Background: Diversity in prion protein oligomerization pathways leads to conformationally distinct assemblies.
Results: Upon pressure perturbation, prion protein oligomers dissociate into monomers, providing new structural and energetic insights into the misfolding process.
Conclusion: Distinct β-sheet-rich oligomers of the prion protein display different void volumes.
Significance: Conformation-enciphered prion strains could be explained in terms of defects in protein packing.
Keywords: oligomer, prion, protein misfolding, protein self-assembly, protein stability, pressure
Abstract
Protein oligomerization has been associated with a wide range of diseases. High pressure approaches offer a powerful tool for deciphering the underlying molecular mechanisms by revealing volume changes associated with the misfolding and assembly reactions. We applied high pressure to induce conformational changes in three distinct β-sheet-rich oligomers of the prion protein PrP, a protein characterized by a variety of infectious quaternary structures that can propagate stably and faithfully and cause diseases with specific phenotypic traits. We show that pressure induces dissociation of the oligomers and leads to a lower volume monomeric PrP state that refolds into the native conformation after pressure release. By measuring the different pressure and temperature sensitivity of the tested PrP oligomers, we demonstrate significantly different void volumes in their quaternary structure. In addition, by focusing on the kinetic and energetic behavior of the pressure-induced dissociation of one specific PrP oligomer, we reveal a large negative activation volume and an increase in both apparent activation enthalpy and entropy. This suggests a transition state ensemble that is less structured and significantly more hydrated than the oligomeric state. Finally, we found that site-specific fluorescent labeling allows monitoring of the transient population of a kinetic intermediate in the dissociation reaction. Our results indicate that defects in atomic packing may deserve consideration as a new factor that influences differences between PrP assemblies and that could be relevant also for explaining the origin of prion strains.
Introduction
The structural conversion of prion proteins (PrPs)2 can be represented in a very simplified way as a process in which a normally well folded constitutive protein (PrPC) is transformed into a misfolded conformer (PrPSc) (1, 2). This misfolding process modifies PrP physico-chemical properties and promotes its self-association into β-sheet-rich oligomers that eventually evolve into large complex deposits called amyloid fibrils (3). It is now clear that these macromolecular complexes play a pivotal role in the pathogenesis of transmissible spongiform encephalopathies, also known as prion diseases (4–6).
However, the mechanisms underlying PrP assembly and the physico-chemical properties that correlate with the toxicity and infectivity of prion diseases remain unclear. This lack of knowledge makes it difficult to identify suitable therapeutic targets and to develop new drugs against these disorders. Because the metastable nature of PrPC (7) and the heterogeneity of the resulting oligomers are intrinsic features of its complex folding energy landscape, a more detailed energetic description of the structural conversion(s) is required.
Elevated temperatures are often used to facilitate in vitro PrP structural conversion. We previously showed that heat treatment of PrP at acidic pH induces various independent assembly pathways, leading to a variety of soluble oligomeric misfolded states with different sizes and conformation-specific antibody reactivity (8, 9). These oligomers are characterized by higher β-sheet content, exposure of hydrophobic clusters, and cytotoxicity, compared with the monomer (10, 11).
Knowing how PrP oligomer diversity is related to the quaternary structure could aid in understanding how multiple prion strains, which cause prion diseases with specific phenotypic traits (12), are generated and stabilized from a unique native fold. Indeed, the main differences between prion strains are due to their alternative conformation that can be stably and faithfully propagated (13, 14).
It has been hypothesized that hydrodynamic/volumetric properties could be a key factor in strain-specific prion replication dynamics (15). Consistent with this finding, PrP misfolding, which exhibits a strong pressure dependence, affects its hydration and packing (16–21), two properties that directly alter the volume of the system. Indeed, the behavior of proteins at different pressures is based on volume differences. As predicted by Le Chatelier's principle, the application of pressure favors conformational states with a smaller specific volume. These observations indicate that determining the pressure-dependent PrP oligomer behavior is crucial for understanding the importance of void volumes or cavities (i.e. defects in atomic packing) and hydration in PrP self-replicating states.
In the present work, we used high pressure and particularly the pressure jump technique (22–25) to provide a new structural dimension that cannot be attained with other experimental approaches: the volumetric comprehension of the kinetic transition state ensemble of PrP oligomer dissociation. We report that the PrP oligomeric energy landscape is strongly influenced by pressure and that different oligomers display different sensitivities. This observation brings new insights into the different oligomer structural changes caused by pressure- and temperature-based treatments and also demonstrates that each oligomer type is associated with a distinct degree of void volumes. We observed that the PrP molecules that constitute the oligomers can attain new conformational coordinates, leading to monomeric PrP that refolds into the native conformation after pressure release. In addition, by focusing on the kinetic and energetic behavior of the pressure-induced dissociation of a PrP oligomer, we reveal a large negative activation volume, together with an increase in both apparent activation enthalpy and entropy. These differences between the transition state ensemble and the starting ground (oligomeric) state suggest a transition state ensemble that is less structured and significantly more hydrated than the oligomeric state.
Experimental Procedures
Recombinant PrPs
Full-length ovine (Ala-136, Arg-154, Gln-171) PrP and its double variant PrPH190K/I208M (26) were produced in Escherichia coli and purified as described previously (27). H190K/I208M amino acid replacements destabilize the C-terminal α-helices H2 and H3 (26, 28) and fine tune the thermally induced oligomerization pathway leading to the formation of O0 and O3 oligomers. Purified PrP monomers were stored lyophilized and recovered in 20 mm sodium citrate, pH 3.4, by elution through a G25 desalting column (GE Healthcare, Orsay, France). The final protein concentration was measured by optical density at 280 nm using an extinction coefficient of 58,718 m−1 cm2.
PrP Oligomerization
The O1 oligomer and the O3 oligomer were produced by heating 100 μm wild type PrP in 20 mm sodium citrate, pH 3.4, at 60 °C for 1 h. The O0 oligomer was obtained by heating 100 μm PrPH190K/I208M in 20 mm sodium citrate, pH 3.4, at 50 °C for 15 min. Homogeneous fractions of each oligomer were collected after separation by size exclusion chromatography.
Size Exclusion Chromatography (SEC)
The oligomerization profiles of recombinant PrPs were analyzed by SEC using a TSK 4000SW gel filtration column (60 × 0.78 cm) (Tosoh Bioscience, Worcestershire, UK), in 20 mm sodium citrate, pH 3.4, at 20 °C. Before each run, the column was equilibrated with at least 4 column volumes of elution buffer. The flow rate was 1 ml/min, and protein elution was monitored by UV absorption at 280 nm. For quantification, the chromatograms were normalized and deconvoluted using Lorentzian and Gaussian fit procedures prior to determining the peak surfaces.
Double Labeling of Oligomers (FRET)
Site-specific labeling was achieved by introducing cysteine residues using QuikChange site-directed mutagenesis (Stratagene) at the N (K29C) or C terminus (S231C) of PrP to enable conjugation with the maleimide of the dyes Oregon Green 488 and Alexa 568 (Life Technologies, Inc.), respectively. Both PrP variants were purified as described above. They were then incubated with a 3-fold molar excess of Oregon Green 488 or Alexa Fluor 568 maleimide at 4 °C overnight, and unconjugated dye was removed by elution through a G25 desalting column (GE Healthcare). Double-labeled O1 oligomers were obtained by heating a mix with equimolar quantities (50 μm) of each labeled protein in 20 mm sodium citrate, pH 3.4, at 60 °C for 1 h. A homogeneous fraction of the oligomer was collected after separation by size exclusion chromatography.
High Pressure Treatment
Purified oligomeric PrP forms were diluted in the same buffer to a final protein concentration of 12 μm (0.7 μm for in situ ANS fluorescence measurements and 4 μm for FRET analysis) and placed in 5-mm diameter quartz cuvettes closed at the top with flexible polyethylene film that was kept in place by a rubber O-ring. Pressure jumps consisted of rapid (<30-s) changes of the atmospheric pressure to obtain a range of final pressures of 200–350 MPa. A pressurization cycle at 300 MPa was performed by maintaining the pressure for 60 min and by decompression of the sample to atmospheric pressure.
