Abstract
U4/U6.U5 tri-snRNP is a 1.5 MDa pre-assembled spliceosomal complex comprising U5 snRNA, extensively base-paired U4/U6 snRNAs and >30 proteins, including the key components Prp8, Brr2 and Snu114. The tri-snRNP combines with a pre-mRNA substrate bound to U1 and U2 snRNPs and transforms into a catalytically active spliceosome following extensive compositional and conformational changes triggered by unwinding of the U4/U6 snRNAs. CryoEM single-particle reconstruction of yeast tri-snRNP at 5.9Å resolution reveals the essentially complete organization of its RNA and protein components. The single-stranded region of U4 snRNA between its 3′-stem-loop and the U4/U6 snRNA stem I is loaded into the Brr2 helicase active site ready for unwinding. Snu114 and the N-terminal domain of Prp8 position U5 snRNA to insert its Loop I, which aligns the exons for splicing, into the Prp8 active site cavity. The structure provides crucial insights into the activation process and the active site of the spliceosome.
The protein coding sequences of most eukaryotic genes are interrupted by non-coding segments called introns. Introns are removed from pre-mRNAs and the flanking coding segments (exons) are spliced together to form mRNAs by two successive trans-esterification reactions within a dynamic multi-megadalton protein-RNA complex known as the spliceosome. This complex comprises five canonical subunits, namely U1, U2, U4, U5 and U6 small nuclear ribonucleoprotein particles (snRNPs) and numerous non-snRNP factors1. Each snRNP contains an snRNA, 7 Sm or LSm proteins and a number of snRNP-specific proteins. During the initial stages of spliceosome assembly, U1 and U2 snRNPs recognize the pre-mRNA 5′ splice site (5′SS) and branch point (BP) forming pre-spliceosomal A complex. The subsequent binding of the preassembled U4/U6.U5 tri-snRNP allows formation of the fully assembled spliceosomal B complex, which is converted to the catalytically active B* complex through extensive structural and compositional remodeling. During this process, U4 and U6 snRNAs, extensively base-paired in tri-snRNP are unwound, U1 and U4 snRNPs are released and many new proteins join the spliceosome1-3. This leads to the formation of a highly structured RNA network between U2, U5 and U6 snRNAs and the 5′SS and BP sequences in the pre-mRNA. The extensively base-paired U2–U6 snRNAs harbour catalytic magnesium ions4 and position the BP and 5′SS for the first trans-esterification reaction, which produces exon 1 and lariat intron–exon 2 intermediates. Further remodeling to C complex enables U5 snRNA loop 1 to align exons 1 and 2 for nucleophilic attack of exon 1 at the 3′-splice site (3′SS), yielding spliced mRNA and lariat intron products5,6. Finally the spliceosome is disassembled before the next round of splicing.
U4/U6.U5 tri-snRNP is the largest preassembled spliceosomal complex, containing U5 snRNA, extensively base-paired U4-U6 snRNAs and over 30 proteins7,8 (Extended Data Table 1). Three key proteins, Prp8, Brr2 and Snu114, play crucial roles in activation of the spliceosome and formation of the active site1. Prp8 forms cross-links with 4-thiouridine introduced at key positions within U5 and U6 snRNAs and the substrate pre-mRNA, showing that Prp8 is involved in substrate positioning and closely associated with the catalytic RNA core9,10. Brr2 helicase, whose activity is regulated by the GTPase Snu114 (ref 11-13), catalyses the unwinding of the U4-U6 snRNA duplex14,15. Interactions between tri-snRNP proteins have been investigated by yeast two-hybrid and in vitro binding assays16,17. Electron cryo-microscopy (cryoEM) reconstruction of cross-linked human tri-snRNP at 21 Å resolution revealed a tetrahedral overall shape with no clear domain separation18. Negative stain microscopy of cross-linked yeast tri-snRNP revealed a tri-angular shape with maximum dimension of 30-34 nm19. The highly biased orientation of tri-snRNP on carbon films precluded full three-dimensional analysis whilst the projection structure revealed three extruding domains termed head, foot and arm; the arm domain adopts variable positions with respect to the rest. Some key proteins were localised within the projection structure using genetically introduced tags 19. Brr2 and U4/U6 snRNP were attributed to the head and arm domains, respectively. Based on this the authors proposed that Brr2 may engage with U4/U6 snRNA for unwinding when Snu114 - mapped in the hinge region - brings the arm and head domains closer.
