Background: Mutations in PAX3 cause Waardenburg syndrome (WS), but the mechanism remains unclear.
Results: PAX3 binds mitotic chromosomes using its homeodomain (HD), a process that depends on arginine methylation and is lost in WS.
Conclusion: PAX3 with mutations in HD causes WS by failing to load on mitotic chromosomes.
Significance: This study pinpoints the molecular defects of a specific group of PAX3 WS mutants.
Keywords: chromosomes, homeobox, mitosis, mutant, protein arginine N-methyltransferase 5 (PRMT5), transcription factor, PAX3, arginine methylation, homeodomain, mitotic chromosome
Abstract
PAX3 is a transcription factor critical to gene regulation in mammalian development. Mutations in PAX3 are associated with Waardenburg syndrome (WS), but the mechanism of how mutant PAX3 proteins cause WS remains unclear. Here, we found that PAX3 loads on mitotic chromosomes using its homeodomain. PAX3 WS mutants with mutations in homeodomain lose the ability to bind mitotic chromosomes. Moreover, loading of PAX3 on mitotic chromosomes requires arginine methylation, which is regulated by methyltransferase PRMT5 and demethylase JMJD6. Mutant PAX3 proteins that lose mitotic chromosome localization block cell proliferation and normal development of zebrafish. These results reveal the molecular mechanism of PAX3s loading on mitotic chromosomes and the importance of this localization pattern in normal development. Our findings suggest that PAX3 WS mutants interfere with the normal functions of PAX3 in a dominant negative manner, which is important to the understanding of the pathogenesis of Waardenburg syndrome.
Introduction
PAX33 is a member of the paired box (PAX) family of transcription factors (1). It contains two DNA binding domains, paired domain (PD) and homeodomain (HD), and an uncharacterized C-terminal domain (CTD) (2, 3). PAX3 binds gene promoters, regulates transcription, and controls differentiation in linages of muscle cells and melanocytes (4–7). In addition to regulating coding genes, PAX3 also targets α-satellite repeats of centromeres and mediates the formation of heterochromatin there (8). This indicates the function of PAX3 can be extended to the regulation of chromatin structure rather than limited to the regulation of expression of protein-coding genes. Moreover, a recent study using an siRNA screen looking for factors important in cell cycle progression through the mitotic phase identified that PAX3 is required for mitotic progression in HeLa cells (9). Loss of PAX3 results in cell cycle arrest at M phase (9). This finding extends the functions of PAX3 from transcriptional regulation in interphase to cell cycle progression through mitosis. Taken together, PAX3 has a critical role in mitosis in addition to being a transcription factor affecting the expression of coding and non-coding genes. However, the function of PAX3 in mitosis remains unknown.
PAX3 gene mutations are associated with Waardenburg syndrome (WS) (10). WS mutations of PAX3 mostly exist in the two DNA binding domains (PD and HD) as nonsense or missense mutations, creating proteins that are truncated or full-length with amino acid substitutions (11). Genetic approaches showed that mice harboring a missense mutation of PAX3 have symptoms of WS (12), further linking the association of PAX3 mutations and WS. However, how mutant PAX3 proteins contribute to the pathogenesis of WS remains disputed. Structural analysis confirmed that amino acid residues of PAX3 mutated in WS patients are the same residues that make contact with DNA or those that affect the folding of these binding domains (13). Therefore, it has been thought that Waardenburg syndrome results from a loss of function in the DNA binding activity of mutant PAX3, which impairs its function as a transcription factor. Using PAX3 WS missense mutants in both PD and HD, Corry et al. (14) found that these PAX3 WS mutants exhibit differences in subnuclear distribution and mobility during interphase of the cell cycle, which is not correlated to their DNA-binding activity. This finding suggests factors in addition to PAX3 DNA binding activity that have direct correlations with WS. However, with a lack of direct and systematic analysis of these mutants, whether there is a direct association between DNA binding activity and function of PAX3 remains unclear. Furthermore, whether PAX3 WS mutants lose functions in cells also remains to be determined. As mentioned above, the recent discovery that PAX3 possesses functions that are unrelated to transcription but critical in mitosis suggests that careful analysis of the consequences of WS mutants in mitosis might clarify the relationship between PAX3 mutants and WS.
In this report we found that PAX3 was located on mitotic chromosomes. PAX3 WS mutants in HD lost the association with mitotic chromosomes. Moreover, loading of PAX3 on mitotic chromosomes required the arginine methyltransferase activity of Protein arginine methyltransferase 5 (PRMT5). Finally, this mitotic loading of PAX3 was required for normal cell cycle progression, cell proliferation, and development of zebrafish embryos, whereas PAX3 WS mutants block these cellular and developmental processes. Our results demonstrate the significance of the mitotic chromosome localization of PAX3 and the association of abnormal localization of PAX3 with WS.
Experimental Procedures
Expression Plasmids
PAX3 cDNA was cloned into pcDNA3-FLAG, pEGFP-C1, and mCherry (Clontech, CA) to make FLAG-tagged, GFP-tagged, and red fluorescent protein-tagged PAX3. FLAG-PAX3(R271G) was made by inserting two fragments from FLAG-PAX3 and PAX3(R271G)-GFP into HindIII/XbaI-digested pcDNA3-FLAG. PAX3 point mutations were created using the QuikChange site-directed mutagenesis kit (Agilent Technologies, CA) and confirmed by sequencing. Deletional mutations of PAX3 were made by PCR and inserted into pEGFP. cDNAs of PRMT5 and PARP1 were cloned into pcDNA3-FLAG and pcDNA3.1-HA. The PRMT5Δ cDNA, which encodes a mutant PRMT5 protein with a deletion spanning amino acids 365–369 (GAGRG) (15), was created by overlap-PCR and then subcloned into pcDNA3-FLAG. cDNA of histone H2A was cloned into pGSTag. cDNAs of JMJD6 and JMJD6(m), which is a triple point-mutant (H187A, D189A, and H273A), were subcloned into pEGFP or pcDNA3-FLAG. cDNAs of JMJD6 and JMJD6(m) were from GST-JMJD6 and GST-JMJD6(m), which were gifts from Richard K. Bruick(16). The plasmids of PAX3(F45L)-GFP, PAX3(S84F)-GFP, PAX3(V265F)-GFP, and PAX3(R271G)-GFP were gifts from D. Alan Underhill (14). The plasmid for expressing GST-PAX3 has been described (17).
