Summary
The first mutation that disrupts BRCA2 mRNA by including a novel, cryptic exon is reported in this issue. The mutation lies deep within an intron and would not have been detected by conventional screening methods. In the future, more mutations may be discovered by direct mRNA analysis.
In this issue of Clinical Cancer Research, Anczuków et al. report the first deep intronic BRCA2 mutation resulting in cryptic exon inclusion, the first such mutation to be found in either the BRCA1 or BRCA2 breast/ovarian tumor suppressor genes (1). Germline mutations in BRCA1 or BRCA2 (collectively BRCA1/2) predict generally high (but highly variable) lifetime risks of breast or ovarian cancer (2, 3). Given a positive BRCA1/2 mutation test result, pre-symptomatic interventions include intensive surveillance or prophylactic surgeries to remove healthy breasts and ovaries. Patients who already have breast or ovarian cancer, along with significant personal or family history of familial breast/ovarian cancer, may seek BRCA1/2 mutation testing, as positive results may inform treatment options and provide potentially life-saving information to family members (4). Thus, genetic testing for germline BRCA1/2 mutations is an important component of comprehensive breast/ovarian cancer care, and biologically informed mutation detection methods are critical to this effort. Conventional genetic testing involves germline sequence analysis of BRCA1/2 exons and flanking intron-exon boundaries, along with molecular assays for some large-scale BRCA1/2 genomic rearrangements. Anczuków et al. (this issue) provide an example of a functionally oriented mutation detection assay that revealed a mutation that would have been missed by more conventional methods. Specifically, they identified a BRCA2 splicing defect caused by inclusion of a cryptic exon, resulting from a deep intronic base change. This work illustrates a practical complement to older mutation detection methods, yet it is only used routinely by a handful of laboratories.
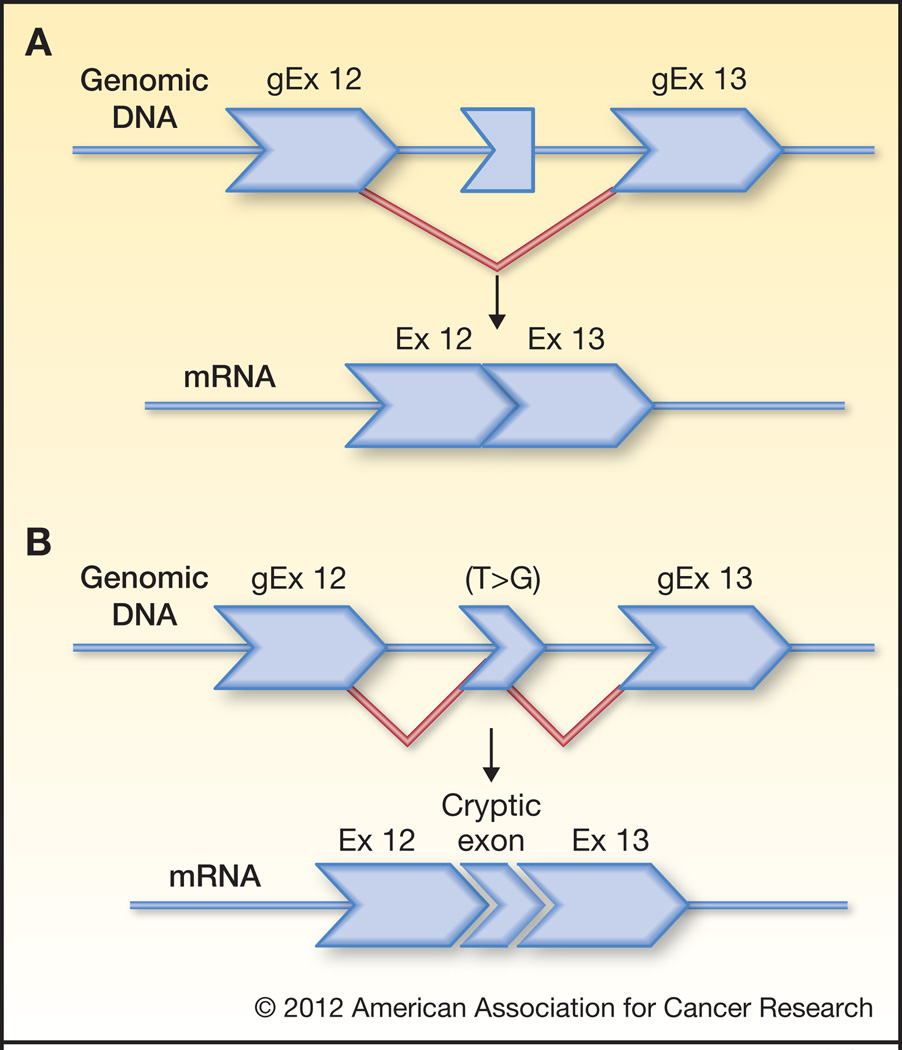
The authors found the cryptic exon by performing RT-PCR analysis of BRCA2 transcripts from Rhône-Alpes area patients referred for genetic testing because of family history of breast/ovarian cancer. After observing the 95 nucleotide sequence inserted between exons 12 and 13 in one family, they performed sequence analysis of the surrounding intronic DNA and found a single base substitution (c.6937+594T>G) that enhances the predicted strength of a 5’ splice site (Fig. 1). Indeed, minigene experiments confirm this single base change alone can confer exon identity on the otherwise-cryptic 95-base exon. To determine the prevalence of this mutation, the authors screened individuals from over 2,000 breast and/or ovarian cancer families and found it in eight additional families. Of these, six families were examined further, and the mutation was found in all 13 affected individuals tested showing that the mutation segregated with cancer in these families in strong support for the deleterious nature of the mutation. However, the frequency of the mutation among unaffected family members was not presented and penetrance was not estimated.
