Background: Many organisms benefit from functional amyloids. Archaea represent the only domain of life without any described functional amyloids.
Results: Biophysical and recombinant techniques show that the sheaths of Methanosaeta thermophila PT are functional amyloid.
Conclusion: Archaea produce functional amyloids.
Significance: The amyloid nature explains many sheath properties that have puzzled researchers for years.
Keywords: amyloid, archaea, cell wall, protein folding, structure-function, functional amyloid, methanogen, sheath
Abstract
Archaea are renowned for their ability to thrive in extreme environments, although they can be found in virtually all habitats. Their adaptive success is linked to their unique cell envelopes that are extremely resistant to chemical and thermal denaturation and that resist proteolysis by common proteases. Here we employ amyloid-specific conformation antibodies and biophysical techniques to show that the extracellular cell wall sheaths encasing the methanogenic archaea Methanosaeta thermophila PT are functional amyloids. Depolymerization of sheaths and subsequent MS/MS analyses revealed that the sheaths are composed of a single major sheath protein (MspA). The amyloidogenic nature of MspA was confirmed by in vitro amyloid formation of recombinant MspA under a wide range of environmental conditions. This is the first report of a functional amyloid from the archaeal domain of life. The amyloid nature explains the extreme resistance of the sheath, the elastic properties that allow diffusible substrates to penetrate through expandable hoop boundaries, and how the sheaths are able to split and elongate outside the cell. The archaeal sheath amyloids do not share homology with any of the currently known functional amyloids and clearly represent a new function of the amyloid protein fold.
Introduction
Archaea form a distinct domain in the tree of life that is separate and unique from bacteria and eukarya (1). Upon their discovery, they were considered extremophiles, although it is now clear that archaea inhabit virtually all environments (2). The ability to thrive in extreme environments is in part due to their cell envelopes, which display distinctive features (3). These features include the use of unique ether lipids in the cell membrane and the replacement of the traditional bacterial peptidoglycan based cell wall with various proteinaceous crystalline layers on the cell surface, often in the form of an S layer (3). However, our understanding regarding the properties and functions of the archaeal cell envelopes remains superficial. Increased knowledge about the constituents of the archaeal cell envelopes is essential for understanding how specific archaea interact with their environment and how they are able to thrive in what we consider extreme environments.
Methanogens are strict anaerobic archaea belonging to the euryarchaeota. They are characterized by their unique ability to produce methane, using carbon dioxide, acetate, isopropanol, and various one-carbon compounds as substrates (2, 4). They play a fundamental role in driving the global biogeochemical carbon cycle, being the only known biological source of methane (4, 5). They also represent the backbone of the anaerobic digestion technology used for production of biogas as energy source, which is considered the most promising sustainable alternative to natural gas (6).
Methanogens of the two genera Methanosaeta and Methanospirillum are characterized by a complex cell wall architecture (3). These microorganisms are occasionally found as single rod-shaped cells encased within a protein layer, which for Methanospirillum has been characterized as an S layer (7, 8). However, more frequently they grow and divide within tubular paracrystalline sheaths to form long filaments. Inside the sheaths, septa or plugs separate the individual cells (8). The sheaths provide resistance against protozoan grazing and cell turgor pressure, while allowing the uptake of substrates like hydrogen, carbon dioxide, and acetate and the release of products, mainly methane (9).
Chemical analyses have shown that the sheaths are proteinaceous; however, the identities of the individual components that comprise them remain unknown (10, 11). Transmission electron microscopy (TEM)2 has previously revealed that the sheaths are composed of circumferential rings or hoops, which can be separated under strongly reducing conditions (10, 12). High angle electron diffraction showed that the sheaths contain components with a cross-β-sheet structure (12, 13). The sheaths furthermore display high stability against chemical (e.g. 6 m urea, 6 m GdmCl, 10 m LiSCN, or 1% SDS) and thermal (>100 °C) denaturation as well as resistance to proteolysis by common proteases (10, 14). Interestingly, most of these features are shared with a group of proteins known as functional amyloids (15).
Amyloids are defined as highly organized proteins or peptide polymers arranged in a cross-β-sheet pattern, in which intermolecular β-sheets are formed along the fibril axis with the β-strands oriented perpendicularly to the fibril axis (16). Functional amyloids were proposed in bacteria in 2002 with the identification of the adhesive curli fimbriae of Escherichia coli (17). The curli system has consequently become the model system for functional bacterial amyloids. However, the TasA fibrils from Bacillus subtilis (18) and Fap fimbria from Pseudomonas spp. (19, 20) also represent important model systems. The functional amyloids are associated with a large variety of functions in bacteria, ranging from structural components of biofilms to spore coat building blocks (15, 21).
Functional amyloids have also been described in eukaryotes. In fungi, they are used to modulate the hydrophobicity of hyphae and as a nongenetic but transmissible regulator of phenotypes (22, 23). Mammals benefit from functional amyloids in the form of scaffolds for melanin synthesis, which provide the basis for our skin pigmentation (24). They also serve as a storage form for peptide hormones (25).
Previous work involving amyloids in archaea is limited. In vitro studies have shown that the acylphosphatase from the hyperthermophile archaea Sulfolobus solfataricus is able to form amyloid-like aggregates directly from an active protein conformation at pH 5.5 in the presence of 15–25% 2,2,2-trifluoroethanol (26–28). However, whether there is any in vivo biological relevance of this observation remains unknown.
Another study has shown that components within the extracellular matrix of Haloferax volcanii biofilms are able to bind the amyloid dyes Congo red and thioflavin T (ThT) (29). This suggests that amyloids may exist within archaeal biofilms. However, an individual functional amyloid and its principle component in association with a particular cellular structure and function have never been shown within the archaeal domain of life.
In this study, we show that the cell wall sheath of the methanogenic archaea Methanosaeta thermophila PT is a functional amyloid assembly composed of the major sheath protein (MspA). The amyloid structure explains the very high resistance of the sheath, the elastic properties that allow diffusible substrates to penetrate the sheath through expandable hoop boundaries, and the ability of the extracellular sheaths to split and elongate to accommodate growth of the archaeal filaments.
Experimental Procedures
Growth Conditions
M. thermophila PT (DSM 6194) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ) and cultured in DSMZ medium 387 according to the instructions provided by DSMZ. The cells were harvested by centrifugation and stored in sealed glass bottles under an oxygen-free atmosphere containing N2 (90%) and CO2 (10%).
Purification of Sheaths
The proteinaceous sheaths were purified using a purification strategy similar to that used for functional amyloids in Pseudomonas (Fap) (19). Briefly, archaea were collected from 300 ml of culture by centrifugation (7500 × g, 15 min) and resuspended in 1 ml of buffer (10 mm Tris-HCl, pH 8.0) containing 0.1 mg/ml RNase A (Sigma-Aldrich, R6513), 0.1 mg/ml Turbo DNase (Ambion, AM2238), 1 mm MgCl2, and 0.1% Triton X-100. The sample was lysed by three cycles of freeze thawing using a −80 °C freezer and a thermo block at 37 °C. The sample was kept at each temperature for at least 15 min. The sample was then incubated (37 °C, 2 h, 750 rpm) for enzymatic degradation of released nucleic acids. SDS was added to 2% and the sampled boiled for 5 min to dissolve the contaminating non-amyloid proteins. Insoluble material was collected by centrifugation (21,000 × g, 10 min), washed in 1 ml of buffer, and boiled once more in 2% SDS. The remaining insoluble material composed of the sheaths was then washed three times in 1 ml of buffer before it was finally resuspended in 500 μl of buffer containing 1 mg/ml sodium azide.
Depolymerization of Sheaths
Purified sheaths were depolymerized using a formic acid procedure. Briefly, 10 μl samples of purified sheaths were mixed with 5 μl of reducing buffer (200 mm Tris-HCl, 200 mm dithiothreitol (DTT), pH 8.0) and incubated at 37 °C for 30 min. The samples were then lyophilized and depolymerized in 100 μl of either 90% or 100% formic acid. The samples were lyophilized again and analyzed by SDS-PAGE.