The development of high-speed direct electron detectors20,21 and powerful maximum likelihood algorithms for classification and particle alignment22 have made it possible to determine the structure of macromolecular assemblies at near atomic resolution by cryoEM23. By applying these new methods we obtained a map of native unstained yeast tri-snRNP at an overall resolution of 5.9 Å in which protein α-helices and RNA double helices are readily discernible. This enabled us to fit the double-stranded helices of U5 snRNA and U4/U6 snRNA as well as previously determined crystal structures or homology models of nearly all the proteins. The structure accounts for a wealth of biochemical and genetic data from yeast and human spliceosomes, and suggests a possible mechanism for B complex formation and the activation of the spliceosome.
Results
CryoEM of the tri-snRNP complex
U4/U6.U5 tri-snRNP was purified from yeast by a gentle procedure without cross-linking, and the sample was subjected to cryoEM analyses (Methods, Extended Data Fig. 1). Using a combination of statistical classification and movie processing22,24 (Extended Data Fig. 2), we obtained a density map with an overall resolution of 5.9 Å by the “gold standard” Fourier shell correlation (FSC) = 0.143 criterion25 with local resolution ranging from 5.0 Å to 20 Å (Extended Data Fig. 3 and 4; Methods). The map revealed clear densities for double-stranded RNA, with protein helices appearing as long tubes and β-sheets as flat densities (Supplementary Video). The density for the Lsm proteins in the flexible arm domain became clearer after using a new multi-body refinement method (Methods; Extended Data Fig. 3).
Overall structure
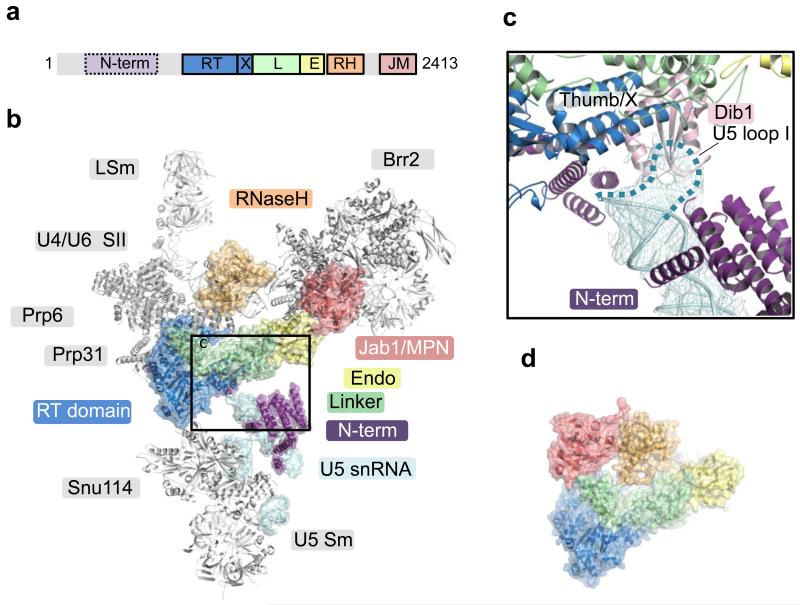
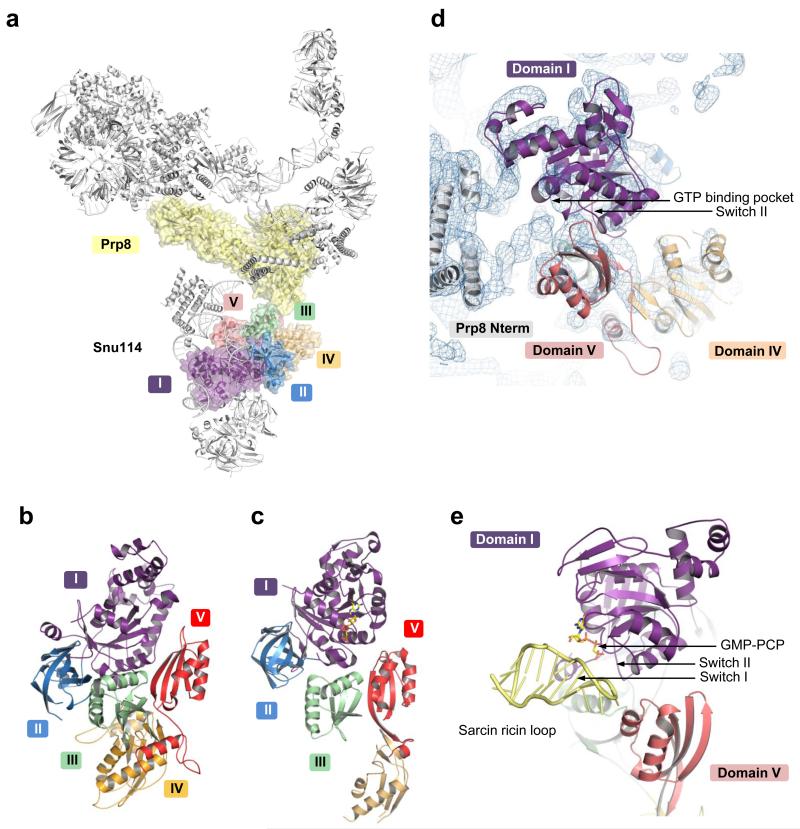
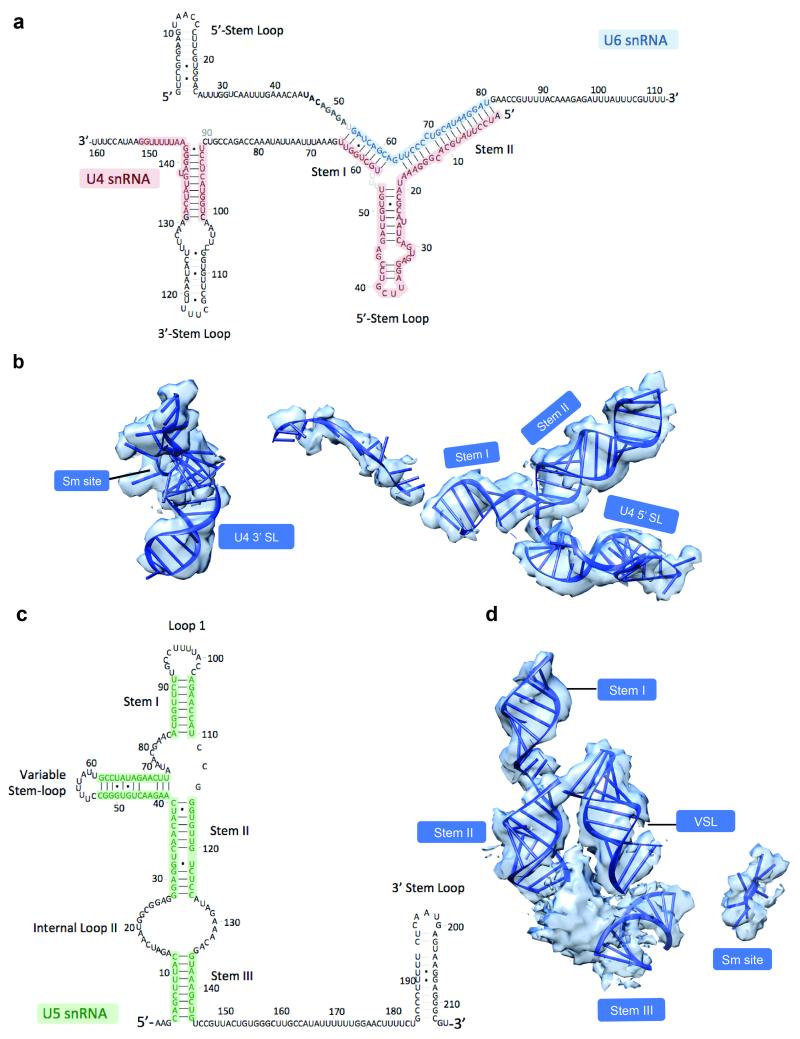
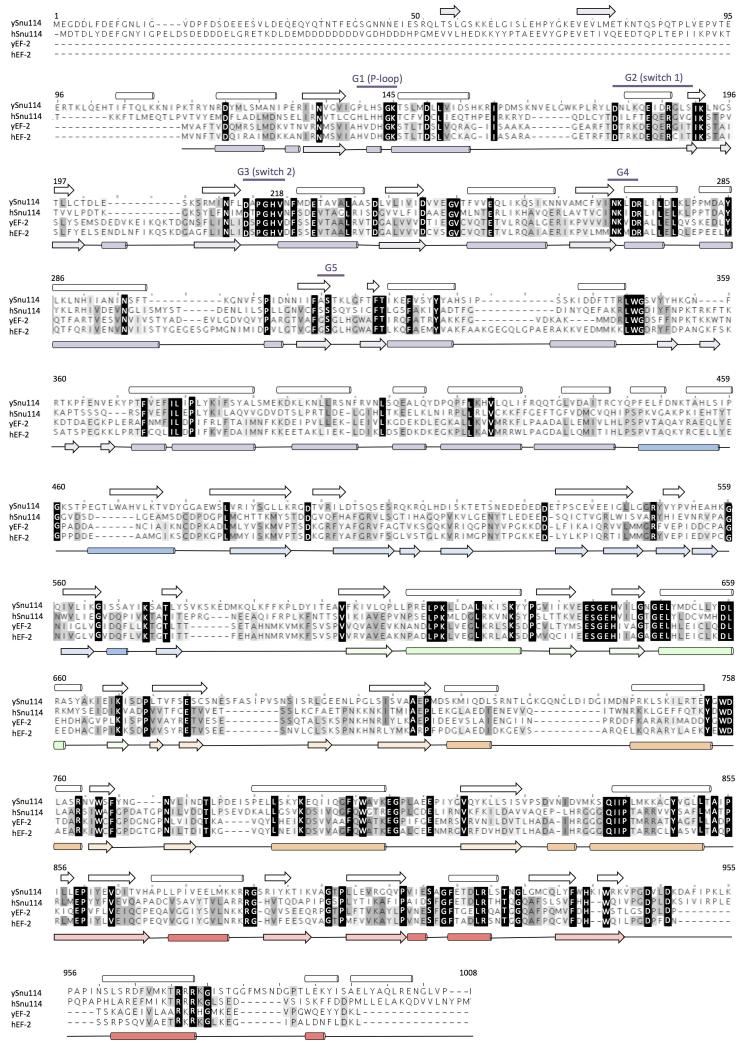
Yeast U4/U6.U5 tri-snRNP has an overall Y-shape with a maximum dimension of approximately 300 Å (Fig. 1). The large domain of Prp8 (residues 885-1824) consisting of the Reverse Transcriptase-like (RT), Linker and type II Endonuclease-like domains10 is located near the centre of the assembly and its crystal structure was fitted into the map as a rigid body10 (Fig. 2; Extended Data Fig. 5a). The orientation of the RNaseH-like domain with respect to the large domain is inverted in tri-snRNP as compared with the Prp8-Aar2 complex10 (Fig. 2d). Three segments of clear double stranded RNA density extending from Prp8 to the foot domain are assigned to coaxially stacked stems I and II, variable stem-loop and stem III of U5 snRNA (Fig. 3; Extended Data Fig. 6) connected to the U5 Sm core (Extended Data Fig. 5e). Snu114 shows a significant sequence similarity to eukaryotic translation elongation factor 2 (EF2)11,26 