Abstract
Surrogates of whole-brain white matter (WM) networks reconstructed using diffusion tensor imaging (DTI) are novel markers of structural brain connectivity. Global connectivity of networks has been found impaired in clinical Alzheimer's disease (AD) compared to cognitively healthy aging. We hypothesized that network alterations are detectable already in preclinical AD and investigated major global WM network properties. Other structural markers of neurodegeneration typically affected in prodromal AD but seeming largely unimpaired in preclinical AD were also examined.
12 cognitively healthy elderly with preclinical AD as classified by florbetapir-PET (mean age 73.4 ± 4.9) and 31 age-matched controls without cerebral amyloidosis (mean age 73.1 ± 6.7) from the ADNI were included. WM networks were reconstructed from DTI using tractography and graph theory. Indices of network capacity and the established imaging markers of neurodegeneration hippocampal volume, and cerebral glucose utilization as measured by fludeoxyglucose-PET were compared between the two groups. Additionally, we measured surrogates of global WM integrity (fractional anisotropy, mean diffusivity, volume).
We found an increase of shortest path length and a decrease of global efficiency in preclinical AD. These results remained largely unchanged when controlling for WM integrity. In contrast, neither markers of neurodegeneration nor WM integrity were altered in preclinical AD subjects.
Our results suggest an impairment of WM networks in preclinical AD that is detectable while other structural imaging markers do not yet indicate incipient neurodegeneration. Moreover, these findings are specific to WM networks and cannot be explained by other surrogates of global WM integrity.
Keywords: Preclinical Alzheimer's disease, DTI, Brain network
Highlights
-
•
We reconstruct WM networks using DTI fiber tractography and graph theory.
-
•
We investigate global network properties in cognitively healthy elderly.
-
•
Global network integration is decreased in subjects with amyloidosis.
-
•
In contrast, no neurodegeneration is found (FDG, hippocampus).
-
•
WM network topology may be altered already in preclinical AD.
1. Introduction
The novel diagnostic concept of Alzheimer's disease (AD) includes subjects with AD dementia, prodromal AD and preclinical AD, i.e. cognitively healthy elderly with positive imaging or neurochemical biomarkers of AD (Albert et al., 2011; McKhann et al., 2011; Sperling et al., 2011). Biomarkers have been arranged along a hypothetical timeline, on which cerebral amyloid (Amyloid-β) deposition is assumed to be a very early event of AD, followed by synaptic dysfunction, hypometabolism and cortical and subcortical atrophy as markers of neurodegeneration (Jack et al., 2013; Jack et al., 2010). Histopathological studies of AD showed demyelination and axonal damage, which are likely to result in functionally relevant disconnections between brain areas in addition to a specific pattern of structural gray matter defects (Delbeuck et al., 2003). Diffusion tensor imaging (DTI) has been used successfully to detect deterioration of white matter (WM) integrity in vivo in AD and prodromal AD (Chua et al., 2008; Damoiseaux et al., 2009). Studies investigating WM in preclinical AD as of yet are rare and show heterogeneous results, reporting regional increases (Racine et al., 2014) and decreases of indices of WM integrity (Chao et al., 2013), or even both (Ryan et al., 2013). However they suggest early alterations of WM in AD.
Surrogates of whole-brain WM networks reconstructed from DTI and assessed by graph theory are a novel imaging marker that integrates microstructural and topological information of WM (Bullmore and Sporns, 2009; Iturria-Medina et al., 2007; Iturria-Medina et al., 2008). Several studies have so far demonstrated impairment of network connectivity both in the prodromal and dementia stages of AD (Daianu et al., 2013; Lo et al., 2010; Wee et al., 2011), thereby supporting the notion of AD as a disconnection disease. Interestingly, Raj et al. demonstrated that AD cortical atrophy and hypometabolism patterns can be predicted from healthy WM network topology (Raj et al., 2012; Raj et al., 2015). Some of the mechanisms suggested, e.g. a prion-like propagation of disease agents along WM tracts, imply very early involvement of the WM network in AD pathology. Furthermore, a recent longitudinal study by Nir et al. showed that WM network architecture not only predicts cortical atrophy but also AD conversion in patients with mild cognitive impairment at risk for developing AD (Nir et al., 2015).
Taken together, these findings led us to hypothesize that WM network alterations might be detectable even at the preclinical stage of AD. In order to test this hypothesis, we examined group differences of network properties between subjects of preclinical AD, i.e. cognitively normal subjects with Amyloid-β deposition (Sperling et al., 2011), and normal controls. For comparison, we also examined other structural imaging markers of neurodegeneration that are known to be affected in prodromal AD, i.e. hippocampal volume and cerebral glucose metabolism. In order to show that WM network alterations are specific to network properties and do not simply reflect global WM integrity, we repeated analyses for WM volume, WM mean fractional anisotropy (FA) and WM mean diffusivity (MD).
2. Materials and methods
The data for the present study were obtained from the database of the Alzheimer's Disease Neuroimaging Initiative (ADNI), available at http://adni.loni.usc.edu. The ADNI was launched in the United States in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early AD. For a more detailed and up to date description of the ADNI please refer to http://www.adni-info.org.