Fluorescence Measurements under High Pressure
Fluorescence measurements were carried out using an Aminco Bowman Series 2 fluorescence-spectrophotometer (SLM Aminco) modified to accommodate a thermostated high pressure optical cell. Protein disaggregation was followed by monitoring the changes in 8-anilino-1-naphthalene sulfonate (ANS; 55 μm final concentration) fluorescence intensity at 490 nm (16-nm slit) and excited at 385 nm (8-nm slit).
Static Light Scattering
Static light scattering kinetic and temperature ramping experiments were performed on a homemade device using a 407-nm laser beam in 2-mm quartz cuvettes. Kinetic experiments were performed after a rapid temperature increase from 15 to 65 °C by placing the cuvette in a preheated thermostated cell holder. Temperature ramping experiments (2 °C/min) were performed starting at 15 °C and up to 90 °C. For both types of experiments, PrP oligomer concentration was fixed at 1 μm (monomer equivalent) in 20 mm sodium citrate, pH 3.4. This technique (multiwavelength mode) was coupled with SEC to estimate the molecular weight of each oligomer type using the relationship between the intensity of light scattered by the molecule and its molecular weight and concentration (equivalent monomer), as described by the Rayleigh theory.
Estimation of the Thermodynamic Apparent Activation Parameters
The relaxation profiles of the oligomer structural reactions were fitted to single or double exponential decays, according to Equations 1 and 2, respectively,
where I(t) and I0 are the fluorescence intensities at time t and at time 0, A and B are the phase amplitudes, and kobs is the measured apparent rate constant at the final pressure P.
The thermodynamic apparent activation parameters ΔH* and ΔS* were determined by fitting NAkBln(kobs/kobs0) = f(1/T) to Equation 3,
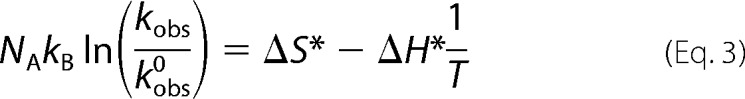 |
where kobs = 1/τ is the observed rate, kobs/kobs0 is the pre-exponential factor for the observed rate corrected for the change in medium viscosity due to the change in pressure, NA is Avogadro's number, kB is the Boltzmann factor, and T is the absolute temperature in Kelvin. From theory, kobs/kobs0 = 1.33kBTcNA(1/η), where c is the molar concentration of PrP protein, and η is the medium viscosity at a given pressure (29).
The change in apparent activation volume of the kinetic transition state was determined by fitting RTln(kobs/kobs0) = ΔG* = f(P) to Equation 4,
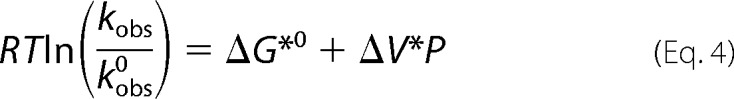 |
where ΔG*0 is the apparent activation free energy change at atmospheric pressure at a given temperature T.
Numerical Simulations of the Experimental Data
The time dependence of the concentration of the individual species A (oligomer), B (intermediate), and C (monomer) in Reaction 1,
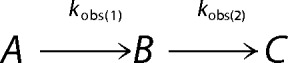 |
was generated by numerical evaluation of Equations 5–7, using a commercial software package (MATLAB version 8.4 (R2014b)/2014, The MathWorks Inc., Natick, MA) and the corresponding measured apparent rate constants.
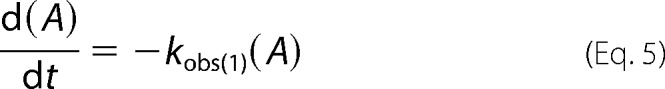 |
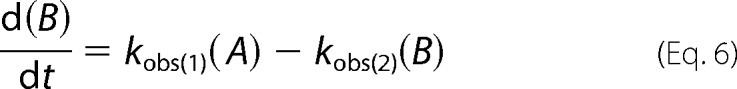 |
 |
Fluorescence Measurements at Atmospheric Pressure
Fluorescence measurements were performed at 20 °C using an FP-6200 fluorimeter (Jasco France, Bouguenais, France). Aliquots of each protein sample (0.4 μm) were incubated with 50 μm ANS at room temperature for 10 min before fluorescence measurements. ANS emission spectra were recorded after excitation at 385 nm. The excitation and emission slit widths were 4 nm.
Circular dichroism (CD) Spectroscopy
CD spectra were recorded at 20 °C using a J810 spectropolarimeter (Jasco). A 0.2-cm optical path length quartz cell was used to record the spectra of each protein sample in the far UV region (195–260 nm). Protein concentration and buffers were those used in the high pressure treatment. Baseline-corrected CD spectra were acquired at a scan speed of 20 nm min−1, 1-nm bandwidth, and 1-s response time. Spectra were signal-averaged over three scans.
Transmission Electron Microscopy
Samples were deposited on Formvar carbon-coated grids, negatively stained with freshly filtered 2% uranyl acetate, dried, and viewed using a JEOL 1200EX2 electron microscope (JEOL USA, Inc., Peabody, MA).
Results
Pressure Inhibits PrP Oligomerization
We previously showed that full-length recombinant PrP can be converted into soluble oligomers upon heating (8, 9). By using this method (as described under “Experimental Procedures”), two discrete and stable oligomers, referred to as O3 and O1, were eluted as separated peaks on a size exclusion column (blue lines in Fig. 1A) and shown to be a ∼12- and ∼36-mer PrP oligomer, respectively (black lines in Fig. 1A). Oligomerization was largely hindered by pressure. Indeed, a single elution peak that corresponded to monomeric PrP was obtained when the pressure at which the heat treatment was performed increased to 150 MPa (red line in Fig. 1B).
FIGURE 1.
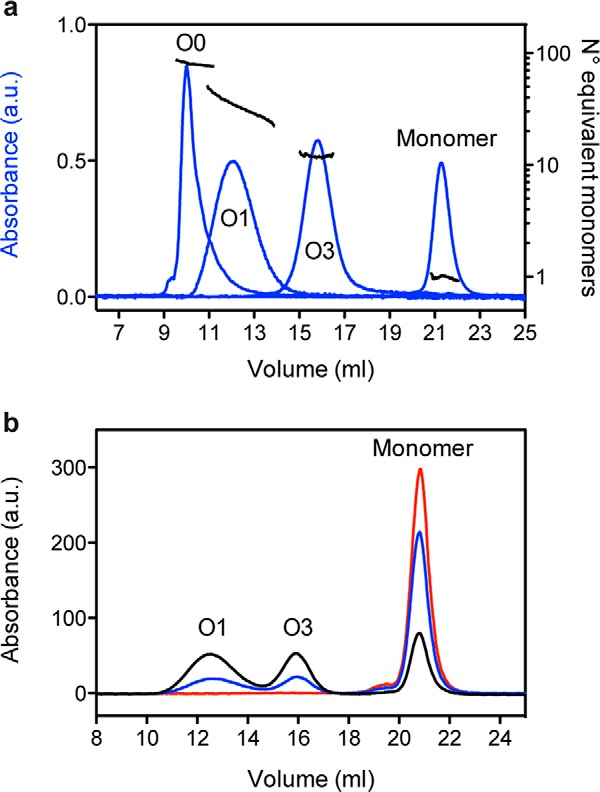
Pressure inhibits heat-induced PrP oligomerization. a, oligomer molecular weight estimation (expressed as the number of equivalent monomers) by multiwavelength light scattering coupled with size exclusion chromatography. b, size exclusion chromatograms obtained after a 1-h incubation (55 °C) of PrP at atmospheric pressure (black line), 50 MPa (blue line), and 150 MPa (red line). The PrP species corresponding to each peak are indicated. a.u., arbitrary units.
These results indicate that the involved oligomerization pathways lead to a population of PrP conformers (i.e. earlier reaction intermediates and/or misfolded oligomers) that are less solvated and more voluminous than the native state protein.