Cell Culture, Transfection, and Treatment
HEK 293 cells were maintained in Dulbecco's modified Eagle's medium (Hyclone) with 10% (v/v) fetal bovine serum (Gibco) and penicillin-streptomycin. Cells were seeded into 60-mm tissue culture dishes 1 day before transfection. Transfection was done through the standard calcium phosphate co-precipitation procedure (18), and the transfection efficiency was at least 60%. To inhibit the enzyme activity of PRMT5, cells were added methylthioadenosine (Sigma) 24 h after transfection to a final concentration of 750 μm (19) and then incubated for another 24 h.
Antibodies
Mouse monoclonal anti-HA (anti-hemagglutinin, H9658) and anti-FLAG M2 (anti-FLAG clone M2, F1804) antibodies were from Sigma. Mouse anti-GFP monoclonal antibody clone 3D8A1B8 (Gm0001-02) was from Abking Biotechnologies (Taipei, Taiwan). Mouse monoclonal antibody specific for PAX3 was obtained from the Developmental Studies Hybridoma Bank, NICHD, National Institutes of Health/University of Iowa, which was developed by C. Ordahl (University of California, San Francisco) (14). Mouse monoclonal antibodies against PAX3 and RNA polymerase II used in fractionated mitotic chromosome preparations were from Enzo Life Sciences and Santa Cruz, respectively. For detection of arginine methylation, anti-mono and -dimethyl arginine antibody (7E6, ChIP Grade, Abcam, MA) was used. For immunostaining, rhodamine (TRITC)-conjugated goat anti-mouse IgG (115-025-003) was from Jackson ImmunoResearch Laboratories.
Co-immunoprecipitation
Co-immunoprecipitation procedure has been described (17). Immunoprecipitation of FLAG-tagged proteins was carried out using anti-FLAG M2 affinity gel (Sigma) following the manufacturer's suggestions. Western blot analyses were performed using standard protocols.
Cell-cycle Analysis
Transfected HEK 293 cells were grown in 100-mm cell culture dishes for 24, 48, 72, and 96 h. Cells were harvested and fixed in ice-cold 70% (v/v) ethanol. On the day of analysis, ethanol was thoroughly removed from the fixed cells. and the cells were stained in 400 μl of propidium iodide staining solution (10 μg/ml propidium iodide, 100 μg/ml RNase A, and 0.1% Triton X-100 in phosphate-buffered saline) in the dark at room temperature for 30 min. Cell suspensions were filtered to obtain single cells, and 10,000 cells were passed through a flow cytometer (FACSCalibur; BD Biosciences). GFP excitation was performed at 488 nm, and emission was measured at 507 nm. Propidium iodide excitation was performed at 536 nm, and emission was measured at 617 nm. Results were analyzed with the Flowjo V9.7.5 software (TreeStar Inc., Ashland, OR) with gating on GFP expression to evaluate the percentages of cells in G0/G1, S, G2/M, and sub-G2/M (ploidy >4n) phases.
Protein Complex Purification and Liquid Chromatography/Tandem Mass Spectrometry Analysis
HEK 293 cells were transfected with FLAG-PAX3. The procedure of protein complex purification has been described elsewhere (20). A mock complex was purified simultaneously from HEK 293 cells transfected with the empty FLAG vector as control. Mass spectrometric analysis was performed essentially as described in Hsieh et al. (21).
Indirect Immunofluorescence and Confocal Laser Microscopy
HEK 293 cells were grown on glass coverslips in 6-cm dishes and transfected as described (18). After 48 h, cells were fixed with 10% formaldehyde or cold ethanol containing 5% (v/v) acetic acid in the case of GFP-HD-expressing cells. For immunofluorescence of endogenous proteins and FLAG-tagged proteins, cells were incubated with primary antibodies and labeled with rhodamine (TRITC)-conjugated secondary antibodies. DAPI (4′,6-diamidino-2-phenylindole; SouthernBiotech, Birmingham, AL) was used to visualize the distribution of mitotic chromosome. Fluorescence microscopy was carried out with an upright epifluorescence microscope, Olympus A BX51, using an UplanFL 100X objective (Tokyo, Japan). Confocal microscopy analysis was carried out with an Olympus FluoView FV1000 laser scanning microscope using a 63 × 1.4 numerical aperture objective (Tokyo, Japan).
Isolation and Fractionation of Mitotic Chromosomes
B16F10 cells were synchronized at M phase with nocodazole and harvested by shake-off. Harvested cells were fractionated into chromatin and supernatant fractions as described (22).
RNA Interference
Small hairpin RNA (shRNA) constructs of pSM2-PRMT5 (RMM1766-9107100), pSM2-YY1 (V2HS_219592), pGIPZ-PARP1 (RHS4430-98912126), and pGIPZ-PAX3 (V3LMM_507115) were obtained from GenDiscovery Biotechnology (Huntsville, AL). The target sequences of PRMT5, YY1, PARP1, and PAX3 shRNA are ACGTTTCAAGAGGGAGTTCATT, CATTAAAGCAGCGTATC, GACCTGATCTGGAACATCA, and AGAAAATTGAGGAATACAA, respectively. shRNA was delivered into cells by calcium phosphate-mediated transfection.
Chromosome Spreading
Transfected HEK 293 cells were synchronized in M phase by treating with nocodazole (100 ng/ml; Sigma) for 24 h. Cells were harvested and suspended in a hypotonic solution (0.2% KCl, 0.2% sodium citrate) at 37 °C for 30 min. Cells were fixed in 10% formaldehyde for 1 min and spread on glass slide. Cells were subsequently fixed for 9 min and mounted with DAPI mounting solution.
In Vitro Arginine Methylation Assay
FLAG-PRMT5 was expressed in HEK 293 cells and purified by anti-FLAG immunoprecipitation. GST-PAX3, GST-H2A, and GST proteins were expressed from Escherichia coli strain BL21. Purified FLAG-PRMT5 was mixed with GST fusion proteins in the presence of S-(5′-Adenosyl)-l-methionine p-toluene sulfonate salt (Sigma), and the reaction was allowed to proceed at 35 °C for 1 h. Reaction mixtures were then immunoprecipitated with anti-mono/dimethyl arginine antibody (7E6) and analyzed by Western blotting.
