Background: Flavonoids are naturally occurring compounds and are known to regulate various transcription factors.
Results: A prenylated flavonoid in hops, xanthohumol, improves diet-induced obesity and fatty liver by suppressing SREBP activation.
Conclusion: Xanthohumol is a novel SREBP inactivator.
Significance: Xanthohumol can be used as a nutraceutical food or pharmacological agent for improving metabolic syndrome.
Keywords: COPII, flavonoid, lipid synthesis, metabolic syndrome, transcription factor, hepatic steatosis, sterol regulatory element-binding proteins, xanthohumol
Abstract
Sterol regulatory element-binding proteins (SREBPs) are key transcription factors that stimulate the expression of genes involved in fatty acid and cholesterol biosynthesis. Here, we demonstrate that a prenylated flavonoid in hops, xanthohumol (XN), is a novel SREBP inactivator that reduces the de novo synthesis of fatty acid and cholesterol. XN independently suppressed the maturation of SREBPs of insulin-induced genes in a manner different from sterols. Our results suggest that XN impairs the endoplasmic reticulum-to-Golgi translocation of the SREBP cleavage-activating protein (SCAP)-SREBP complex by binding to Sec23/24 and blocking SCAP/SREBP incorporation into common coated protein II vesicles. Furthermore, in diet-induced obese mice, dietary XN suppressed SREBP-1 target gene expression in the liver accompanied by a reduction of the mature form of hepatic SREBP-1, and it inhibited the development of obesity and hepatic steatosis. Altogether, our data suggest that XN attenuates the function of SREBP-1 by repressing its maturation and that it has the potential of becoming a nutraceutical food or pharmacological agent for improving metabolic syndrome.
Introduction
Metabolic syndrome is characterized by a group of risk factors for cardiovascular disease, including obesity, hyperglycemia, hypertension, dyslipidemia, insulin resistance, and fatty liver (1). This syndrome has been increasing worldwide to become one of the most common health problems. A modern lifestyle and diet contribute to excessive energy intake and low energy expenditure, leading to visceral lipid accumulation. Increasing evidence indicates that poor regulation and control of lipid metabolism promote metabolic syndrome (2).
Lipid biosynthesis in mammals is tightly regulated by a family of transcription factors called sterol regulatory element-binding proteins (SREBPs)3 (3). SREBPs are synthesized as inactive precursors inserted into the endoplasmic reticulum (ER) membrane. To function as transcription factors, these precursors must undergo proteolytic cleavage in the Golgi apparatus, and this process is regulated by sterols. In cells with low sterol levels, SREBP cleavage-activating protein (SCAP) binds to Sec23/24, which clusters the SCAP/SREBP complex into common coated protein II (COP II) vesicles (4). These vesicles then transport the SCAP/SREBP complex from the ER to the Golgi, wherein two proteases, site-1 protease (S1P) and site-2 protease (S2P), sequentially cleave SREBPs (5, 6). Once the mature forms of SREBPs are released from the Golgi, they activate the expression of genes in the nucleus important for fatty acid and cholesterol biosynthesis. When cellular sterol levels are sufficient, SCAP associates with insulin-induced genes (Insigs), which facilitate the retention of the SCAP/SREBP complex in the ER, suppressing the proteolytic processing of SREBPs (7).
There are three major SREBP isoforms in mammals, SREBP-1a, -1c, and -2 (8). SREBP-1a and -1c mainly activate the expression of genes involved in fatty acid synthesis, whereas SREBP-2 preferentially stimulates the expression of genes involved in cholesterol synthesis. Up-regulation of the expression and processing of SREBP-1c has been observed in the liver of obese mice (9, 10), and liver-specific SREBP-1c transgenic mice have displayed hepatic steatosis, hypertriglyceridemia, and insulin resistance (11). These studies indicate that the dysregulation of SREBP-1c contributes to the pathogenesis of these diseases.
In this study, we screened for SREBP modulators in natural food constituents and identified xanthohumol (XN) as an inactivator. XN is the most abundant prenylated flavonoid in hops (12) and is known as a putative cancer chemopreventive agent because of its antioxidant (13) and anti-inflammatory properties (14). Moreover, XN has been shown to inhibit triglyceride synthesis and apolipoprotein B secretion associated with the low activity of diacylglycerol acyltransferase 1 and microsomal triglyceride transfer protein (15). Several reports have recently shown that XN exerts anti-metabolic syndrome effects in vivo (16–19); however, detailed mechanisms contributing to these effects remain to be clarified.
In this study, we demonstrated that XN affects SREBP processing; it interacted with Sec23/24 and blocked the sorting of the SCAP/SREBP complex into COP II vesicles. XN reduced the mature forms of SREBPs and de novo biosynthesis of fatty acid and cholesterol in cultured cells. Furthermore, XN ameliorated obesity and fatty liver in mice fed a high fat diet (HFD), and this is associated with the down-regulation of hepatic SREBP processing.
Experimental Procedures
Materials
Cholesterol, 25-hydroxycholesterol (25-HC), fluvastatin, lipoprotein-deficient serum (LPDS), dialyzed fetal bovine serum (FBS), 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), and brefeldin A (BFA) were purchased from Sigma. Dulbecco's modified Eagle's medium (DMEM), DMEM/Ham's F-12 medium, thapsigargin (Tg), and 8-prenylnaringenin (8-PN) were from Wako (Osaka, Japan). Blasticidin S was from Invitrogen. XN (92.4% pure) and isoxanthohumol (IXN) were from Hopsteiner (Mainburg, Germany). Naringenin (NG) was from LKT Laboratories (St. Paul, MN).
Antibodies
Monoclonal anti-SREBP-1 (2A4), anti-SREBP-1 (H-160), anti-SCAP (9D5), and anti-Sec23 (E-19) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). Monoclonal anti-FLAG (M2) and anti-β-actin (AC-15) antibodies were purchased from Sigma. Monoclonal anti-GM130 (35) antibody was from BD Biosciences. Monoclonal anti-activating transcription factor 6 (ATF6) antibody was from Bio Academia (Osaka, Japan). Furthermore, polyclonal anti-Sec24C, anti-phospho-Akt (Ser-473), anti-phospho-Akt (Thr-308), anti-Akt, anti-phospho-S6K (Thr-389), anti-S6K, anti-phospho-AMP-activated protein kinase (AMPK) (Thr-172), and anti-AMPK antibodies were from Cell Signaling Technology (Beverly, MA). Polyclonal anti-Sec61α antibody was from Millipore (Billerica, MA). Polyclonal anti-SREBP-2 (RS004) antibody has been previously described (20). Peroxidase-conjugated affinity-purified donkey anti-mouse IgG, peroxidase-conjugated affinity-purified donkey anti-rabbit IgG, and Cy3-conjugated affinity-purified donkey anti-mouse IgG were purchased from Jackson ImmunoResearch (West Grove, PA).
Media and Buffers
Medium A contained DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% (v/v) FBS. Medium B contained DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 10% FBS, 50 μm sodium mevalonate, and 12.5 μm fluvastatin. Furthermore, medium C contained DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 5% (v/v) LPDS, 50 μm sodium mevalonate, and 12.5 μm fluvastatin. Medium D contained DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% LPDS. Medium E contained DMEM/Ham's F-12 supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. Medium F contained DMEM/Ham's F-12 supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 5% LPDS, 50 μm sodium mevalonate, and 12.5 μm fluvastatin. Buffer A contained 50 mm Tris-HCl (pH 7.5) and 150 mm NaCl. Buffer B was Buffer A supplemented with a protease inhibitor mixture (Nacalai Tesque, Kyoto, Japan).
Plasmid Constructs
An expression plasmid for SREBP-1c was constructed by inserting fragments coding amino acids 2–463 of human SREBP-1c into pCMV-3×FLAG (Sigma). Expression plasmids for SREBP-1a and SREBP-2 (pCMV-3×FLAG-SREBP-1a(2–487) and pCMV-3×FLAG-SREBP-2 (2–481)) were previously described (21).
Cell Culture
Huh-7 (a human hepatoma cell line) cells were maintained in medium A. Huh-7/FAS-luc (a stable cell line of Huh-7 expressing a luciferase reporter driven by a sterol regulatory element-containing fatty-acid synthase (FAS) promoter) (22) cells were maintained in medium A containing 2 μg/ml blasticidin S. CHO-7 (a Chinese hamster ovarian cell line adapted to grow in LPDS medium), SRD-15 (a CHO-7 cell line deficient in Insig-1 and -2) (23), and CHO/pGFP-SCAP (a stable cell line of SCAP-deficient CHO-7 cells expressing GFP-SCAP) (24) cells were maintained in medium E.
Luciferase Assays
Huh-7/FAS-luc cells were plated in 12-well plates at a density of 1.0 × 105 cells/well and cultured with medium A for 24 h. The cells were then switched to medium B for 16 h. After incubation for another 24 h in the absence or presence of 10 or 30 μm XN, luciferase activity was measured as described previously (25). XN was dissolved in DMSO. The final DMSO concentration of the cultured medium was 0.1%. Normalized luciferase values were determined by dividing luciferase activity by the protein content in cell extracts quantified using the BCA protein assay (Pierce).
Small Interfering RNA (siRNA) Experiments
siRNAs (40 pmol/6-well plate) for human Insig-1 and Insig-2 (sc-44432 and sc-45781, respectively, Santa Cruz Biotechnology) and control (pGL2 luciferase; Bonac) were transfected using Lipofectamine RNAiMAX (Invitrogen) into Huh-7 cells according to the manufacturer's instructions.
Real Time Quantitative PCR
Total RNA was extracted from Huh-7 cells or mouse livers using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. RNA was reverse-transcribed using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Real time quantitative PCR (TaqMan probe and SYBR Green) analysis was performed on StepOnePlus real time PCR systems. Expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or 18S rRNA control. The TaqMan identification numbers for the genes analyzed are as follows: human (h) FAS, Hs00188012_m1; human stearoyl-CoA desaturase 1 (SCD1), Hs00748952_s1; hInsig-1, Hs01650977_g1; hInsig-2, Hs00379223_m1; hGAPDH, 4352934; mouse (m) acetyl-CoA carboxylase 1 (ACC1), Mm01304286_m1; mFAS, Mm00662319_m1; mHMG-CoA synthase (HMGCS), Mm00524111_m1; mSREBP-2, Mm01306283_m1; mouse (ATGL), Mm00503040_m1; and mAdiponectin, Mm00456425_m1. The sequences of the primer sets used were as follows: hACC1, 5′-TGGGCCTCAAGAGGATTTGT-3′ and 5′-TCCACTGTTGGCTGATACATAGATG-3′; hHMGCS, 5′-GACTTGTGCATTCAAACATAGCAA-3′ and 5′-GCTGTAGCAGGGAGTCTTGGTACT-3′; hHMG-CoA reductase (HMGCR), 5′-TACCATGTCAGGGGTACGTC-3′ and 5′-CAAGCCTAGAGACATAATCATC-3′; human squalene synthase, 5′-ATGACCATCAGTGTGGAAAAGAAG-3′ and 5′-CCGCCAGTCTGGTTGGTAA-3′; mSCD1, 5′-CCGGAGACCCCTTAGATCGA-3′ and 5′-TAGCCTGTAAAAGATTTCTGCAAACC-3′ (26); mSREBP-1a, 5′-GAACAGACAGTGGCCGAGAT-3′ and 5′-GGGAGTCACTGTCTTGGTTG-3′; mSREBP-1c, 5′-GAGCCATGGATTGCACATTT-3′ and 5′-CGGGAAGTCACTGTCTTGGT-3′; mHMGCR, 5′-CCGGCAACAACAAGATCTGTG-3′ and 5′-ATGTACAGGATGGCGATGCA-3′; mSQS, 5′-CACACTGGCTGCCTGTTACAA-3 and 5′-CCCCTTCCGAATCTTCACTACTC-3′; mouse ATP-binding cassette subfamily A member 1 (ABCA1), 5′-TCCTCATCCTCGTCATTCAAA-3′ and 5′-GGACTTGGTAGGACGGAACCT-3′; mABCG5, 5′-TGTCCTACAGCGTCAGCAACC-3′ and 5′-GGCCACTCTCGATGTACAAGG-3′; mABCG8, 5′-AGAGTTGCATCCCCCTAGCC-3′ and 5′-TCCTTGACACAGGCATGAAGC-3′; mouse cholesterol 7α-hydroxylase (CYP7A1), 5′-AGCAACTAAACAACCTGCCAGTACTA-3′ and 5′-GTCCGGATATTCAAGGATGCA-3′; mouse peroxisome proliferator-activated receptor (PPAR) α, 5′-CTCGCGTGTGATAAAGC-3′ and 5′-CGATGCTGTCCTCCTTG-3′; mouse acyl-CoA oxidase, 5′-CAGCACTGGTCTCCGTCATG-3′ and 5′-CTCCGGACTACCATCCAAGATG-3′; mouse carnitine palmitoyltransferase 1a (CPT-1a), 5′-TGGGCTACTCAGAGGATGG-3′ and 5′-AAGGTGTCAAATGGGAAGG-3′; mouse hormone-sensitive lipase, 5′-GCTGGGCTGTCAAGCACTGT-3′ and 5′-GTAACTGGGTAGGCTGCCAT-3′; mPPARγ2, 5′-CTCTGTTTTATGCTGTTATGGGTGA-3′ and 5′-GGTCAACAGGAGAATCTCCCAG-3′; mouse tumor necrosis factor (TNF) α, 5′-CAGCCGATGGGTTGTACCTT-3′ and 5′-GGCAGCCTTGTCCCTTGA-3′; mouse uncoupling protein 1 (UCP1), 5′-GGAGGTGTGGCAGTGTTCATTGG-3′ and 5′-AGCATTGTAGGTCCCCGTGTAGCG-3′; mouse deiodinase, iodothyronine, type II (Dio2), 5′-CCACCTTCTTGACTTTGCCA-3′ and 5′-GGTGAGCCTCATCAATGTATAC-3′; and mouse PPARγ coactivator-1 (PGC1α), 5′-TTCTGGGTGGATTGAAGTGGTG-3′ and 5′-TGTCAGTGCATCAAATGAGGGC-3′.