comprising Domains I-V (Fig. 4; Extended Data Fig. 5c and 7). Homology models of each domain of Snu114 (residues 120-1008) were fitted individually into the density adjacent to Prp8 and U5 snRNA revealing a contact between Domain III of Snu114 and the RT domain of Prp8 (Fig. 4). The structure of the N-terminal 884 residues of Prp8 is still unknown. The N-terminal helix of the RT domain of Prp8 (RTα1)10 extends further in tri-snRNP towards a bundle of four long helices. Another cluster of long helices, which makes close contact with the coaxially stacked stems I and II of U5 snRNA, is found in the vicinity (Figure 3b; Extended data Fig. 6c). The region containing residues 420-542 of Prp8 is known to interact with the N-terminal half of Snu114 (ref. 27) and 4-thiouridine introduced at C79 of U5 snRNA cross-links with both Prp8 and Snu114 (ref. 28), suggesting that the density adjacent to Snu114 and U5 snRNA is part of the Prp8 N-terminus. At the tip of stem I the density assigned to the U5 loop I extends towards the RT Thumb/X domain of Prp8 and makes close contact with a thioredoxin-like fold of Dib1 (Fig. 2c; Extended Data Fig. 5f) (ref. 29). This is in good agreement with the fact that a 16 kDa protein is cross-linked to 4-thiouridine incorporated at U97 in the U5 snRNA loop I (ref. 28). The binding of Dib1 is further stabilized by the N-terminal helices of Prp8 (Fig. 2c).