Zebrafish Maintenance and Microinjection
Zebrafish (Danio rerio) AB strain was acquired from Taiwan Zebrafish Core Facility at Academia Sinica (TZCAS) and maintained in 20-liter aquariums with filtered fresh water and aeration at 28 °C. The photoperiod is 14 h of light followed by 10 h of darkness in standard laboratory condition (23). On the day of injection, 100 ng/μl DNA were mixed with 0.05% phenol red of working solutions. Embryos were obtained by natural mating and injecting 4∼5 nl of the working solution containing DNA into the egg yolk of the zygote immediately after fertilization (24). A typical group of microinjection contained 100 embryos and maintained in 96-well plates for individual observation. The morphology and the expression fluorescence of the embryos were recorded at 2 day post-fertilization.
Results
PAX3 Associated with Mitotic Chromosomes Using the Homeodomain
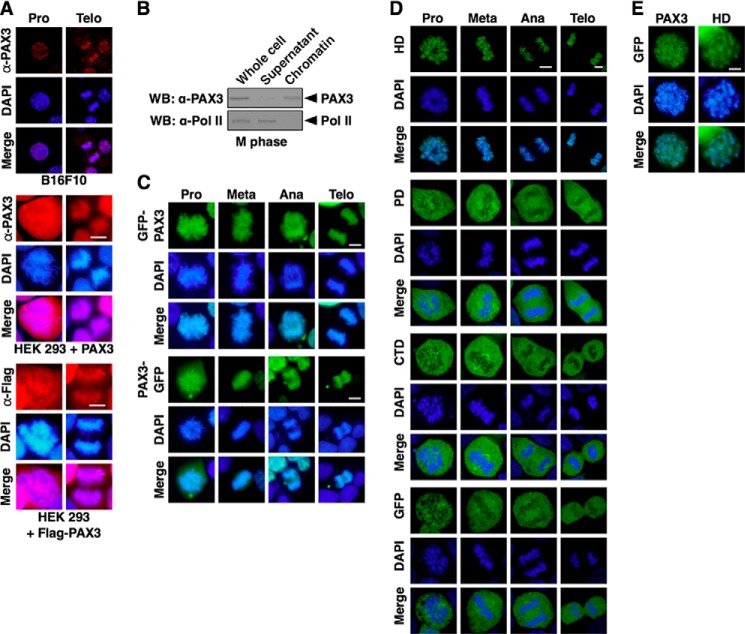
To test if PAX3 associates with the mitotic chromosome, we visually determined the localization of PAX3 by immunofluorescence. As shown in Fig. 1A, endogenous PAX3 was detected on mitotic chromosomes in B16F10 cells using an anti-PAX3 antibody. In HEK 293 cells, which express little endogenous PAX3, exogenously expressed PAX3 was also detected on mitotic chromosomes using the same anti-PAX3 antibody. This association with mitotic chromosomes was further confirmed using FLAG-tagged PAX3 in cells (Fig. 1A) to eliminate the possibility of cross-reactivity with the anti-PAX3 antibody. Using PAX3 tagged with GFP at either the N terminus or the C terminus, we confirmed that PAX3 consistently located on mitotic chromosomes in each stage of mitosis (Fig. 1B). Furthermore, we determined the domain of PAX3 responsible for the association with mitotic chromosomes was the HD, not the PD or the C-terminal domain (Fig. 1C). HD alone exhibited the same localization pattern as the full-length PAX3 in chromatin spreading assay (Fig. 1D). These results demonstrate that PAX3 associates with mitotic chromosomes through its DNA binding domain, HD.
FIGURE 1.
PAX3 used the HD to load on mitotic chromosomes. A, PAX3 binds mitotic chromosomes. Subcellular localization of endogenous PAX3 in B16F10 cells was assessed by confocal microscopy after staining with anti-PAX3 antibodies (red) and DAPI. Subcellular localization of exogenously expressed PAX3 as a non-tagged protein or a FLAG-tagged protein was also determined by fluorescence microscopy in HEK 293 cells, which express little endogenous PAX3. Pro, prophase; Telo, telophase. B, PAX3 associates with mitotic chromosomes. B16F10 cells were synchronized at M phase and collected. Whole cell lysate was fractionated into chromatin and supernatant fractions. Endogenous PAX3 was analyzed by Western blotting (WB) using anti-PAX3 antibody. RNA polymerase II (Pol II) served as the negative control. of the total whole cell lysate was loaded in the lane marked Whole cell. C, GFP-PAX3 and PAX3-GFP load on mitotic chromosome. HEK 293 cells were transfected with the GFP-tagged PAX3 expression vectors in which GFP was fused at the N terminus or C terminus of PAX3. Mitotic chromosome localization in asynchronous cells was determined by fluorescence microscopy. Images were taken from the cells in four subphases of mitosis (Pro, prophase; Meta, metaphase; Ana, anaphase; Telo, telophase). All of the cells that expressed GFP-tagged PAX3 and were in the same subphase showed the same localization pattern. D, HD of PAX3, but not PD or CTD, loaded on mitotic chromosomes. HEK 293 cells were transfected with GFP-HD, GFP-PD, GFP-CTD, or GFP vector. Localization to mitotic chromosomes was determined by confocal microscopy, and all of the cells examined showed the same localization pattern. E, PAX3 and HD coat mitotic chromosomes evenly. HEK 293 cells transfected with GFP-PAX3 or GFP-HD were synchronized at M phase. Localization of PAX3 or HD was examined in chromosome spreading assay.
PAX3 HD Mutants Found in Waardenburg Syndrome Fail to Bind Mitotic Chromosomes
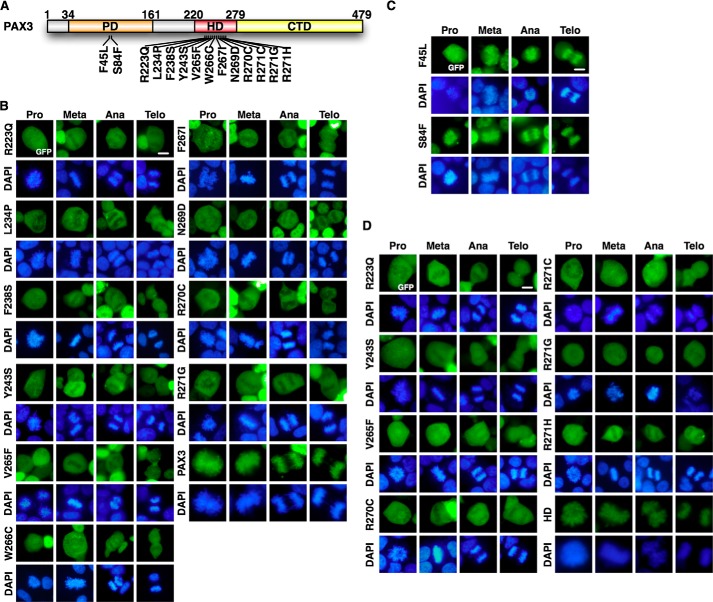
Mutant PAX3 proteins found in patients of WS have mutations in the PD or the HD (Fig. 2A). To determine if the ability of PAX3 to load on mitotic chromosomes correlated with WS, we examined the mitotic localization of PAX3 WS mutants. As shown in Fig. 2B, PAX3 WS mutants that had mutations in HD lost the ability to bind mitotic chromosomes. However, PAX3 WS mutants that had mutations in PD retained their association with mitotic chromosomes (Fig. 2C). Using PAX3 HD alone to rule out potential interference from other domains of PAX3, we confirmed that mutations in HD found in WS prevented binding of PAX3 to the mitotic chromosome (Fig. 2D). These results demonstrate that abnormal PAX3 proteins from Waardenburg syndrome that have mutations in HD lose the ability to load on mitotic chromosomes.