Pressure-induced PrP Oligomer Dissociation
We then asked whether the quaternary, tertiary, and secondary structural features of PrP oligomers were affected by pressure and whether pressure sensitivity was dependent on the oligomer type. With this aim, O1 and O3 oligomers were generated from the wild type protein, and O0, another oligomer with higher molecular weight, was generated from the H190K/I208M PrP variant (26). O0 eluted as a separate peak in the void volume of the column and, on the basis of its scattering intensity, was considered to be an ∼82-mer PrP oligomer (Fig. 1A). The effects of pressure on these assemblies (O0, O1, and O3) were then assessed by applying a pressure of 550 MPa. After a cycle of compression (15 °C for 1 h) and decompression to atmospheric pressure, the extent of dissociation of each purified oligomer was analyzed by SEC (Fig. 2). The loss of each oligomer population (quaternary structure) was concomitant with the gain of monomeric conformers, suggesting that pressurization led to irreversible dissociation without any oligomeric intermediate forms. The O1 oligomer was completely dissociated after pressure treatment; conversely, the ratio of monomeric PrP rescued from the O3 and O0 assemblies was about 41 and 78%, respectively. This indicates that the barostability of the O1 oligomer is lower than that of the O0 and O3 assemblies.
FIGURE 2.
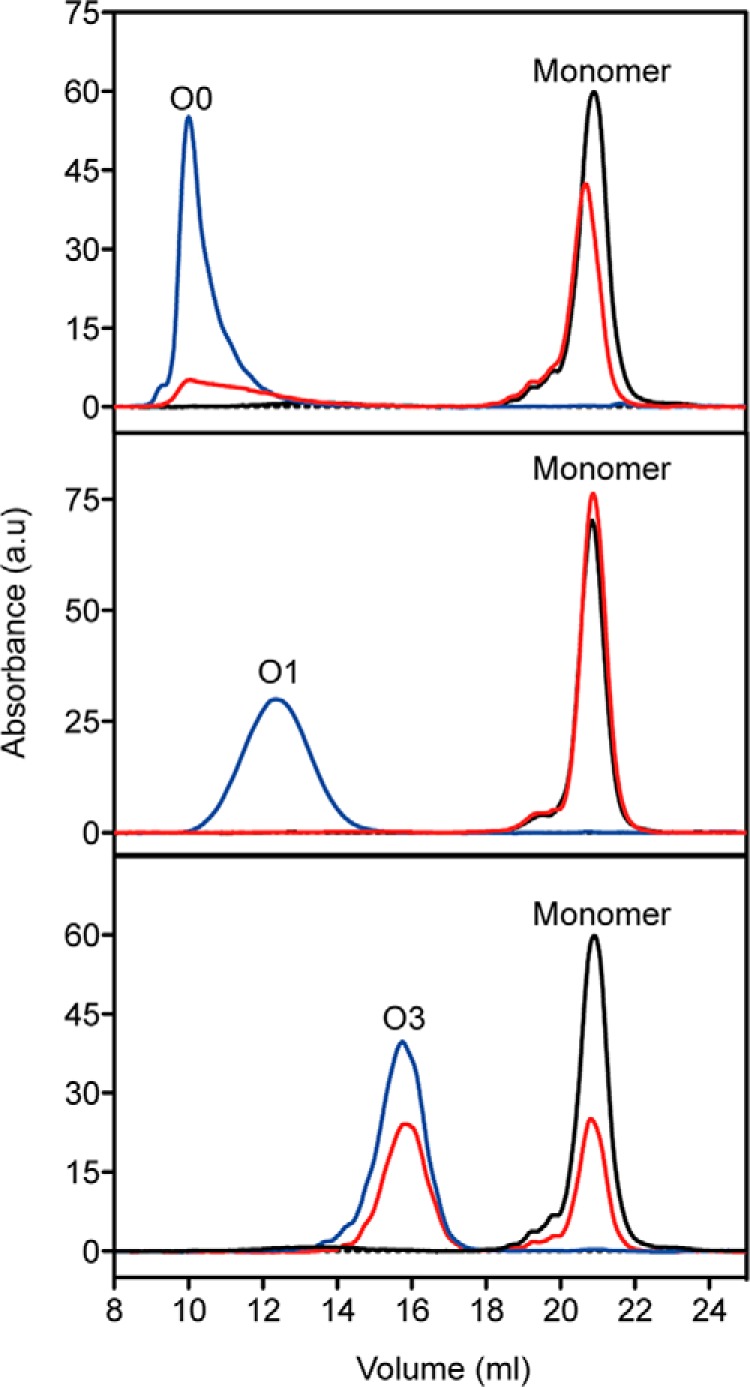
Pressure induces PrP oligomer dissociation. Overlay of size exclusion chromatograms of purified O0 (top), O1 (middle), and O3 (bottom) PrP oligomers untreated (at 15 °C for 1 h; blue line) or after a cycle (1 h) of compression (550 MPa)/decompression at 15 °C (red line). The profile of monomeric PrP (black line) is shown in each panel to estimate the amount of resolubilized PrP rescued from each oligomeric form after pressure treatment. The PrP species corresponding to each peak are indicated. a.u., arbitrary units.
Similar results were obtained when changes in the PrP solvent-exposed hydrophobic clusters and secondary structure that accompany oligomer dissociation were assessed by ANS fluorescence (Fig. 3A) and CD spectroscopy (Fig. 3B), respectively. The ANS fluorescence emission spectrum of pressure-treated O1 oligomers was comparable with that of monomeric PrP (Fig. 3A, middle panel), demonstrating the complete and irreversible loss of its solvent-exposed hydrophobic clusters. In line with the previous results, pressure-treated O0 and O3 oligomers showed an intermediate spectrum (Fig. 3A, top and bottom) that mirrored the mixture of oligomers and monomers present in the sample after the compression/decompression cycle. Concerning the secondary structure, the major contribution in the native protein spectrum was from the α-helical component, in accordance with its structural prevalence (Fig. 3B, black line) (30). In contrast, both O1 and O3 oligomers showed a strong signal characteristic of β-sheet structures. The blue-shifted profile of the O0 oligomer could be assigned to enrichment in loop components with a possible contribution by β-turns and a disordered structure. The observed spectral changes upon pressure treatment reflected the refolding of dissociated monomeric PrP into the native α-helix-rich conformation after pressure release. Therefore, the specific gain in α-helical forms observed for each oligomer mirrors its instability toward pressure.
FIGURE 3.
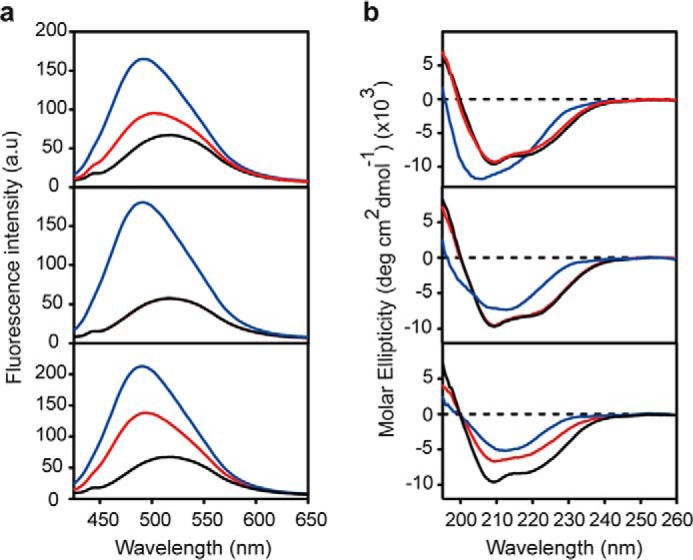
Pressure leads to irreversible loss of ANS binding to PrP oligomers and modifies their secondary structure. Top panels, O0; middle panels, O1; bottom panels, O3. a, ANS fluorescence emission spectra of untreated (1 h at 15 °C) PrP oligomers (blue line) and after a cycle (1 h) of compression (550 MPa)/decompression at 15 °C (red line). For comparison, the ANS spectrum of monomeric PrP (black line) is shown in each panel. b, CD spectra of untreated (1 h at 15 °C) PrP oligomers (blue line) and after a cycle (1 h) of compression (550 MPa)/decompression at 15 °C (red line). For comparison, the CD spectrum of monomeric PrP (black line) is shown in each panel. a.u., arbitrary units.