Figure 1.
1A. A sequence within intron 12 of BRCA2 is somewhat exon-like, but does not have a sufficiently strong 5’ exon donor site to be efficiently included as a high frequency exon among BRCA2 splice variatnts. 1B. A T>G transversion generates a much stronger 5’ exon donor site consensus sequence, resulting in high-frequency inclusion of the cryptic exon.
The authors further speculate on the interesting idea that the deleterious cryptic exon could be a target for antisense oligonucleotide therapy. Using their minigene system, they demonstrate that an oligonucleotide spanning the 5’ cryptic splice donor and the flanking intron can prevent incorporation of the cryptic exon and restore normal splicing of BRCA2 exons 12 and 13. It will be interesting to see whether such an oligonucleotide will have the same effect in the context of splicing all 27 BRCA2 exons.
While this particular mutation was seen in only 9/2,000 (0.5%) of families examined, it is likely that mRNA-based mutation detection methods will identify other previously unidentified deleterious mutations. Will such mutations be significant contributors to breast/ovarian cancer risk? That is difficult to know, but it is worth considering that only 44% of patients predicted to carry a BRCA1 or BRCA2 mutation test positive for recognized mutations in either gene. (5). Additionally, only 63% of familial breast cancers that map to the BRCA1 locus on chromosome 17 carry detectable mutations in the BRCA1 DNA sequence (6). Collectively, these observations suggest there may be large numbers of mutations in BRCA1 or BRCA2 that are either not detected by conventional sequencing analysis, or are not recognized as deleterious even when they are detected.
There are several types of mutations seen in other disease-associated genes that would also be potentially missed by current screening strategies, and these have never been systematically sought in BRCA1 or BRCA2. These include promoter mutations, mutations in 5’ and 3’ untranslated mRNA that could affect message stability and/or processing, deep intronic mutations that promote exon-skipping, spurious “exonization” of genomic repeat elements, or other types of splicing defect (7).
Importantly, Anczuków et al. show, using isoform specific RT-PCR, that the cryptic exon inclusion associated with the intronic base change can also be detected at low levels among wild type BRCA2 mRNA products. As the relative abundance of mutation-associated alternate splice variants may not be “all or none,” it is critical to characterize the penetrance of the mutation. Events resulting in increased levels of different splice variants will not necessarily always be phenotypically deleterious.