Figure 1. Overview of the U4/U6.U5 tri-snRNP structure with its protein and RNA components modeled into cryo-EM density.
a, Front view, facing concave surface; b, back view; c, top view. d, 2D class average showing the different domains of tri-snRNP: head, body, arm and foot.
Figure 2. Prp8 in tri-snRNP.
a, Domain organization of Prp8. The structure of the N-terminal domain (residues 1-884) is unknown. RT, Reverse transcriptase-like domain; X, Thumb/X; L, linker, RH, RNaseH-like; JM, Jab1/MPN domains. b, The large domain of Prp8 is located at the centre of tri-snRNP. The Jab1/MPN domain is bound to Brr2 (ref. 31,32). c, Inset, Loop 1 of U5 snRNA is inserted to the active site cavity and in contact with Dib1. d, Prp8 in the Prp8/Aar2 complex10 is shown with its large domain in the same orientation as in b. In tri-snRNP, the RNaseH domain is inverted while the Jab1/MPN domain in complex with Brr2 is located at the opposite end of the large domain.
Figure 3. The snRNA components of U4/U6.U5 tri-snRNP.
a, Secondary structures of U4/U6 and U5 snRNAs. b, Double-stranded regions of U4/U6 and U5 snRNAs modeled into the cryo-EM map.
Figure 4. Structure of Snu114 in tri-snRNP.
a, Location of Snu114 in the U4/U6.U5 tri-snRNP. b, Arrangement of Domains (I-V) in Snu114 (see Extended Data Fig. 7). c, Domain arrangement in EF-G bound to the ribosome44. d, The interface between the N-terminal domain of Prp8 and Snu114. Some of the uninterpreted density at the interface may be attributed to the unmodeled switch I loop. e, The interaction of the switch region of EF-G with the sarcin-ricin loop44 for GTPase activation.
Brr2 forms a stable complex with the Jab1/MPN domain of Prp8 (ref. 30-32) and its characteristic shape was recognized in the less ordered head domain (Fig. 1 and 5; Extended Data Fig. 5b). Although this part of the map is lower in resolution the individual domains of yeast Brr2 were fitted into the density together with the Jab1/MPN domain31. This revealed a widening of the gap between the two RecA domains of the N-terminal cassette (Fig. 5c). Coaxially stacked stems I and II of U4/U6 snRNAs and the 5′-stem-loop (5′SL) of U4 snRNA branching from the 3-way junction are unambiguously identified near the N-terminal helicase cassette of Brr2 and Prp8 (Fig. 1 and 3; Extended Data Fig. 6). Snu13 and the Nop domain of Prp31 bind to the kink-turn at the tip of the U4 snRNA 5′SL33. Initially the crystal structures of Snu13 and Prp31 were fitted individually around the kink-turn but the density clearly showed that the coiled-coil domain of Prp31 is rotated by approximately 60° with respect to the Nop domain in tri-snRNP (Fig. 1; Extended Data Fig. 5g). The four-helix bundle of the coiled-coil domain is in contact with the RT domain of Prp8 (Fig. 1). Furthermore, Prp31 used for crystallization was a truncated form 33 and in our map clear α-helical density extends from both N- and C-termini (Extended Data Fig. 5g). The C-terminal helix extends from the 5′SL to the three-way junction, in agreement with previous foot-printing experiments34 (Fig. 1a). Prp4 contains seven WD repeats at the C-terminus35 and its characteristic seven-bladed β-propeller domain is packed against Snu13 and the junction between the Nop and coiled-coil domain of Prp31 (Fig. 1 and 5, Extended Data Fig. 5j). Four additional α-helices, likely belonging to the N-terminus of Prp4, were built into the rod-like density on top of the β-propeller. Prp3 is predicted to have a ferredoxin-like domain at its C-terminus36 and interacts with Prp4 and U4/U6 stem II (ref. 37). Density sandwiched between the Prp4 WD40 domain and U4/U6 stem II, two long helices lying along U4/U6 stem II and a number of connected helices nearby are likely to belong to Prp3 (Fig. 1; Extended Data Fig. 5i). The U4 core domain is wedged between the tandem helicase cassettes of Brr2 (Fig. 1a, 1c and 5b; Extended Data Fig. 6a). The 3′SL of U4 snRNA contacts the helix-loop-helix domain of the N-terminal helicase cassette (Fig. 5b) which contains several lysine/arginine residues close to the RNA backbone of the 3′SL of U4 snRNA. Based on previous labeling data19 U6 LSm proteins are fitted into the flexible arm region in the multi-body refined map (Fig. 1a, Extended Data Fig. 3c and 5l).