The macrostructural changes induced by pressure were also investigated (Fig. 4). Analysis of the starting PrP assemblies by transmission electron microscopy revealed their precise oligomer size and morphology, ranging from round-shaped particles (O3 oligomer) to wormlike structures (O0 oligomer). After pressure treatment, the structure of O1 and O0 was strongly modified, supporting the observed loss of quaternary structure. Only the amount of O3 oligomers remained almost unchanged.
FIGURE 4.
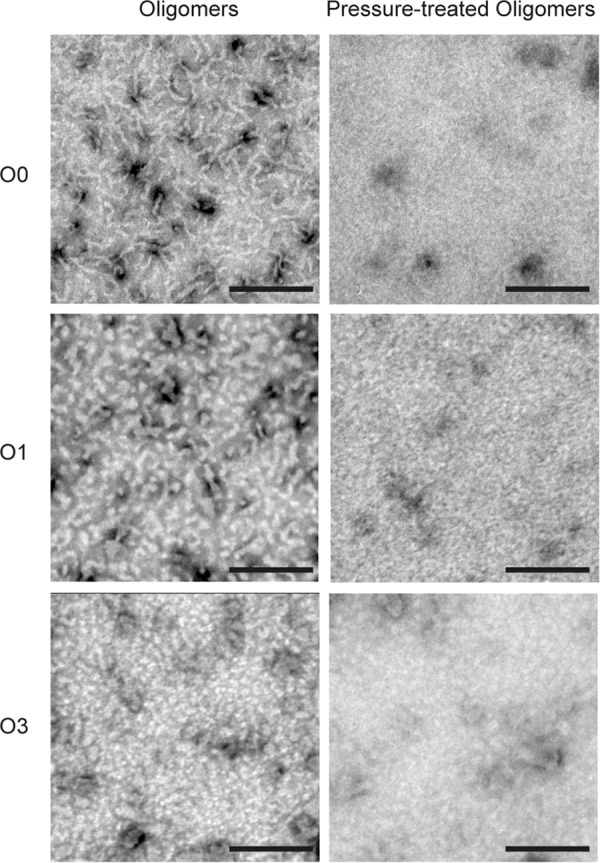
Pressure alters the macrostructure of PrP oligomers. Top panels, O0; middle panels, O1; bottom panels, O3. Shown are negative stained transmission electron microscopy micrographs of untreated (1 h at 15 °C) PrP oligomers (left) and after a cycle of compression (550 MPa)/decompression at 15 °C for 1 h. Scale bars, 200 nm.
Kinetics of Pressure Jump-induced Oligomer Dissociation
The previous results showed that the pressure treatment of oligomers led to a significant decrease in the β-sheet content and a large decrease in the ANS fluorescence yield. This last feature turned out to be a suitable intrinsic probe for kinetic measurements. To better understand the structural changes induced by pressure, the kinetics of the structural changes of each oligomer type was measured by monitoring ANS fluorescence after a fast increase (within 30 s) of pressure to 300 MPa at 37 °C (Fig. 5). A single exponential kinetics was obtained for the O1 oligomer. Conversely, higher order processes were observed for O3 and O0. The obtained kinetic profiles were in good agreement with the specific barostability of the three oligomers: O1 the most sensitive and O3 the most resistant to pressure dissociation.
FIGURE 5.
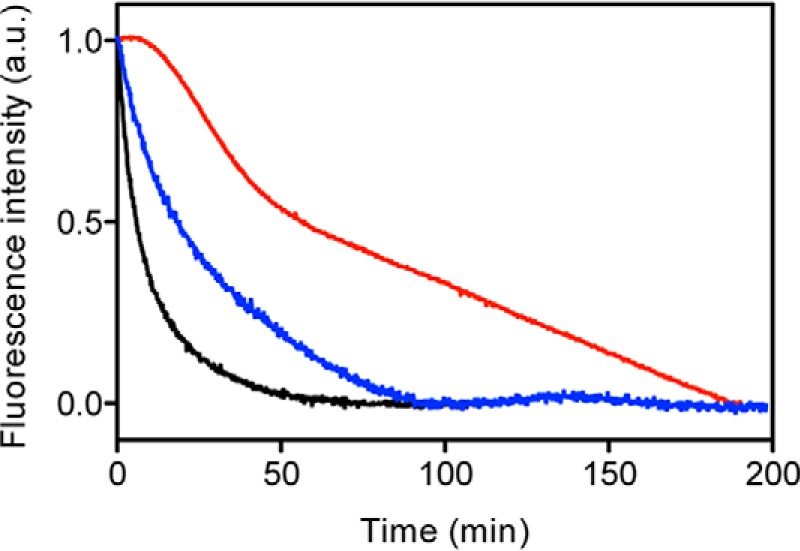
Pressure-induced PrP oligomer dissociation after a rapid increase of pressure to 300 MPa. The structural change kinetics for O0 (blue line), O1 (black line), and O3 (red line) were recorded as a decrease in ANS fluorescence emission intensity. The temperature was 37 °C. a.u., arbitrary units.
Kinetics of Temperature Jump-induced Oligomer Dissociation
To determine whether there was a direct relationship between the oligomer barostability and thermal stability, static light scattering measurements for each oligomer type were performed during temperature ramping (Fig. 6A). The loss in scattering intensity observed when the temperature increased from 15 to 90 °C indicated a decrease in the average molecular weight of the sample, denoting the irreversible dissociation of the oligomers to a native monomeric PrP at the protein concentrations used (data not shown). This finding is in agreement with our previous observations (8, 28). In contrast to what was observed during pressure treatment, the O3 oligomer was the most unstable, whereas O1 and O0 thermal stabilities were almost identical. The different pressure- and temperature-driven oligomer dissociation behaviors were supported by the relaxation kinetics observed after a rapid temperature increase to 75 °C (Fig. 6B).
FIGURE 6.
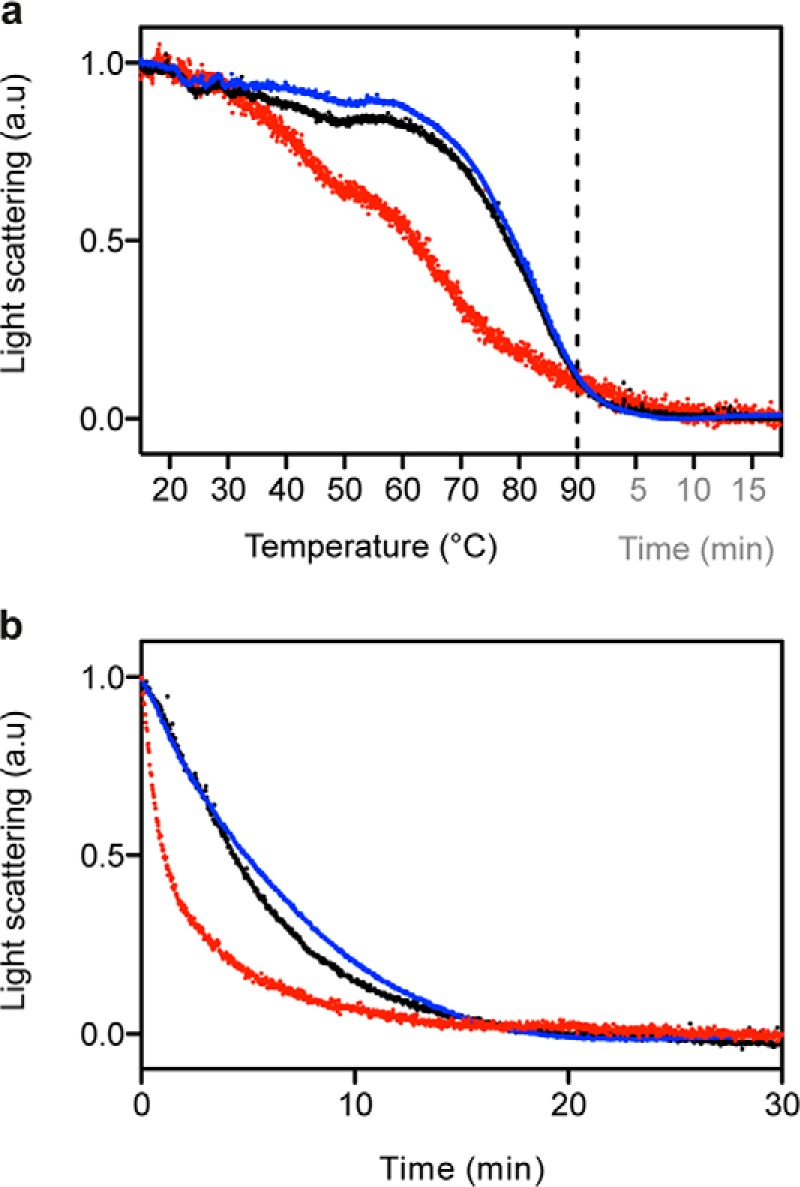
Temperature-induced PrP oligomer dissociation after a gradual or rapid temperature increase. The extent of structural changes for O0 (blue line), O1 (black line), and O3 (red line) was recorded as a decrease in light scattering intensity. The pressure was 0.1 MPa. a.u., arbitrary units. a, temperature ramping experiments (2 °C/min) were performed starting at 15 °C up to 90 °C. The signal stability at 90 °C was monitored for another 20 min. b, kinetic experiments were performed after a rapid temperature increase from 15 to 75 °C.