With the wealth of genomic sequence data now available, it is worth asking whether this deep intronic variant would have been recognized as potentially deleterious had it been seen during sequence analysis. Certainly bioinformatics will play an increasingly important role in assessing the true contribution of BRCA1 and BRCA2 mutations to breast cancer risk. In the current paper, the authors make use of splice site prediction algorithms to determine why a single base change promotes efficient inclusion of the cryptic exon. In the future, we expect bioinformatics to be used to predict mutation-associated alternate splicing events that can then be tested with molecular analysis. Prediction models will incorporate advancing but still incomplete understanding of the molecular events that select and correctly splice exons, including chromatin density, co-transcriptional events, secondary structures, and trans acting factors, coupled with the technologies of exon arrays used for alternate spicing events (8, 9)
Certainly bioinformatics will play an increasingly important role in assessing the true contribution of BRCA1 and BRCA2 mutations and for that matter other yet uncharacterized genes to breast cancer risk. Genome Wide Association Studies (GWAS) have successfully contributed to provide large numbers of intronic SNPs with significant associations to diseases or traits. Analysis of high throughput sequencing data (10), have advanced on identifying SNPs that primarily affect alterative splicing (AS). However, the molecular mechanisms by which intronic SNPs affect AS and increase risk of disease remain largely unknown. Thus, there is great need to initiate a new era of research that investigates how intronic SNPs affect alternative splicing, including activation of cryptic exons, in clinically relevant genes such as described in this report. Bioinformatics tools developed for splice site predictions (11) have successfully contributed to identification of novel cryptic exons in dystrophy research ((12). While bioinformatics may make increasingly powerful predictions about splicing defects in the future, such predictions will always require molecular testing. And, as the number of rare mutant alleles of BRCA1 and BRCA2 and other cancer susceptibility genes grows, the need for creative methods for assessing the mutation status in individual families and their clinical significance becomes more pressing.
Footnotes
The authors have no conflict of interests.
References
- 1.Anczuków O, Buisson M, Léoné M, Coutanson C, Lasset C, Calender A, et al. BRCA2 deep intronic mutation causing activation of a cryptic exon: opening towards a new preventive therapeutic strategy. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-12-1100. (this issue). [DOI] [PubMed] [Google Scholar]
- 2.Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4:174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrucelli N, Daly MB, Feldman GL. Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet Med. 2010;12:245–259. doi: 10.1097/GIM.0b013e3181d38f2f. [DOI] [PubMed] [Google Scholar]
- 4.Domchek SM, Friebel TM, Neuhausen SL, Wagner T, Evans G, Isaacs C, et al. Mortality after bilateral salpingo-oophorectomy in BRCA1 and BRCA2 mutation carriers: a prospective cohort study. Lancet Oncol. 2006;7:223–229. doi: 10.1016/S1470-2045(06)70585-X. [DOI] [PubMed] [Google Scholar]
- 5.Nanda R, Schumm LP, Cummings S, Fackenthal JD, Sveen L, Ademuyiwa F, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294:1925–1933. doi: 10.1001/jama.294.15.1925. [DOI] [PubMed] [Google Scholar]
- 6.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomassen M, Blanco A, Montagna M, Hansen TV, Pedersen IS, Gutierrez-Enriquez S, et al. Characterization of BRCA1 and BRCA2 splicing variants: a collaborative report by ENIGMA consortium members. Breast Cancer Res Treat. 2012;132:1009–1023. doi: 10.1007/s10549-011-1674-0. [DOI] [PubMed] [Google Scholar]
- 8.Jin Y, Yang Y, Zhang P. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol. 2011;8:450–457. doi: 10.4161/rna.8.3.15388. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz S, Ast G. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. Embo J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 12.Malueka RG, Takaoka Y, Yagi M, Awano H, Lee T, Dwianingsih EK, et al. Categorization of 77 dystrophin exons into 5 groups by a decision tree using indexes of splicing regulatory factors as decision markers. BMC genetics [electronic resource] 2012;13:23. doi: 10.1186/1471-2156-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]