SDS-PAGE of Sheath Monomers
The lyophilized samples were dissolved in 20 μl of reducing SDS-PAGE loading buffer (75 mm M Tris, 0.6% (w/w) SDS, 15% (v/v) glycerol, 7.5% (v/v) β-mercaptoethanol, 0.9 mg/liter bromphenol blue, 8 m urea at pH 6.8). Urea was required to avoid precipitation of the sheath proteins. 10 μl were loaded to an AnyKD gel (Bio-Rad), and electrophoresis was done at 140 V for 40 min. Gels were stained with Coomassie Brilliant Blue G250.
Mass Spectrometry
Gel bands were excised and cut into pieces. The gel plugs were then washed with 50 mm ammonium bicarbonate and 30% (v/v) acetonitrile, followed by reduction and alkylation with DTT and iodoacetamide, respectively, in consecutive incubation steps. The gel plugs were dried and rehydrated with 10 ng/μl sequencing grade trypsin (Promega). Trypsin digestion was done overnight at 37 °C. The supernatant and the following organic extract containing the digested peptides were combined and dried. The digests were reconstituted in 30 μl of LC-MS sample buffer (2% acetonitrile, 0.1% formic acid).
A sample volume of 10 μl was injected onto a Dionex Ultimate 3000 nanoLC system that was connected to a Quadrupole Orbitrap (Q Exactive) mass spectrometer equipped with a NanoSpray Flex ion source. Both instruments were from Thermo Scientific. The flow settings were 8 μl/min for the sample loading onto a trapping column, which was an Acclaim PepMap100 C18, 5-μm column from Thermo Scientific. The nanoflow was set to 300 nl/min for the peptide separation on the analytical column, which was a 50-cm Acclaim PepmapRSLC, 75-μm column connected with nanoviper fittings. The nano-electrospray was done using a Picotip Silicatip emitter from New Objectives. The LC buffers were buffer A (0.1% formic acid, 0.005% heptafluorobutyric acid) and buffer B (90% acetonitrile, 0.1% formic acid, 0.005% heptafluorobutyric acid). The applied gradient was from 12 to 40% buffer B over 30 min.
The mass spectrometer was operated in data-dependent acquisition mode. A full MS scan in the mass range of 350–1850 m/z was acquired at a resolution of 70,000 with an AGC target of 3 × 106 and maximum fill time set to 250 ms. Instrument lock mass correction was applied using the contaminant ion at 391.28429 m/z. In each cycle, the mass spectrometer will trigger up to 12 MS/MS acquisitions on abundant peptide precursors ions. The MS/MS scans were acquired with a dynamic mass range at a resolution of 17,500 and with an AGC target of 2 × 105 and a maximum fill time of 60 ms. The precursor ions were isolated using a quadruple isolation window of 3 m/z and the fragmented in the HCD trap with a normalized collision energy set to 30. The under-fill ratio was set to 3.5% with the intensity threshold at 1.2 × 105. Apex triggering was 3–10 s with charge and exclude isotopes exclusion on, and dynamic exclusion set to 15 s.
Peak lists were generated from the raw data files using ProteomeDiscoverer version 1.3 (ThermoScientific). Mascot server version 2.3 (Matrix Sciences) was used for searching against the Uniprot complete proteome databases for M. thermophila PT (METTP, released November 14, 2006). The search criteria were mass tolerances of 5 and 15 ppm on precursor and fragment level, respectively. No enzyme specificity and two missed cleavages were allowed. Cysteine carboxyamidomethylation was set as fixed, and methionine oxidation and lysine formylation were set as variable modifications. The peptide probability p value was set 0.01. The Mascot scores, protein sequence coverage, spectral counts, and emPAI scores were recorded from the Mascot search log.
Transmission Electron Microscopy
Suspension culture samples, purified sheaths and in vitro formed fibrils were mounted on 400-mesh carbon-coated, glow discharged nickel grids for 30 s. Grids were washed with one drop of double distilled water (ddH2O) and stained with three drops of 1% (w/v) phosphotungstic acid at pH 7.2. Samples were inspected in a transmission electron microscope (JEOL, 1010) at 60 kV. Images were obtained using an electron-sensitive CCD camera (Olympus, KeenView). For size determination, a cross-line carbon replica grid (2160 lines/mm) was used.
Binding of Amyloid-specific Antibodies
The presence of amyloid epitopes was analyzed using the amyloid conformational specific antibody WO1 as previously described (30). Briefly, cells from 500 μl of culture of purified sheaths corresponding to an optical density at 600 nm of 1 were pelleted by centrifugation (21,000 × g, 30 min), washed in 1 ml of PBS, and then resuspended in 1 ml of PBS containing 1% (w/v) gelatin as a blocking buffer. The sample was preincubated (1 h, 37 °C), after which antibodies WO1 and Tween 20 were added to a final concentration of 10 nm and 0.05% (w/v), respectively. The sample was incubated (2 h, 37 °C). The primary antibody was removed by centrifugation, and the pellets were washed three times in 400 μl of PBS containing 1% (w/v) gelatin and 0.1% (v/v) Triton X-100. The sample was then incubated (1 h, 37 °C) with a 1:256 dilution of the Alexa Fluor® 488 goat anti-mouse IgM (μ chain) (Molecular Probes) and 0.025% Tween 20 in PBS containing 1% (w/v) gelatin. Finally, the samples were washed three times as described above but without gelatin in the washing buffer. After the final washing step, the samples were resuspended in 100 μl of PBS and stored at 4 °C until microscopy.
Images were obtained using a LSM 510 META confocal laser scanning microscope (Carl Zeiss) equipped with an argon ion laser (458 and 488 nm) and two helium-neon lasers (543 and 633 nm). The settings for the CLSM were calibrated using an activated sludge sample containing both amyloid-positive and -negative cells as previously described (30). All samples were also prepared in the absence of the primary antibody. These samples served as controls to evaluate unspecific binding of the secondary antibody.
FTIR
FTIR was carried out using a Tensor 27 (Bruker) FTIR spectrophotometer equipped with a DTGS Mid-infrared detector and a Golden Gate single reflection diamond attenuated total reflectance (ATR) cell (Specac). Purified sheaths or in vitro polymerized sheath proteins were pelleted by centrifugation and dried on the ATR crystal using dry nitrogen. In vitro polymerized sheath proteins were pelleted (100,000 × g, 30 min, 4 °C) in a MAX XP Ultracentrifuge (Optima) with a TLA 110 rotor, resuspended in ddH2O, and pelleted again. Pellets were resuspended in a small volume of ddH2O and dried on the ATR crystal. ATR spectra were recorded from 4000–1000 cm−1 using a nominal resolution of 2 cm−1 and 128 accumulations. The resulting spectra were baseline-corrected and interfering signals from H2O and CO2 were removed using the atmospheric compensation filter in the OPUS 5.5 system (Bruker). Different components of the amide I region were identified by second derivative analysis in the OPUS 5.5 system.
Circular Dichroism
CD spectra from 250–200 nm were collected on a Jasco J-810 spectropolarimeter using 0.2-nm steps, scan speed of 50 nm/min, bandwidth of 2 nm, and a response time of 2 s. A 1-mm quartz cuvette (Hellma) was used, and the temperature was kept constant at 20 °C with a thermostatically controlled cell holder (Jasco PTC 423S). The protein concentration was diluted to 0.25 mg/ml with ddH2O, and all spectra were baseline-corrected with respect to the buffer. To improve the signal to noise ratio, five scans were averaged on each sample. Data points with a HT value higher than 600 V were removed. The results were expressed as mean residue ellipticity. All samples were sonicated in a Bandelin Sonorex Digitec water bath sonicator for 2 min to minimize light scattering and settling of the insoluble amyloids.
X-ray Fiber Diffraction
Fiber diffraction specimens were prepared by suspending 10 μl of 5 mg/ml purified sheaths or 1 mg/ml in vitro fibrillated MspA between two capillary glass tubes. Samples were dried at room temperature resulting in fibers with a length of 2–3 mm. The data were collected using a sealed tube x-ray source (wavelength of 1.5418 Å) in a Compact HomeLab system (Rigaku), equipped with a PILATUS3 R 200K hybrid pixel array detector (Dectris). The sample to detector distance was 60 mm, and an exposure time of 20–30 min was used. The images were evaluated and Bragg distances measured using Adxv 1.9.10.