2.1. Subjects
Following the study design, the selection criteria for the database were classification as cognitively healthy and the availability of T1-weighted, florbetapir (AV45) and DTI imaging at baseline, yielding 47 subjects from the ADNI 2 phases of the ADNI project. One of these had to be excluded because of data corruption of the DTI scan and another subject was excluded because of extensive white matter pathology, defined as a visual Fazekas scale rating of 3 (Fazekas et al., 1987), all in all yielding 45 subjects that were included in the present study. The sample was dichotomized using a cutoff point for AV45-imaging standardized uptake value ratio (SUVR) of 1.1 (Joshi et al., 2012). 13 subjects with an SUVR ≥1.1 were classified as preclinical AD. 32 with an SUVR <1.1 were classified as normal controls (NC). One subject from each group later had to be excluded due to classification as outliers (see Section 3). For descriptive statistics of the subjects' demographical data, please refer to Table 1.
Table 1.
Descriptive statistics of demographical data.
| Total sample | NC | Preclinical AD | p-Value | |
|---|---|---|---|---|
| N | 43 | 31 | 12 | |
| Gender (f/m) | 23/20 | 13/18 | 10/2 | .015a |
| Age | 73.1 ± 6.2 | 73.1 ± 6.7 | 73.4 ± 4.9 | .886 |
| Years of education | 16.2 ± 2.7 | 16.7 ± 2.7 | 14.9 ± 2.4 | .052 |
NC, normal controls. p-Values of between group differences were calculated using the chi-square test for gender, and t-test for all other variables.
Considered statistically significant.
In ADNI, subjects undergo several neuropsychological examinations. For this study, we chose to report the well-known mini mental state examination (MMSE) and Alzheimer's disease assessment score — cognitive section (ADAS-Cog) in order to allow the reader to judge the cognitive state of the sample. Detailed information about neuropsychological testing and diagnostic criteria are available at the ADNI website (http://adni.loni.usc.edu/methods).
2.2. Imaging data acquisition
DTI and inversion-recovery spoiled gradient recalled (IR-SPGR) T1-weighted imaging data were acquired on several General Electric 3 T scanners using scanner specific protocols. Briefly, DTI data were acquired with a voxel size of 1.372 × 2.70 mm3, 41 diffusion gradients and a b-value of 1000 s/mm2. IR-SPGR data were acquired with a voxel size of 1.022 × 1.20 mm3.
AV45 and fludeoxyglucose (FDG-PET) imaging data were acquired on several types of scanners using different acquisition protocols. In order to increase data uniformity, the data underwent a standardized preprocessing procedure at the ADNI project.
All imaging protocols and preprocessing procedures are available at the ADNI website (http://adni.loni.usc.edu/methods/).
2.3. AV45 and FDG-PET data processing
Subject AV45 and FDG standardized uptake value ratios (SUVRs) were calculated at ADNI core laboratories following a standardized pipeline that is available at the ADNI website (http://adni.loni.usc.edu/methods/pet-analysis/). Briefly, AV45 SUVR was calculated as the average of the uptake values of the frontal, angular/posterior cingulate, lateral parietal and temporal cortices divided by the mean uptake values of the cerebellum. FDG SUVR was calculated as the mean uptake of the left and right angular, bilateral posterior cingular and inferior temporal gyri normalized by the uptake of the pons/cerebellar vermis region.
2.4. DTI and T1-weighted imaging data processing
For detailed information on T1-weighted data processing, please refer to Appendix A — section Data processing. T1-weighted IR-SPGR data were automatically segmented using Freesurfer (https://surfer.nmr.mgh.harvard.edu/) in order to calculate white matter hypointensity volume (WMHV) and hippocampal volume. Additionally, IR-SPGR data were tissue segmented using SPM8 (http://www.fil.ion.ucl.ac.uk/spm/) in order to extract total gray matter (GM) and WM volume.
For detailed information on DTI data processing, please refer to Appendix A — section Data processing. DTI data were processed as previously described (Fischer et al., 2014). Briefly, diffusion tensors were fitted to the data using CAMINO (http://cmic.cs.ucl.ac.uk/camino/) and FA and MD maps were calculated and coregistered to IR-SPGR data in order to extract total WM mean FA and MD.
2.5. Network reconstruction
For detailed information on brain network reconstruction please refer to Appendix A — section Network reconstruction. Briefly, we defined the network nodes as the 111 cortical and subcortical brain areas defined by the Harvard–Oxford probabilistic brain atlas. Deterministic streamline tractography was conducted between each possible pair of the 111 brain regions and the number of resulting streamlines was considered the respective edge weight (Fischer et al., 2014; Hagmann et al., 2008; Li et al., 2009) (for exemplary tractography results please see Supplementary Fig. 1 of Appendix B). The resulting undirected and weighted brain network graphs were then thresholded to contain at least three fibers per connection in order to reduce the likelihood of false positives (Li et al., 2009; Lo et al., 2010; Shu et al., 2009).
2.6. Network properties
For a detailed description of how the network property indices were calculated, please refer to Appendix A — section Quantification of brain networks. We assessed the clustering coefficient CC and the normalized clustering coefficient CCnorm, shortest paths L and normalized shortest paths Lnorm as well as global efficiency Eglob, number of nodes N, number of edges E as well as total cost C in order to quantify topological brain network properties. These properties were chosen for the sake of comparability with other studies investigating brain networks in patients of AD (Tijms et al., 2013). An introduction and overview to graph theoretical methods in neuroimaging can be found in Kaiser (2011).