FIGURE 2.
PAX3 HD mutants found in WS lost the ability to load on mitotic chromosomes. A, schematic diagram of PAX3 mutations found in WS patients. B, PAX3 WS mutants in HD do not load on mitotic chromosomes. The expression vectors for GFP-PAX3 WS mutants in HD were transfected in HEK 293 cells. Localization to the mitotic chromosome was determined by fluorescence microscopy. GFP-PAX3 served as control. Meta, metaphase; Ana, anaphase; Telo, telophase. C, PAX3 WS mutants in PD retained the ability to load on mitotic chromosomes. The expression vectors for GFP-PAX3 WS mutants in PD were transfected in HEK 293 cells. Localization to the mitotic chromosome was determined by fluorescence microscopy. D, HD of PAX3 with WS mutations lost mitotic chromosome localization. GFP-tagged HDs harboring mutations found in WS patients were expressed and analyzed for chromosomal localization as described in panels B and C. GFP-HD served as the positive control.
Loading of PAX3 on Mitotic Chromosomes Required the Arginine Methyltransferase Activity of PRMT5
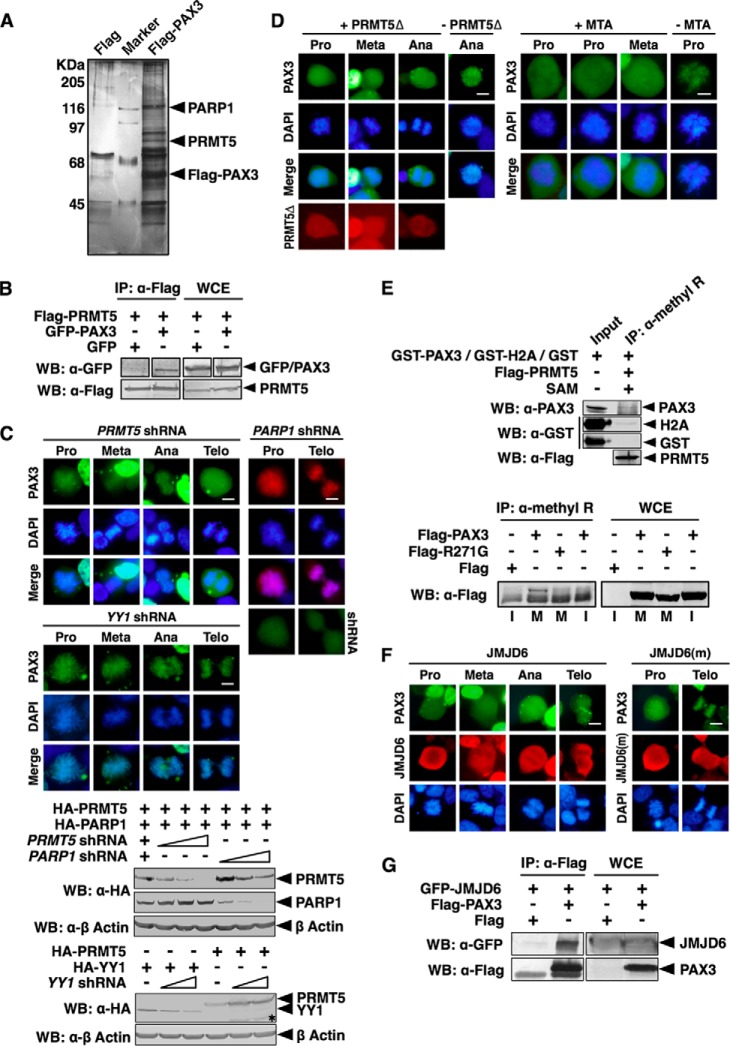
To understand the mechanism of how PAX3 loaded on mitotic chromosome, we used an immunoaffinity purification strategy to look for proteins that form a complex with PAX3. FLAG-PAX3 expressed in HEK 293 cells was immobilized on an anti-FLAG affinity column. Proteins that stably associated with FLAG-PAX3 were eluted and identified by mass spectrometry. PRMT5, which catalyzes methylation of arginine residues in proteins, was identified as a member of the PAX3 complex (Fig. 3A). Poly(ADP-ribose) polymerase 1 (PARP1) was another member of the PAX3 complex (Fig. 3A) (25). PRMT5 was confirmed to specifically interact with PAX3 in co-immunoprecipitation assay (Fig. 3B). Knocking down PRMT5 using shRNA blocked the mitotic chromosome localization of PAX3, whereas knocking down PARP1, a member of the PAX3 protein complex (Fig. 3A), or YY1, an unrelated transcription factor, had no effect (Fig. 3C), suggesting that PRMT5 is specifically required for loading of PAX3 on mitotic chromosomes. A dominant negative mutant of PRMT5 (15) that lost the methyltransferase activity blocked the mitotic chromosome localization of PAX3 (Fig. 3D). Methylthioadenosine (MTA), an inhibitor of PRMT5, also blocked the mitotic chromosome localization of PAX3 (Fig. 3D), confirming that the methyltransferase activity of PRMT5 is required for loading PAX3 on mitotic chromosomes. In a modified in vitro methylation assay using purified FLAG-PRMT5 as enzyme and S-adenosylmethionine as methyl donor, GST-PAX3 could be immunoprecipitated by an antibody specific for arginine methylation (Fig. 3E, upper panel). Using the same anti-methylarginine antibody, we were able to pull down PAX3 from the lysate of cells synchronized at M phase but were not able to do so from cell lysates at interphase (Fig. 3E, lower panel). R271G, one of the PAX3 mutants found in WS that had mutations in HD, could not be pulled down by the anti-methylarginine antibody from M phase lysates (Fig. 3E), further suggesting that arginine methylation is required for mitotic chromosome loading of PAX3. Jumonji domain-containing 6 (JMJD6), which is an arginine demethylase, blocked the mitotic chromosome localization of PAX3, but a mutant of JMJD6 (16) without the demethylase activity had no effect on the localization of PAX3 (Fig. 3F), confirming the importance of methylation to PAX3 localization in M phase. We also confirmed that JMJD6 interacted with PAX3 (Fig. 3G). However, JMJD6 is not a component of the PAX3 protein complex (Fig. 3A), suggesting that the interaction between JMJD6 and PAX3 is not stable enough to survive immunoaffinity purification. Taken together, these results demonstrate that PAX3 is a mitotic phase-specific protein, and arginine methylation is required for the mitotic chromosome localization of PAX3, which is regulated by PRMP5 and JMJD6.
