O2 and induction of defense enzymes have been considered crucial for pathogens that have to fight against H2O2 assault by macrophages or neutrophils (35, 36). However, the perR null mutant S. aureus strain, which is more resistant to H2O2 than the wild type by constitutive expression of H2O2 defense enzymes, exhibits attenuated virulence in our C. elegans model of infection (Fig. 7C) as observed with other models of infection (11, 27, 28). Moreover, the H2O2-sensitive S. aureus strain by the expression of PerRBS (Fig. 7, A and B) is not attenuated in C. elegans (Fig. 7C), consistent with the previous finding that neither KatA nor AhpC are required for resistance to neutrophil-dependent killing or virulence of S. aureus (14). Instead, the expression of PerRBS, which is less sensitive to H2O2 compared with PerRSA, increases the virulence suggesting that the activity of PerR positively correlates with the virulence of S. aureus. This may imply that the inactivation of PerR by H2O2, rather than the direct poisoning of bacteria by H2O2, can be exploited by phagocytic cells that wish to reduce the virulence of S. aureus. It is not clear why inactivation of PerR activity reduces the virulence of S. aureus. One possible explanation would be poor growth of S. aureus in the iron-limited host environment. Derepression of PerR-regulon is likely to lead to Fe2+ deficiency due to the elevated expression of KatA, which consumes Fe2+, and Fur, which represses Fe2+ uptake, as observed with B. subtilis (37). Alternatively, or in addition, active PerR may be involved in the induction of the virulence factor, either directly or indirectly. Indeed it has been shown that Streptococcus pyogenes PerR regulates an extracellular virulence factor, MF3, directly (38). All together, our findings that the activity of PerR is directly linked to the virulence of S. aureus suggests that PerRSA can be an attractive target for a novel approach to design new drugs for S. aureus treatment (39).
In contrast to virulence, H2O2 resistance is inversely correlated with the activity of PerR because the H2O2 defense systems are derepressed by H2O2-mediated PerR inactivation. Our results clearly show this relationship (Fig. 7, A and B). As reported previously the perR null mutant S. aureus exhibits increased resistance to H2O2, whereas S. aureus expressing PerRBS exhibits an increased sensitivity to H2O2 than that expressing PerRSA, presumably due to the hyper-repression of PerRSA-regulated genes by PerRBS. Although KatA and AhpC are not required for virulence, they are known to play important roles for survival under aerobic conditions and especially for colonization at the anterior nares, which are the primary ecological niche for S. aureus (14). Considering that most of the PerRSA is fully oxidized and no significant further derepression of PerR regulon is triggered by H2O2 treatment under aerobic conditions, it is likely that S. aureus, as a facultative anaerobic bacterium, has evolved PerRSA to sense low levels of endogenous H2O2 normally encountered under aerobic environment, rather than to sense higher levels of external H2O2 produced by microbial competitor or by host immune system.
Author Contributions
C. J., J. K., J. H., and J. L. conceived and designed the experiments. C. J., J. K., Y. W., Y. L., T. C., and S. J. performed the experiments. C. J., J. K., H. Y., J. H., and J. L. analyzed the data and wrote the paper.
Acknowledgments
We thank Dr. Joohong Ahnn for help with the C. elegans killing assay and Dr. Cheolju Lee for help with mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grant GM059323 (to J. D. H.) and National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIP) Grants NRF-2011-0017199 and NRF-2009-0068133 (to J.-W. L.). The authors declare that they have no conflict of interest.
- PerRBS
- B. subtilis PerR
- PerRSA
- S. aureus PerR
- MLMM
- metal-limited minimal medium
- PAR
- 4-(2-pyridylazo)resorcinol
- FA
- fluorescence anisotropy
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1. Imlay J. A. (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Imlay J. A. (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toledano M. B., Delaunay A., Monceau L., Tacnet F. (2004) Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem. Sci. 29, 351–357 [DOI] [PubMed] [Google Scholar]
- 4. Dubbs J. M., Mongkolsuk S. (2012) Peroxide-sensing transcriptional regulators in bacteria. J. Bacteriol. 194, 5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee J. W., Helmann J. D. (2007) Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499 [DOI] [PubMed] [Google Scholar]
- 6. Zuber P. (2009) Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 63, 575–597 [DOI] [PubMed] [Google Scholar]
- 7. Lee J. W., Helmann J. D. (2006) The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367 [DOI] [PubMed] [Google Scholar]
- 8. Traoré D. A., El Ghazouani A., Jacquamet L., Borel F., Ferrer J. L., Lascoux D., Ravanat J. L., Jaquinod M., Blondin G., Caux-Thang C., Duarte V., Latour J. M. (2009) Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 5, 53–59 [DOI] [PubMed] [Google Scholar]