Immunoblotting
Cells and mouse liver were lysed in RIPA buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% (v/v) Triton X-100, 0.5% (w/v) deoxycholate, and 0.1% (w/v) SDS) supplemented with a protease inhibitor mixture. The lysates were subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA), and probed with the antibodies indicated in the figure legends. The immunoreactive proteins were visualized using ECL (GE Healthcare) or Immobilon (Millipore, Billerica, MA) immunoblotting detection reagents. The signals on the membrane were detected by ImageQuant LAS 4000 mini (GE Healthcare).
Measurement of de Novo Fatty Acid and Cholesterol Synthesis
Huh-7 cells were plated in 12-well plates at a density of 2 × 105 cells/well and cultured with medium A for 24 h. The cells were then switched to medium D for 16 h. After incubation for another 18 h in the absence or presence of 10 or 30 μm XN, the cells were placed in fresh medium supplemented with 1.6 μCi/ml [14C]acetate and incubated for 6 h. Furthermore, the cells were washed with phosphate-buffered saline (PBS), scraped, and collected by centrifugation. The cell pellets were mixed with 1 ml of 8 n potassium hydrate and 1 ml of ethanol. The mixture was heated at 100 °C for 2 h and extracted two times with 2 ml of petroleum ether (cholesterol extract). Moreover, 2 ml of the lower aqueous layer was mixed with 1 ml of 12 n hydrochloric acid and then extracted two times with 3 ml of petroleum ether (fatty acid extract). The lipid extracts were then purified by thin layer chromatography on Silica Gel 60 (Merck) and quantified with a BAS2000 image analysis system (Fujifilm, Tokyo, Japan). Normalized fatty acid and cholesterol synthesis rates were determined by dividing the signals of 14C-labeled fatty acid and [14C]cholesterol by the amounts of total cellular protein quantified using the BCA protein assay.
Immunofluorescence Staining and Fluorescence Microscopy
CHO/pGFP-SCAP cells were plated on a poly-d-lysine Cellware 4-well culture slide (BD Biocoat) and cultured with medium E for 48 h. Moreover, the cells were switched to medium F for 3 h. After incubation for another 3 h in the absence or presence of 30 μm XN, the cells were fixed with 3% (w/v) paraformaldehyde in PBS. The cells were permeabilized with 0.2% (v/v) Triton X-100 in PBS, blocked with 2% (w/v) bovine serum albumin in PBS, treated with primary antibody against GM130, and subjected to reaction with Cy3-conjugated secondary antibody. Then the cells were mounted on glass slides with Gel Mount aqueous mounting medium (Sigma). Images were obtained using an FV500 confocal laser scanning microscope (Olympus, Tokyo, Japan).
Cell Fractionation
Cell fractionation was performed according to a previously published procedure (27), with minor modifications. CHO-7 cells were plated in 100-mm dishes at a density of 2 × 106 cells/dish and cultured with medium E for 24 h. The cells were then switched to medium F for 16 h. After incubation for another 3 h in the absence or presence of 30 μm XN, the cells were washed with PBS, scraped, and collected by centrifugation. The cell pellets were resuspended in buffer B supplemented with 15% (w/v) sucrose and passed through a 25-gauge needle 20 times. The disrupted cell suspension was centrifuged at 3,000 × g for 10 min, and the supernatant was diluted to 3 ml with buffer A supplemented with 15% sucrose. A discontinuous sucrose density gradient was prepared in a 13 PET tube (Hitachi Koki, Tokyo, Japan) by layering the following sucrose density solutions in buffer A: 3 ml of 45% sucrose, 5 ml of 30% sucrose, 3 ml of diluted supernatant on 15% sucrose, and 2 ml of 7.5% sucrose. The gradient solution was centrifuged at 24,000 rpm in a Himac CP85β (Hitachi Koki, Tokyo, Japan) for 1 h, and whole cells were separated into two fractions, light (14–18%) and heavy (34–38%). The light fraction contained the Golgi apparatus, plasma membrane, and endosome. The heavy fraction contained the ER and peroxisome. Aliquots of each fraction were subjected to immunoblotting to examine the protein levels of SCAP and SREBPs.
Detection of Binding Proteins for XN Beads
The preparation of XN beads and binding reactions were performed as described previously (28). CHO-7 cells were plated in 100-mm dishes at a density of 2 × 106 cells/dish and cultured with medium E for 24 h. The cells were then switched to medium F for 16 h. After incubation for another 3 h in the absence or presence of 50 μm XN, the cells were washed with PBS, scraped, and collected by centrifugation. The cell pellets were resuspended in binding buffer (10 mm Tris-HCl (pH 7.6), 50 mm KCl, 5 mm MgCl2, and 1 mm EDTA) supplemented with a protease inhibitor mixture and passed through a 25-gauge needle 20 times. The disrupted cell suspension was centrifuged at 15,000 × g for 1 min, and the supernatant was collected. The cell lysate was precleared by incubation with control beads at 4 °C for 1 h and then incubated with control or XN beads at 4 °C for 12 h. The pelleted beads were washed three times with binding buffer, and the bound proteins were eluted with SDS sample buffer and were subjected to immunoblotting.
Immunoprecipitation
CHO-7 cells were plated in 100-mm dishes at a density of 2 × 106 cells/dish and cultured with medium E for 24 h. The cells were then switched to medium F for 16 h. After incubation for another 3 h in the absence or presence of 30 μm XN, the cells were harvested and lysed with Nonidet P-40 lysis buffer (50 mm Hepes-KOH (pH 7.6), 100 mm NaCl, 1.5 mm MgCl2, 1% (v/v) Nonidet P-40, 1 mm DTT) supplemented with a protease inhibitor mixture. The cell lysate was passed through a 25-gauge needle 20 times, rotated at 4 °C for 1.5 h, and centrifuged at 20,000 × g for 30 min. The supernatant was rotated with anti-SCAP (9D5) at 4 °C for 1.5 h, and then protein A-Sepharose beads (GE Healthcare) were added. After rotation for 1.5 h, the pelleted Sepharose was washed three times with Nonidet P-40 lysis buffer, and the bound proteins were eluted with SDS sample buffer and were subjected to immunoblotting.
Mice and Diet
All experiments were performed according to the guidelines of the Animal Usage Committee of The University of Tokyo. Six-week-old C57BL/6J mice purchased from Clea Japan (Tokyo, Japan) were housed in the animal care facility under controlled conditions (12-h light/dark cycle, 23 °C). The mice were provided free access to water and a standard chow diet (Labo MR Stock; Nosan Corp. Bio Department, Kanagawa, Japan) for the 1st week following their arrival.
Oral Administration of XN to Mice
Mice were randomized into three groups (n = 5/group). Vehicle, 75 or 150 mg/kg body weight of XN, was administered to the mice by oral gavage once daily. After a 3-day treatment, livers were isolated, frozen in liquid nitrogen, and stored at −80 °C until they were analyzed.
Administration of XN-supplemented HFD to Mice
HFD (60% energy supplied by fat; D12492) was purchased from Research Diet (New Brunswick, NJ). After feeding the HFD for 10 weeks, the mice were divided into three groups with similar average body weight and blood glucose levels (n = 10–11/group). The mice were fed a HFD, 0.2% (w/w) or 0.4% (w/w) XN-supplemented HFD for 50 days. The body weight and food intake were measured every 2 days. Mice were then sacrificed, and their blood and liver were collected. Blood was collected in tubes with 0.1 m EDTA and centrifuged at 3,000 rpm for 30 min to obtain plasma. Livers were rapidly excised, frozen in liquid nitrogen, and stored at −80 °C until further analysis.
Plasma Measurements
Plasma glucose, triglyceride (TG), and cholesterol concentrations were determined using kits purchased from Wako (Osaka, Japan). Plasma insulin and C-peptide concentrations were determined using an ELISA kit (Shibayagi, Gunma, Japan). Plasma lipoprotein profile was analyzed by LipoSEARCH (Skylight Biotech, Tokyo, Japan).
Measurement of TG and Cholesterol in Liver
Mouse livers (100–150 mg) were homogenized in 4-ml of ice-cold chloroform/methanol (2:1, v/v) and incubated for 30 min at room temperature. Then 1 ml of 50 mm NaCl was added, and the mixture was centrifuged at 1,500 × g for 30 min. The lower organic layer was collected, and 1-ml 0.36 m CaCl2/methanol (1:1, v/v) was added. This mixture was then centrifuged at 1,500 × g for 10 min, and the upper aqueous layer was removed. The lower organic layer was washed again with 1-ml 0.36 m CaCl2/methanol (1:1, v/v) and diluted up to 5 ml with chloroform. Aliquots of the lipid extract (20 μl for TG determination and 500 μl for cholesterol) contained Triton X-100/chloroform (1:1, v/v) and were then air-dried. TG and cholesterol concentrations were determined using kits purchased from Wako (Osaka, Japan).
Measurement of TG in Feces
Dried mouse feces were homogenized and mixed with 25 ml of ethanol. The mixture was heated at 65 °C for 2 h and filtered. The filtrate was diluted up to 25 ml with ethanol, and 5 ml of the fecal extract was evaporated to dryness under reduced pressure to remove ethanol. Isopropyl alcohol (2 ml) was added to the pellet (lipid extract), and TG concentration was determined using kits purchased from Wako.
Computed Tomography (CT) Scan Analysis
Mice were anesthetized with isoflurane and scanned with an x-ray CT scanner designed for experimental animals (LaTheta LCT-200; Hitachi Aloka Medical, Tokyo, Japan). Contiguous slice images were obtained at 2-mm intervals in the abdominal region (from the diaphragm to the bottom edge of the pubic bone) and at 0.1-mm intervals in the liver. The percentage of fat area in the abdomen and liver and the abdominal visceral and subcutaneous fat volumes were calculated using LaTheta software (version 3.00).
Oral Glucose Tolerance Test (OGTT)
After a 16-h fasting, glucose (2 g/kg body weight) was administered to mice by oral gavage. Blood glucose levels were measured from a nick in a tail vein before and at 15, 30, 60, 90, and 120 min after administration using a hand-held glucometer (Ascensia Breeze 2; Bayer Yakuhin, Osaka, Japan).
Statistical Analysis
All data are represented as mean ± S.E. Statistical analysis was performed using the Ekuseru-Toukei Version 2.0 (Social Survey Research Information). One-way analysis of variance followed by the Bonferroni procedure was used to compare more than two groups. Differences were considered significant at p < 0.05.