For the next analyses, we focused on the O1 oligomer because it was the only assembly for which the simple two-state pressure- and temperature-induced kinetics were represented by a single exponential curve. Because the dissociation process was irreversible, we assumed that the individual rate constant for the backward reaction was negligibly small, such that the relaxation time predominantly reflected the inverse of the individual rate constant for the forward reaction.
Temperature Dependence of the Pressure-driven Oligomer-Monomer Relaxation Kinetics
The observed pressure-induced structural changes in the O1 oligomer accelerated when the temperature at which pressure jumps (of 300 MPa) were applied increased from 15 to 45 °C (Fig. 7A). Within our experimental temperature range, the Arrhenius plot of the observed rate constants was linear (Fig. 7A, inset). The free energy barrier associated with the pressure-induced process was estimated from the temperature dependence of the observed rate constant. With this aim, the relative contribution of the apparent activation enthalpy and entropy to the rate constants was calculated using Equation 2 (see “Experimental Procedures”). The resulting apparent activation parameters (Table 1) indicate that the pressure-induced reaction kinetics was controlled by competition between the apparent activation enthalpy and entropy. Furthermore, the pressure-induced structural transition involved the high energy barrier ΔG* that accounted for the stability of PrP oligomers in physiological conditions.
FIGURE 7.
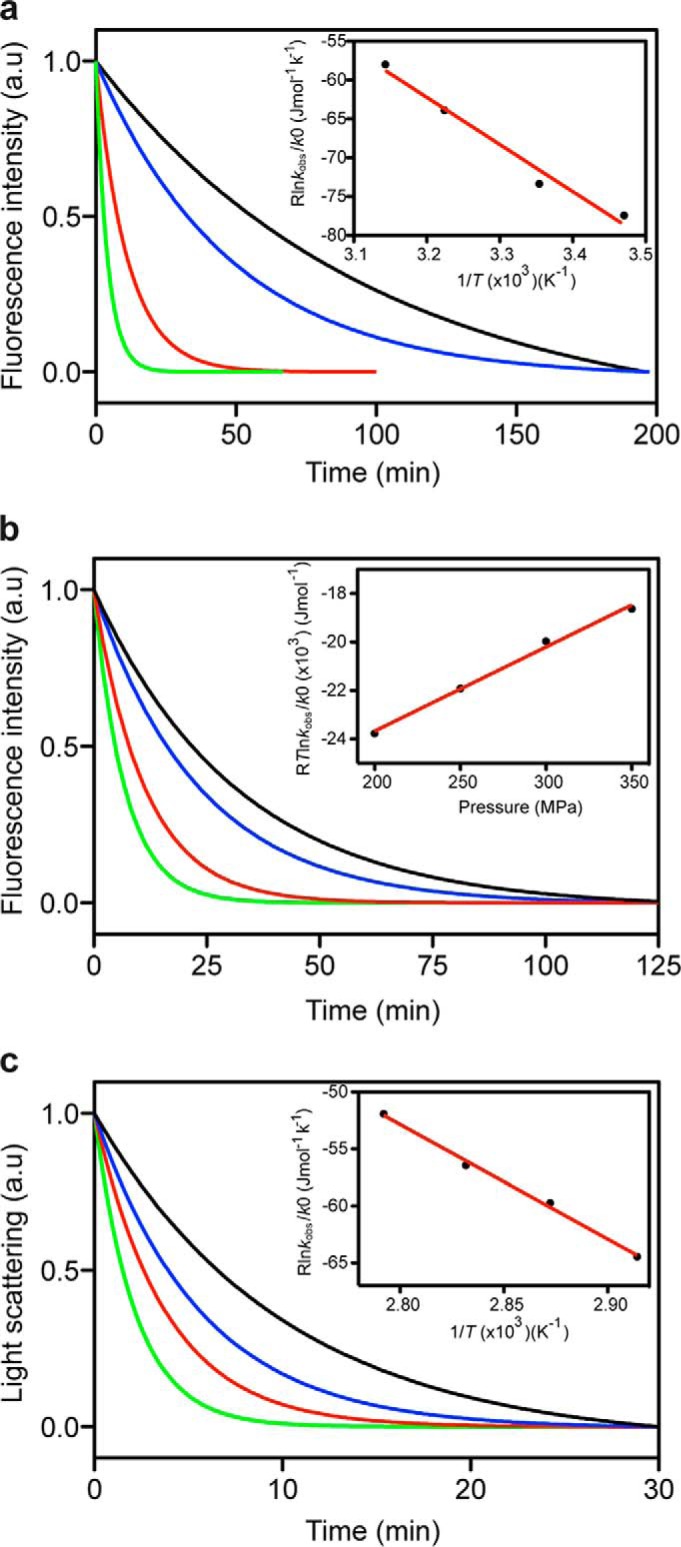
Kinetics of the pressure- and temperature-induced dissociation of the O1 PrP oligomer. Temperature (a) and pressure (b) dependence of the pressure-induced O1 structural kinetics. a, pressure jumps (from 0.1 to 300 MPa) were performed at different temperatures: 15 °C (black line), 25 °C (blue line), 37 °C (red line), and 45 °C (green line). b, pressure jumps (at 37 °C) were performed starting at 0.1 MPa to obtain different final pressures: 200 MPa (black line), 250 MPa (blue line), 300 MPa (red line), and 350 MPa (green line). c, temperature dependence of the temperature-induced O1 structural kinetics. Rapid temperature increases of different magnitudes (starting at 15 °C and always at 0.1 MPa) were performed to reach different final temperatures: 70 °C (black line), 75 °C (blue line), 80 °C (red line), and 85 °C (green line). Insets show the pressure and temperature dependence of the observed rate constants (see “Experimental Procedures” for further details). Solid lines, linear fits to the data. a.u., arbitrary units.
TABLE 1.
Thermodynamic activation parameters of O1 oligomer
| Parameter | Value |
|---|---|
| Temperature dependence (pressure-induced kinetics) (300 MPa) | |
| ΔH* (kJ mol−1) | 60.6 ± 6.5 |
| ΔS* (J mol−1 K−1) | 131.9 ± 21.4 |
| TΔS*310 K(kJ mol−1) | 40.9 ± 6.6 |
| ΔG*310 K,300 MPa (kJ mol−1) | 19.7 ± 9.3 |
| Pressure dependence (pressure-induced kinetics) (310 K) | |
| ΔG*0310 K (kJ mol−1) | 30.6 ± 0.5 |
| ΔV* (ml mol−1) | -34.7 ± 1.9 |
| ΔG*310 K, 300 MPa (kJ mol−1) | 20.2 ± 0.5 |
| Temperature dependence (thermally induced kinetics) (0.1 MPa) | |
| ΔH* (kJ mol−1) | 100.4 ± 4.6 |
| ΔS* (J mol−1 K−1) | 228.4 ± 13.2 |
| TΔS*310 K (kJ mol−1) | 70.8 ± 4.1 |
| ΔG*310 K, 0.1 MPa (kJ mol−1) | 29.6 ± 6.2 |
Pressure Dependence of the Pressure-driven Oligomer-Monomer Relaxation Kinetics
Next, pressure jumps of different magnitude with final pressures ranging from 200 to 350 MPa were performed. The relaxation profile of the O1 oligomer structural changes was faster at high pressure, and the plot of the observed rate constant as a function of pressure was linear (Fig. 7B). The thermodynamic apparent activation parameters derived from the analysis are shown in Table 1. The negative apparent activation volume (ΔV* = −34.7 ± 1.9 ml mol−1) indicates that the oligomer in its ground state was more voluminous than in its kinetic transition state. Although the pressure-induced conformational change of oligomers was apparently prohibited by relatively large apparent activation free energies (ΔG*), these energetic barriers were compensated by the negative contribution from ΔV*, leading to an acceleration of the dissociation at increasing pressure.