- 9. Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 10. Horsburgh M. J., Ingham E., Foster S. J. (2001) In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183, 468–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horsburgh M. J., Clements M. O., Crossley H., Ingham E., Foster S. J. (2001) PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69, 3744–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morrissey J. A., Cockayne A., Brummell K., Williams P. (2004) The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect. Immun. 72, 972–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horsburgh M. J., Wharton S. J., Cox A. G., Ingham E., Peacock S., Foster S. J. (2002) MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44, 1269–1286 [DOI] [PubMed] [Google Scholar]
- 14. Cosgrove K., Coutts G., Jonsson I. M., Tarkowski A., Kokai-Kun J. F., Mond J. J., Foster S. J. (2007) Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J. W., Helmann J. D. (2006) Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J. Biol. Chem. 281, 23567–23578 [DOI] [PubMed] [Google Scholar]
- 16. Arnaud M., Chastanet A., Débarbouillé M. (2004) New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L., Keramati L., Helmann J. D. (1995) Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. U.S.A. 92, 8190–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luong T. T., Lee C. Y. (2007) Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods 70, 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Novick R. P. (1991) Genetic systems in staphylococci. Methods Enzymol. 204, 587–636 [DOI] [PubMed] [Google Scholar]
- 20. Charpentier E., Anton A. I., Barry P., Alfonso B., Fang Y., Novick R. P. (2004) Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 70, 6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Z., Lee J. W., Helmann J. D. (2011) Identification of altered function alleles that affect Bacillus subtilis PerR metal ion selectivity. Nucleic Acids Res. 39, 5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Won Y. B., Ji C. J., Cho J. H., Lee J. W. (2010) Mutational analysis of the metal-binding sites of peroxide sensor PerR. B Korean Chem. Soc. 31, 1573–1576 [Google Scholar]
- 23. Sifri C. D., Begun J., Ausubel F. M., Calderwood S. B. (2003) Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71, 2208–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang J. S., Nam H. J., Seo M., Han S. K., Choi Y., Nam H. G., Lee S. J., Kim S. (2011) OASIS: online application for the survival analysis of lifespan assays performed in aging research. PloS One 6, e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacquamet L., Traoré D. A., Ferrer J. L., Proux O., Testemale D., Hazemann J. L., Nazarenko E., El Ghazouani A., Caux-Thang C., Duarte V., Latour J. M. (2009) Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73, 20–31 [DOI] [PubMed] [Google Scholar]
- 26. Traoré D. A., El Ghazouani A., Ilango S., Dupuy J., Jacquamet L., Ferrer J. L., Caux-Thang C., Duarte V., Latour J. M. (2006) Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 61, 1211–1219 [DOI] [PubMed] [Google Scholar]
- 27. Needham A. J., Kibart M., Crossley H., Ingham P. W., Foster S. J. (2004) Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology 150, 2347–2355 [DOI] [PubMed] [Google Scholar]
- 28. Prajsnar T. K., Cunliffe V. T., Foster S. J., Renshaw S. A. (2008) A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell. Microbiol. 10, 2312–2325 [DOI] [PubMed] [Google Scholar]
- 29. Fillat M. F. (2014) The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546, 41–52 [DOI] [PubMed] [Google Scholar]
- 30. Bsat N., Herbig A., Casillas-Martinez L., Setlow P., Helmann J. D. (1998) Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29, 189–198 [DOI] [PubMed] [Google Scholar]
- 31. Seaver L. C., Imlay J. A. (2004) Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 279, 48742–48750 [DOI] [PubMed] [Google Scholar]
- 32. Stone J. R. (2004) An assessment of proposed mechanisms for sensing hydrogen peroxide in mammalian systems. Arch. Biochem. Biophys. 422, 119–124 [DOI] [PubMed] [Google Scholar]
- 33. Aslund F., Zheng M., Beckwith J., Storz G. (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A. 96, 6161–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim S., Bang Y. J., Kim D., Lim J. G., Oh M. H., Choi S. H. (2014) Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Mol. Microbiol. 93, 992–1009 [DOI] [PubMed] [Google Scholar]
- 35. Spaan A. N., Surewaard B. G., Nijland R., van Strijp J. A. (2013) Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu. Rev. Microbiol. 67, 629–650 [DOI] [PubMed] [Google Scholar]
- 36. van Kessel K. P., Bestebroer J., van Strijp J. A. (2014) Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front. Immunol. 5, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Faulkner M. J., Ma Z., Fuangthong M., Helmann J. D. (2012) Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J. Bacteriol. 194, 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wen Y. T., Tsou C. C., Kuo H. T., Wang J. S., Wu J. J., Liao P. C. (2011) Differential secretomics of Streptococcus pyogenes reveals a novel peroxide regulator (PerR)-regulated extracellular virulence factor mitogen factor 3 (MF3). Mol. Cell. Proteomics 10, M110.007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duarte V., Latour J. M. (2013) PerR: a bacterial resistance regulator and can we target it? Future Med. Chem. 5, 1177–1179 [DOI] [PubMed] [Google Scholar]
- 40. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Varghese S., Wu A., Park S., Imlay K. R., Imlay J. A. (2007) Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol. Microbiol. 64, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]