FIGURE 3.
Loading of PAX3 on mitotic chromosomes required arginine methyltransferase activity of PRMT5. A, PRMT5 is a component of the PAX3 protein complex. A FLAG-PAX3 protein complex was purified from HEK 293 cells as described under “Experimental Procedures.” Components of the PAX3 protein complex were separated by SDS-PAGE, visualized by silver staining, and identified by LC-MS/MS. B, PAX3 interacts with PRMT5 in a coimmunoprecipitation assay. + indicates the plasmids transfected into HEK 293 cells. Whole cell extracts (WCE) and anti-FLAG immunoprecipitates (IP: α-FLAG) were immunoblotted by anti-FLAG and anti-GFP as indicated. WB, Western blot. C, knocking down PRMT5 prevents localization of PAX3 to the mitotic chromosome. PRMT5 or YY1 shRNA was transfected into HEK 293 cells with GFP-PAX3 as indicated. Mitotic chromosome localization of GFP-PAX3 was determined by fluorescence microscopy. Cells expressing PARP1 shRNA and FLAG-PAX3 served as the control, and the expression of the PARP1 shRNA was confirmed by the fluorescence from the GFP tag of the shRNA construct (right panels). Knockdown efficiency of shRNAs was determined by Western blotting. An asterisk indicates nonspecific protein bands. Meta, metaphase; Ana, anaphase; Telo, telophase. D, inhibition of the methyltransferase activity of PRMT5 prevents PAX3 from binding to the mitotic chromosome. Left panels, FLAG-PRMT5Δ (a PRMT5 dominant negative mutant) and GFP-PAX3 were transfected into HEK 293 cells. Mitotic chromosome localization of GFP-PAX3 was determined by fluorescence microscopy. Right panels, HEK 293 cells expressing GFP-PAX3 were treated with the PRMT enzyme inhibitor methylthioadenosine (MTA) for 24 h before analysis of localization of PAX3. E, PAX3 can be detected by an anti-methylarginine antibody specifically during mitosis. Upper panel, in vitro methylation reactions containing GST fusion proteins (GST-PAX3, GST-H2A, or GST) purified from HEK 293 cells, FLAG-PRMT5 (also purified from HEK 293 cells), and S-(5′-adenosyl)-l-methionine (SAM) were immunoprecipitated with an anti-methylarginine antibody and then blotted with anti-PAX3, anti-GST, or anti-FLAG antibodies. Lower panel, HEK 293 cells were transfected with expression vectors indicated and then synchronized. Anti-methylarginine immunoprecipitates from transfected cells synchronized at mitotic phase (M) or interphase (I) was blotted with the anti-FLAG antibody. FLAG-R271G, FLAG-tagged PAX3 with the R271G mutation in HD. F, arginine demethylase JMJD6 pulls PAX3 from mitotic chromosomes. FLAG-JMJD6 or FLAG-JMJD6(m) (a mutant JMJD6 without enzyme activity) was transfected with GFP-PAX3 into HEK 293 cells. Mitotic chromosome localization of GFP-PAX3 was determined by fluorescence microscopy. G, JMJD6 interacted with PAX3. HEK 293 cells were transfected with plasmids as indicated. Anti-FLAG immunoprecipitates (α-FLAG) and whole cell extracts were immunoblotted with anti-FLAG and anti-GFP antibodies as indicated.
Cell Proliferation Required Proper Loading of PAX3 on Mitotic Chromosomes
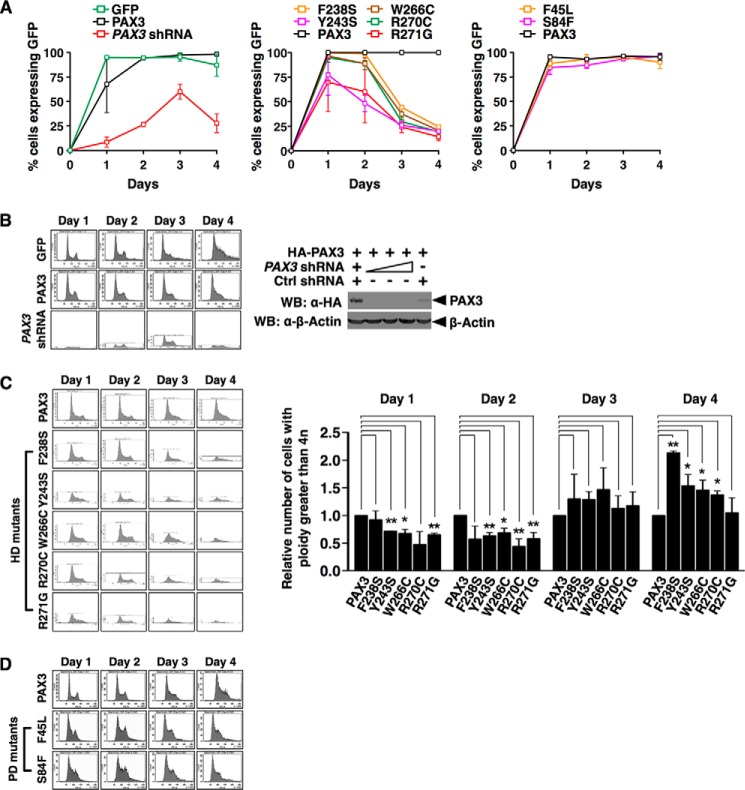
We found that cells expressing PAX3 shRNA or PAX3 WS mutants in HD proliferated slower than cells expressing wild type PAX3, but PAX3 WS mutations in PD had no effect on the proliferation of cells (Fig. 4A). To determine if prevention of mitotic chromosome retention of PAX3 caused a decrease in cell proliferation due to abnormalities in cell cycle progression, we counted the number of cells expressing different PAX3 mutants at different cell cycle stages. Loss of PAX3 in HEK 293 caused an accumulation of cells at S/G2/M phases 4 days after transfection of the shRNA, whereas expression of GFP or GFP-PAX3 had no effect (Fig. 4B), suggesting PAX3 is required for cell cycle progression through mitosis. Expression of PAX3 WS mutants in HD (F238S, Y243S, W266C, R270C, and R271G) also blocked cell cycle progression through S/G2/M phases (Fig. 4C, left panel) and resulted in an increased proportion of cells having a ploidy greater than 4n (Fig. 4C, right panel), which demonstrated that mutations in HD possess a dominant negative effect on cell cycle progression. However, expression of PAX3 WS mutants in PD (F45L and S84F) had no effect compared with expression of wild type PAX3 or R271G (Fig. 4D). Our results show that binding of PAX3 to the mitotic chromosome is required for cellular proliferation and proper cell cycle progression.