For supplementary regional analyses, we also calculated the nodal weighted degree WD in each subject, as well as the mean nodal betweenness centrality BC across all subjects.
2.7. Statistics
The significance threshold was set to p ≤ .05 for all analyses.
All continuous variables were tested for normal distribution within subgroups using the Kolmogorov–Smirnov test. The Grubbs-test was used to detect outliers (Grubbs, 1950).
Differences between preclinical AD subjects and NC regarding demographical data (age, sex, years of education) and WMHV were calculated as follows: the chi-square test was used for categorical variables, the t-test was used for continuous normally distributed variables and the Mann–Whitney test was used for continuous not normally distributed variables.
Simple linear regression was used to investigate whether the potential confounding variables age, gender, education and WM hypointensity volume (WMHV) had a significant influence on any of the network indices. In order to investigate possible group differences of WM network indices between NC and preclinical AD as well as markers of neurodegeneration (hippocampal volume, cerebral glucose metabolism), global GM and WM properties (mean FA, mean MD, volume), we used analysis of covariance (ANCOVA), wherein all confounding variables that had a significant influence on at least one network index were used as covariates. In an additional analysis we investigated regional network alterations by repeating ANCOVA on the WD of each node controlling for age, education, gender and WMHV. Group differences of mean BC between nodes with altered WD in preclinical AD were calculated using the Mann–Whitney test.
Residuals were tested for normal distribution using the Kolmogorov–Smirnov test throughout ANCOVA analyses.
3. Results
All continuous variables except WMHV were normally distributed. Two outliers were found for global efficiency (standard deviation 3.1 and 2.5) and excluded from all subsequent analyses, one within each subgroup. Significant differences of demographic data between NC and preclinical AD were found for gender (please see Table 1 for details). WMHV did not differ significantly between groups (NC: 3.2 ± 1.9; preclinical AD: 4.0 ± 2.5; p = .285). No group differences in cognitive performance were found for MMSE (NC: 28.7 ± 1.6; preclinical AD: 28.9 ± 1.3; p = .621) or ADAS-cog (NC: 8.4 ± 4.1; preclinical AD: 9.4 ± 4.5; p = .457).
Linear regression revealed a significant influence of age, gender, education and WMHV on at least one network index and they were thus used as covariates for ANCOVA. Residuals of all ANCOVA were normally distributed.
3.1. Group differences
Regarding WM network properties, analyses revealed an increase of shortest paths and a decrease of global efficiency in the preclinical AD group. Please see Table 2 for an overview of network property group differences and Fig. 1 for boxplots of the data.
Table 2.
Descriptive statistics and group differences of network properties controlling for age, gender, education and white matter hypointensities.
| Normal controls | Preclinical AD | p-Values of ANCOVA | |
|---|---|---|---|
| CC | .0192 ± .0046 | .0182 ± .0035 | .365 |
| L | .0599 ± .0141 | .0723 ± .0085 | .036a |
| Eglobal | 38.1 ± 5.3 | 32.5 ± 3.2 | .015a |
| CCnorm | 5.2 ± 0.3 | 5.1 ± 0.2 | .982 |
| Lnorm | 2.0 ± 0.3 | 2.1 ± 0.2 | .253 |
| N | 108.4 ± 0.9 | 108.1 ± 1.4 | .339 |
| E | 701.4 ± 65.2 | 676.7 ± 71.8 | .282 |
| C | 85,116.1 ± 12,691.8 | 75,084.0 ± 10,925.1 | .109 |
Mean ± standard deviation. ANCOVA, analysis of covariance. Group differences are controlled for age, gender and white matter hypointensity volume. CC, clustering coefficient, L, shortest paths, Eglobal, global efficiency, CCnorm, normalized clustering coefficient, Lnorm, normalized shortest paths, N, number of connected nodes, E, number of edges, C, total sum of edge weights.
Considered statistically significant.
Fig. 1.
Boxplots of network properties, global white matter properties and markers of neurodegeneration. A–C: Global white matter network properties. D–F: Global white matter properties. FA, fractional anisotropy. MD, mean diffusivity. G and H: Markers of neurodegeneration.
No differences were found for the markers of early neurodegeneration hippocampal volume and cerebral glucose metabolism as measured by FDG-uptake. Likewise, total GM and WM volume as well as FA and MD were unchanged in preclinical AD. Please see Table 3 for details.
Table 3.
Descriptive statistics and group differences of markers of neurodegeneration, and global properties of gray matter and white matter controlling for age, gender, education and white matter hypointensities.
| Normal controls | Preclinical AD | p-Values of ANCOVA | |
|---|---|---|---|
| Hippocampal volume | 7.11 ± 1.1 | 7.3 ± 0.9 | .505 |
| FDG-PET | 6.5 ± 0.6 | 6.5 ± 0.8 | .409 |
| GM volume | 724.8 ± 62.2 | 691.1 ± 56.6 | .807 |
| WM volume | 454.2 ± 43.6 | 428.5 ± 34.5 | .334 |
| WM mean FA | 0.37 ± 0.02 | 0.36 ± 0.01 | .083 |
| WM mean MD | 0.83 ± 0.05 | 0.83 ± 0.03 | .186 |
Mean ± standard deviation. Group differences are controlled for age, gender and white matter hypointensity volume. ANCOVA, analysis of covariance. FDG-PET, fludeoxyglucose positron emission tomography. GM, gray matter. WM, white matter. FA, fractional anisotropy. MD, mean diffusivity.