ThT Binding
Fluorescence spectra were recorded on a Varian Cary Fluorimeter (Varian, Palo Alto, CA) with a built-in Peltier element for temperature regulation. Proteins were dissolved in PBS buffer to a final protein concentration of 50 μg/ml. The final concentration of ThT was 40 μm. The samples were excited at 440 nm, and emission was measured from 450 to 600 nm with a slit with of 5 nm for both excitation and emission.
Preparation of Recombinant Sheath Monomers
A synthetic M. thermophila PT mspA gene (Mthe_1069), optimized for the E. coli codon usage corresponding to the mature protein (without signal sequence) flanked by XbaI and XhoI restriction sites, were designed using CLC main workbench 6.0 and synthesized by GenScript (Germany). The synthetic gene was cloned into the pET32b(+) vector (Novagen) using the XbaI and XhoI restriction sites. This adds a C-terminal His tag to the construct. The plasmid was transformed into E. coli BL21 (DE3) (Life Technologies). Protein expression was carried out in 500 ml of LB medium (37 °C, 200 rpm). Induction was done with 1 mm isopropyl β-d-thiogalactopyranoside (IPTG) at A600 nm = 0.7–1.2, and cells were harvested 3 h after induction by centrifugation (10,000 × g, 30 min). The cell pellet was resuspended in 25 ml of extraction buffer (6 m GdmCl, 20 mm sodium phosphate, 500 mm NaCl, 20 mm imidazole, pH 7.4) and sonicated three times for 1 min using a rod sonicator. The sample was placed 1 min on ice between sonications. The lysate was then incubated overnight at 4 °C with gentle shaking. Insoluble material was pelleted by centrifugation (20,000 × g, 30 min). The His-tagged MspA protein was isolated from the supernatant using a 5-ml HisTrap HP column (GE Healthcare) and an elution buffer, corresponding to the extraction buffer, containing 500 mm imidazole.
In Vitro Fibrillation
Purified monomers in extraction buffer were desalted using a PD-10 desalting column (GE Healthcare) equilibrated with ddH2O. Protein concentration was estimated by UV absorbance using the following extinction coefficients for the recombinant MspA from M. thermophila PT: ϵ280 nm = 1.885 cm−1·(g/liter)−1. The proteins were diluted in ddH2O to 1 mg/ml and mixed with an equal amount of 40 mm buffer (see below). 200 μl samples were loaded into a 96-well Nunc optical bottom plate (catalog no. 265301; Thermo Scientific). Immediately afterward, the plate was transferred to a Tecan GENios Pro plate reader and incubated at 37 °C overnight with 30 s of linear shaking at 180 rpm applied every 84 s. The following buffers were used: citric acid (pH 3.0 and 4.0), acetic acid (pH 5.0), MES (pH 6.0), imidazole (pH 7.0), Tris (pH 8.0), and glycine (pH 9.0). All buffers were adjusted to an ionic strength of 60 and 300 mm with NaCl.
Stopped Flow Kinetics
5 ml of purified monomers in extraction buffer was dialyzed against 1 liter of GdmCl buffer (6 m GdmCl, 5 mm phosphate, pH 7.0) twice to remove imidazole from the purification. α-synuclein was purified as previously described (31) and solubilized in GdmCl buffer. Concentration was determined by UV absorption. Fibrillation kinetics was measured on a stopped flow apparatus SX18.MV rapid reaction microanalyzer from Applied Photophysics (Leatherhead, Surrey, UK) with the settings: photomultiplier, 400 V; filter, 1 ms; excitation, 450 nm with 4.7-nm slit width; emission, 470-nm filter; time, 200 s; 25 °C and with a 1:9 dilution into low ionic strength buffers as above. Final GdmCl and protein concentrations were 0.6 m and 0.46 mg/ml, respectively. ThT was added to the buffer solution resulting in a final ThT concentration of 40 μm. All experiments were done in triplicate. The first 30 ms was removed because of mixing effects.
Bioinformatic Analysis
The blastP algorithm (32) was used with standard settings to identify sheath protein homologs within and the NCBI Reference Sequence Database (RefSeq). Repeat regions were identified using the Rapid automatic detection and alignment of repeats in protein sequences (Radar) tool (33). Amyloid propensity was determined using the AmylPred2 algorithm using the specified output option (34).
Results
The cell wall sheaths of filamentous methanogens share many traits with functional amyloids, but there is currently insufficient evidence to conclude that the sheaths are amyloid structures (16). We therefore analyzed the sheaths of M. thermophila PT using methods derived from the functional amyloid field. M. thermophila PT was chosen as the model organism because its genome has been sequenced, and it represents an important genus of acetoclastic methanogens in relation to anaerobic digestion (35–37).
The Sheaths of Intact Filaments Display Amyloid Morphology and Are Targeted by Amyloid Conformational Antibodies
TEM was used to visualize the morphology of the cell wall sheaths on intact filaments (Fig. 1A). The sheaths displayed 17-nm wide striations perpendicular to the filament length, similar to those previously observed on the sheaths of Methanospirillum hungatei and Methanosaeta concilii (10, 12). The sheaths of M. thermophila PT are therefore proposed also to be composed of stacked hoops. The width of these hoops is consistent with the common widths of mature amyloid fibrils, which are in the range of 6–20 nm (38). This led us to hypothesize that the hoops represent circulated amyloid fibrils. Clusters of gas vesicles were also observed within the filaments (Fig. 1A).
FIGURE 1.
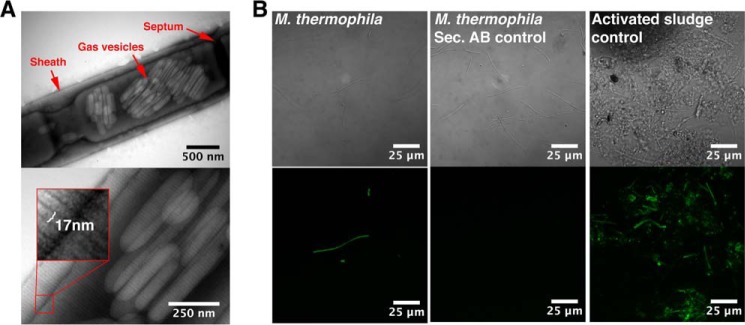
The sheaths on intact filaments display regular striations perpendicular to the filament length and bind the amyloid-specific conformational antibody WO1. A, transmission electron micrographs of intact filaments. The width of the striations is highlighted in the magnified inset. B, binding of the amyloid specific antibody (WO1) to the intact filaments. Differential interference contrast images are shown in the top row, and the green fluorescence signals from the antibodies are shown in the bottom row. Activated sludge from a wastewater treatment plant known to contain both amyloid-positive and -negative cells (40) was used to adjust the settings of the microscope. Secondary controls with only the secondary antibody were included to rule out unspecific binding of the secondary antibody.
The amyloid conformational specific antibody WO1 has previously proved useful for the detection of amyloid-like epitopes in complex samples (39–41). It was therefore investigated whether the WO1 antibody was able to target the sheaths of intact M. thermophila PT filaments. The filaments showed strong binding of WO1 (Fig. 1B), confirming the presence of exposed amyloid-like epitopes on the outer surfaces of the sheaths. Addition evidence is, however, required to determine whether these epitopes reflect a true amyloid structure.
Purified Sheaths Display Amyloid Properties
Confirming a novel functional amyloid is challenging and requires validation of the fibrillar morphology by high resolution microscopy techniques and specific binding of amyloid dyes (Congo red or ThT) or amyloid-conformational antibodies (WO1). In addition, the amyloid β-sheet secondary structure must be confirmed for purified amyloids (15, 16). The sheaths were therefore isolated using a purification strategy that exploits the high structural stability of functional amyloids (20). This purification strategy involved cell lysis by freeze-thawing, enzymatic hydrolysis of nucleic acids, and solubilization of contaminating non-amyloid protein by boiling in 2% SDS. Successful sheath purification was confirmed by TEM analysis (Fig. 2A). The 17-nm-wide striations perpendicular to the filament length were intact after purification, confirming the ability of the sheaths to withstand the harsh purification procedure.