Results
Identification of XN as a New SREBP Inactivator
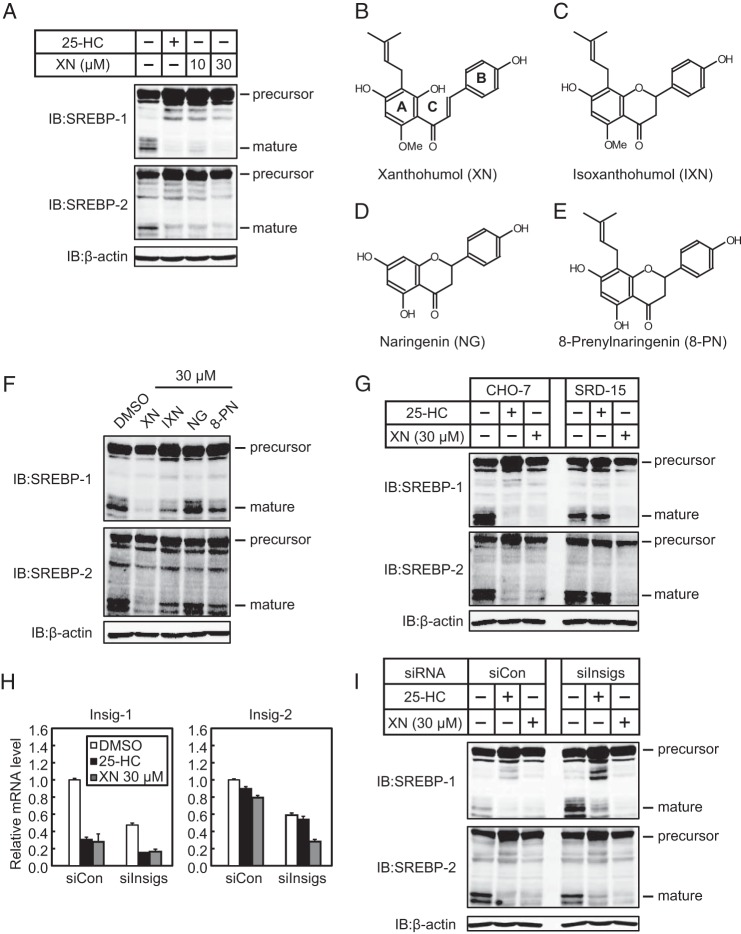
To identify SREBP inactivators, we used a human hepatoma Huh-7 cell line that stably expressed a luciferase reporter gene driven by an SRE-containing FAS promoter (Huh-7/FAS-luc). To increase SREBP activity, the cells were depleted of sterols by incubating them with LPDS and fluvastatin, an agent that inhibits HMGCR and subsequently blocks cholesterol biosynthesis. The cells were then treated with ∼100 naturally occurring food components for identifying potential SREBP inactivators. As a positive control, 25-HC, a well known inhibitor of SREBP processing, was tested and was shown to markedly lower luciferase activity (Fig. 1A). Using this screening system, we identified XN as a potent SREBP inactivator (Fig. 1, A and B).
FIGURE 1.
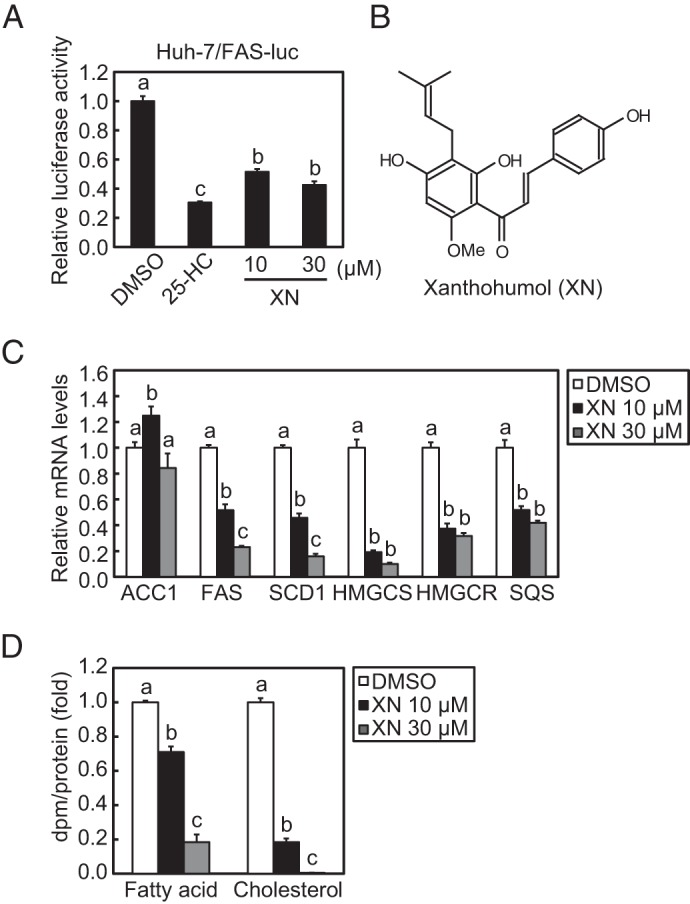
XN reduces SREBP activity and de novo synthesis of fatty acid and cholesterol. A, Huh-7/FAS cells were depleted of sterols by incubating in medium B for 16 h. The cells were then switched to medium B in the presence of vehicle, 1 μg/ml (2.5 μm) 25-HC, 10 μm XN, or 30 μm XN. After incubation for 24 h, luciferase assay was performed, and relative luciferase activity was obtained by normalization to the activity in the presence of the vehicle. B, structure of XN. C, Huh-7 cells were depleted of sterols by incubating in medium C for 16 h. The cells were then switched to medium C in the presence of vehicle, 10 μm XN, or 30 μm XN. After incubation for 24 h, total RNA was isolated from the cells. Real time quantitative PCR was performed, and relative mRNA levels were obtained by normalization to GAPDH mRNA. D, Huh-7 cells were cultured with medium D for 16 h. The cells were then switched to medium D in the presence of vehicle, 10 μm XN, or 30 μm XN. After incubation for 18 h, the cells were treated with 1.6 μCi/ml [14C]acetate and cultured for an additional 6 h. Fatty acid and cholesterol were extracted, and synthesis rates were determined. All data are presented as means ± S.E. (n = 3). Different superscript letters denote statistical significance (p < 0.05).
Quantitative real time PCR analyses in Huh-7 cells revealed that XN treatment reduced mRNA levels of two SREBP-1 target genes, FAS and SCD1, involved in fatty acid synthesis, but not of ACC1 (Fig. 1C). Similarly, three SREBP-2 target genes, HMGCS, HMGCR, and SQS, involved in cholesterol synthesis, were down-regulated by XN (Fig. 1C). Consistent with these changes in gene expression, XN decreased the de novo synthesis of both fatty acid and cholesterol (Fig. 1D). Altogether, these results indicate that XN suppresses SREBP activity and reduces lipid synthesis.
XN Reduces Level of Mature Forms of SREBPs Independently of Insigs
SREBPs are synthesized as inactive precursors. When cellular cholesterol is low, the inactive precursors are proteolytically processed to the mature forms that function as active transcription factors. To determine whether XN affects cellular levels of mature SREBPs, Huh-7 cells were treated with XN for 3 h under low cholesterol conditions; the cell lysates were subjected to immunoblotting. Similar to 25-HC, XN reduced the level of the mature forms of both SREBP-1 and -2 (Fig. 2A). We also examined the effect of three flavonoids with structures similar to XN, IXN, NG, and 8-PN, on cellular levels of mature SREBPs (Fig. 2, B–E). Although not to the extent of XN, IXN and 8-PN markedly decreased the level of the mature forms of SREBPs, whereas NG had little effect (Fig. 2F).
FIGURE 2.
XN decreases production of mature SREBPs. A, Huh-7 cells were depleted of sterols by incubating in medium C for 16 h. The cells were then switched to medium C in the presence of vehicle, 1 μg/ml 25-HC, 10 μm XN, or 30 μm XN. After incubation for 3 h, whole cell extracts underwent immunoblotting (IB) with anti-SREBP-1 (2A4), anti-SREBP-2, or anti-β-actin antibodies. B–E, structures of XN (B), IXN (C), NG (D), and 8-PN (E). F, Huh-7 cells were depleted of sterols by incubating in medium C for 16 h. The cells were then switched to medium C in the presence of 30 μm XN, IXN, NG, or 8-PN. After incubation for 3 h, whole cell extracts were subjected to immunoblotting with the antibodies listed in A. G, CHO-7 and SRD-15 cells were depleted of sterols by incubating in medium F for 16 h. The cells were then switched to medium F in the presence of vehicle, 1 μg/ml 25-HC, or 30 μm XN. After incubation for 3 h, whole cell extracts underwent immunoblotting with the antibodies listed in A. H and I, Huh-7 cells were transfected with either control (siCon) or Insig-1 and -2 siRNA oligonucleotides (siInsigs), cultured with medium A for 24 h, then cultured with medium C for 16 h, and re-fed the medium containing 1 μg/ml 25-HC or 30 μm XN for 3 h before harvest. H, real time PCR analysis was performed, and relative mRNA levels were obtained by normalization to GAPDH mRNA. I, whole cell extracts underwent immunoblotting with anti-SREBP-1 (2A4), anti-SREBP-2, or anti-β-actin antibodies. The same results were obtained in more than three separate experiments.
To further analyze this effect, we used a previously established Insig-1- and -2-deficient Chinese hamster ovary cell line (SRD-15). Previous studies have reported that 25-HC blocks SREBP processing by binding to Insigs, triggering their binding to SCAP and retaining the SCAP/SREBP complex in the ER (29). Consistent with these findings, we found that 25-HC did not affect the level of the mature forms of SREBPs in SRD-15 cells, whereas it suppressed SREBP processing in parental CHO-7 cells (Fig. 2G). XN reduced the level of the mature forms of SREBPs in both CHO-7 and SRD-15 cells (Fig. 2G). When endogenous Insig-1 and Insig-2 expressions in Huh7 cells were reduced with gene-specific siRNA (Fig. 2H), XN-mediated suppression of mature forms of SREBPs was still observed, whereas 25-HC-mediated suppressions were partly suppressed (Fig. 2I). It should be noted that treatment with 25-HC and XN for 3 h decreased the expression of the Insig-1 gene possibly due to the suppression of SREBP activity. In addition, treatment with XN unexpectedly decreased the expression of the Insig-2 gene. These results indicate that the decrease in mature SREBPs by XN occurs without Insigs, acting in a manner different from 25-HC.
XN Suppresses SREBP Processing by Blocking ER-to-Golgi Transport of the SCAP/SREBP Complex
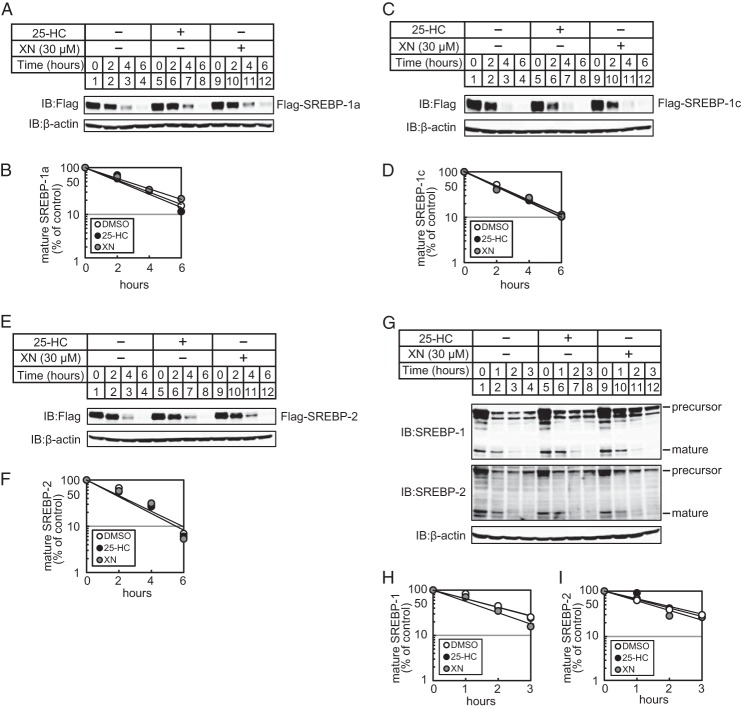
Next, we investigated the mechanism by which XN reduces the level of mature forms of SREBPs. One possibility is that XN promotes the protein degradation of mature SREBPs. To examine this, CHO-7 cells were transfected with an expression plasmid encoding a mature form of human SREBP-1a (amino acids 2–487; pCMV-3×FLAG-SREBP-1a(2–487), and a turnover of exogenous SREBP-1a was measured. In the presence of the translation inhibitor cycloheximide, the amount of mature SREBP-1a gradually reduced (Fig. 3A, lanes 1–4). 25-HC did not promote the degradation of mature SREBP-1a (Fig. 3A, compare lanes 1–4 and 5–8), consistent with a previous report (30). Moreover, XN did not promote the degradation of mature SREBP-1a (Fig. 3A, compare lanes 1–4 and 9–12). The quantification of these signals is shown in Fig. 3B. Similar results were obtained when cells were transfected with an expression plasmid coding a FLAG-tagged mature form of human SREBP-1c (amino acids 2–463) (Fig. 3, C and D) or that of human SREBP-2 (amino acids 2–481) (Fig. 3, E and F). Next, we examined whether XN promotes the degradation of endogenous mature SREBPs. Sterol-depleted CHO-7 cells were preincubated with the serine protease inhibitor, AEBSF, that inhibits S1P, thereby inhibiting SREBP cleavage and maturation (31). In the presence of AEBSF, the level of mature SREBPs decreased in a time-dependent manner, and 25-HC and XN had no effect on the degradation (Fig. 3, G, compare lanes 1–4, 5–8, and 9–12, and quantification in H and I). These data indicate that XN does not accelerate the degradation of mature SREBPs.