Supplementary regional analyses revealed decreased WD in the preclinical AD group for the following 13 nodes defined by the Harvard–Oxford Atlas: left frontal pole (p = .018), posterior division of the left inferior temporal gyrus (p = .04), posterior division of the left (p = .027) and right cingulate gyri (p = .020), left precuneus cortex (p = .033), anterior division of the left parahippocampal gyrus (p = .05), left central opercular cortex (p = .019), left planum polare (p = .025), left hippocampus (p = .047), right insular cortex (p = .007), superior (p = .045) and inferior (p = .029) divisions of the right lateral occipital cortex and frontal medial cortex (p = .029). The mean BC of these nodes across subjects was higher than the remaining nodes showing increased importance for global network integration of the nodes impaired in preclinical AD (p = .016). Please see Supplementary Table 1 Appendix C for details.
4. Discussion
The main finding of this study was that WM network alterations previously reported in clinical AD could also be detected in preclinical AD. Notably, preclinical AD patients with WM network alterations showed no signs of neurodegeneration such as atrophy or reduced cortical glucose utilization. Moreover, these findings were specific to network properties and could not be explained by global WM integrity surrogates (FA, MD, volume).
Alterations of structural and functional brain network properties in AD have been demonstrated repeatedly (Tijms et al., 2013). Using structural MRI, DTI, EEG or magnetoencephalography (MEG) in combination with graph theory, these studies generally reported an increase of shortest path length. One study assessed WM networks reconstructed using DTI fiber tractography in a manner comparable to the present study (Lo et al., 2010). They reported decreased global efficiency and increased mean shortest path length in AD. The findings of the present study may be interpreted as an extension of these findings to preclinical AD. This means that an impairment of the global structural integration of brain regions is already measurable at the preclinical stage of AD. Notably this is in contrast to normal aging, where global efficiency was found to be preserved (Gong et al., 2009).
Assessing the total brain WM has two main advantages over ROI based, voxel-wise or morphometric approaches. First, the networks and their properties integrate more of the information available from imaging scans than either of the approaches cited above. Specifically, microstructural tissue composition (e.g. FA), morphometry (fiber thickness) as well as topology (fiber orientation, interconnection of GM regions) are all condensed into one WM network surrogate and its properties. Second, in addition to information integration, the network property indices provide a straightforward and objective way of quantifying, characterizing and interpreting the data contained in the network. Network property indices such as clustering coefficient, shortest paths and global efficiency intuitively model the efficiency of local and global information traversal and processing and (in the case of DTI networks) have been found to be associated with cognition, age and gender (Fischer et al., 2014; Gong et al., 2009; Li et al., 2009) as well as sensitive to WM pathology (Lo et al., 2010). These views are supported further by the findings of the present study. The integration of more information available from the imaging data than other approaches is beneficial with respect to detecting alterations in preclinical AD: neither global FA and MD nor total WM volume showed group differences in ANCOVA. We additionally performed voxelwise analyses for group differences between HC and preclinical AD for FA and MD using tract based spatial statistics (TBSS) and simple t-test, but none were found (data not shown).
Cerebral Amyloid-β deposition plays an early key role in the preclinical pathogenesis of AD. However, it is not sufficient by itself for the clinical manifestation of AD. The prodromal phase of AD is instead marked by neurodegenerative processes, whose association with Amyloid-β deposition is an ongoing area of research (Jack et al., 2013). One of these neurodegenerative processes is an impairment of WM connectivity measurable by DTI. Studies in mouse models have histologically confirmed a negative association of Amyloid-β deposition and WM connectivity (Song et al., 2004; Sun et al., 2005). However, more recent studies in mice reported contradicting results showing both negative and positive associations (Muller et al., 2013; Qin et al., 2013; Shu et al., 2013; Zerbi et al., 2013). Studies in asymptomatic humans with Amyloid-β deposition are rare, of limited comparability and heterogeneous (Chao et al., 2013; Racine et al., 2014; Ryan et al., 2013). Thus the relationship between amyloid and WM integrity does not seem to be straight forward. Our finding of unaltered FA and MD of total WM as well as at the voxel level (data not shown, please see above) may thus be due to unknown modifying factors, or due to the small sample size.