FIGURE 4.
Loading of PAX3 on mitotic chromosomes was required for proper progression of the cell division cycle and cell proliferation. A, mutations in PAX3 HD, but not in PD, block cell proliferation. HEK 293 cells were transfected with expression vectors indicated. The PAX3 shRNA construct also expressed GFP. The percentages of cells expressing the GFP tag were shown as the means ± S.D. from three sets of experiments. B, knocking down PAX3 blocks cell cycle progression. Left panel, transfected cells were stained with propidium iodide and analyzed for their DNA content using FACS analysis. Results presented here are representatives from at least three independent experiments. Right panel, immunoblot showing PAX3 shRNA effectively knocked down expression of PAX3. WB, Western blot. C, mutations in HD of PAX3 block cell cycle progression. Left panel, FACS analysis was performed on HEK 293 cells expressing GFP-PAX3 HD mutants (F238S, Y243S, W266C, R270C, or R271G) as in B. Right panel, percentages of cells expressing GFP-PAX3 or GFP-PAX3 mutants that had a ploidy greater than 4n were plotted. Statistical analysis was performed using Student's t test. *, p < 0.05; **, p < 0.01; error bar, S.D.; n = 3. D, mutations in PD of PAX3 had no effect on cell cycle progression. FACS analysis was performed on HEK 293 cells expressing GFP-PAX3 PD mutants (F45L or S84F) as in B.
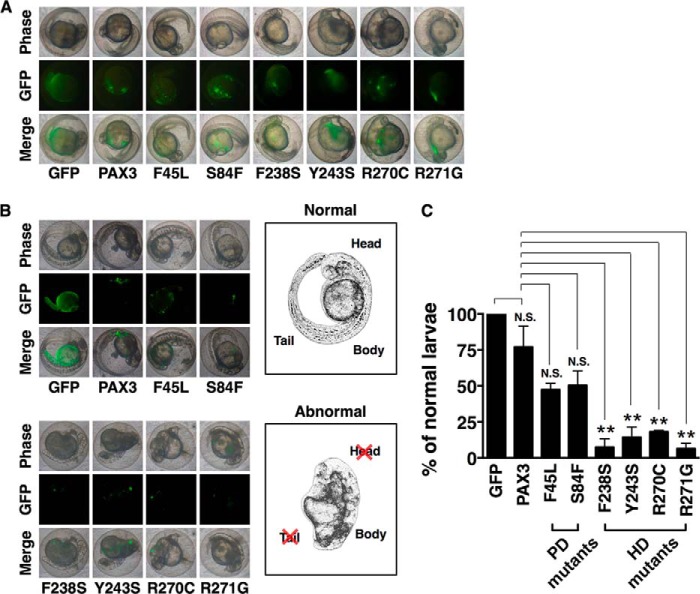
PAX3 Mutants in HD Caused Abnormal Development in Zebrafish Larvae
To determine if proper localization of PAX3 to the mitotic chromosome was essential for normal larval development in zebrafish, we injected expression vectors for wild type and mutant PAX3 into one-cell embryos of zebrafish and observed the developmental progress (Fig. 5, A and B). Larvae that had been injected with wild type PAX3 or the GFP control developed normally (Fig. 5B, upper left panels). Larvae injected with PAX3 PD mutants (F45L or S84F) also developed normally (Fig. 5B, upper left panels), although the number of viable larvae was reduced compared with that of larvae expressing PAX3 or GFP control (data not shown). On the other hand, the head and the tail of the larvae injected with PAX3 HD mutants (F238S, Y243S, R270C, or R271G) were underdeveloped (Fig. 5B, lower left panels; also see drawings on the right for comparison of morphology), and the number of normal larvae was significantly reduced (Fig. 5C). This result demonstrates that PAX3 mutations in HD undermine the normal development of zebrafish larvae.
FIGURE 5.
Expression of PAX3 HD mutants impeded normal development of zebrafish larvae. A, expression plasmids for GFP, GFP-PAX3, GFP-PAX3 HD mutants, or GFP-PAX3 PD mutants were microinjected into 100 zebrafish zygotes at one-cell stage. Bright field and fluorescence microscopic images were taken at 24 h post fertilization. B, one-cell zebrafish embryos were microinjected as described in A. Images from bright field and fluorescence microscopy were taken to show the morphology of larva at 2 days post fertilization. Sketches of a normal and an abnormal larva are presented at the right for comparison. C, the percentage of normal larvae that had been microinjected with PAX3 HD mutants was significantly lower than those injected with PAX3 or PAX3 PD mutants. From B, statistical analysis was performed using Student's t test. **, p < 0.01; N.S., non-significant; error bar, S.D.; n = 3.
Discussion
Mitotic Localization of PAX3 and the Association with Waardenburg Syndrome
Our results here demonstrate that PAX3 associates with the mitotic chromosome, but PAX3 mutants found in WS patients with mutations in HD lose the association with the mitotic chromosome (Figs. 1 and 2). The association with mitotic chromosomes is mediated specifically by the HD region, although both PD and HD have been characterized as DNA binding domains. Consistent with this localization pattern, expression of these HD mutants causes a delay in cell cycle progression, but mutants in PD have no effect on cell cycle progression. These results support the hypothesis that a subset of pathogenic mutants of PAX3 causes Waardenburg syndrome through aberrant mitotic localization.