FIGURE 3.
XN does not promote the degradation of mature SREBPs. A, C, and E, CHO-7 cells were transfected with pCMV-3×FLAG-SREBP-1a (2–487) (A), pCMV-3×FLAG-SREBP-1c(2–463) (C), or pCMV-3×FLAG-SREBP-2 (2–481) (E) and cultured in medium E for 24 h. The cells were trypsinized and seeded in 6-well plates and cultured further under the same conditions for 24 h. After incubation with 50 μm cycloheximide for 0.5 h, the cells were switched to medium E supplemented with 50 μm cycloheximide in the presence of vehicle, 1 μg/ml 25-HC, or 30 μm XN. G, CHO-7 cells were depleted of sterols by incubating in medium F for 16 h. After incubation with 300 μm AEBSF for 0.5 h, the cells were switched to medium F supplemented with 300 μm AEBSF in the presence of vehicle, 1 μg/ml 25-HC, or 30 μm XN. After incubation for the indicated period of time, whole cell extracts underwent immunoblotting (IB) with anti-FLAG, anti-SREBP-1 (2A4), anti-SREBP-2, or anti-β-actin antibodies. B, D, F, H, and I, The signals detected on the membrane in (A, C, E, and G) were quantified, and data were plotted as the percentage of FLAG-SREBP-1a (B), FLAG-SREBP-1c (D), FLAG-SREBP-2 (E), mature SREBP-1 (H), and -2 (I) protein remaining. The same results were obtained in more than three separate experiments.
Another possibility in which XN may reduce the level of mature SREBPs is by inhibiting S1P and S2P activities. S1P and S2P are proteases that cleave the precursors of SREBPs in the Golgi apparatus, releasing the mature forms. To examine whether XN inhibits the proteolytic cleavage of SREBPs in the Golgi, we pretreated CHO-7 cells with BFA, a fungal metabolite that blocks the anterograde transport of proteins from the ER to the Golgi. BFA causes SREBPs to be proteolytically cleaved in the ER independently of sterols by translocating S1P and S2P from the Golgi back to the ER (32). Furthermore, the mature forms of SREBPs were present in cells treated with BFA, even in the presence of 25-HC or XN (Fig. 4). These data suggest that XN does not inhibit the activity of S1P or S2P.
FIGURE 4.
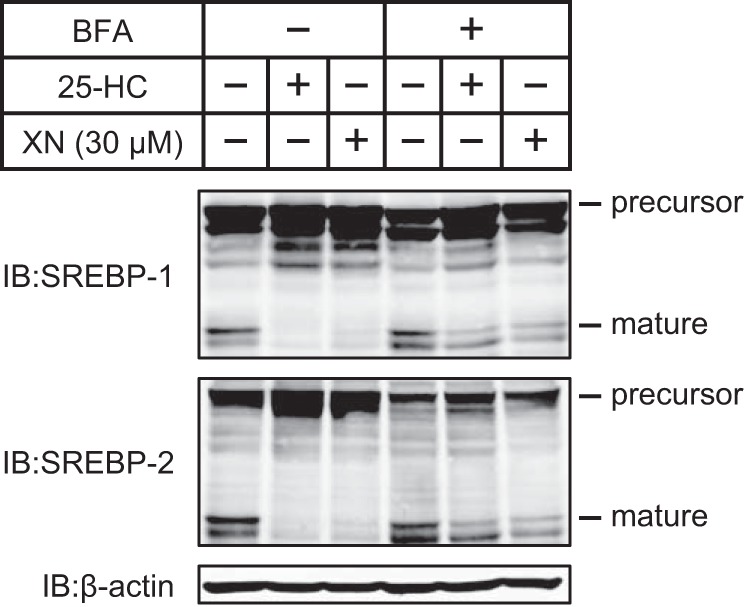
XN does not inhibit the proteolysis of SREBPs. CHO-7 cells were depleted of sterols through incubation in medium F for 16 h. After incubation with or without 1 μg/ml BFA for 0.5 h, the cells were switched to medium F in the presence of vehicle, 1 μg/ml BFA plus 1 μg/ml 25-HC, or 1 μg/ml BFA plus 30 μm XN. After incubation for 3 h, whole cell extracts were subjected to immunoblotting (IB) with anti-SREBP-1 (2A4), anti-SREBP-2, or anti-β-actin antibodies. The same results were obtained in more than three separate experiments.
The precursors of SREBPs undergo two sequential cleavage events; however, before this, the SCAP/SREBP complex embedded in the ER must be transported to the Golgi. Therefore, we hypothesized that XN suppresses the ER-to-Golgi translocation of the SCAP/SREBP complex. To investigate this, we used two types of assays. First, we used immunofluorescence microscopy to examine SCAP-deficient cells stably expressing GFP-SCAP (CHO/pGFP-SCAP). When CHO/pGFP-SCAP cells were incubated under sterol-depleted conditions, GFP-SCAP exhibited bright juxtanuclear fluorescence (Fig. 5A). The colocalization of GFP-SCAP with the Golgi marker GM130 indicated the localization of GFP-SCAP in the Golgi (Fig. 5, A–C). In contrast, when the cells were incubated in the presence of sterols, the GFP signal was diffuse and reticular, suggesting ER localization (Fig. 5D). There was little colocalization of GFP and GM130 staining under this condition (Fig. 5, D–F). Importantly, a similar diffuse, reticular signal was observed when the cells were treated with XN under sterol-depleted conditions (Fig. 5, G–I), indicating that XN blocks the transport of the SCAP/SREBP complex to the Golgi.
FIGURE 5.
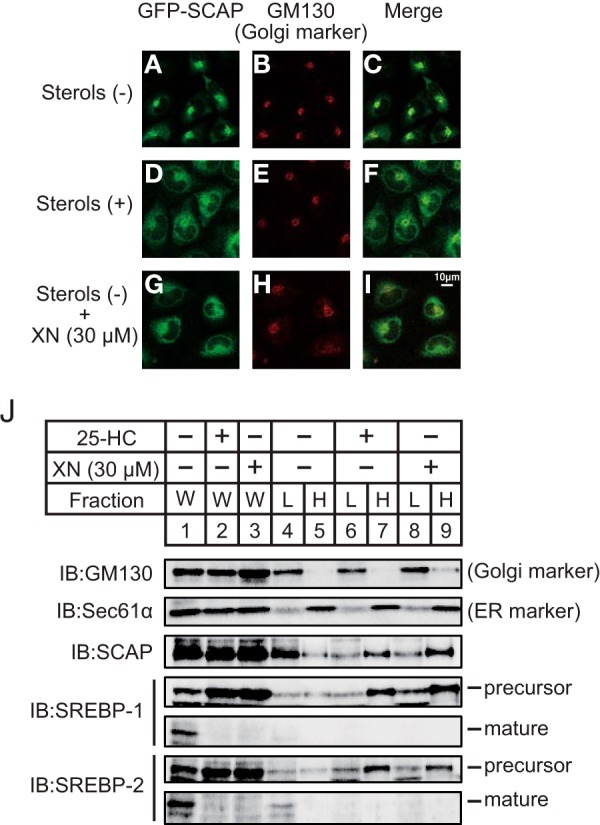
XN blocks the ER-to-Golgi translocation of the SCAP/SREBP complex. A–I, CHO/pGFP-SCAP cells were depleted of sterols by incubating in medium F for 3 h. The cells were then switched to medium F in the absence or presence of sterols (10 μg/ml cholesterol plus 1 μg/ml 25-HC) or 30 μm XN. After incubation for 3 h, the cells were fixed and then stained with primary antibody against GM130 followed by Cy3-conjugated secondary antibody. The cells were then imaged for GFP-SCAP (A, D, and G) and GM130 (B, E, and H). C, F, and I are merged images of GFP-SCAP and GM130. J, CHO-7 cells were depleted of sterols by incubating in medium F for 16 h. The cells were then switched to medium F in the presence of vehicle, 1 μg/ml 25-HC, or 30 μm XN. After incubation for 3 h, the cells were fractionated into a light fraction, containing the Golgi, and a heavy fraction, containing the ER, by sucrose density-gradient centrifugation, as described under “Experimental Procedures.” Equal volumes of each fraction were subjected to immunoblotting (IB) with anti-GM130, anti-Sec61α, anti-SCAP, anti-SREBP-1 (2A4), or anti-SREBP-2 antibodies. The same results were obtained in more than three separate experiments. W, whole cell lysates; L, light fraction; H, heavy fraction.
Second, we performed cell fractionation to examine the protein levels of SCAP and SREBPs in both the ER and Golgi of the XN-treated cells. Before cell fractionation, aliquots of protein were divided, and the suppressive effects of XN and 25-HC on SREBP maturation were confirmed (Fig. 5J, lanes 1–3). The lysates from CHO-7 cells were then subjected to centrifugation through a discontinuous sucrose gradient, generating two discrete fractions, light and heavy. The light fraction contained the Golgi and expressed GM130 (Fig. 5J, lanes 4, 6, and 8), and the heavy fraction contained the ER and expressed the ER-resident protein Sec61α (Fig. 5J, lanes 5, 7, and 9). When the cells were cultured under sterol-depleted conditions, SCAP was localized to the light fraction (Fig. 5J, lanes 4 and 5), and SREBPs were not detected in either the light or heavy fractions. This may have occurred if SREBPs were proteolytically cleaved in the Golgi in response to sterol depletion and released from the Golgi. In contrast, in the presence of 25-HC, SCAP, and SREBP precursors were found in the heavy fraction (Fig. 5J, lanes 6 and 7). These results are consistent with previous reports, wherein the SCAP/SREBP complex was anchored to the ER in sterol-loaded cells but transported to the Golgi in sterol-depleted cells. Similar to 25-HC, XN treatment led to SCAP and SREBPs found in the heavy fraction (Fig. 5J, lanes 8 and 9). These data indicate that XN contributes to the tethering of the SCAP/SREBP complex to the ER membrane and suppresses the transport of the SCAP/SREBP complex to the Golgi.
XN Inhibits Incorporation of the SCAP/SREBP Complex into COP II Vesicles
Considering our findings from previous experiments, we hypothesized that XN interacts with proteins involved in the ER-to-Golgi transport of the SCAP/SREBP complex. When cells are depleted of sterols, Sec23/24 binds to SCAP, mediating the sorting of the SCAP/SREBP complex into COP II vesicles. Furthermore, SREBPs are transported via the vesicles to the Golgi apparatus. To examine whether XN interacts with SCAP, SREBPs, or COP II proteins, we prepared XN-immobilized agarose beads (XN beads). The lysates from sterol-depleted CHO-7 cells were incubated with XN beads, and the coprecipitated proteins were subjected to immunoblotting. As shown in Fig. 6A, the proteins coprecipitated with XN beads contained Sec23 and Sec24 but not SCAP and SREBP-1, whereas the control beads contained trace levels of proteins (Fig. 6A, lanes 3 and 5). Moreover, when cells were pretreated with additional XN, the amount of Sec23 and Sec24 associated with XN beads reduced (Fig. 6A, lanes 5 and 6), indicating that the XN used in the pretreatment competed with the XN beads for associating with Sec23 and Sec24. These results suggest that XN binds to Sec23/24.
FIGURE 6.