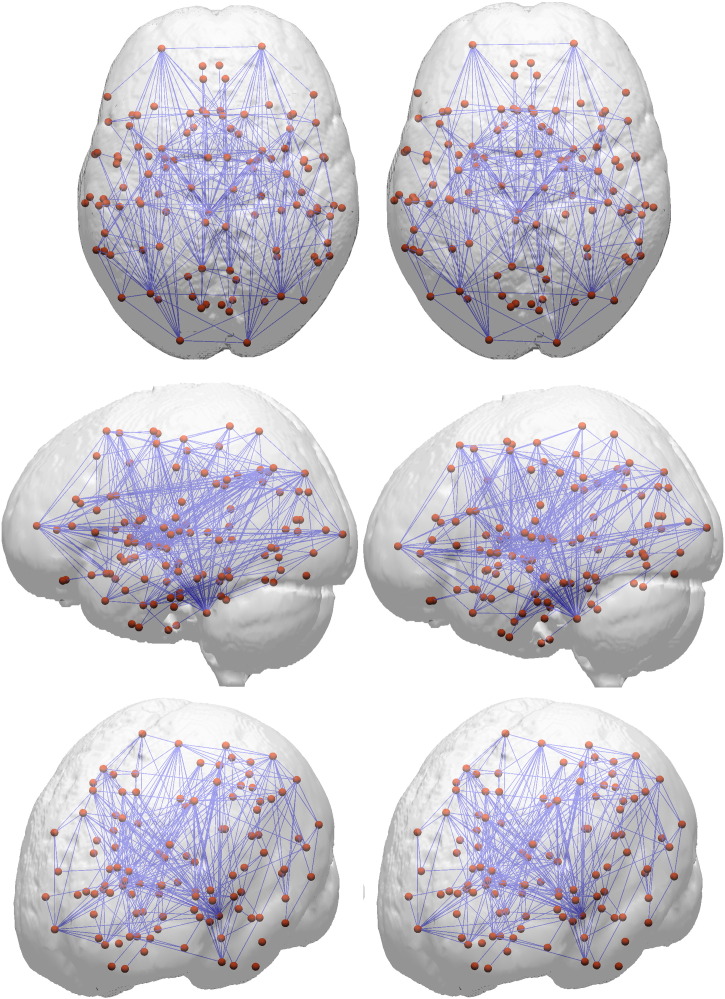
The human brain network as reconstructed from various imaging modalities has consistently been shown to exhibit small world organization, where several regions form locally densely connected clusters (Bullmore and Sporns, 2009). These clusters are in turn interconnected by network hubs that provide “shortcuts” for efficient information traversal between any region in the network. This network layout is robust to random elimination of nodes, since the number of these network hubs is relatively small compared to overall network size (He et al., 2008). The finding of the present study where shortest paths were increased and global efficiency decreased while network reference values such as size, number of edges and total cost remained unaltered in preclinical AD thus implies degradation of hub regions in preclinical AD (please see Fig. 2 for network plots). Indeed, there is mounting evidence that hubs are focal areas of pathology in neurodegenerative diseases (Buckner et al., 2009; Crossley et al., 2014; Raj et al., 2015). In a supplementary regional analysis, we were able to confirm this view: network nodes with locally impaired connectivity in preclinical AD (i.e. decreased weighted degree) also showed a significantly higher contribution to global network integration (i.e. betweenness centrality, see Appendix A, Quantification of brain networks) than nodes with unaltered connectivity and may thus be regarded as hub regions (Bullmore and Sporns, 2009).
Fig. 2.
For this figure, we averaged individual networks in the groups of NC (left) and preclinical AD (right). A per-connection threshold of at least 3 streamlines was applied. Red spheres symbolize gray matter regions and blue lines white matter connections among them. Please note that networks were only averaged for this figure; in all statistical analyses individual networks were considered.
Unlike shortest path length, normalized shortest path length did not show an increase between NC and preclinical AD. This constellation is analogous to findings of other studies that showed consistent increase of shortest path length in patients of AD but heterogeneous results for normalized shortest path length (Tijms et al., 2013). This apparent discrepancy may be due to different effects of the normalization depending on underlying network topology (van Wijk et al., 2010). However, as topology could be altered in AD (Sanz-Arigita et al., 2010), this might lead to underestimation of effects.
Atrophy and hypometabolism have been detected in preclinical AD before by other studies, particularly in the hippocampus (Chetelat et al., 2010; Mosconi et al., 2008). However, neither the hippocampus nor the other structural markers of neurodegeneration investigated in this study, i.e. brain volume and FDG uptake, were altered in preclinical AD. This finding implies that WM network impairment may occur before significant neurodegeneration is detectable.
Integration of different structures spread across the brain is vital for many cognitive functions. It follows that connectivity between these regions has an impact on how well they can synchronize and cooperate to work on a specific task. Indeed, properties of structural and functional whole brain networks have been shown to be associated with cognitive performance in healthy subjects and patients of AD (Li et al., 2009; van den Heuvel and Hulshoff Pol, 2010; Wen et al., 2011). Furthermore, other studies have shown an association of amyloid deposition and cognitive performance (Rodrigue et al., 2012; Storandt et al., 2009, 2009; Villemagne et al., 2011). Considering these prior findings together with the altered network indices in preclinical AD as reported by the present study, one might assume cognitive differences between the preclinical AD group and NC that are driven by an association with the networks. However, in an exploratory analysis, we were unable to show either (data not shown). Possible explanations are the small sample size as well as the cognitive tests available with ADNI, which put more emphasis on diagnosis and disease progression monitoring than sensitivity to subtle cognitive differences within the healthy range (http://adni.loni.use.edu/methods/documents/).
This study has several limitations. First, due to the cross sectional design of the study, group differences between HC and preclinical AD may be due to selection bias. However, we controlled for age, education and sex in all analyses of covariance. Second, the sample size of this study is relatively small and it may not adequately represent the population. Third, the data were acquired at several different centers. However, major efforts for the standardization of data acquisition were undertaken during the design and execution of the ADNI project. Fourth, we refrained from using correction for multiple comparisons in light of the exploratory nature of this study.
5. Conclusion
Using DTI tractography and graph theory we were able to demonstrate that alterations of whole-brain white matter network properties are detectable in preclinical AD even before structural markers of brain atrophy show significant neurodegeneration. Moreover, WM network alterations could not be explained by global WM integrity.