Figure 5. Brr2 mode of unwinding.
a, Domain organisation of Brr2 N-terminal helicase cassette (NHC). WH, Winged-helix; HLH, helix-loop-helix; and FN3, fibronectin3-like domains. The inactive C-terminal helicase cassette (CHC) has the same domain organisation. b, U4/U6 di-snRNP and its interaction with Brr2 in tri-snRNP. The domains of Brr2 NHC are coloured as in a. The single-stranded RNA between U4/U6 stem I and U4 3′SL is already loaded in the active site of Brr2. When the Hel308 structure43 is overlaid onto the NHC of Brr2, its 10-nucleotide DNA substrate coincide with the density in the Brr2 active site, which extends to U4 snRNA 3′SL (red dotted line). The helix-loop-helix domain of Brr2 interacts with U4 snRNA 3′SL (inset). c, Superposition of the RecA1 domain of Brr2 in the crystal structure31 (PDB ID 4BGD, in grey) and in tri-snRNP (domains coloured as in a) shows the opening of the gap between the RecA1 and RecA2 domains (indicated by the red arrow) to accommodate the RNA substrate.
A striking elongated curved α-solenoid density bridging the RNaseH-like domain of Prp8 and the WD40 domain of Prp4 is assigned to the tetratricopeptide (TPR) motifs of Prp6 (Fig. 1b-c; Extended Data Fig. 5d). Prp6 is required for the accumulation of tri-snRNP38 and is proposed to act as a bridge between U5 and U4/U6 snRNPs16,39. Prp6 contains up to 19 predicted TPR motifs, each comprising a helix-loop-helix motif39 and 37 connected idealized poly-Ala helices were built into the map. Nine canonical tandem TPR motifs at the C-terminus of the protein form a highly curved alpha-helical solenoid-like structure, which contacts Snu13, U4 snRNA 5′SL and the Prp4 WD40 domain in tri-snRNP (Fig. 1b-c). This is consistent with the fact that antibodies against the C-terminal fragment of human Prp6 immunoprecipitate U5 snRNP but not tri-snRNP, as the C-terminal domain in our structure is in close contact with U4/U6 snRNP, which presumably occludes the epitope39.
Central role of Prp8 in tri-snRNP assembly
Our single particle cryoEM reconstruction of yeast U4/U6.U5 tri-snRNP has revealed a nearly complete organization of its RNA and protein components although some densities remain unassigned, and Snu66, Snu23, Prp38 and possibly substoichiometric Spp381 are yet to be located (Extended Data Table 1; Extended Data Fig. 4f and 8d-e). Prp8 positioned at the centre of the assembly functions as a hub of protein-protein and protein-RNA interactions holding the whole assembly together (Fig. 1 and 2b). In yeast, a stable Prp8-Snu114-Aar2-U5 core domain complex is imported into the nucleus40, where Brr2 replaces Aar2. The Jab1/MPN and RNaseH-like domains, held tightly onto the Prp8 large domain by Aar2 (Fig. 2d), are released in tri-snRNP wherein the Jab1/MPN domain forms a stable complex with Brr2 as in the crystal structure31,32 (Fig. 1b and 2b; Extended Data Fig. 5b). The tri-snRNP structure provides the first glimpse of the interaction between Snu114, the U5 core domain and the N-terminal domain of Prp8 which holds the co-axially stacked stems I and II, and variable stem-loop of U5 snRNA (Fig. 3). On the opposite side of Prp8, Snu13 and Prp31 firmly bound to 5′SL of U4 snRNA33, the U4 Sm protein assembly, the Brr2-Jab1/MPN domain complex, the Prp3/Prp4 complex, the RNaseH domain of Prp8 and the U4/U6 snRNA duplex assemble together (Fig. 1 and 2).