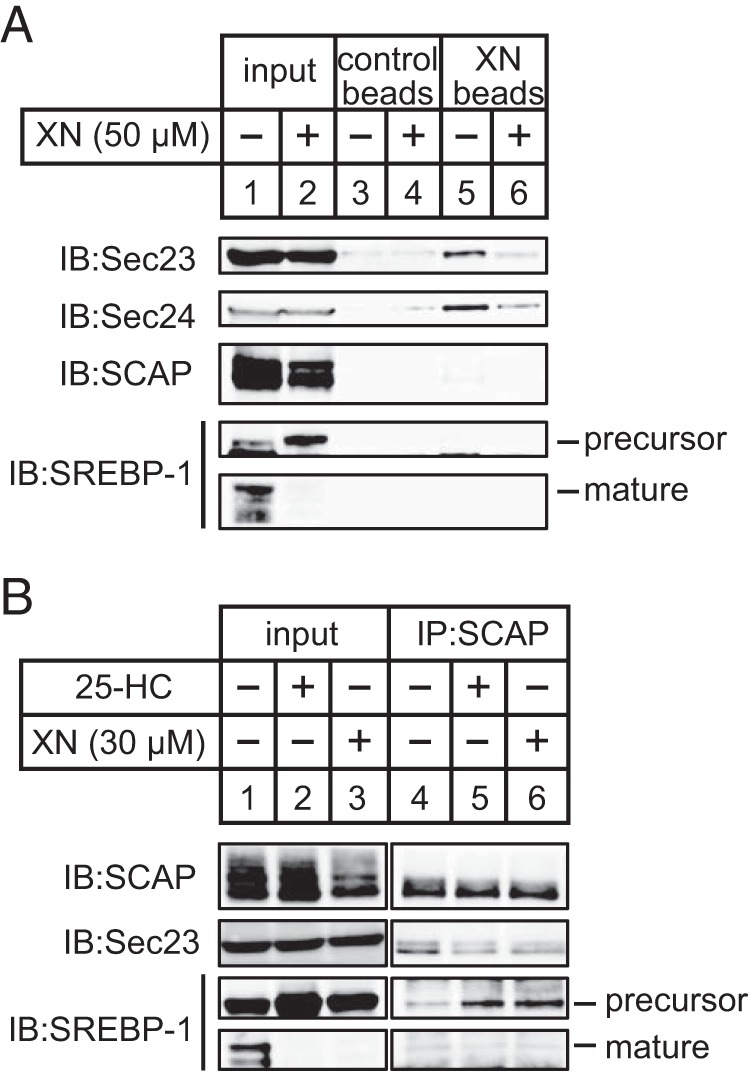
XN suppresses clustering of the SCAP/SREBP complex into COP II vesicles. A, CHO-7 cells were depleted of sterols by incubating in medium F for 16 h. The cells were then switched to medium F in the presence or absence of 50 μm XN. After incubation for 3 h, the cell lysates were precleared with control beads and then incubated with control or XN beads. The pelleted beads were washed, and the eluted proteins were subjected to immunoblotting with anti-Sec23 (recognize Sec23A and Sec23B), anti-Sec24C, anti-SCAP, or anti-SREBP-1 (2A4) antibodies. B, CHO-7 cells were depleted of sterols by incubating in medium F for 16 h. The cells were then switched to medium F in the presence of vehicle, 1 μg/ml 25-HC, or 30 μm XN. After incubation for 3 h, cell lysates were subjected to immunoprecipitation with anti-SCAP antibody. The immunoprecipitates were subjected to immunoblotting (IB) with anti-Sec23, anti-SCAP, or anti-SREBP-1 (2A4) antibodies. The same results were obtained in more than three separate experiments.
The association of XN with Sec23/24 and its inhibitory effect on the translocation of the SCAP/SREBP complex indicate that XN impairs sorting of the SCAP/SREBP complex into COP II vesicles. To examine this, we investigated whether the association between Sec23 and SCAP is attenuated by XN. CHO-7 cells were treated with XN under sterol-depleted conditions; the cell lysates were then coimmunoprecipitated with SCAP and subjected to immunoblotting with Sec23. Although SCAP associated with Sec23 proteins, this association was attenuated in cells treated with 25-HC (Fig. 6B, lanes 4 and 5). We also found that the association between SCAP and SREBP-1 precursor was promoted in cells treated with 25-HC (Fig. 6B, lanes 4 and 5); this is consistent with an increase in the amount of SREBP-1 precursor in the cell lysates before immunoprecipitation because of the suppression of proteolytic processing (Fig. 6B, lanes 1 and 2). Furthermore, these observations are consistent with a previous report that found that sterols cause a conformational change in SCAP, rendering it inaccessible to COP II proteins (29). XN also reduced the amount of Sec23 associated with SCAP and increased the amount of SREBP-1 associated with SCAP (Fig. 6B, lanes 4 and 6). These data suggest that XN blocks the incorporation of SCAP/SREBP into COP II vesicles by binding to Sec23/24.
ATF6 is also transported from the ER to the Golgi by COP II vesicles (33) and is cleaved by S1P and S2P (34). Therefore, we examined whether XN suppresses ATF6 processing. Huh-7 cells were pretreated with Tg, which causes ER stress by blocking the ER Ca2+-ATPase and stimulates ATF6 processing; the cells were then treated with XN. ATF6 processing stimulated by Tg was inhibited in cells treated with AEBSF but not in those treated with XN (Fig. 7). In contrast, SREBP processing was suppressed in both AEBSF- and XN-treated cells (Fig. 7). Therefore, XN appears to block the COP II-dependent transport of specific cargo proteins.
FIGURE 7.
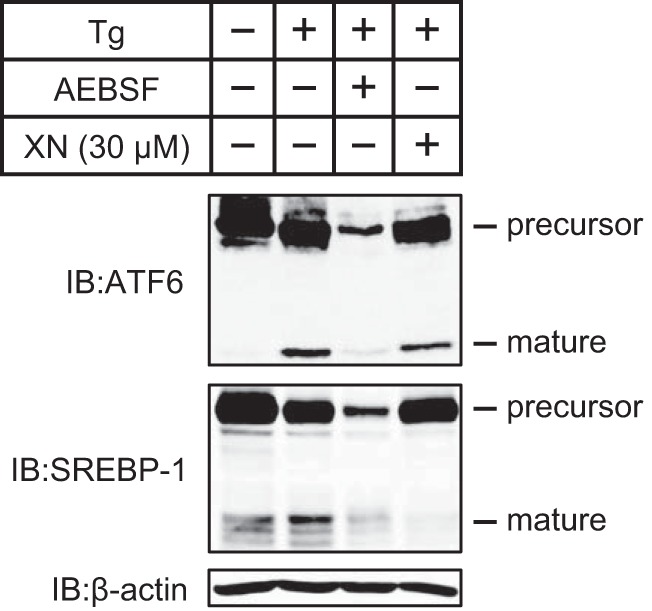
XN does not inhibit ATF6 processing. Huh-7 cells were cultured with medium A for 48 h and then incubated with or without 250 nm Tg for 5 h. Cells incubated in Tg were then switched to medium A in the presence of 250 nm Tg alone or Tg plus 300 μm AEBSF or 30 μm XN. After incubation for 3 h, whole cell extracts were subjected to immunoblotting (IB) with anti-ATF6, anti-SREBP-1 (2A4), or anti-β-actin antibodies. The same results were obtained in more than three separate experiments.
XN Reduces the Mature Form of SREBP-1 in Mouse Liver
Next, we investigated the effect of XN in mice in vivo. First, to determine whether XN suppresses SREBP processing in mouse liver, 75 or 150 mg/kg body weight of XN was administered to C57BL/6J mice by oral gavage once daily for 3 days. The food intake and body weight were not affected by XN (Fig. 8, A and B), whereas the level of hepatic mature SREBP-1 was reduced (Fig. 8C). SCD1 gene expression decreased in the livers of mice treated with XN; however, other SREBP target genes, such as ACC1, FAS, HMGCS, HMGCR, and SQS, were not affected by XN (Fig. 8, D and E). These results indicate that XN inhibits the proteolytic activation of SREBP in mouse liver as it does in cultured cells.
FIGURE 8.
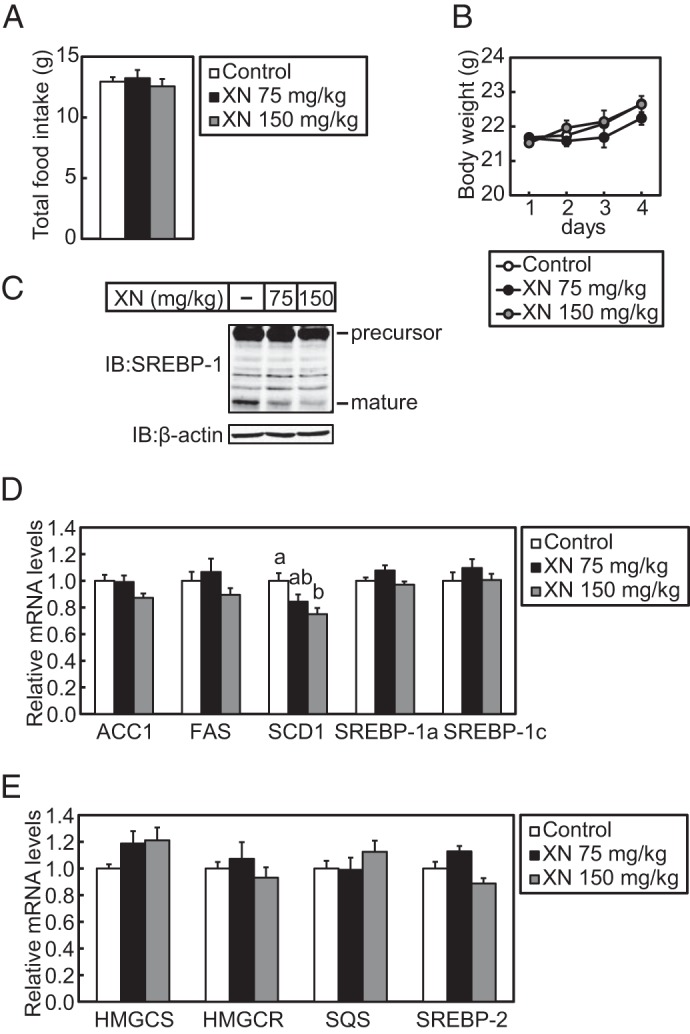
XN blocks SREBP processing in vivo. Male 6-week-old C57BL/6J mice received oral administration of vehicle or XN (75 or 150 mg/kg body weight) once daily. The mice had ad libitum access to water and a standard chow diet. After a 3-day treatment, livers were harvested from the mice. A, average mouse body weight for each treatment group during the 4-day experiment. B, total average mouse food intake for each treatment group during the 4-day experiment. C, immunoblotting (IB) analysis of liver extracts with anti-SREBP-1 (H-160) or anti-β-actin antibodies. D and E, real time quantitative PCR analysis of gene expression in the liver. Relative mRNA levels were obtained by normalization to 18S rRNA. All data are presented as mean ± S.E. (n = 5). Different superscript letters denote statistical significance (p < 0.05).
XN Ameliorates Diet-induced Obesity and Fatty Liver in Mice
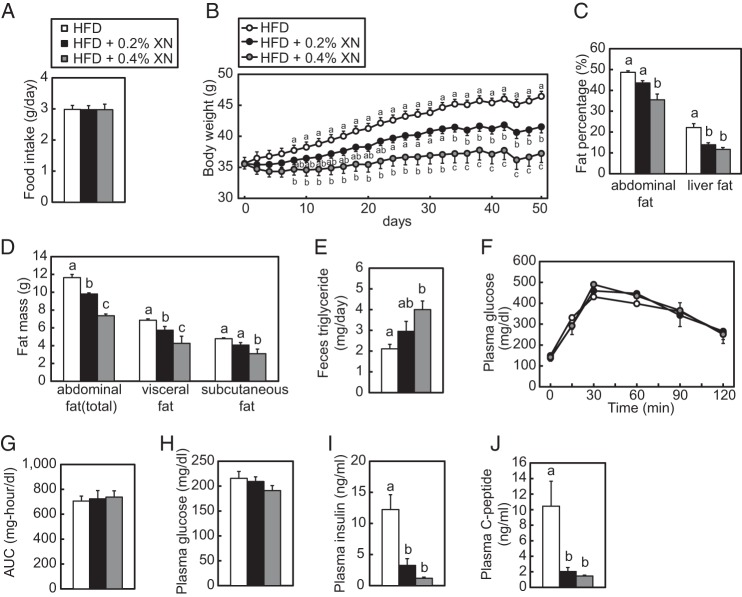
Next, to verify the effect of XN on diet-induced obesity, C57BL/6J mice were fed a HFD or HFD supplemented with XN (0.2 or 0.4%) for 50 days. During the feeding experiment, the average daily food intake was similar in all groups (Fig. 9A). Considering daily food intake, dietary supplementation of 0.2% XN is estimated ∼150 mg/kg/day XN. The mice fed XN-supplemented HFD exhibited lesser weight gain than those fed a HFD alone, in a concentration-dependent manner (Fig. 9B). Moreover, the weight of the liver, epididymal, mesenteric, subcutaneous, and inguinal white fat pads, as well as interscapular brown fat pads, was reduced in the mice fed a HFD supplemented with XN (Table 1). Consistent with this, CT scan analysis demonstrated that XN reduced the percentage of fat area in the abdomen and liver (Fig. 9C). In this analysis, the abdominal visceral and subcutaneous fat volumes were separately calculated, and XN decreased both volumes (Fig. 9D). These results suggest that XN can ameliorate diet-induced obesity. To determine whether dietary XN reduces fat absorption from the intestine, we analyzed lipid levels in feces. The triglyceride levels in feces showed a tendency to increase in the HFD + 0.2% XN group and significantly increased in the HFD + 0.4% XN group (Fig. 9E), suggesting that dietary XN reduced lipid absorption from the intestine.