FIGURE 2.
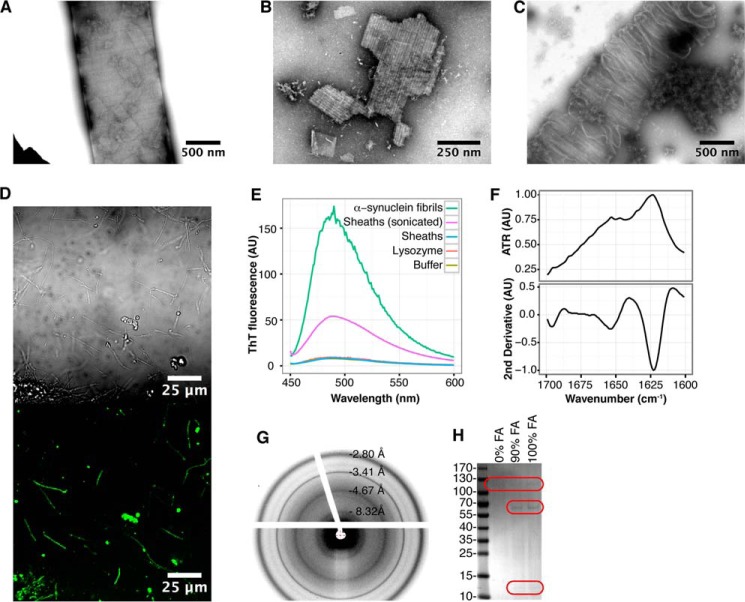
Purified sheaths display amyloid-like properties. A–C, transmission electron micrographs of purified sheaths that were untreated (A), sonicated (B), or partly depolymerized (C) using DTT and formic acid. D, binding of the amyloid specific antibody (WO1) to purified sheaths. A differential interference contrast image is shown in the top panel, and the green fluorescence signal from the antibody is shown in the bottom panel. The setting of the microscope was the same as used for obtaining the images in Fig. 1B. E, fluorescence emission of ThT excited at 440 nm when mixed with 50 μg/ml untreated and sonicated sheaths. α-Synuclein fibrils and lysozyme monomers were used as positive and negative controls, respectively. F, FTIR spectra of purified sheaths. G, x-ray fiber diffraction pattern of purified sheaths. H, SDS-PAGE of sheaths depolymerized by DTT and formic acid (FA) at various concentrations. Red boxes highlight the faint protein bands. The numbers to the left of the SDS-PAGE gel indicate the molecular mass (kDa) of the marker proteins.
The gas vesicles observed within the intact filaments (Fig. 1A) were lysed during the purification procedure and remained as empty shells within the purified sheaths (Fig. 2A). The shells of gas vesicles are proteinaceous and display many similarities to the archaeal sheaths, including a high content of cross-β-sheet structure (42, 43). These shells may consequently represent a second amyloid structure within the Archaea. Fragmentation of the purified sheath by sonication often occurred between individual hoops (Fig. 2B), suggesting a stronger stability of the sheaths perpendicular to the filament length. Brief treatment of the sheaths with the reducing agent DTT and formic acid resulted in sheath disassembly into bundles of a few hoops (Fig. 2C) or occasionally individual hoops. This strongly supports a sheath structure in which disulfide bonds are used to interlink the individual hoops.
Binding of the amyloid-specific antibody WO1 to the purified sheaths were evaluated to confirm the in situ data (Fig. 2D). A strong binding of WO1 to the sheaths was observed, confirming the presence of amyloid-like epitopes on the sheaths. Although amyloid-specific antibodies are useful for the identification of amyloid-like epitopes in situ, they are not completely specific. The binding of the amyloid dye ThT to the sheaths was therefore also evaluated (Fig. 2E). The ThT emission spectrum of sonicated sheaths was comparable with that of α-synuclein amyloid fibrils. A strong fluorescence emission at 485 nm showed that ThT binds specifically to the purified sheaths, supporting an amyloid-like structure (44, 45). The negative controls (buffer and lysozyme monomers) did not show any binding of ThT. Untreated sheaths were not able to bind ThT either, suggesting that the ThT binding epitopes were buried within the compact sheath structure.
Fourier transform infrared spectroscopy (FTIR) was used to evaluate the secondary structure of the sheaths (Fig. 2F). The presence of an intense amide I peak at 1618 cm−1 demonstrated the presence of extended β-sheet structures, supporting an amyloid structure of the sheaths (46).
Although the FTIR data provide strong support for an amyloid structure, these data cannot be used to prove the defining cross-β-sheet of amyloids. The sheaths were therefore subjected to x-ray fiber diffraction analysis (Fig. 2C). This revealed a clear reflection at 4.66 Å and a weaker reflection at 8.35 Å, indicative of a cross-β-sheet conformation (47). Additional strong reflections were observed at 2.82 and 3.42 Å. However, the structural basis of these reflections is unknown. Combined, the data clearly show that the sheaths are amyloid structures.
Sheaths Are Composed of the Major Protein Subunit (MspA)
The purified sheaths were treated with DTT and formic acid to release the sheath monomers. A sample lacking the formic acid treatment was included to distinguish the amyloid proteins from contaminants. The latter are equally abundant in treated and nontreated samples. The samples were subsequently separated by SDS-PAGE optimized for amyloid proteins (Fig. 2H) and analyzed by mass spectrometry (Table 1) (20). Mass spectrometry revealed that the sheaths were composed of a major protein subunit of hitherto unassigned function, which we named MspA (major sheath protein A). The presence of gas vesicle shell proteins (GvpA) was furthermore observed (Table 1). Genomic analyses showed that the open reading frame encoding MspA was not part of an operon structure. A 22-amino acid residue signal peptide was predicted in the N terminus of MspA using the SignalP algorithm (48), suggesting secretion of the protein. However, it was not possible to assign the secretion to a specific secretion system. MspA contained three internal cysteine residues, which may allow the formation of intermolecular disulfide bridges between adjacent hoops, either directly or by the help of linker or glue peptides as previously suggested (10).
TABLE 1.
Proteins identified by MS/MS after depolymerization of purified sheaths with DTT and formic acid
A sample lacking the formic acid treatment was included to distinguish the amyloid proteins from contaminants. The latter are equally abundant in treated and untreated samples. The major sheath protein (MspA) is highlighted in bold type and is identified by its increased abundance in the formic acid-treated sample as evident from the emPAI score. The major contaminating proteins are gas vesicle proteins, which are known to form amyloid-like structures on the surface of gas vesicles (42). An abundant S layer-related protein, which shows similar abundance in the formic acid treated and untreated samples, is furthermore observed. FA, formic acid.
| RefSeq accession nb | Protein description | Molecular massa | Mascot score |
Protein sequence coverage |
Spectral MS/MS counts (Mascot) |
emPAI (Mascot)b |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0% FA | 100% FA | 0% FA | 100% FA | 0% FA | 100% FA | 0% FA | 100% FA | |||
| kDa | % | |||||||||
| YP_843493 | Putative uncharacterized protein | 61.1 | 164 | 2534 | 18 | 77 | 17 | 158 | 0.11 | 7.51 |
| YP_842502 | Gas vesicle structural protein | 8 | 0 | 496 | 0 | 98 | 0 | 20 | 0 | 11.75 |
| YP_842503 | Gas vesicle structural protein | 7.8 | 0 | 498 | 0 | 98 | 0 | 23 | 0 | 12.16 |
| YP_843107 | S layer-related duplication domain | 79.8 | 1031 | 1441 | 24 | 24 | 125 | 139 | 1.72 | 5.58 |
| YP_842724 | Acetyl-CoA decarbonylase/synthase gamma subunit | 53.3 | 86 | 43 | 39 | 46 | 12 | 14 | 0.05 | 0.06 |
| YP_842729 | Acetyl-CoA decarbonylase/synthase complex subunit α | 91.1 | 323 | 431 | 28 | 39 | 80 | 109 | 0.47 | 0.88 |
| YP_843615 | Acetyl-coenzyme A synthetase | 74.5 | 405 | 560 | 29 | 37 | 59 | 87 | 0.67 | 1.26 |
| YP_843000 | Methyl-coenzyme M reductase, α subunit | 61.8 | 127 | 180 | 47 | 54 | 26 | 23 | 0.36 | 0.76 |
| YP_843003 | Methyl-coenzyme M reductase, β subunit | 46 | 167 | 204 | 36 | 54 | 35 | 34 | 0.41 | 0.41 |
a The gel migration generally correlated well with the molecular weights of the identified proteins.
b The Exponentially Modified Protein Abundance Index (emPAI) offers approximate, label-free, relative quantitation of the proteins in a mixture based on protein coverage by the peptide matches in a database search result (73).