Temperature Dependence of the Temperature-driven Oligomer-Monomer Relaxation Kinetics
To study the temperature dependence of the thermally induced oligomer dissociation reaction rate, temperature increases of different magnitudes were performed at atmospheric pressure, starting at the same temperature (15 °C) and reaching different final temperatures (70–85 °C). Fig. 7C shows the typical monoexponential kinetic profiles of O1 dissociation after heating. The thermodynamic apparent activation parameters were then calculated (Table 1) based on the linear dependence from temperature of the observed rate constants (Fig. 7C, inset).
The absolute value of the Gibbs free energy of activation for O1 thermal dissociation was estimated to be 29.6 ± 6.2 kJ mol−1. This barrier, assessed at atmospheric pressure and physiological temperature, was comparable with the value obtained after pressure treatment (30.6 ± 0.5 kJ mol−1). In addition, in both dissociation processes, the reaction was entropically favorable but enthalpically unfavorable. Because the oligomer-monomer transition depends mainly on enthalpy, a conformational remodeling must occur from the ground state to the activated state.
Site-specific Monitoring of Oligomer Dissociation by FRET
PrP monomers were N-terminally labeled with Oregon Green 488 (Exmax/Emmax ≈ 496/524 nm) or C-terminally labeled with Alexa 568 (Exmax/Emmax ≈ 578/603 nm). After purification, equimolar quantities of each labeled protein were mixed to obtain a double-labeled O1 oligomer. The fluorescent behavior of the purified O1 species depended on its oligomeric state. When excited at 496 nm, the oligomer fluoresced with Imax ∼520 and ∼605 nm (black line in Fig. 8A). In the dissociated state (after a 1-h pressure treatment at 300 MPa, 25 °C), the same incident light resulted in a markedly different emission spectrum, with a simultaneous decrease in emission at 605 nm and a slight increase at 525 nm (red line in Fig. 8A).
FIGURE 8.
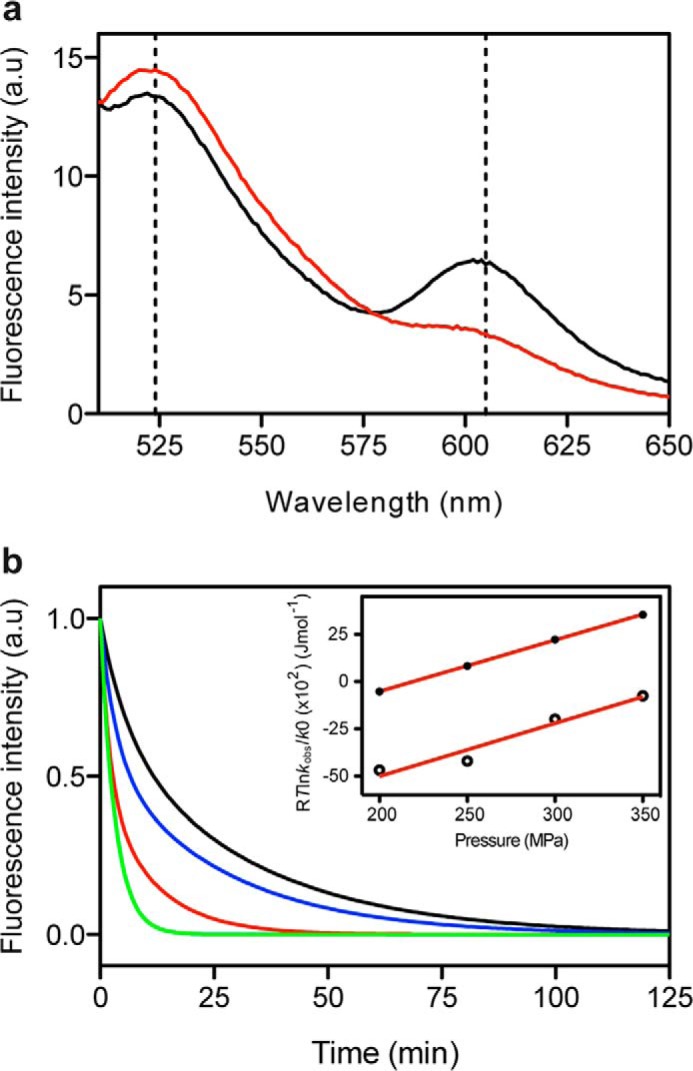
Kinetics of the pressure-induced dissociation monitored by loss of FRET activity of the double-labeled O1 PrP oligomer. a, fluorescence emission spectrum of the FRET pair Oregon Green 488/Alexa 568 covalently linked to the protomers of O1 oligomers (black line). Loss of FRET activity after a cycle of compression (300 MPa)/decompression at 25 °C (red line). b, pressure jumps (at 25 °C) were performed starting at 10 MPa to obtain different final pressures: 200 MPa (black line), 250 MPa (blue line), 300 MPa (red line), and 350 MPa (green line). Inset, pressure dependence of the observed rate constants (see “Experimental Procedures” for further details) calculated from the fast phase (solid circles) and from the slow phase (open circles). Solid lines, linear fits to the data. a.u., arbitrary units.
Next, the kinetics of pressure-induced oligomer dissociation was monitored by following the FRET efficiency loss. Pressure jumps of different magnitudes were performed with final pressures ranging from 200 to 350 MPa. Each pressure jump produced biphasic kinetics with faster relaxation profiles at high pressure (Fig. 8B). Each phase encompassed about half of the total signal change at pressures of >300 MPa. The measured apparent rate constants for the slower decay at each final pressure corresponded to those measured using ANS as extrinsic fluorescent reporter. These results indicate that O1 dissociation does not directly lead to the PrP native state but rather through an ANS-silent precursor that still retains significant hydrophobic patches exposed to the solvent. As illustrated in the inset to Fig. 8B, the plot of the observed rate constants as a function of pressure was linear. The thermodynamic apparent activation parameters derived from the analysis (Table 2) differed considerably from those determined by using ANS fluorescence. These results provide strong evidence that region-localized (monitored by FRET) and global (ANS fluorescence) conformational changes within the O1 oligomer do not exhibit a concerted behavior.
TABLE 2.
Thermodynamic activation parameters obtained from pressure-dependent kinetics of a double-labeled O1 oligomer
| Fast phase | Slow phase | |
|---|---|---|
| ΔG*0298 K (kJ mol−1) | 6.0 ± 0.1 | 10.61 ± 1.3 |
| ΔV* (ml mol−1) | −27.2 ± 0.2 | −28.00 ± 4.5 |
| ΔG*298 K, 300 MPa (kJ mol−1) | −2.2 ± 0.1 | 2.2 ± 1.3 |
Discussion
The structural changes involved in the formation of PrP assemblies, such as during PrP oligomerization (8, 31, 32), are not well understood. Little is known about the changes in hydration properties and intrinsic packing that directly affect the volume of the system during the native to oligomer transition. However, knowledge of the volumetric properties of PrP assemblies is crucial because they might govern strain-specific prion replication dynamics (15) and also because they might dictate the gains in biological functions, such as binding to other molecules and cytotoxicity (10, 11).
Rather than rely on structural data alone, in this work, we investigated the pressure-induced PrP structural transformations. Specifically, we focused on the effects of high pressure on three distinct non-fibrillar β-sheet-rich oligomers formed from full-length PrP, after their purification from a heterogeneous mixture of species. The obtained experimental results allow us to discuss how packing and hydration may account for multiple stable and conformationally distinct oligomers.