FIGURE 9.
XN inhibits the development of obesity and fatty liver in HFD-fed mice. A–D and F–J, 6-week-old C57BL/6J male mice were fed a HFD for 10 weeks and subsequently fed a HFD, 0.2% XN, or 0.4% XN-supplemented HFD for 50 days (n = 10–11). The mice were given free access to water and their diet. CT scans and OGTT were performed at 35 and 44 days after the initiation of XN supplementation, respectively. The mice were sacrificed and subjected to various analyses as described below. A, total daily average mouse food intake for each treatment group during the 50-day experiment. B, average mouse body weight for each treatment group during the 50-day experiment. C, fat percentages in the abdomen and liver after 35 days of XN feeding. Mice were scanned using an experimental animal x-ray CT system (LaTheta LCT-200). D, abdominal fat mass after 35 days of XN feeding. Mice were analyzed as in C. Whole abdominal fat, visceral fat, and subcutaneous fat masses were determined using CT analysis. E, feces triglyceride levels for each treatment group. Six-week-old C57BL/6J male mice (n = 5) were fed a HFD for 1 week and subsequently fed a HFD, 0.2% XN, or 0.4% XN-supplemented HFD for 8 days. Feces were collected for last 4 days. F, effect of XN on glucose tolerance in HFD-fed mice determined by OGTT after 44 days of XN feeding. OGTT was performed by oral administration of 2 g/kg body weight glucose to mice fasted for 16 h. Blood glucose levels were measured after the indicated period of time. G, quantification of the area under the curve (AUC) from OGTT in F. H–I, plasma glucose (H), insulin (I), and C-peptide (J) levels after the 50-day experiment. All data are presented as mean ± S.E. Different superscript letters denote statistical significance (p < 0.05).
TABLE 1.
Weights of liver and adipose tissue in C57BL/6J mice fed HFD + XN for 50 days
Data are presented as mean ± S.E. (n = 10–11). Different superscript letters denote statistical significance (p < 0.05).
| HFD | HFD + 0.2% XN | HFD + 0.4% XN | |
|---|---|---|---|
| Liver (g) | 2.10 ± 0.49a | 1.53 ± 0.23b | 1.36 ± 0.23b |
| Epididymal WAT (g) | 2.35 ± 0.44a | 2.43 ± 0.27a | 1.83 ± 0.53b |
| Mesenteric WAT (g) | 1.35 ± 0.25a | 0.87 ± 0.28b | 0.55 ± 0.19c |
| Subcutaneous WAT (g) | 1.55 ± 0.25a | 1.19 ± 0.24b | 0.77 ± 0.29c |
| Inguinal WAT (g) | 1.03 ± 0.16a | 0.93 ± 0.28ab | 0.74 ± 0.26b |
| Interscapular BAT (g) | 0.25 ± 0.05a | 0.19 ± 0.08b | 0.08 ± 0.06c |
As shown in Table 2, the plasma total TG levels were lower in the mice fed a HFD supplemented with XN than in mice fed a HFD alone, and there was a marked decrease in cholesterol levels in the mice fed a HFD supplemented with XN. Table 2 also summarizes the plasma lipoprotein profiles in each group, as measured by HPLC. TG and cholesterol levels in chylomicron and very low density lipoprotein (VLDL) fractions tended to decrease, and levels in low density lipoprotein (LDL) and high density lipoprotein (HDL) fractions markedly decreased in the mice fed a HFD supplemented with XN. Consistent with the decreases in liver weight and fat percentage in the mice fed a HFD supplemented with XN, the livers of these mice had lower TG and cholesterol accumulation than those of the mice fed a HFD alone.
TABLE 2.
Plasma lipoprotein profile and hepatic lipid accumulation in C57BL/6J mice fed HFD + XN for 50 days
Data are presented as mean ± S.E. (n = 10–11). Different superscript letters denote statistical significance (p < 0.05). CM is chylomicron.
| HFD | HFD + 0.2% XN | HFD + 0.4% XN | |
|---|---|---|---|
| Plasma | |||
| Triglyceride (mg/dl) | |||
| Total | 56.8 ± 10.0 | 59.2 ± 27.6 | 34.1 ± 11.0 |
| CM | 7.7 ± 2.0 | 8.0 ± 5.0 | 3.8 ± 1.2 |
| VLDL | 30.4 ± 6.4 | 37.6 ± 20.7 | 19.9 ± 7.8 |
| LDL | 15.7 ± 3.6a | 11.7 ± 3.0ab | 8.7 ± 2.0b |
| HDL | 3.0 ± 0.5a | 2.0 ± 0.4b | 1.7 ± 0.4b |
| Cholesterol (mg/dl) | |||
| Total | 180.4 ± 5.5a | 143.3 ± 17.0b | 120.9 ± 13.0c |
| CM | 0.9 ± 0.2 | 1.0 ± 0.6 | 0.6 ± 0.3 |
| VLDL | 5.4 ± 1.6 | 4.9 ± 2.3 | 3.2 ± 1.0 |
| LDL | 40.0 ± 1.8a | 27.0 ± 5.7b | 20.7 ± 3.1b |
| HDL | 134.1 ± 6.0a | 110.5 ± 11.1b | 96.4 ± 9.5b |
| Liver | |||
| Triglyceride (mg/g) | 130.8 ± 31.2a | 46.2 ± 19.5b | 29.6 ± 8.8b |
| Cholesterol (mg/g) | 2.1 ± 0.9a | 1.3 ± 0.3b | 1.1 ± 0.3b |
To evaluate whether XN affects glucose metabolism, we performed the OGTT 44 days after the initiation of XN supplementation. There was no marked difference in the plasma glucose level between the groups during OGTT (Fig. 9, F and G). However, the average glucose levels did trend to lower values in the mice fed a HFD supplemented with XN (Fig. 9H). Moreover, there was a significant decrease in insulin and C-peptide levels in the mice fed a HFD supplemented with XN at the end of the 50-day experiment (Fig. 9, I and J). This suggests that XN improves insulin sensitivity in mice experiencing diet-induced obesity. Next, to assess the activity of hepatic proteins involved in insulin signaling, we analyzed their phosphorylation state. The phosphorylation of hepatic Akt was not altered by XN (Fig. 10A). Phosphorylated S6K increased in the liver of mice fed a HFD supplemented with XN; however, the amount of total S6K was also elevated by XN (Fig. 10A). In contrast, phosphorylated AMPK levels were increased by XN (Fig. 10A), indicating that XN activated AMPK in mouse liver. However, XN did not change the phosphorylation state of AMPK in CHO-7 cells (Fig. 10B).
FIGURE 10.
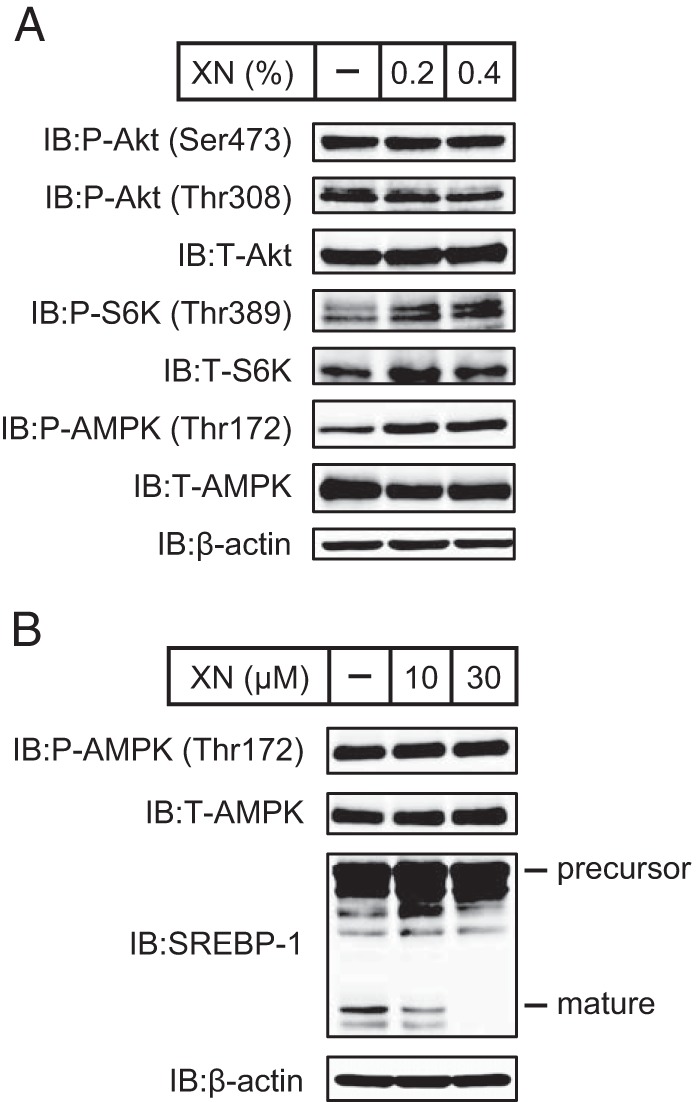
Phosphorylation states of proteins involved in insulin signaling in livers of mice fed a HFD supplemented with XN and XN do not affect AMPK phosphorylation in cells. A, immunoblot (IB) analysis of liver extracts with anti-phospho-Akt (Ser-473), anti-phospho-Akt (Thr-308), anti-Akt, anti-phospho-S6K (Thr-389), anti-S6K, anti-phospho-AMPK (Thr-172), anti-AMPK, or anti-β-actin antibodies. The mice used here were the same as those used in Fig. 9. B, CHO-7 cells were depleted of sterols by incubating in medium F for 16 h. The cells were then switched to medium F in the presence of vehicle, 10 μm XN, or 30 μm XN. After incubation for 3 h, whole cell extracts were subjected to immunoblotting with anti-phospho-AMPK (Thr-172), anti-AMPK, anti-SREBP-1 (2A4), or anti-β-actin antibodies. The same results were obtained in more than three separate experiments.
XN reduced the mature form of hepatic SREBP-1 (Fig. 11A) and mRNA levels of its target genes involved in fatty acid synthesis, such as ACC1, FAS, and SCD1 (Fig. 11B). This suggests that XN lowered lipogenesis by inhibiting SREBP maturation in the mouse liver. In contrast, SREBP-2 target genes involved in cholesterol synthesis, such as HMGCS, HMGCR, and SQS, were not affected in the liver of mice fed a HFD supplemented with XN (Fig. 11C). However, mRNA levels of two other genes involved in cholesterol metabolism, ATP-binding cassette subfamily G member 8 (ABCG8) and CYP7A1, increased in mice fed a HFD supplemented with XN (Fig. 11D), suggesting that XN accelerated the catabolism of cholesterol. The expression of genes involved in fatty acid β-oxidation, such as ACO and CPT-1a, was not altered by XN (Fig. 11E). As shown in Fig. 12A, XN did not reduce the mature form of SREBP-1, although it reduced the precursor form of SREBP-1 in the subcutaneous white adipose tissue (WAT). FAS gene expression decreased in the subcutaneous WAT of mice treated with XN; however, other SREBP target genes, such as ACC1 and SREBP-1c, were not affected by XN (Fig. 12B). These results indicate that XN does not inhibit the proteolytic activation of SREBP in WAT. XN increased the expression of ATGL, adiponectin, and UCP1 genes, although it did not affect the expression of HSL, PPARγ2, and TNFα gene in the subcutaneous WAT (Fig. 12B). XN did not alter the expression of UCP1, Dio2, and PGC1α gene in brown adipose tissue (BAT) (Fig. 12C). Altogether, these results suggest that although XN has different effects on several metabolic parameters, the suppression of hepatic SREBP activation by XN contributed to the amelioration of obesity and hepatic steatosis in HFD-fed mice.
FIGURE 11.