Homology searches within the NCBI Reference Sequence Database (RefSeq) using either the BlastP or BlastN algorithm were not able to identify any homologs to MspA from M. thermophila PT. Low sequence similarity between MspA proteins from the closely related species combined with the small number of sequenced archaea genomes may explain why no homologs could be identified in other Methanosaeta strains.
Evolutionary studies have shown that the E. coli curli (CsgA) Pseudomonas Fap (FapC) functional amyloids display low sequence similarity between homologs from closely related species (49, 50). The TasA functional amyloid of B. subtilis is another example of a genus-specific amyloid protein (18). These observations suggest that the amyloid properties can be maintained despite large changes in the primary structure of amyloid proteins.
MspA Shares Structural Features with Known Functional Amyloids
Only a few functional amyloid proteins are presently known, and most of them contain partially conserved repeat units in their primary structure (15). Such repeat units could also be found in MspA, but they were substantially less conserved than those of the curli and Fap amyloid proteins in E. coli and pseudomonads, respectively (results not shown). Interestingly, MspA displayed an increased frequency in the use of tiny or small residues (63.7% versus 49.8%) compared with the average for the proteins in M. thermophila PT and a decrease in the frequency of large aliphatic (8.2% versus 16.8%) and positively charged (7.7% versus 13.3%) residues. Similar usage of residues is seen for other functional amyloids. This allows for a more compact amyloid core, which could in part explain the high structural stability of functional amyloids (17, 19, 51–53).
A variety of bioinformatic tools has been developed to identify amyloidogenic hotspots within proteins. The AmylPred2 algorithm determines amyloidogenic regions based on the consensus score of 11 of such tools and thus provides a reliable estimate of the amyloid propensity (34). AmylPred2 confirmed the presence of amyloidogenic regions within MspA (Fig. 3). The amyloid propensity was especially pronounced in the C terminus of the protein, suggesting that this region is critical for amyloid formation.
FIGURE 3.
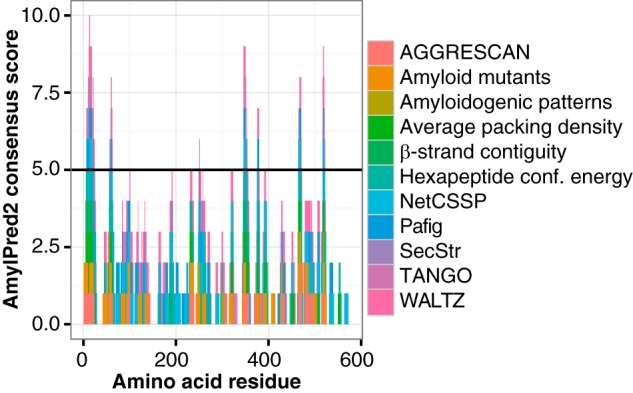
Identification of amyloidogenic hotspots within the major sheath protein MspA using the AmylPred2 algorithm. Predictions from individual methods are included, and positive predictions of at least five methods are considered a positive hit (horizontal line).
MspA Is Highly Amyloidgenic
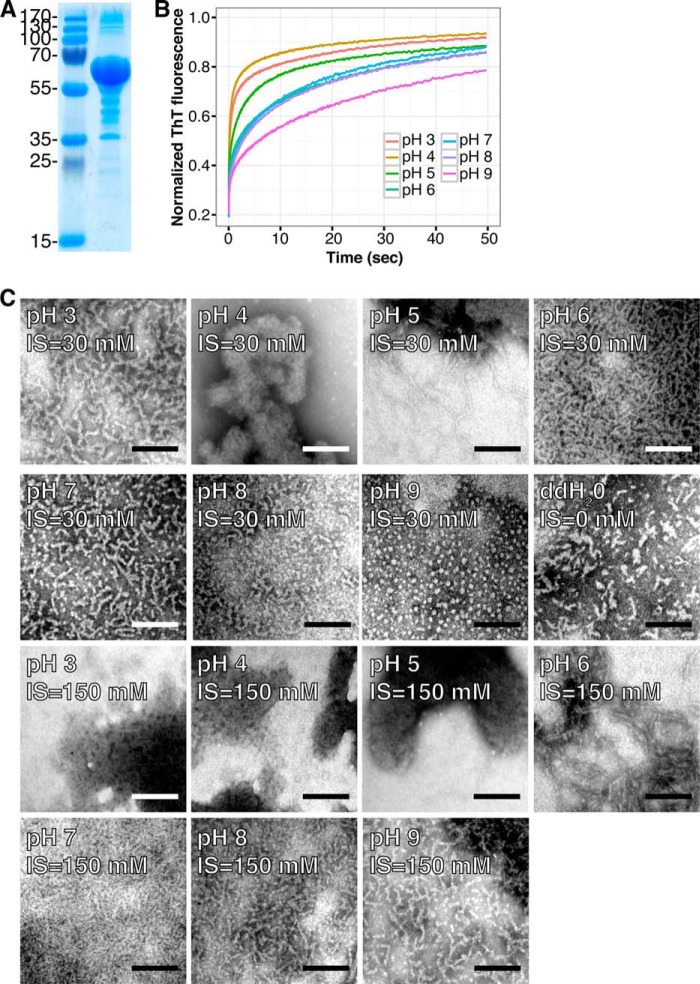
Functional amyloid proteins are often able to self-assemble into amyloid fibrils in vitro under physiological conditions, a feature rarely seen for non-amyloid proteins (54–56). MspA was therefore cloned and recombinantly expressed in E. coli to determine its amyloidic propensity (Fig. 4A). Initial attempts to asses the fibrillation kinetic with ThT in a plate reader setup, similar to that previously used to study fibrillation of the curli amyloid protein CsgA (56), were unsuccessful. A high initial ThT signal indicated that the fibrillation process could be extremely rapid and might have occurred already before data could be collected. This was confirmed by stopped flow experiments (Fig. 4B) showing that MspA was able to fibrillate within 1 min after a 10-fold dilution from a stock solution in 6 m GdmCl into the appropriated GdmCl-free buffer. Similar rapid fibrillation kinetics have previously been shown for the functional amyloid Pmel-17, which is involved in melanin biosynthesis (24, 57). MspA was able to fibrillate at all examined pH values, ranging from pH 3 to pH 9, and the fibrillation kinetics in general increased with decreasing pH of the buffer used. The ThT fluorescence of the MspA monomer in 6 m GdmCl was similar to that of 6 m GdmCl alone, confirming that there were no fibril seeds in the monomer stock solution. The disease-related amyloid protein α-synuclein, which was included as a control, did not fibrillate in the time frame analyzed.
FIGURE 4.
Recombinant MspA monomers are extremely amyloidogenic and form amyloids in vitro under a wide range of conditions. A, SDS-PAGE of purified recombinant MspA protein. The gel was overloaded to visualize impurities. These accounted for <5% of the total proteins. The numbers to the left indicate the molecular mass (kDa) of the marker proteins. B, fibrillation kinetics evaluated by stopped flow and ThT fluorescence at various pH values. C, transmission electron micrographs of MspA amyloids formed in vitro at various pH values with low (30 mm) and high (150 mm) ionic strength (IS). The scale bars represent 100 nm.