Author contributions
All statistical analyses were carried out by the corresponding author.
Conflict of interest
The authors have nothing to disclose. For funding sources of ADNI please see the Acknowledgment section.
Acknowledgments
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; EISAI Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F Hoffmann-La Roche Ltd. and it's affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research and Development, LLC; Johnson and Johnson Pharmaceutical Research and Development LLC; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2015.06.007.
Contributor Information
Florian Udo Fischer, Email: florian.fischer@unimedizin-mainz.de.
Dominik Wolf, Email: dominik.wolf@unimedizin-mainz.de.
Armin Scheurich, Email: armin.scheurich@unimedizin-mainz.de.
Andreas Fellgiebel, Email: andreas.fellgiebel@unimedizin-mainz.de.
Appendix A. Supplementary data
Supplementary material.
References
- Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C., Gamst A., Holtzman D.M., Jagust W.J., Petersen R.C., Snyder P.J., Carrillo M.C., Thies B., Phelps C.H. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging–Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. 21514249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner R.L., Sepulcre J., Talukdar T., Krienen F.M., Liu H., Hedden T., Andrews-Hanna J.R., Sperling R.A., Johnson K.A. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J. Neurosci. 2009;29(6):1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. 19211893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10(3):186–198. doi: 10.1038/nrn2575. 19190637 [DOI] [PubMed] [Google Scholar]
- Chao L.L., Decarli C., Kriger S., Truran D., Zhang Y., Laxamana J., Villeneuve S., Jagust W.J., Sanossian N., Mack W.J., Chui H.C., Weiner M.W. Associations between white matter hyperintensities and beta amyloid on integrity of projection, association, and limbic fiber tracts measured with diffusion tensor MRI. PLOS One. 2013;8(6):e65175. doi: 10.1371/journal.pone.0065175. 23762308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chételat G., Villemagne V.L., Bourgeat P., Pike K.E., Jones G., Ames D., Ellis K.A., Szoeke C., Martins R.N., O'Keefe G.J., Salvado O., Masters C.L., Rowe C.C., Australian Imaging Biomarkers and Lifestyle Research Group Relationship between atrophy and beta-amyloid deposition in Alzheimer disease. Ann. Neurol. 2010;67(3):317–324. doi: 10.1002/ana.21955. 20373343 [DOI] [PubMed] [Google Scholar]
- Chua T.C., Wen W., Slavin M.J., Sachdev P.S. Diffusion tensor imaging in mild cognitive impairment and Alzheimer's disease: a review. Curr. Opin. Neurol. 2008;21(1):83–92. doi: 10.1097/WCO.0b013e3282f4594b. 18180656 [DOI] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Scott J., Carletti F., Fox P.T., McGuire P., Bullmore E.T. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137(8):2382–2395. doi: 10.1093/brain/awu132. 25057133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daianu M., Jahanshad N., Nir T.M., Toga A.W., Jack C.R., Jr., Weiner M.W., Thompson P.M., Alzheimer's Disease Neuroimaging Initiative Breakdown of brain connectivity between normal aging and Alzheimer's disease: a structural k-core network analysis. Brain Connect. 2013;3(4):407–422. doi: 10.1089/brain.2012.0137. 23701292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux J.S., Smith S.M., Witter M.P., Sanz-Arigita E.J., Barkhof F., Scheltens P., Stam C.J., Zarei M., Rombouts S.A. White matter tract integrity in aging and Alzheimer's disease. Hum. Brain Mapp. 2009;30(4):1051–1059. doi: 10.1002/hbm.20563. 18412132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbeuck X., Van der Linden M., Collette F. Alzheimer's disease as a disconnection syndrome? Neuropsychol. Rev. 2003;13(2):79–92. doi: 10.1023/a:1023832305702. 12887040 [DOI] [PubMed] [Google Scholar]
- Fazekas F., Chawluk J.B., Alavi A., Hurtig H.I., Zimmerman R.A. MR signal abnormalities at 1.5 T in Alzheimer's dementia and normal aging. A.J.R. Am. J. Roentgenol. 1987;149(2):351–356. doi: 10.2214/ajr.149.2.351. 3496763 [DOI] [PubMed] [Google Scholar]
- Fischer F.U., Wolf D., Scheurich A., Fellgiebel A. Association of structural global brain network properties with intelligence in normal aging. PLOS One. 2014;9(1):e86258. doi: 10.1371/journal.pone.0086258. 24465994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G., Rosa-Neto P., Carbonell F., Chen Z.J., He Y., Evans A.C. Age- and gender-related differences in the cortical anatomical network. J. Neurosci. 2009;29(50):15684–15693. doi: 10.1523/JNEUROSCI.2308-09.2009. 20016083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs F.E. Sample criteria for testing outlying observations. Ann. Math. Stat. 1950;21(1):27–58. [Google Scholar]