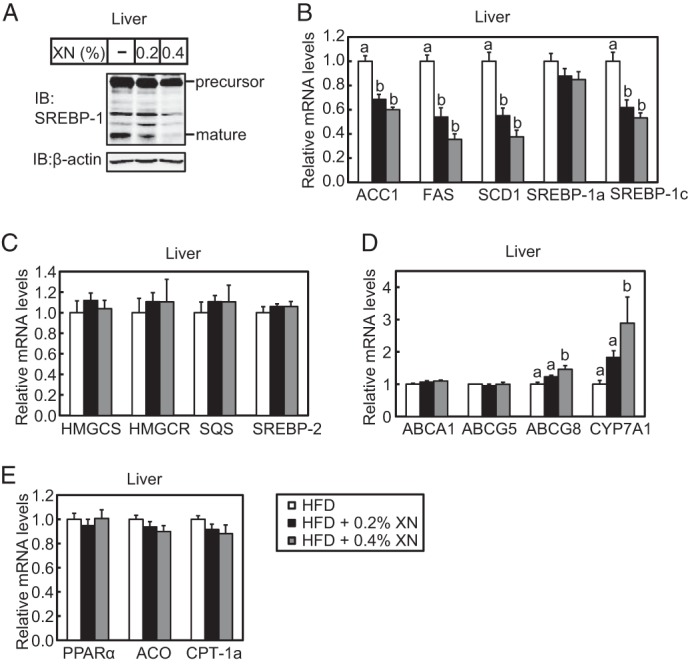
XN inhibits SREBP processing in the liver of HFD-fed mice. A, immunoblot (IB) analysis of liver extracts with anti-SREBP-1 (H-160) or anti-β-actin antibodies. B–E, real time quantitative PCR analysis of gene expression in the liver. Genes involved in fatty acid synthesis (B), cholesterol synthesis (C), cholesterol efflux and catabolism (D), and fatty acid β-oxidation (E) were analyzed. Relative mRNA levels were obtained by normalization to 18S rRNA. All data are presented as mean ± S.E. (n = 10–11). Different superscript letters denote statistical significance (p < 0.05). The mice used here were the same as those used in Fig. 9.
FIGURE 12.
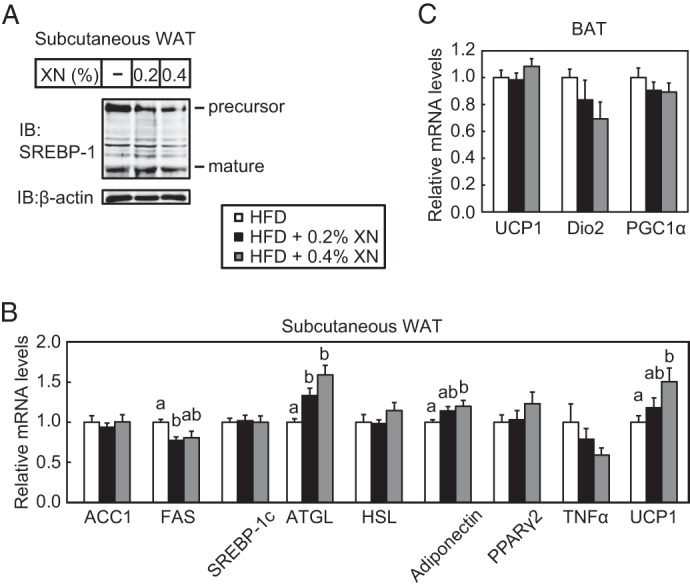
Effect of XN on SREBP processing and gene expression in the subcutaneous WAT and BAT in HFD-fed mice. A, immunoblot (IB) analysis of subcutaneous WAT extracts with anti-SREBP-1 (H-160) or anti-β-actin antibodies. B and C, real time quantitative PCR analysis of gene expression in the subcutaneous WAT (B) and BAT (C). Relative mRNA levels were obtained by normalization to 18S rRNA. All data are presented as mean ± S.E. (n = 10–11). Different superscript letters denote statistical significance (p < 0.05). The mice used here were the same as those used in Fig. 9.
Discussion
In this study, we identified XN as a new SREBP inactivator by screening numerous food components. XN suppresses SREBP processing by binding to Sec23/24 and attenuating the association between SCAP and Sec23/24, causing the SCAP/SREBP complex to remain anchored to the ER. Furthermore, XN reduces mRNA levels of SREBP target genes involved in fatty acid and cholesterol synthesis and inhibits de novo lipid synthesis.
It has been reported that several naturally derived small molecules inhibit SREBP processing. Betulin, a pentacyclic triterpene isolated from birch bark, impairs the ER-to-Golgi translocation of SREBPs by promoting the association of SCAP with Insig, a step also regulated by sterols (35). Polyunsaturated fatty acids suppress the proteasomal degradation of Insig-1 by triggering the dissociation of Insig-1 from UBX domain-containing protein 8, a protein that mediates the extraction of ubiquitinated Insig-1 from the ER membrane (36). It is noteworthy that these small molecules affect the function of Insig, inhibiting SREBP processing. In contrast, in this study, we analyzed the regulation of SREBPs in the absence of Insigs. We demonstrated that XN inhibits the transport of the SCAP/SREBP complex from the ER to the Golgi independently of Insigs and in a manner different from betulin, polyunsaturated fatty acids, and sterols (Fig. 2G); in contrast, XN acts on the COP II components Sec23/24. The Sec23/24 complex associates with cargo proteins that have sequences recognized by Sec24 and clusters these proteins into COP II transport vesicles. Because COP II vesicles transport various proteins, it is possible that the association between XN and Sec23/24 blocks the translocation of some proteins besides SREBPs. However, we found that XN does not affect the transport of ATF6, a cargo normally transported in COP II vesicles (Fig. 7). This indicates that XN may inhibit only a specific set of cargo proteins. Sec24 has multiple independent sites of cargo recognition (37); therefore, XN may bind near a specific site of Sec24, making it inaccessible to specific cargo proteins. It should be noted that because we used whole cell lysates to determine XN-binding proteins (Fig. 6A), but it is still unclear whether XN directly binds to Sec23/24. It is conceivable that XN indirectly associates with Sec23/24 through unidentified factors. Further investigations are required for determining the direct targets of XN, the binding pattern of SCAP to Sec23/24, and molecular mechanisms by which XN suppresses the association between SCAP and Sec23/24.
As shown in Fig. 2, XN, IXN, and 8-PN reduced the mature forms of SREBPs in Huh-7 cells. XN has an open C-ring, whereas IXN and 8-PN lack the open C-ring, suggesting that this component is not essential for decreasing the observed mature SREBPs. However, the nonprenylated analog of 8-PN, NG, had little effect on the level of mature SREBPs. Therefore, the prenyl group at the A-ring in XN, IXN, and 8-PN may play an important role in reducing the level of mature forms of SREBPs. Prenylation has also been shown to enhance the membrane permeability and cellular uptake of compounds by introducing hydrophobic properties (38). This may assist XN and its derivatives in regulating SREBPs within cells. In vivo, XN is metabolized into IXN and 8-PN (39–42). Therefore, the effects of these XN metabolites on mice fed a HFD supplemented with XN must also be considered. Because IXN and 8-PN decrease the mature forms of SREBPs in Huh-7 cells, when XN is consumed in the diet, its metabolites, such as IXN and 8-PN, may also exert an inhibitory effect on SREBP activity and metabolic disorders.
To evaluate and validate the effect of XN in vivo, we performed an experiment wherein XN was included in the diet of mice with diet-induced obesity. XN inhibited the development of obesity and fatty liver in a concentration-dependent manner (0.2 and 0.4%). Consistent with the results in cultured cells, XN decreased mature SREBP-1 and down-regulated the expression of its target genes in mouse liver. In contrast, mRNA levels of hepatic SREBP-2 target genes were not reduced (Fig. 11C), indicating that XN does not decrease mature SREBP-2 in the mouse liver. Although we could not detect SREBP-2 protein with immunoblotting in this experiment, it is important to analyze SREBP-2 processing in the livers of mice fed a HFD supplemented with XN. A previous study reported that in contrast to the marked increase in mature SREBP-1c, the amount of mature SREBP-2 changes very little in the livers of obese mice (9). Considering this, one possibility is that XN had little effect on SREBP-2 because of insufficient activation of SREBP-2 in the livers of mice fed a HFD. Alternatively, the down-regulation of SREBP-2 processing and its target genes by XN did occur; however, the subsequent decrease in cholesterol may have positively stimulated and restored the production of mature SREBP-2.
In the mice fed a HFD supplemented with XN, plasma insulin levels were markedly reduced (Fig. 9I). Because insulin activates the proteolytic processing of SREBP-1c, the predominant SREBP-1 isoform in the liver (43), an effect of decreased plasma insulin levels on hepatic SREBP-1c processing in the mice fed a HFD supplemented with XN may have occurred. Furthermore, the stimulation of SREBP-1c processing by insulin has been shown to require the activation of the Akt/mTOR/S6K pathway (44). However, the phosphorylation of Akt and S6K did not decrease in the livers of mice fed a HFD supplemented with XN (Fig. 10A). This suggests that the decrease in mature hepatic SREBP-1 by XN is not attributable to the reduction in plasma insulin levels. Despite the remarkable decline in plasma insulin levels, plasma glucose levels and glucose tolerance did not markedly improve by XN (Fig. 9, F–I). In the mice fed a HFD supplemented with XN, lower insulin levels produced nearly the same effect on glucose removal as higher insulin levels did in mice fed a HFD alone. This indicates that insulin sensitivity was improved by XN.
It has been previously demonstrated that AMPK phosphorylation is induced in the liver of ApoE−/− mice fed a western-type diet supplemented with XN (45). Our results also demonstrate that dietary XN increases phosphorylated AMPK in the livers of mice fed a HFD (Fig. 10A). Because phosphorylation activates AMPK and inhibits the proteolytic processing of SREBPs (46), in this study the decrease in the mature forms of SREBPs caused by XN may be partly attributable to AMPK activation. However, XN treatment did not affect the phosphorylation of AMPK in CHO-7 cells (Fig. 10B), suggesting that AMPK activation is not required for the suppression of SREBP processing by XN.
Consistent with our result, several reports indicate that dietary or oral administration of XN exhibits an anti-obesity effect in obese model animals (16, 18, 19). Yui et al. (19) reported that a dietary XN-rich extract from the hop suppressed fat absorption from the intestine and reduced the activity of fatty acid synthesis enzyme, such as FAS, glucose-6-phosphate dehydrogenase, and malic enzyme, in the liver accompanied by the reduction of SREBP-1c mRNA expression. It has been reported that oral administration of XN reduced plasma levels of dicarboxylic fatty acids and acylcarnitines, markers of dysfunctional β-oxidation, indicating that XN promotes fatty acid oxidation (17). Thus, XN exhibit anti-obesity effect through multiple functions. Given that SREBP inactivators, such as betulin and fatostatin, exhibit anti-obesity effect (35, 47), the suppression of SREBP activation by XN is, at least in part, attributable to the anti-obesity effect.
SREBPs function as master regulators of fatty acid and cholesterol biosynthesis, and aberrant SREBP activity may be linked to metabolic disease states. The inhibition of SREBP activation is a promising therapeutic approach for the treatment of metabolic disorders. Here, we examined the regulation of SREBP activity and demonstrated that XN suppresses SREBP processing not only in cultured cells but also in the livers of mice. This contributed to the amelioration of diet-induced obesity and fatty liver. Although XN is the principal prenylated flavonoid in hops, drinks made from hops, beers, for example, contain very small amounts of XN (1–100 μg/liter) (48). Thus, supplemental intake of XN is suitable to get its metabolic improvement. Altogether, our results indicate that XN is an effective nutritional and pharmacological agent that can be used for improving metabolic syndrome, including obesity and fatty liver.
Author Contributions
J. I. and R. S. conceived the project. S. M. and J. I. designed the study. S. M. performed the experiments. S. M. J. I., M. S., and R. S. analyzed the data. S. M. and J. I. wrote the manuscript. All authors read and approved the manuscript.
Acknowledgments
We thank Drs. Joseph L. Goldstein, Michael S. Brown, Andrew J. Brown, and Russell A. DeBose-Boyd for generously sharing their valuable tools. We also thank Kaori Honda and Drs. Yasumitsu Kondoh, Tamio Saito, and Hiroyuki Osada for providing xanthohumol-conjugated agarose beads and Dr. Hisanori Kato for the use of LCT-200. We are grateful to Kirin Co., Ltd., for providing XN. We also thank Enago for the English language review.