The ability of PAX3 to bind mitotic chromosomes might be a result of changes in protein conformation. When we examined the localization of endogenous PAX3 in B16F10 cells during mitosis, we could see signals consistently in prophase and telophase but not in metaphase and anaphase. In addition, Corry et al. (14) reported that PAX3 is not located on mitotic chromosomes. These inconsistencies most likely arose from failure of particular antibodies to detect PAX3 in immunostaining experiments because PAX3 has a particular conformation. Nevertheless, when we used PAX3 tagged with GFP either on the N terminus or the C terminus, we confirmed that PAX3 associates with mitotic chromosomes. Furthermore, all of the HD mutants we analyzed failed to bind mitotic chromosomes at all times except for full-length PAX3(V265F). Depending on the precise subphase during M phase, 17–67% of the full-length V265F mutant failed to bind mitotic chromosomes. The number rose to 100% when the mutant PAX3 is the HD domain with the V265F mutation, which further suggests that a particular conformation or conformational change is required for the mitotic localization of PAX3. The necessary changes in the conformation of PAX3 to bind mitotic chromosomes might come from protein methylation (see “Discussion” below).
Our results in Fig. 4 show that PAX3 HD mutants exhibit defects in cell proliferation, possibly through a subtle delay in cell cycle progression. Our findings are consistent with a previous report that PAX3 is required for the progression of the cell cycle in HeLa cells (9). In patients of Waardenburg syndrome, usually only one of the two PAX3 alleles is mutated. Considering the fact that one mutant allele is sufficient to cause WS, we reason that PAX3 HD mutations behave in a dominant negative fashion in WS patients. Moreover, exogenous expression of PAX3 HD mutants also cause developmental abnormalities in zebrafish larvae (Fig. 5), which is consistent with the dominant negative characteristics of these mutants from WS. Taken together, our results demonstrate that one mechanism of the pathogenesis of WS is the loss of wild type PAX3 association with mitotic chromosomes due to interference from PAX3 HD mutants. However, whether these effects stem solely from the failure to bind mitotic chromosomes remain unclear. Because PAX3 also regulates gene expression during interphase, further experiments using the Degron system to express PAX3 only in interphase might help answer this question (26). Moreover, the function of PD during mitosis remains unclear. Further structural analysis might be necessary to reveal the answer.
Our finding that failure of mutant PAX3 to bind mitotic chromosomes associates with Waardenburg syndrome by causing cellular and developmental abnormalities offers a new understanding of the mechanism of the disease. Before our finding, it was thought that PAX3 mutants contribute to WS through aberrant regulation of gene expression during interphase. Our finding stresses the importance of proper loading of PAX3 on mitotic chromosomes. Whether PAX3 functions as a true bookmark protein for the maintenance of lineage identity when exiting mitosis or as a cell cycle regulator (27) requires further experimental support and will be investigated in the near future.
The Mechanism of Loading PAX3 onto Mitotic Chromosomes
Our results show that loading of PAX3 onto mitotic chromosomes is regulated dynamically by arginine methylation and demethylation. Furthermore, PAX3 is methylated specifically during mitosis. Preliminary results in our laboratory indicate a conformational change in PAX3 when cells enter mitosis,4 suggesting that arginine methylation changes the conformation of PAX3 during mitosis. Whether this conformation change is through arginine methylation by PRMT5 remains to be further confirmed. On the other hand, it is possible that PRMT5 alters the chromatin structure by modifying histone proteins, allowing PAX3 to bind during mitosis. PRMT5 is known to methylate histone H3, H4, and H2A (28). However, no specific reader domain in PAX3 is required in this scenario, and the binding with chromatin could be mediated by interactions with other proteins. Our results indicate a specific requirement for HD in the loading of PAX3 on mitotic chromosomes, which does not support this hypothesis.
A recent finding that the binding specificity of FOXA1 to chromatin changes from high specificity in interphase to low specificity in mitosis (29) confirms that global binding specificity of proteins like PAX3 or FOXA1 could change according to cell cycle stages. We propose that arginine methylation, which takes place specifically during mitosis, causes the conformation of PAX3 to change, exposing the HD of PAX3 to bind mitotic chromosomes. Further investigation on whether PAX3 is a direct target of PRMT5 and which arginine residue of PAX3 is methylated in mitosis will confirm this model.
Author Contributions
Y.-L. Y. and W.-M. Y. conceived and coordinated the study and wrote the paper. T.-F. W. designed, performed, and analyzed the experiments shown in Figs. 1, 2, 4, and 5. T.-F. W., Y.-L. Y., I.-L. L., C.-C. L., and P.-L. L. designed, performed, and analyzed the experiments shown in Fig. 3. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We greatly appreciate expression plasmids from Richard K. Bruick and D. Alan Underhill. We thank Mei-Ju Hsieh, Yi-Chung Lin, Ya-Chen Liang, Feng-Shu Hsieh, Yi-Ping Kuo, Han-Yu Wang, Yi-Zu Lee, and Tung-Ping Lin for making tagged-expression plasmids. We thank Taiwan Zebrafish Core Facility at Academia Sinica (TZCAS) for providing zebrafish (AB strain). We especially thank Hui-Ying Hsu of School of Medicine, China Medical University and Mei-Chun Liu of Instrument Technology Research Center, Taichung Veterans General Hospital, as well as Hsin-Chi Lan for technical assistance. We also thank Hsin-Chi Lan, Ya-Chen Liang, and Feng-Shu Hsieh for helpful discussion and critical reading of this manuscript.
This work was supported by grants from the Ministry of Science and Technology (NSC 99-2311-B-005-005-MY3 and NSC 102-2311-B-005-005 (to W.-M. Y.); NSC 98-2311-B-468-001-MY3 (to Y.-L. Y.)) and Asia University (103-asia-05; to Y.-L. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
T.-F. Wu, Y.-L. Yao, I.-L. Lai, and W.-M. Yang, unpublished results.
- PAX3
- paired box 3
- WS
- Waardenburg syndrome
- PD
- paired domain
- HD
- homeodomain
- CTD
- C-terminal domain
- PRMT5
- protein arginine methyltransferase 5
- JMJD6
- Jumonji domain-containing 6
- TRITC
- tetramethylrhodamine isothiocyanate
- PARP1
- poly(ADP-ribose) polymerase 1.