- Hagmann P., Cammoun L., Gigandet X., Meuli R., Honey C.J., Wedeen V.J., Sporns O. Mapping the structural core of human cerebral cortex. PLOS Biol. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. 18597554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Chen Z., Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J. Neurosci. 2008;28(18):4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. 18448652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y., Canales-Rodríguez E.J., Melie-García L., Valdés-Hernández P.A., Martínez-Montes E., Alemán-Gómez Y., Sánchez-Bornot J.M. Characterizing brain anatomical connections using diffusion weighted MRI and graph theory. Neuroimage. 2007;36(3):645–660. doi: 10.1016/j.neuroimage.2007.02.012. 17466539 [DOI] [PubMed] [Google Scholar]
- Iturria-Medina Y., Sotero R.C., Canales-Rodríguez E.J., Alemán-Gómez Y., Melie-García L. Studying the human brain anatomical network via diffusion-weighted MRI and graph theory. Neuroimage. 2008;40(3):1064–1076. doi: 10.1016/j.neuroimage.2007.10.060. 18272400 [DOI] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S., Shaw L.M., Vemuri P., Wiste H.J., Weigand S.D., Lesnick T.G., Pankratz V.S., Donohue M.C., Trojanowski J.Q. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–216. doi: 10.1016/S1474-4422(12)70291-0. 23332364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W., Petersen R.C., Trojanowski J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9(1):119–128. doi: 10.1016/S1474-4422(09)70299-6. 20083042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.D., Pontecorvo M.J., Clark C.M., Carpenter A.P., Jennings D.L., Sadowsky C.H., Adler L.P., Kovnat K.D., Seibyl J.P., Arora A., Saha K., Burns J.D., Lowrey M.J., Mintun M.A., Skovronsky D.M., Florbetapir F 18 Study Investigators Performance characteristics of amyloid PET with florbetapir F 18 in patients with Alzheimer's disease and cognitively normal subjects. J. Nucl. Med. 2012;53(3):378–384. doi: 10.2967/jnumed.111.090340. 22331215 [DOI] [PubMed] [Google Scholar]
- Kaiser M. A tutorial in connectome analysis: topological and spatial features of brain networks. Neuroimage. 2011;57(3):892–907. doi: 10.1016/j.neuroimage.2011.05.025. 21605688 [DOI] [PubMed] [Google Scholar]
- Li Y., Liu Y., Li J., Qin W., Li K., Yu C., Jiang T. Brain anatomical network and intelligence. PLOS Comput. Biol. 2009;5(5) doi: 10.1371/journal.pcbi.1000395. 19492086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.Y., Wang P.N., Chou K.H., Wang J., He Y., Lin C.P. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer's disease. J. Neurosci. 2010;30(50):16876–16885. doi: 10.1523/JNEUROSCI.4136-10.2010. 21159959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging–Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. 21514250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L., Pupi A., De Leon M.J. Brain glucose hypometabolism and oxidative stress in preclinical Alzheimer's disease. Ann. N. Y. Acad. Sci. 2008;1147:180–195. doi: 10.1196/annals.1427.007. 19076441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.P., Kassubek J., Vernikouskaya I., Ludolph A.C., Stiller D., Rasche V. Diffusion tensor magnetic resonance imaging of the brain in APP transgenic mice: a cohort study. PLOS One. 2013;8(6):e67630. doi: 10.1371/journal.pone.0067630. 23840754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir T.M., Jahanshad N., Toga A.W., Bernstein M.A., Jack C.R., Jr., Weiner M.W., Thompson P.M., Alzheimer's Disease Neuroimaging Initiative Connectivity network measures predict volumetric atrophy in mild cognitive impairment. Neurobiol. Aging. 2015;36(Suppl 1):S113–S120. doi: 10.1016/j.neurobiolaging.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y.Y., Li M.W., Zhang S., Zhang Y., Zhao L.Y., Lei H., Oishi K., Zhu W.Z. In vivo quantitative whole-brain diffusion tensor imaging analysis of APP/PS1 transgenic mice using voxel-based and atlas-based methods. Neuroradiology. 2013;55(8):1027–1038. doi: 10.1007/s00234-013-1195-0. 23644540 [DOI] [PubMed] [Google Scholar]
- Racine A.M., Adluru N., Alexander A.L., Christian B.T., Okonkwo O.C., Oh J., Cleary C.A., Birdsill A., Hillmer A.T., Murali D., Barnhart T.E., Gallagher C.L., Carlsson C.M., Rowley H.A., Dowling N.M., Asthana S., Sager M.A., Bendlin B.B., Johnson S.C. Associations between white matter microstructure and amyloid burden in preclinical Alzheimer's disease: a multimodal imaging investigation. Neuroimage Clin. 2014;4:604–614. doi: 10.1016/j.nicl.2014.02.001. 24936411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Kuceyeski A., Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73(6):1204–1215. doi: 10.1016/j.neuron.2011.12.040. 22445347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., LoCastro E., Kuceyeski A., Tosun D., Relkin N., Weiner M., Alzheimer's Disease Neuroimaging Initiative Network diffusion model of progression predicts longitudinal patterns of atrophy and metabolism in Alzheimer's disease. Cell Rep. 2015:S2211. doi: 10.1016/j.celrep.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue K.M., Kennedy K.M., Devous M.D., Sr., Rieck J.R., Hebrank A.C., Diaz-Arrastia R., Mathews D., Park D.C. Beta-amyloid burden in healthy aging: regional distribution and cognitive consequences. Neurology. 