This work was supported by research grants from Japan Science and Technology Agency (Research for Promoting Technological Seeds) (to J. I.), The Tojuro Iijima Foundation for Food Science and Technology (to J. I.), the Japanese Council for Science, Technology and Innovation, and Cross-ministerial Strategic Innovation Promotion Program SIP Project ID 14533567. The authors declare that they have no conflicts of interest with the contents of this article.
- SREBP
- sterol regulatory element-binding protein
- AEBSF
- 4-(2-aminoethyl) benzenesulfonyl fluoride
- BFA
- brefeldin A
- COP II
- coated protein II
- CT
- computed tomography
- ER
- endoplasmic reticulum
- FAS
- fatty-acid synthase
- 25-HC
- 25-hydroxycholesterol
- HMGCR
- HMG-CoA reductase
- HMGCS
- HMG-CoA synthase
- HSL
- hormone-sensitive lipase
- Insigs
- insulin-induced genes
- IXN
- isoxanthohumol
- LPDS
- lipoprotein-deficient serum
- NG
- naringenin
- OGTT
- oral glucose tolerance test
- 8-PN
- 8-prenylnaringenin
- PPAR
- peroxisome proliferator-activated receptor
- SCAP
- SREBP cleavage-activating protein
- S1P
- site-1 protease
- S2P
- site-2 protease
- Tg
- thapsigargin
- XN
- xanthohumol
- h
- human
- m
- mouse
- BAT
- brown adipose tissue
- WAT
- white adipose tissue
- S6K
- S6 kinase
- AMPK
- AMP-activated protein kinase.
References
- 1. Zimmet P., Magliano D., Matsuzawa Y., Alberti G., Shaw J. (2005) The metabolic syndrome: a global public health problem and a new definition. J. Atheroscler. Thromb. 12, 295–300 [DOI] [PubMed] [Google Scholar]
- 2. Evans R. M., Barish G. D., Wang Y. X. (2004) PPARs and the complex journey to obesity. Nat. Med. 10, 355–361 [DOI] [PubMed] [Google Scholar]
- 3. Brown M. S., Goldstein J. L. (1999) A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. U.S.A. 96, 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Espenshade P. J., Li W. P., Yabe D. (2002) Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. U.S.A. 99, 11694–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duncan E. A., Brown M. S., Goldstein J. L., Sakai J. (1997) Cleavage site for sterol-regulated protease localized to a Leu-Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J. Biol. Chem. 272, 12778–12785 [DOI] [PubMed] [Google Scholar]
- 6. Rawson R. B., Zelenski N. G., Nijhawan D., Ye J., Sakai J., Hasan M. T., Chang T. Y., Brown M. S., Goldstein J. L. (1997) Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1, 47–57 [DOI] [PubMed] [Google Scholar]
- 7. Yabe D., Brown M. S., Goldstein J. L. (2002) Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horton J. D., Goldstein J. L., Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimomura I., Bashmakov Y., Horton J. D. (1999) Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 274, 30028–30032 [DOI] [PubMed] [Google Scholar]
- 10. Kammoun H. L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T., Ferré P., Foufelle F. (2009) GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 119, 1201–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knebel B., Haas J., Hartwig S., Jacob S., Köllmer C., Nitzgen U., Muller-Wieland D., Kotzka J. (2012) Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PLoS ONE 7, e31812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stevens J. F., Taylor A. W., Nickerson G. B., Ivancic M., Henning J., Haunold A., Deinzer M. L. (2000) Prenylflavonoid variation in Humulus lupulus: distribution and taxonomic significance of xanthogalenol and 4′-O-methylxanthohumol. Phytochemistry 53, 759–775 [DOI] [PubMed] [Google Scholar]
- 13. Miranda C. L., Stevens J. F., Ivanov V., McCall M., Frei B., Deinzer M. L., Buhler D. R. (2000) Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J. Agric. Food Chem. 48, 3876–3884 [DOI] [PubMed] [Google Scholar]
- 14. Lupinacci E., Meijerink J., Vincken J. P., Gabriele B., Gruppen H., Witkamp R. F. (2009) Xanthohumol from hop (Humulus lupulus L.) is an efficient inhibitor of monocyte chemoattractant protein-1 and tumor necrosis factor-α release in LPS-stimulated RAW 264.7 mouse macrophages and U937 human monocytes. J. Agric. Food Chem. 57, 7274–7281 [DOI] [PubMed] [Google Scholar]
- 15. Casaschi A., Maiyoh G. K., Rubio B. K., Li R. W., Adeli K., Theriault A. G. (2004) The chalcone xanthohumol inhibits triglyceride and apolipoprotein B secretion in HepG2 cells. J. Nutr. 134, 1340–1346 [DOI] [PubMed] [Google Scholar]
- 16. Nozawa H. (2005) Xanthohumol, the chalcone from beer hops (Humulus lupulus L.), is the ligand for farnesoid X receptor and ameliorates lipid and glucose metabolism in KK-A(y) mice. Biochem. Biophys. Res. Commun. 336, 754–761 [DOI] [PubMed] [Google Scholar]
- 17. Kirkwood J. S., Legette L. L., Miranda C. L., Jiang Y., Stevens J. F. (2013) A metabolomics-driven elucidation of the anti-obesity mechanisms of xanthohumol. J. Biol. Chem. 288, 19000–19013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Legette L. L., Luna A. Y., Reed R. L., Miranda C. L., Bobe G., Proteau R. R., Stevens J. F. (2013) Xanthohumol lowers body weight and fasting plasma glucose in obese male Zucker fa/fa rats. Phytochemistry 91, 236–241 [DOI] [PubMed] [Google Scholar]
- 19. Yui K., Kiyofuji A., Osada K. (2014) Effects of xanthohumol-rich extract from the hop on fatty acid metabolism in rats fed a high-fat diet. J. Oleo Sci. 63, 159–168 [DOI] [PubMed] [Google Scholar]
- 20. Sato R., Miyamoto W., Inoue J., Terada T., Imanaka T., Maeda M. (1999) Sterol regulatory element-binding protein negatively regulates microsomal triglyceride transfer protein gene transcription. J. Biol. Chem. 274, 24714–24720 [DOI] [PubMed] [Google Scholar]
- 21. Hirano Y., Murata S., Tanaka K., Shimizu M., Sato R. (2003) Sterol regulatory element-binding proteins are negatively regulated through SUMO-1 modification independent of the ubiquitin/26 S proteasome pathway. J. Biol. Chem. 278, 16809–16819 [DOI] [PubMed] [Google Scholar]
- 22. Miyata S., Inoue J., Shimizu M., Sato R. (2012) 4′-Hydroxyflavanone suppresses activation of sterol regulatory element-binding proteins and de novo lipid synthesis. FEBS Lett. 586, 1778–1782 [DOI] [PubMed] [Google Scholar]
- 23. Lee P. C., Sever N., Debose-Boyd R. A. (2005) Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J. Biol. Chem. 280, 25242–25249 [DOI] [PubMed] [Google Scholar]
- 24. Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. (2000) Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102, 315–323 [DOI] [PubMed] [Google Scholar]
- 25. Sato R., Inoue J., Kawabe Y., Kodama T., Takano T., Maeda M. (1996) Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 271, 26461–26464 [DOI] [PubMed] [Google Scholar]
- 26. Kalaany N. Y., Gauthier K. C., Zavacki A. M., Mammen P. P., Kitazume T., Peterson J. A., Horton J. D., Garry D. J., Bianco A. C., Mangelsdorf D. J. (2005) LXRs regulate the balance between fat storage and oxidation. Cell Metab. 1, 231–244 [DOI] [PubMed] [Google Scholar]
- 27. Radhakrishnan A., Goldstein J. L., McDonald J. G., Brown M. S. (2008) Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawatani M., Okumura H., Honda K., Kanoh N., Muroi M., Dohmae N., Takami M., Kitagawa M., Futamura Y., Imoto M., Osada H. (2008) The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I. Proc. Natl. Acad. Sci. U.S.A. 105, 11691–11696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun L. P., Seemann J., Goldstein J. L., Brown M. S. (2007) Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hirano Y., Yoshida M., Shimizu M., Sato R. (2001) Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J. Biol. Chem. 276, 36431–36437 [DOI] [PubMed] [Google Scholar]
- 31. Colgan S. M., Tang D., Werstuck G. H., Austin R. C. (2007) Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int. J. Biochem. Cell Biol. 39, 1843–1851 [DOI] [PubMed] [Google Scholar]
- 32. DeBose-Boyd R. A., Brown M. S., Li W. P., Nohturfft A., Goldstein J. L., Espenshade P. J. (1999) Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99, 703–712 [DOI] [PubMed] [Google Scholar]
- 33. Schindler A. J., Schekman R. (2009) In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc. Natl. Acad. Sci. U.S.A. 106, 17775–17780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ye J., Rawson R. B., Komuro R., Chen X., Davé U. P., Prywes R., Brown M. S., Goldstein J. L. (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 35. Tang J. J., Li J. G., Qi W., Qiu W. W., Li P. S., Li B. L., Song B. L. (2011) Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 13, 44–56 [DOI] [PubMed] [Google Scholar]
- 36. Lee J. N., Zhang X., Feramisco J. D., Gong Y., Ye J. (2008) Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J. Biol. Chem. 283, 33772–33783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C., Hamamoto S., Orci L., Schekman R. (2003) Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509 [DOI] [PubMed] [Google Scholar]
- 38. Mukai R., Fujikura Y., Murota K., Uehara M., Minekawa S., Matsui N., Kawamura T., Nemoto H., Terao J. (2013) Prenylation enhances quercetin uptake and reduces efflux in Caco-2 cells and enhances tissue accumulation in mice fed long-term. J. Nutr. 143, 1558–1564 [DOI] [PubMed] [Google Scholar]
- 39. Wunderlich S., Zürcher A., Back W. (2005) Enrichment of xanthohumol in the brewing process. Mol. Nutr. Food Res. 49, 874–881 [DOI] [PubMed] [Google Scholar]
- 40. Guo J., Nikolic D., Chadwick L. R., Pauli G. F., van Breemen R. B. (2006) Identification of human hepatic cytochrome P450 enzymes involved in the metabolism of 8-prenylnaringenin and isoxanthohumol from hops (Humulus lupulus L.). Drug Metab. Dispos. 34, 1152–1159 [DOI] [PubMed] [Google Scholar]
- 41. Possemiers S., Heyerick A., Robbens V., De Keukeleire D., Verstraete W. (2005) Activation of proestrogens from hops (Humulus lupulus L.) by intestinal microbiota; conversion of isoxanthohumol into 8-prenylnaringenin. J. Agric. Food Chem. 53, 6281–6288 [DOI] [PubMed] [Google Scholar]
- 42. Possemiers S., Bolca S., Grootaert C., Heyerick A., Decroos K., Dhooge W., De Keukeleire D., Rabot S., Verstraete W., Van de Wiele T. (2006) The prenylflavonoid isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8-prenylnaringenin in vitro and in the human intestine. J. Nutr. 136, 1862–1867 [DOI] [PubMed] [Google Scholar]
- 43. Hegarty B. D., Bobard A., Hainault I., Ferré P., Bossard P., Foufelle F. (2005) Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc. Natl. Acad. Sci. U.S.A. 102, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Owen J. L., Zhang Y., Bae S. H., Farooqi M. S., Liang G., Hammer R. E., Goldstein J. L., Brown M. S. (2012) Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl. Acad. Sci. U.S.A. 109, 16184–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Doddapattar P., Radoví B., Patankar J. V., Obrowsky S., Jandl K., Nusshold C., Kolb D., Vujić N., Doshi L., Chandak P. G., Goeritzer M., Ahammer H., Hoefler G., Sattler W., Kratky D. (2013) Xanthohumol ameliorates atherosclerotic plaque formation, hypercholesterolemia, and hepatic steatosis in ApoE-deficient mice. Mol. Nutr. Food Res. 57, 1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Xu S., Mihaylova M. M., Zheng B., Hou X., Jiang B., Park O., Luo Z., Lefai E., Shyy J. Y., Gao B., Wierzbicki M., Verbeuren T. J., Shaw R. J., Cohen R. A., Zang M. (2011) AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 13, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kamisuki S., Mao Q., Abu-Elheiga L., Gu Z., Kugimiya A., Kwon Y., Shinohara T., Kawazoe Y., Sato S., Asakura K., Choo H. Y., Sakai J., Wakil S. J., Uesugi M. (2009) A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16, 882–892 [DOI] [PubMed] [Google Scholar]
- 48. Stevens J. F., Page J. E. (2004) Xanthohumol and related prenylflavonoids from hops and beer: to your good health! Phytochemistry 65, 1317–1330 [DOI] [PubMed] [Google Scholar]