Samples of MspA that were incubated under the wide range of environmental conditions were analyzed by TEM to confirm the formation of amyloid fibrils (Fig. 4C). Small fibrils were evident at physiological conditions (pH 6–8) at low and high ionic strength. These fibrils resembled those formed in vitro by the SC3 hydrophobin from the fungus Schizophyllum commune and of chaplin proteins of Streptomyces coelicolor (58, 59). MspA also formed fibrils at pH 3 and 5 in low ionic strength buffers, at pH 9 in high ionic strength buffers, and in deionized water. MspA showed a tendency to form amorphous aggregates close to its isoelectric point (pI = 4.66). This was especially evident at high ionic strength, where large aggregates were evident at pH 3, 4, and 5. Interestingly, regularly sized oligomers were observed at pH 9 in the low ionic strength buffer. The relatively slow fibrillation kinetics at pH 9 could be a requirement for the formation of the ordered structures.
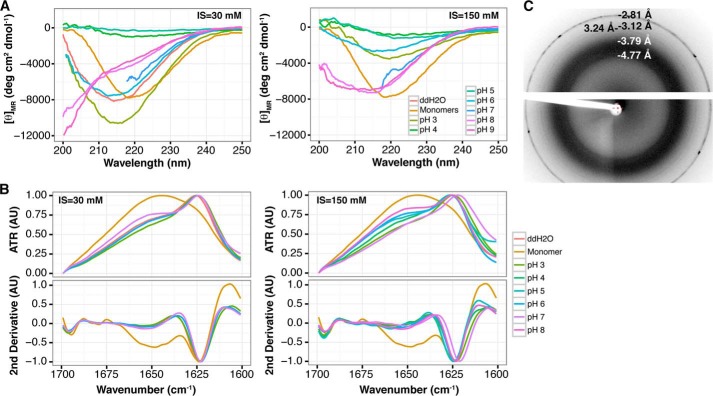
The secondary structure of soluble and suspended MspA was determined for all samples using CD (Fig. 5A). The CD spectra confirmed that the MspA fibrils in suspension had a high proportion of β-sheet structure as evident from the single minimum at ∼217 nm (60). The samples prepared using buffers with pH values close to the pI of MspA produced weak signals because of sample precipitation. The secondary structure of the insoluble material was determined by FTIR (Fig. 5B). A major peak between 1622 and 1625 cm−1 was seen for all samples, confirming the presence amyloid-like extended β-sheet structures (46). A few samples were analyzed by x-ray fiber diffraction to confirm the cross-β-sheet (Fig. 5C). However, the sample heterogeneity made it impossible to obtain high quality diffraction patterns. However, a faint band at 4.77 Å was observed, supporting a cross-β-sheet structure. Combined, the TEM, CD, FTIR, and x-ray fiber diffraction results strongly support our hypothesis that MspA is an amyloidogenic protein.
FIGURE 5.
In vitro polymerized MspA has amyloid-like secondary structure. A, CD spectra of samples containing MspA amyloids formed in vitro at various pH values with low (30 mm) and high (150 mm) ionic strength (IS). CD does not measure large amyloid aggregates that have precipitated. B, FTIR spectra of insoluble MspA amyloids formed in vitro at the same conditions as above. The insoluble amyloids were isolated by centrifugation prior to the FTIR analysis. C, x-ray fiber diffraction pattern of MspA polymerized in vitro at pH 7 at low ionic strength.
Discussion
The special tubular cell wall sheaths of the methanogenic archaea M. thermophila was shown to be functional amyloid assemblies composed of the major sheath protein (MspA). This is the first report about a functional amyloid from archaea.
Gas vesicles were also isolated from M. thermophila PT using the applied purification strategy. MS analysis furthermore showed that these required treatment with formic acid to be depolymerized. Accordingly, it might be hypothesized that these may represent another example of archaeal amyloids. This is supported by previous solid state NMR correlation spectroscopy data that provided evidence for an extended cross-β structure in gas vesicles from cyanobacteria (42, 61).
Although MspA is able to form amyloid fibrils in vitro, they are not able to spontaneously assemble into ordered tubular sheaths. This suggests the requirement of axillary factors for sheath biogenesis. Such factors could include the presence of scaffolds as is known from the formation of the highly organized rodlet layer on the surface of aerial hyphae and spores of S. coelicolor, which depends upon the interplay between rodlin proteins and the amyloidogenic chaplins (62). The sheaths of M. hungatei GP1 was previously shown to contain phenol-soluble polypeptides of unknown structure that conferred rigidity to the sheath (63). Such polypeptides were not observed in this study, but because of their unique physiochemical properties, they might not be applicable for the MS analyses. The axillary factors could also be chaperones that guide the amyloid formation as is seen for the curli system (21) or glue peptides that bind the individual hoops together as previously suggested (10). However, in the current study we were not able to identify such factors. Previously, sheaths of filamentous methanogens have been viewed as impermeable barriers, and consequently substrates and waste products had to transverse the sheaths though pores located between sheath subunits (13).
We propose a model of the sheaths (Fig. 6) that is based on the primary protein structure of the MspA proteins combined with previously published electron diffraction data (13). The large size and the primary structure of MspA suggest that the protein forms a β-solenoid structure in analogy to the amyloid structure of E. coli curli (64–66) and Het-s fibers (67, 68). The amyloid cores of circulated MspA fibrils form individual hoops, and dual intermolecular disulfide bridges between adjacent hoops are proposed formed from cysteine residues found within the more flexible noncore regions of the fibrils, either directly or via small glue peptides. The presence of disulfide bridges between hoops is supported by the observation that isolated sheaths are disassembled to the individual hoops upon treatment with the reducing agent DTT and formic acid. A similar observation was previously made after treatment of sheaths from M. hungatei GP1 with β-mercaptoethanol at 90 °C (10).
FIGURE 6.
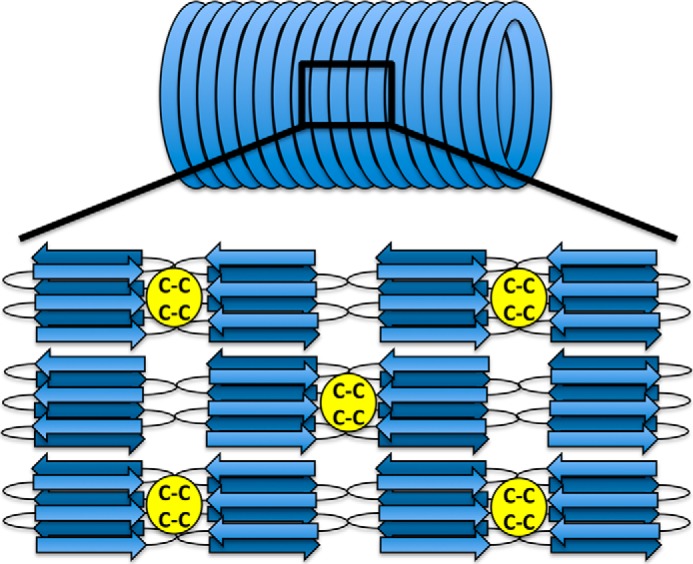
A hypothetical model of the organization of MspA molecules within the sheath. The structure is based on our data and previous electron diffraction data (13). Two closely spaced cysteine residues found within the middle of the MspA sequence may allow for the formation of intermolecular disulfide bonds that links the hoops together. The linkage could either be direct or mediated by small linker peptides. The exact number of β-strands formed by each MspA subunit is not known.
In this model, diffusible substrates are able to penetrate the sheaths through expandable hoop boundaries. This hypothesis is in agreement with studies on the elastic properties of archaeal sheaths, which show that the sheaths are able to creep with increasing pressure (69). Consequently, the sheaths may act as pressure regulators, allowing gas to escape only above a certain pressure.