Prp8 has a surface on which are exposed the 5′SS-binding ACAGAGA sequence of U6 snRNA, the U6 sequences which pair with U2 snRNA, and U5 snRNA loop I which interacts with exon 1 and exon 2. This surface is partly occluded by a highly conserved protein Dib1 (Fig. 2c and 6), suggesting its potential role in regulating the incorporation of RNA components into the active site cavity during spliceosome assembly and activation. When U4 and U6 snRNAs are unwound, releasing U4 snRNA from the spliceosome together with Snu13, Prp31, Prp3/Prp4 (ref. 3), Brr2/Jab1MPN are no longer held in place and could be repositioned during catalysis and spliceosome disassembly (Fig. 5; Extended Data Fig. 8d).
Figure 6. Insights into activation mechanism and the active site of the spliceosome.
a, Mapping of the U4-cs1 suppressor mutations on the surface of Prp8. Three clusters of mutations are found in close proximity to the key elements of spliceosomal activation: Prp31/U4snRNA 5′SL, Snu114, and ACAGAGA box of the U6 snRNA. b, A model of the catalytic core of group II intron docked into the active site cavity by superposition of the EBS1 stem of group II intron (PDB ID 3IGI) and stem I of the U5 snRNA.
Brr2 mode of action during activation
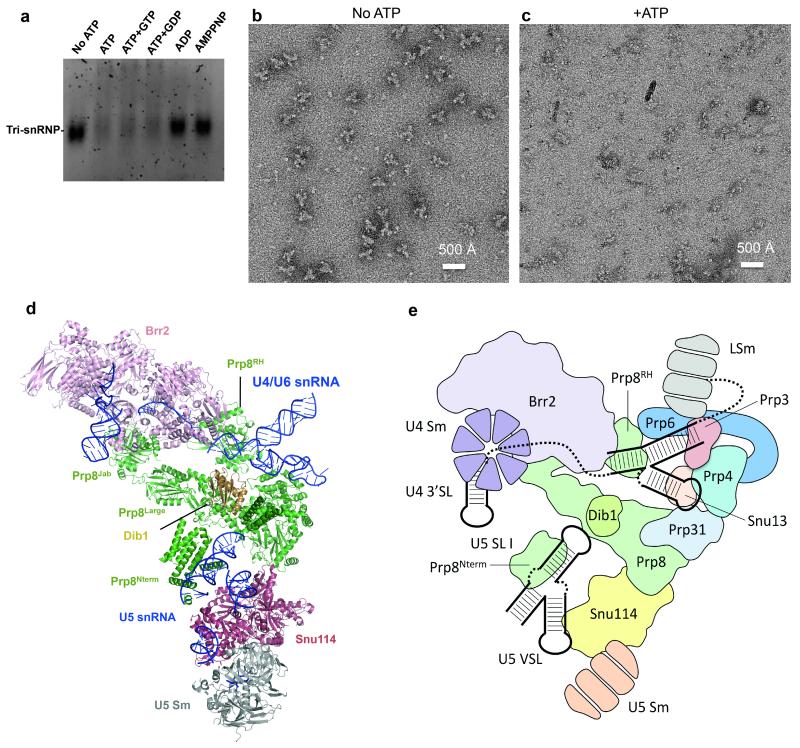
The unwinding of the U4/U6 snRNA duplex is an essential step in spliceosomal activation and is catalyzed by Brr2 (ref. 14,15). Like other Ski2-like helicases, Brr2 unwinds any RNA duplex with 3′ overhangs41. The U4/U6 snRNA duplex has 3′ overhangs on both ends (Fig. 3a; Extended Data Fig. 6a) and it has been suggested that Brr2 binds to the single stranded region of U4 snRNA and translocates along U4 snRNA41,42. Our structure shows that Brr2 is pre-loaded onto the single-stranded region between U4 snRNA 3′SL and stem I of the U4/U6 duplex, showing definitively that it translocates along U4 snRNA. The gap between the two RecA domains is widened in tri-snRNP and the prominent separator β-hairpin is located adjacent to stem I of the U4/U6 snRNA duplex (Fig. 5b-c). Our purified U4/U6.U5 tri-snRNP disintegrates upon addition of ATP regardless of the presence of GTP or GDP but remains intact after addition of ADP or AMPPNP, a non-hydrolyzable ATP analogue (Extended Data Fig 8). This shows that Brr2 is in an active state in our purified U4/U6.U5 tri-snRNP in perfect agreement with the structure. In vitro the RNaseH domain binds to the forked region of the U4/U6 snRNA duplex adjacent to stem I and inhibits Brr2 binding to the substrate RNA41. In tri-snRNP the RNaseH domain fails to prevent substrate loading onto Brr2 or unwinding. The helix-loop-helix domain of the Brr2 N-terminal helicase cassette interacts with the 3′SL of U4 snRNA and this interaction may be important for positioning U4 snRNA in the Brr2 active site41,42. Because of the limited resolution we cannot model the single-stranded region of U4 snRNA de novo. However, when we superpose the N-terminal helicase cassette of Brr2 onto the Hel308 structure with partially unwound DNA duplex43, ten nucleotides of the substrate DNA coincide with the extra density in the Brr2 active site and six additional nucleotides can be accommodated in the density extending further to the 5′ end of the U4 3′ SL (Fig. 5b).