Void Volumes in Oligomers Dictate Their Distinct Pressure Versus Temperature Lability
We show that pressure can prevent PrP association into oligomers and even reverse their formation. The dissociation reaction leads to monomeric PrP molecules that refold into the native conformation after pressure release. This finding contributes crucial knowledge about PrP structural changes that accompany the assembly process. Particularly, it implies that the volume of the system differs in the monomeric and oligomeric PrP conformers. Because a reaction is favored under pressure when the volume change of the system is negative (33), the observed pressure-induced oligomer destabilization must be dictated by a decrease in volume. On the basis of a previous study on the molecular determinants of the decrease in volume upon protein unfolding (34), the present observations are explained by the collapse of cavities and not just by a disruption of the weak bond network and hydration of newly exposed residues.
We also provide conclusive evidence that the pressure-induced dissociation of PrP oligomers occurs without populating any lower molecular weight oligomer, in agreement with the effects of temperature (9). However, pressure and temperature affect each oligomer type in a specific manner. Indeed, whereas thermostability was O0 ≥ O1 > O3, barostability was O3 > O0 > O1. High molecular weight complexes, which are more prone to be enriched in void volumes in the intermolecular boundaries, are preferentially destabilized by pressure due to the larger volume decrease upon dissociation, compared with smaller oligomers. O0 arises through the homoassociation of O1 via conformational rearrangements (8). This could collapse potential packing defects or even create favorable interface interactions, explaining the higher barostability of O0 than O1. Thus, the O0 pathway forces the PrP structure to attain new conformational coordinates that lead to a quaternary structure that, as described in the present work, contains a different amount of misfolded β-sheets and possibly more hydrogen bonds, reminiscent of a protofibrillar structure. Because hydrogen bonds are not significantly dependent on pressure or even stabilized (35), their presence can indeed contribute to O0 stabilization at increasing pressure, compared with O1.
These pressure-induced perturbations are different from those induced by temperature, the effects of which also depend on the total energy of the system. PrP assemblies with a better shielding of backbone-backbone hydrogen bonds from the solvent are more protected against temperature-induced dissociation, regardless of their structural defects (cavities). Therefore, the experimental use of pressure and temperature ranges provides alternative insights into the oligomer structure.
The Transition State Ensemble of O1 Oligomer Dissociation
In complex energy landscapes, such as those of protein misfolding and oligomerization, the reaction rate (e.g. oligomer dissociation) might depend on the pathway. To verify experimentally this hypothesis and to investigate the energetics of protein conformation, we focused on the pressure- and temperature-induced dissociation of O1. By analyzing the dissociation kinetic data of this oligomer, obtained with experiments performed at different final temperatures and pressures, we reveal a large negative activation volume, with an increase in the apparent activation enthalpy (ΔH*) and entropy (ΔS*). This suggests a substantial structure difference between the transition state ensemble and the starting ground state (oligomer). Whereas ΔH* is clearly unfavorable to dissociation, ΔS* favors the reaction. The apparent activation free energies (ΔG*) are dominated by the enthalpic term and may be explained by the unfavorable solvation energy of newly exposed surfaces and by the disruption of atomic packing interactions in the oligomeric boundaries. Several factors may contribute to the increased ΔS* of the activated state, such as partial PrP unfolding and release of bound water molecules. The observed decrease in ΔV* in the dissociation transition state can be explained by a more hydrated state, particularly the hydration of voids and the electrostriction of water surrounding charged residues.
We obtained virtually identical apparent activation free energies (ΔG*) in both pressure and temperature dissociation experiments (30.6 ± 0.5 and 29.6 ± 6.2 kJ mol−1, respectively), with the same starting O1 oligomer dissociation conditions (310 K and 0.1 MPa). In addition, both pressure and temperature dependence of the pressure-induced kinetics led to identical ΔG* values (20.2 ± 0.5 and 19.7 ± 9.3 kJ mol−1, respectively), at 310 K and 300 MPa. Although this suggests comparable rate-limiting free energy barriers between the pressure and temperature dissociation reactions, the divergent associated enthalpic/entropic costs reveal distinct reaction pathways.
O1 Oligomer Dissociation Occurs through the Formation of a Transient Kinetic Intermediate
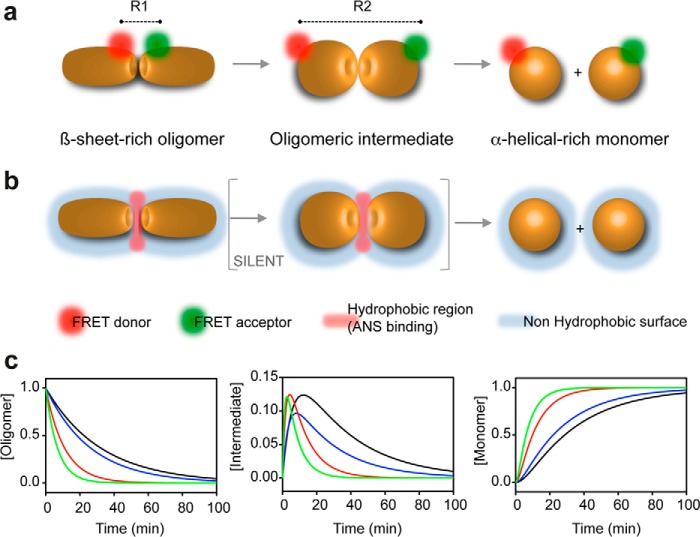
The results of the ANS fluorescence experiments allowed us to mechanistically characterize O1 dissociation; however, they only monitor the overall behavior of a protein rather than providing more local information. Conversely, with the FRET-based assay, we could follow oligomer dissociation locally. The biphasic kinetics obtained using this approach could readily be explained by the transient population of a kinetic intermediate. Indeed, the pressure-induced dissociation of O1 can be schematized as indicated in Fig. 9, A (FRET analysis) and B (ANS fluorescence).
FIGURE 9.
Schematic representation of PrP oligomer dissociation induced by pressure. The oligomer is depicted as a dimer for clarity. This model is based on the kinetic data obtained using FRET (a) and ANS fluorescence (b). R, distance between donor and acceptor. c, plot of the simulated time dependence of the concentration of individual species (oligomer, intermediate, and monomer) for the O1 oligomer dissociation reaction, obtained after pressure jumps from 10 to 200 MPa (black line), 250 MPa (blue line), 300 MPa (red line), and 350 MPa (green line).
The simulated time dependence of the concentration of individual species (Fig. 9C), obtained using the assumptions described under “Experimental Procedures” and the observed rate constants (data not shown), suggests a predissociation conformational change leading to a transient kinetic intermediate that retains its partially exposed nonpolar core, to which ANS can bind. Because some regions of the protomer/oligomer can be more poorly packed than others, the high pressure sensitivity of the transition state ensembles of the double-labeled O1 oligomer, compared with the transition state ensemble monitored using ANS, could be ascribed to a large amount of void volume in the immediate vicinity of the fluorescent dyes of the PrP N and C termini.
Conclusion
Here, we demonstrate significantly different void volumes in the quaternary structure of distinct β-sheet-rich oligomers of the prion protein PrP. Further investigations on the volumetric properties that characterize the plethora of structurally different quaternary folds of PrP are important for thoroughly understanding how multiple prion strains are generated and stabilized from a unique native fold. This will provide information not only on prion self-perpetuating structural states but also on the aberrant protein folds associated with many proteinopathies (36).
Author Contributions
J. T. conceived and coordinated the study and wrote the paper. J. T., R. L., and H. R. performed and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
J. T. and R. L. are grateful to INSERM for institutional support. We thank Pascal Wodling for assistance.
The authors declare that they have no conflicts of interest with the contents of this article.
- PrP
- prion protein
- PrPC
- PrP cellular isoform
- PrPSc
- PrP pathogenic isoform
- ANS
- 8-anilino-1-naphthalene sulfonate
- SEC
- size exclusion chromatography
- MPa
- megapascals.