Growth of the methanogenic filaments by cell expansion and division is accompanied by sheath splitting and elongation (70). This is only possible when the sheath precursors are able to integrate with the existing surface macromolecules (63). The autocatalytic or nucleated formation of amyloid structures explains how the sheaths can split and elongate by incorporation of additional MspA molecules.
Extreme structural stability is a common feature for functional amyloids, because they are stabilized by extensive hydrogen bonds besides internal salt bridges and hydrophobic interaction (19, 71, 72). The physical properties of the amyloid structure therefore also explain the high structural stability of the sheaths.
Our discovery of the methanogenic sheaths as amyloid structures represents the first documentation of functional amyloids within the archaeal domain of life. It also denotes the first example of a functional amyloid for a strict anaerobe. The amyloid sheath protein MspA is evolutionary unrelated to all currently known amyloid systems and represents a new function of the amyloid protein fold. The discovery of archaeal functional amyloids suggests that the amyloid in general has proved useful to organisms from all three domains of life because of their common useful biophysical properties.
Author Contributions
M. S. D., P. L., D. E. O., and P. H. N. designed the study. KF cultured the archaea. M. S. D. and K. F. purified the cell wall sheaths. M. S. D. and P. L. performed antibody staining and confocal laser scanning microscopy. G. C. performed electron microscopy. M. S. D., M. R. S., B. S. V., and A. B. performed biophysical characterization of the sheaths. M. S. D. depolymerized the sheaths and performed the MS/MS analysis. M. S. D. designed and constructed vectors for expression of recombinant MspA. M. S. D., M. R. S., B. S. V., and A. B. performed biophysical characterization of the recombinant MspA. M. S. D. and P. H. N. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to the Lundbeck Foundation for supporting our research on bacterial amyloid. We also thank Anne Stentebjerg for valuable help in the laboratory.
This work was supported in part by funds from the Villum Foundation and Innovation Fund Denmark (to M. S. D.) and is in part supported by the Sino-Danish Centre for Education and Research (to M. R. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- TEM
- transmission electron microscopy
- ThT
- thioflavin T
- ddH2O
- double distilled water
- ATR
- attenuated total reflectance
- GdmCl
- guanidinium chloride
- FTIR
- Fourier transform infrared spectroscopy
- DTT
- dithiothreitol.
References
- 1. Woese C. R., Kandler O., Wheelis M. L. (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87, 4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cavicchioli R. (2011) Archaea: timeline of the third domain. Nat. Rev. Microbiol. 9, 51–61 [DOI] [PubMed] [Google Scholar]
- 3. Albers S.-V., Meyer B. H. (2011) The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426 [DOI] [PubMed] [Google Scholar]
- 4. Liu Y., Whitman W. B. (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N.Y. Acad. Sci. 1125, 171–189 [DOI] [PubMed] [Google Scholar]
- 5. Hedderich R., Whitman W. B. (2013) Physiology and biochemistry of the methane-producing archaea. In The Prokaryotes (Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. eds) pp. 635–662, Springer, Berlin [Google Scholar]
- 6. Weiland P. (2010) Biogas production: current state and perspectives. Appl. Microbiol. Biotechnol. 85, 849–860 [DOI] [PubMed] [Google Scholar]
- 7. Firtel M., Southam G., Harauz G., Beveridge T. J. (1993) Characterization of the cell wall of the sheathed methanogen Methanospirillum hungatei GP1 as an S layer. J. Bacteriol. 175, 7550–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaw P. J., Hills G. J., Henwood J. A., Harris J. E., Archer D. B. (1985) Three-dimensional architecture of the cell sheath and septa of Methanospirillum hungatei. J. Bacteriol. 161, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walsby A. E. (1994) Gas vesicles. Microbiol. Rev. 58, 94–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sprott G. D., Beveridge T. J., Patel G. B., Ferrante G. (1986) Sheath disassembly in Methanospirillum hungatei strain GP1. Can. J. Microbiol. 32, 847–854 [Google Scholar]
- 11. Sprott G. D., McKellar R. C. (1980) Composition and properties of the cell wall of Methanospirillum hungatii. Can. J. Microbiol. 26, 115–120 [DOI] [PubMed] [Google Scholar]
- 12. Patel G. B., Sprott G. D., Humphrey R. W., Beveridge T. J. (1986) Comparative analyses of the sheath structures of Methanothrix concilii GP6 and Methanospirillum hungatei strains GP1 and JF1. Can. J. Microbiol. 32, 623–631 [Google Scholar]
- 13. Stewart M., Beveridge T. J., Sprott G. D. (1985) Crystalline order to high resolution in the sheath of Methanospirillum hungatei: a cross-β structure. J. Mol. Biol. 183, 509–515 [DOI] [PubMed] [Google Scholar]
- 14. Beveridge T. J., Stewart M., Doyle R. J., Sprott G. D. (1985) Unusual stability of the Methanospirillum hungatei sheath. J. Bacteriol. 162, 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dueholm M. S., Nielsen P. H., Chapman M., Otzen D. (2013) Functional amyloids in bacteria. In Amyloid Fibrils and Prefibrillar Aggregates (Otzen D. E. ed), pp. 411–438, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 16. Fändrich M. (2007) On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell Mol. Life Sci. 64, 2066–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., Hultgren S. J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Romero D., Aguilar C., Losick R., Kolter R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dueholm M. S., Petersen S. V., Sønderkaer M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., Otzen D. E. (2010) Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 20. Dueholm M. S., Søndergaard M. T., Nilsson M., Christiansen G., Stensballe A., Overgaard M. T., Givskov M., Tolker-Nielsen T., Otzen D. E., Nielsen P. H. (2013) Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. MicrobiologyOpen 2, 365–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blanco L. P., Evans M. L., Smith D. R., Badtke M. P., Chapman M. R. (2012) Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20, 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bayry J., Aimanianda V., Guijarro J. I., Sunde M., Latgé J.-P. (2012) Hydrophobins: unique fungal proteins. PLoS Pathog. 8, e1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wickner R. B., Edskes H. K., Shewmaker F., Nakayashiki T. (2007) Prions of fungi: inherited structures and biological roles. Nat. Rev. Microbiol. 5, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGlinchey R. P., Shewmaker F., McPhie P., Monterroso B., Thurber K., Wickner R. B. (2009) The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc. Natl. Acad. Sci. U.S.A. 106, 13731–13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Plakoutsi G., Taddei N., Stefani M., Chiti F. (2004) Aggregation of the acylphosphatase from Sulfolobus solfataricus: the folded and partially unfolded states can both be precursors for amyloid formation. J. Biol. Chem. 279, 14111–14119 [DOI] [PubMed] [Google Scholar]
- 27. Bemporad F., Chiti F. (2009) “Native-like aggregation” of the acylphosphatase from Sulfolobus solfataricus and its biological implications. FEBS Lett. 583, 2630–2638 [DOI] [PubMed] [Google Scholar]
- 28. Pagano K., Bemporad F., Fogolari F., Esposito G., Viglino P., Chiti F., Corazza A. (2010) Structural and dynamics characteristics of acylphosphatase from Sulfolobus solfataricus in the monomeric state and in the initial native-like aggregates. J. Biol. Chem. 285, 14689–14700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chimileski S., Franklin M. J., Papke R. T. (2014) Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC Biol. 12, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larsen P., Nielsen J. L., Otzen D., Nielsen P. H. (2008) Amyloid-like adhesins produced by floc-forming and filamentous bacteria in activated sludge. Appl. Environ. Microbiol. 74, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lorenzen N., Lemminger L., Pedersen J. N., Nielsen S. B., Otzen D. E. (2014) The N-terminus of α-synuclein is essential for both monomeric and oligomeric interactions with membranes. FEBS Lett. 588, 497–502 [DOI] [PubMed] [Google Scholar]