2012;78(6):387–395. doi: 10.1212/WNL.0b013e318245d295. 22302550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan N.S., Keihaninejad S., Shakespeare T.J., Lehmann M., Crutch S.J., Malone I.B., Thornton J.S., Mancini L., Hyare H., Yousry T., Ridgway G.R., Zhang H., Modat M., Alexander D.C., Rossor M.N., Ourselin S., Fox N.C. Magnetic resonance imaging evidence for presymptomatic change in thalamus and caudate in familial Alzheimer's disease. Brain. 2013;136(5):1399–1414. doi: 10.1093/brain/awt065. 23539189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Arigita E.J., Schoonheim M.M., Damoiseaux J.S., Rombouts S.A., Maris E., Barkhof F., Scheltens P., Stam C.J. Loss of ‘small-world’ networks in Alzheimer's disease: graph analysis of FMRI resting-state functional connectivity. PLOS One. 2010;5(11):e13788. doi: 10.1371/journal.pone.0013788. 21072180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu N., Liu Y., Li J., Li Y., Yu C., Jiang T. Altered anatomical network in early blindness revealed by diffusion tensor tractography. PLOS One. 2009;4(9):e7228. doi: 10.1371/journal.pone.0007228. 19784379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X., Qin Y.Y., Zhang S., Jiang J.J., Zhang Y., Zhao L.Y., Shan D., Zhu W.Z. Voxel-based diffusion tensor imaging of an APP/PS1 mouse model of Alzheimer's disease. Mol. Neurobiol. 2013;48(1):78–83. doi: 10.1007/s12035-013-8418-6. 23877934 [DOI] [PubMed] [Google Scholar]
- Song S.K., Kim J.H., Lin S.J., Brendza R.P., Holtzman D.M. Diffusion tensor imaging detects age-dependent white matter changes in a transgenic mouse model with amyloid deposition. Neurobiol. Dis. 2004;15(3):640–647. doi: 10.1016/j.nbd.2003.12.003. 15056472 [DOI] [PubMed] [Google Scholar]
- Sperling R.A., Aisen P.S., Beckett L.A., Bennett D.A., Craft S., Fagan A.M., Iwatsubo T., Jack C.R., Jr., Kaye J., Montine T.J., Park D.C., Reiman E.M., Rowe C.C., Siemers E., Stern Y., Yaffe K., Carrillo M.C., Thies B., Morrison-Bogorad M., Wagster M.V., Phelps C.H. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging–Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. 21514248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storandt M., Mintun M.A., Head D., Morris J.C. Cognitive decline and brain volume loss as signatures of cerebral amyloid-beta peptide deposition identified with Pittsburgh compound B: cognitive decline associated with Abeta deposition. Arch. Neurol. 2009;66(12):1476–1481. doi: 10.1001/archneurol.2009.272. 20008651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.W., Song S.K., Harms M.P., Lin S.J., Holtzman D.M., Merchant K.M., Kotyk J.J. Detection of age-dependent brain injury in a mouse model of brain amyloidosis associated with Alzheimer's disease using magnetic resonance diffusion tensor imaging. Exp. Neurol. 2005;191(1):77–85. doi: 10.1016/j.expneurol.2004.09.006. 15589514 [DOI] [PubMed] [Google Scholar]
- Tijms B.M., Wink A.M., de Haan W., van der Flier W.M., Stam C.J., Scheltens P., Barkhof F. Alzheimer's disease: connecting findings from graph theoretical studies of brain networks. Neurobiol. Aging. 2013;34(8):2023–2036. doi: 10.1016/j.neurobiolaging.2013.02.020. 23541878 [DOI] [PubMed] [Google Scholar]
- van den Heuvel M.P., Hulshoff Pol H.E. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 2010;20(8):519–534. doi: 10.1016/j.euroneuro.2010.03.008. 20471808 [DOI] [PubMed] [Google Scholar]
- van Wijk B.C., Stam C.J., Daffertshofer A. Comparing brain networks of different size and connectivity density using graph theory. PLOS One. 2010;5(10):e13701. doi: 10.1371/journal.pone.0013701. 21060892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne V.L., Pike K.E., Chételat G., Ellis K.A., Mulligan R.S., Bourgeat P., Ackermann U., Jones G., Szoeke C., Salvado O., Martins R., O'Keefe G., Mathis C.A., Klunk W.E., Ames D., Masters C.L., Rowe C.C. Longitudinal assessment of Abeta and cognition in aging and Alzheimer disease. Ann. Neurol. 2011;69(1):181–192. doi: 10.1002/ana.22248. 21280088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C.Y., Yap P.T., Li W., Denny K., Browndyke J.N., Potter G.G., Welsh-Bohmer K.A., Wang L., Shen D. Enriched white matter connectivity networks for accurate identification of MCI patients. Neuroimage. 2011;54(3):1812–1822. doi: 10.1016/j.neuroimage.2010.10.026. 20970508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Zhu W., He Y., Kochan N.A., Reppermund S., Slavin M.J., Brodaty H., Crawford J., Xia A., Sachdev P. Discrete neuroanatomical networks are associated with specific cognitive abilities in old age. J. Neurosci. 2011;31(4):1204–1212. doi: 10.1523/JNEUROSCI.4085-10.2011. 21273405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbi V., Kleinnijenhuis M., Fang X., Jansen D., Veltien A., Van Asten J., Timmer N., Dederen P.J., Kiliaan A.J., Heerschap A. Gray and white matter degeneration revealed by diffusion in an Alzheimer mouse model. Neurobiol. Aging. 2013;34(5):1440–1450. doi: 10.1016/j.neurobiolaging.2012.11.017. 23273575 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.