References
- 1. Blake J. A., Ziman M. R. (2014) Pax genes: regulators of lineage specification and progenitor cell maintenance. Development 141, 737–751 [DOI] [PubMed] [Google Scholar]
- 2. Vogan K. J., Gros P. (1997) The C-terminal subdomain makes an important contribution to the DNA binding activity of the Pax-3 paired domain. J. Biol. Chem. 272, 28289–28295 [DOI] [PubMed] [Google Scholar]
- 3. Lang D., Powell S. K., Plummer R. S., Young K. P., Ruggeri B. A. (2007) PAX genes: roles in development, pathophysiology, and cancer. Biochem. Pharmacol. 73, 1–14 [DOI] [PubMed] [Google Scholar]
- 4. Maroto M., Reshef R., Münsterberg A. E., Koester S., Goulding M., Lassar A. B. (1997) Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell 89, 139–148 [DOI] [PubMed] [Google Scholar]
- 5. Ridgeway A. G., Skerjanc I. S. (2001) Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J. Biol. Chem. 276, 19033–19039 [DOI] [PubMed] [Google Scholar]
- 6. Lang D., Lu M. M., Huang L., Engleka K. A., Zhang M., Chu E. Y., Lipner S., Skoultchi A., Millar S. E., Epstein J. A. (2005) Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433, 884–887 [DOI] [PubMed] [Google Scholar]
- 7. Kubic J. D., Young K. P., Plummer R. S., Ludvik A. E., Lang D. (2008) Pigmentation PAX-ways: the role of Pax3 in melanogenesis, melanocyte stem cell maintenance, and disease. Pigment Cell Melanoma Res. 21, 627–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bulut-Karslioglu A., Perrera V., Scaranaro M., de la Rosa-Velazquez I. A., van de Nobelen S., Shukeir N., Popow J., Gerle B., Opravil S., Pagani M., Meidhof S., Brabletz T., Manke T., Lachner M., Jenuwein T. (2012) A transcription factor-based mechanism for mouse heterochromatin formation. Nat. Struct. Mol. Biol. 19, 1023–1030 [DOI] [PubMed] [Google Scholar]
- 9. Neumann B., Walter T., Hériché J. K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., Cetin C., Sieckmann F., Pau G., Kabbe R., Wünsche A., Satagopam V., Schmitz M. H., Chapuis C., Gerlich D. W., Schneider R., Eils R., Huber W., Peters J. M., Hyman A. A., Durbin R., Pepperkok R., Ellenberg J. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tassabehji M., Newton V. E., Liu X. Z., Brady A., Donnai D., Krajewska-Walasek M., Murday V., Norman A., Obersztyn E., Reardon W. (1995) The mutational spectrum in Waardenburg syndrome. Hum. Mol. Genet. 4, 2131–2137 [DOI] [PubMed] [Google Scholar]
- 11. Pingault V., Ente D., Dastot-Le Moal F., Goossens M., Marlin S., Bondurand N. (2010) Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 31, 391–406 [DOI] [PubMed] [Google Scholar]
- 12. Epstein D. J., Vekemans M., Gros P. (1991) splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell 67, 767–774 [DOI] [PubMed] [Google Scholar]
- 13. Birrane G., Soni A., Ladias J. A. (2009) Structural basis for DNA recognition by the human PAX3 homeodomain. Biochemistry 48, 1148–1155 [DOI] [PubMed] [Google Scholar]
- 14. Corry G. N., Hendzel M. J., Underhill D. A. (2008) Subnuclear localization and mobility are key indicators of PAX3 dysfunction in Waardenburg syndrome. Hum. Mol. Genet. 17, 1825–1837 [DOI] [PubMed] [Google Scholar]
- 15. Branscombe T. L., Frankel A., Lee J. H., Cook J. R., Yang Z., Pestka S., Clarke S. (2001) PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276, 32971–32976 [DOI] [PubMed] [Google Scholar]
- 16. Chang B., Chen Y., Zhao Y., Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 17. Hsieh M. J., Yao Y. L., Lai I. L., Yang W. M. (2006) Transcriptional repression activity of PAX3 is modulated by competition between corepressor KAP1 and heterochromatin protein 1. Biochem. Biophys. Res. Commun. 349, 573–581 [DOI] [PubMed] [Google Scholar]
- 18. Lai I. L., Lin T. P., Yao Y. L., Lin C. Y., Hsieh M. J., Yang W. M. (2010) Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J. Biol. Chem. 285, 7187–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richard S., Morel M., Cléroux P. (2005) Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem. J. 388, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yao Y. L., Yang W. M. (2003) The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. J. Biol. Chem. 278, 42560–42568 [DOI] [PubMed] [Google Scholar]
- 21. Hsieh F. S., Chen N. T., Yao Y. L., Wang S. Y., Chen J. J., Lai C. C., Yang W. M. (2014) The transcriptional repression activity of STAF65γ is facilitated by promoter tethering and nuclear import of class IIa histone deacetylases. Biochim. Biophys. Acta 1839, 579–591 [DOI] [PubMed] [Google Scholar]
- 22. Burke L. J., Zhang R., Bartkuhn M., Tiwari V. K., Tavoosidana G., Kurukuti S., Weth C., Leers J., Galjart N., Ohlsson R., Renkawitz R. (2005) CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 24, 3291–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Westerfield M. (2000) The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th Ed., University of Oregon Press, Eugene, OR [Google Scholar]
- 24. Jeon H. Y., Lee H. (2013) Depletion of Aurora-A in zebrafish causes growth retardation due to mitotic delay and p53-dependent cell death. FEBS J. 280, 1518–1530 [DOI] [PubMed] [Google Scholar]
- 25. Wu T. F., Yao Y. L., Lai I. L., Lee T. H., Underhill D. A., Yang W. M. (2014) PAX3 Loads onto pericentromeric heterochromatin during S phase through PARP1. Anticancer Res. 34, 4717–4722 [PubMed] [Google Scholar]
- 26. Kadauke S., Udugama M. I., Pawlicki J. M., Achtman J. C., Jain D. P., Cheng Y., Hardison R. C., Blobel G. A. (2012) Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150, 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zaidi S. K., Young D. W., Montecino M. A., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S. (2010) Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat. Rev. Genet. 11, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Lorenzo A., Bedford M. T. (2011) Histone arginine methylation. FEBS Lett. 585, 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caravaca J. M., Donahue G., Becker J. S., He X., Vinson C., Zaret K. S. (2013) Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 27, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]