- 32. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 33. Heger A., Holm L. (2000) Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41, 224–237 [DOI] [PubMed] [Google Scholar]
- 34. Tsolis A. C., Papandreou N. C., Iconomidou V. A., Hamodrakas S. J. (2013) A consensus method for the prediction of “aggregation-prone” peptides in globular proteins. PLoS One 8, e54175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kamagata Y., Kawasaki H., Oyaizu H., Nakamura K., Mikami E., Endo G., Koga Y., Yamasato K. (1992) Characterization of three thermophilic strains of Methanothrix (“Methanosaeta”) thermophila sp. nov., and rejection of Methanothrix (“Methanosaeta”) thermoacetophila. Int. J. Syst. Bacteriol. 42, 463–468 [DOI] [PubMed] [Google Scholar]
- 36. Smith K. S., Ingram-Smith C. (2007) Methanosaeta, the forgotten methanogen? Trends Microbiol. 15, 150–155 [DOI] [PubMed] [Google Scholar]
- 37. Welte C., Deppenmeier U. (2011) Membrane-bound electron transport in Methanosaeta thermophila. J. Bacteriol. 193, 2868–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goldsbury C., Baxa U., Simon M. N., Steven A. C., Engel A., Wall J. S., Aebi U., Müller S. A. (2011) Amyloid structure and assembly: insights from scanning transmission electron microscopy. J. Struct. Biol. 173, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Nuallain B., Wetzel R. (2002) Conformational Abs recognizing a generic amyloid fibril epitope. Proc. Natl. Acad. Sci. U.S.A. 99, 1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larsen P., Nielsen J. L., Dueholm M. S., Wetzel R., Otzen D., Nielsen P. H. (2007) Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9, 3077–3090 [DOI] [PubMed] [Google Scholar]
- 41. Jordal P. B., Dueholm M. S., Larsen P., Petersen S. V., Enghild J. J., Christiansen G., Højrup P., Nielsen P. H., Otzen D. E. (2009) Widespread abundance of functional bacterial amyloid in mycolata and other Gram-positive bacteria. Appl. Environ. Microbiol. 75, 4101–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bayro M. J., Daviso E., Belenky M., Griffin R. G., Herzfeld J. (2012) An amyloid organelle, solid-state NMR evidence for cross-β assembly of gas vesicles. J. Biol. Chem. 287, 3479–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pfeifer F. (2012) Distribution, formation and regulation of gas vesicles. Nat. Rev. Microbiol. 10, 705–715 [DOI] [PubMed] [Google Scholar]
- 44. Ban T., Hamada D., Hasegawa K., Naiki H., Goto Y. (2003) Direct observation of amyloid fibril growth monitored by thioflavin T fluorescence. J. Biol. Chem. 278, 16462–16465 [DOI] [PubMed] [Google Scholar]
- 45. Fändrich M., Forge V., Buder K., Kittler M., Dobson C. M., Diekmann S. (2003) Myoglobin forms amyloid fibrils by association of unfolded polypeptide segments. Proc. Natl. Acad. Sci. U.S.A. 100, 15463–15468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zandomeneghi G., Krebs M. R., McCammon M. G., Fändrich M. (2004) FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 13, 3314–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sunde M., Serpell L. C., Bartlam M., Fraser P. E., Pepys M. B., Blake C. C. (1997) Common core structure of amyloid fibrils by synchrotron x-ray diffraction. J. Mol. Biol. 273, 729–739 [DOI] [PubMed] [Google Scholar]
- 48. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 49. Dueholm M. S., Albertsen M., Otzen D., Nielsen P. H. (2012) Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One 7, e51274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dueholm M. S., Otzen D., Nielsen P. H. (2013) Evolutionary insight into the functional amyloids of the pseudomonads. PLoS One 8, e76630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sunde M., Kwan A. H., Templeton M. D., Beever R. E., Mackay J. P. (2008) Structural analysis of hydrophobins. Micron. 39, 773–784 [DOI] [PubMed] [Google Scholar]
- 52. Claessen D., Rink R., de Jong W., Siebring J., de Vreugd P., Boersma F. G., Dijkhuizen L., Wosten H. A. (2003) A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17, 1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Elliot M. A., Karoonuthaisiri N., Huang J., Bibb M. J., Cohen S. N., Kao C. M., Buttner M. J. (2003) The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17, 1727–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shokri M. M., Ahmadian S., Bemporad F., Khajeh K., Chiti F. (2013) Amyloid fibril formation by a normally folded protein in the absence of denaturants and agitation. Amyloid 20, 226–232 [DOI] [PubMed] [Google Scholar]
- 55. Jahn T. R., Parker M. J., Homans S. W., Radford S. E. (2006) Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 13, 195–201 [DOI] [PubMed] [Google Scholar]
- 56. Dueholm M. S., Nielsen S. B., Hein K. L., Nissen P., Chapman M., Christiansen G., Nielsen P. H., Otzen D. E. (2011) Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry 50, 8281–8290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E., Kelly J. W. (2006) Functional amyloid formation within mammalian tissue. PLoS Biol. 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zykwinska A., Guillemette T., Bouchara J.-P., Cuenot S. (2014) Spontaneous self-assembly of SC3 hydrophobins into nanorods in aqueous solution. Biochim. Biophys. Acta 1844, 1231–1237 [DOI] [PubMed] [Google Scholar]
- 59. Sawyer E. B., Claessen D., Haas M., Hurgobin B., Gras S. L. (2011) The assembly of individual chaplin peptides from Streptomyces coelicolor into functional amyloid fibrils. PLoS One 6, e18839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miles A. J., Wallace B. A. (2006) Synchrotron radiation circular dichroism spectroscopy of proteins and applications in structural and functional genomics. Chem. Soc. Rev. 35, 39–51 [DOI] [PubMed] [Google Scholar]
- 61. Ezzeldin H. M., Klauda J. B., Solares S. D. (2012) Modeling of the major gas vesicle protein, GvpA: from protein sequence to vesicle wall structure. J. Struct. Biol. 179, 18–28 [DOI] [PubMed] [Google Scholar]
- 62. Claessen D., Stokroos I., Deelstra H. J., Penninga N. A., Bormann C., Salas J. A., Dijkhuizen L., Wösten H. A. (2004) The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol. Microbiol. 53, 433–443 [DOI] [PubMed] [Google Scholar]
- 63. Southam G., Beveridge T. J. (1992) Characterization of novel, phenol-soluble polypeptides which confer rigidity to the sheath of Methanospirillum hungatei GP1. J. Bacteriol. 174, 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. White A. P., Collinson S. K., Banser P. A., Gibson D. L., Paetzel M., Strynadka N. C., Kay W. W. (2001) Structure and characterization of AgfB from Salmonella enteritidis thin aggregative fimbriae. J. Mol. Biol. 311, 735–749 [DOI] [PubMed] [Google Scholar]
- 65. Shewmaker F., McGlinchey R. P., Thurber K. R., McPhie P., Dyda F., Tycko R., Wickner R. B. (2009) The functional curli amyloid is not based on in-register parallel β-sheet structure. J. Biol. Chem. 284, 25065–25076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tian P., Boomsma W., Wang Y., Otzen D. E., Jensen M. H., Lindorff-Larsen K. (2015) Structure of a functional amyloid protein subunit computed using sequence variation. J. Am. Chem. Soc. 137, 22–25 [DOI] [PubMed] [Google Scholar]
- 67. Wasmer C., Lange A., Van Melckebeke H., Siemer A. B., Riek R., Meier B. H. (2008) Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science 319, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 68. Van Melckebeke H., Wasmer C., Lange A., Ab E., Loquet A., Böckmann A., Meier B. H. (2010) Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid-state NMR spectroscopy. J. Am. Chem. Soc. 132, 13765–13775 [DOI] [PubMed] [Google Scholar]
- 69. Xu W., Mulhern P. J., Blackford B. L., Jericho M. H., Firtel M., Beveridge T. J. (1996) Modeling and measuring the elastic properties of an archaeal surface, the sheath of Methanospirillum hungatei, and the implication of methane production. J. Bacteriol. 178, 3106–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Beveridge T. J., Harris B. J., Sprott G. D. (1987) Septation and filament splitting in Methanospirillum hungatei. Can. J. Microbiol. 33, 725–732 [Google Scholar]
- 71. Collinson S. K., Emody L., Muller K. H., Trust T. J., Kay W. W. (1991) Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173, 4773–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wosten H., De Vries O., Wessels J. (1993) Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell 5, 1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]