References
- 1. Prusiner S. B. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 2. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kelly J. W. (1998) The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 8, 101–106 [DOI] [PubMed] [Google Scholar]
- 4. Deleault N. R., Walsh D. J., Piro J. R., Wang F., Wang X., Ma J., Rees J. R., Supattapone S. (2012) Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. U.S.A. 109, E1938–E1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Makarava N., Kovacs G. G., Bocharova O., Savtchenko R., Alexeeva I., Budka H., Rohwer R. G., Baskakov I. V. (2010) Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 119, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang F., Wang X., Yuan C. G., Ma J. (2010) Generating a prion with bacterially expressed recombinant prion protein. Science 327, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thirumalai D., Klimov D. K., Dima R. I. (2003) Emerging ideas on the molecular basis of protein and peptide aggregation. Curr. Opin. Struct. Biol. 13, 146–159 [DOI] [PubMed] [Google Scholar]
- 8. Eghiaian F., Daubenfeld T., Quenet Y., van Audenhaege M., Bouin A. P., van der Rest G., Grosclaude J., Rezaei H. (2007) Diversity in prion protein oligomerization pathways results from domain expansion as revealed by hydrogen/deuterium exchange and disulfide linkage. Proc. Natl. Acad. Sci. U.S.A. 104, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rezaei H., Eghiaian F., Perez J., Doublet B., Choiset Y., Haertle T., Grosclaude J. (2005) Sequential generation of two structurally distinct ovine prion protein soluble oligomers displaying different biochemical reactivities. J. Mol. Biol. 347, 665–679 [DOI] [PubMed] [Google Scholar]
- 10. Chich J. F., Chapuis C., Henry C., Vidic J., Rezaei H., Noinville S. (2010) Vesicle permeabilization by purified soluble oligomers of prion protein: a comparative study of the interaction of oligomers and monomers with lipid membranes. J. Mol. Biol. 397, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 11. Simoneau S., Rezaei H., Salès N., Kaiser-Schulz G., Lefebvre-Roque M., Vidal C., Fournier J. G., Comte J., Wopfner F., Grosclaude J., Schätzl H., Lasmézas C. I. (2007) In vitro and in vivo neurotoxicity of prion protein oligomers. PLoS Pathog. 3, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aguzzi A., Heikenwalder M., Polymenidou M. (2007) Insights into prion strains and neurotoxicity. Nat. Rev. Mol. Cell Biol. 8, 552–561 [DOI] [PubMed] [Google Scholar]
- 13. Peretz D., Williamson R. A., Legname G., Matsunaga Y., Vergara J., Burton D. R., DeArmond S. J., Prusiner S. B., Scott M. R. (2002) A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 34, 921–932 [DOI] [PubMed] [Google Scholar]
- 14. Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. (1998) Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 4, 1157–1165 [DOI] [PubMed] [Google Scholar]
- 15. Laferrière F., Tixador P., Moudjou M., Chapuis J., Sibille P., Herzog L., Reine F., Jaumain E., Laude H., Rezaei H., Béringue V. (2013) Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog. 9, e1003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cordeiro Y., Kraineva J., Ravindra R., Lima L. M., Gomes M. P., Foguel D., Winter R., Silva J. L. (2004) Hydration and packing effects on prion folding and β-sheet conversion. High pressure spectroscopy and pressure perturbation calorimetry studies. J. Biol. Chem. 279, 32354–32359 [DOI] [PubMed] [Google Scholar]
- 17. De Simone A., Dodson G. G., Verma C. S., Zagari A., Fraternali F. (2005) Prion and water: tight and dynamical hydration sites have a key role in structural stability. Proc. Natl. Acad. Sci. U.S.A. 102, 7535–7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El Moustaine D., Perrier V., Acquatella-Tran Van Ba I., Meersman F., Ostapchenko V. G., Baskakov I. V., Lange R., Torrent J. (2011) Amyloid features and neuronal toxicity of mature prion fibrils are highly sensitive to high pressure. J. Biol. Chem. 286, 13448–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Silva J. L., Vieira T. C., Gomes M. P., Bom A. P., Lima L. M., Freitas M. S., Ishimaru D., Cordeiro Y., Foguel D. (2010) Ligand binding and hydration in protein misfolding: insights from studies of prion and p53 tumor suppressor proteins. Acc. Chem. Res. 43, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Torrent J., Alvarez-Martinez M. T., Harricane M. C., Heitz F., Liautard J. P., Balny C., Lange R. (2004) High pressure induces scrapie-like prion protein misfolding and amyloid fibril formation. Biochemistry 43, 7162–7170 [DOI] [PubMed] [Google Scholar]
- 21. Torrent J., Alvarez-Martinez M. T., Heitz F., Liautard J. P., Balny C., Lange R. (2003) Alternative prion structural changes revealed by high pressure. Biochemistry 42, 1318–1325 [DOI] [PubMed] [Google Scholar]
- 22. Font J., Torrent J., Ribó M., Laurents D. V., Balny C., Vilanova M., Lange R. (2006) Pressure-jump-induced kinetics reveals a hydration dependent folding/unfolding mechanism of ribonuclease A. Biophys. J. 91, 2264–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Torrent J., Font J., Herberhold H., Marchal S., Ribó M., Ruan K., Winter R., Vilanova M., Lange R. (2006) The use of pressure-jump relaxation kinetics to study protein folding landscapes. Biochim. Biophys. Acta 1764, 489–496 [DOI] [PubMed] [Google Scholar]
- 24. Vidugiris G. J., Markley J. L., Royer C. A. (1995) Evidence for a molten globule-like transition state in protein folding from determination of activation volumes. Biochemistry 34, 4909–4912 [DOI] [PubMed] [Google Scholar]
- 25. Woenckhaus J., Köhling R., Thiyagarajan P., Littrell K. C., Seifert S., Royer C. A., Winter R. (2001) Pressure-jump small-angle x-ray scattering detected kinetics of staphylococcal nuclease folding. Biophys. J. 80, 1518–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cavaliere P., Torrent J., Prigent S., Granata V., Pauwels K., Pastore A., Rezaei H., Zagari A. (2013) Binding of methylene blue to a surface cleft inhibits the oligomerization and fibrillization of prion protein. Biochim. Biophys. Acta 1832, 20–28 [DOI] [PubMed] [Google Scholar]
- 27. Rezaei H., Marc D., Choiset Y., Takahashi M., Hui Bon Hoa G., Haertlé T., Grosclaude J., Debey P. (2000) High yield purification and physico-chemical properties of full-length recombinant allelic variants of sheep prion protein linked to scrapie susceptibility. Eur. J. Biochem. 267, 2833–2839 [DOI] [PubMed] [Google Scholar]
- 28. Chakroun N., Prigent S., Dreiss C. A., Noinville S., Chapuis C., Fraternali F., Rezaei H. (2010) The oligomerization properties of prion protein are restricted to the H2H3 domain. FASEB J. 24, 3222–3231 [DOI] [PubMed] [Google Scholar]
- 29. Först P., Werner F., Delgado A. (2000) The viscosity of water at high pressures: especially at subzero degrees centigrade. Rheologica Acta 39, 566–573 [Google Scholar]
- 30. James T. L., Liu H., Ulyanov N. B., Farr-Jones S., Zhang H., Donne D. G., Kaneko K., Groth D., Mehlhorn I., Prusiner S. B., Cohen F. E. (1997) Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl. Acad. Sci. U.S.A. 94, 10086–10091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baskakov I. V., Legname G., Baldwin M. A., Prusiner S. B., Cohen F. E. (2002) Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 277, 21140–21148 [DOI] [PubMed] [Google Scholar]
- 32. Hornemann S., Glockshuber R. (1998) A scrapie-like unfolding intermediate of the prion protein domain PrP(121–231) induced by acidic pH. Proc. Natl. Acad. Sci. U.S.A. 95, 6010–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silva J. L., Foguel D., Royer C. A. (2001) Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem. Sci. 26, 612–618 [DOI] [PubMed] [Google Scholar]
- 34. Roche J., Caro J. A., Norberto D. R., Barthe P., Roumestand C., Schlessman J. L., García A. E., Garcia-Moreno B. E., Royer C. A. (2012) Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 6945–6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mozhaev V. V., Heremans K., Frank J., Masson P., Balny C. (1996) High pressure effects on protein structure and function. Proteins 24, 81–91 [DOI] [PubMed] [Google Scholar]
- 36. Carrell R. W., Lomas D. A. (1997) Conformational disease. Lancet 350, 134–138 [DOI] [PubMed] [Google Scholar]