Role of Snu114 in spliceosome activation
Snu114 shows substantial sequence similarity to EF2 (Fig. 4; Extended Data Fig. 7) suggesting that it might induce conformational change in the spliceosome upon GTP binding or hydrolysis and regulate spliceosomal activation11-13. EF-G, bacterial counterpart of EF2, enters the ribosome in the GTP-bound form. Its GTPase is activated when switch regions I and II are remodeled upon interacting with the sarcin-ricin loop of 23S rRNA44 (Fig. 4e) and GTP hydrolysis leads to translocation45. The activation process of the spliceosome has not been dissected in detail and it is not known at what stage GTP is hydrolysed or how Snu114 GTPase is activated. Snu114 and EF2 share highly similar switch I and II sequences, including the critical His residue, which places a water molecule adjacent to the γ-phosphate in EF-G. Unassigned density connecting the junction between stem I and II of U5 snRNA and the switch I and II loops coincides with the position of the sarcin-ricin loop (Fig. 4d). This is likely to be the N-terminal domain of Prp8 which may play a role in the activation of GTP hydrolysis.
Prior to the unwinding of the U4/U6 duplex, the 5′SS sequence pairs with the ACAGAGA sequence in U6 snRNA2. The U4-cs1 cold sensitive mutation, which extends U4/U6 stem I at the restrictive temperature and sequesters the ACAGAGA box from the 5′SS, stalls the spliceosome prior to unwinding46 (Extended Data Fig. 6a). A suppressor of U4-cs1 has a duplication of the ACAGA sequence in U6 snRNA47. This shows that pairing of the ACAGAGA sequence with the 5′SS is a checkpoint to ensure proper assembly of Complex B prior to the unwinding of the U4/U6 snRNA duplex. Interestingly suppressors of U4-cs1 in Prp8 form three clusters on the surface of the large domain of Prp8 (Fig. 6a; Extended Data Table 2)10. In tri-snRNP one of these clusters is located at the interface between the RT domain and Domain III of Snu114, and another is at the interface with Prp31 and the junction between the RT and N-terminal domains of Prp8, showing this checkpoint can be bypassed when these subunit interfaces are tampered with (Fig. 6a). This suggests that the interactions between these components undergo allosteric changes, which possibly couple the guanine-nucleotide binding state of Snu114 and the pairing between 5′SS and the ACAGAGA sequence to the activation of the U4/U6 duplex unwinding. Understanding the activation process will require extensive interplay between structural and biochemical work; the tri-snRNP structure provides an important structural framework for further investigation of this process.
The structural resemblance between the Group II intron active site48 and the catalytic RNA core of the spliceosome4,49 endorsed the hypothesis that they evolved from a common ancestor50. Based on the similarity of the domain architecture between the group II intron encoded protein (IEP) and Prp8, we proposed that Prp8 evolved from IEP and recruited more domains and interacting proteins to assemble spliceosomal snRNAs10, which derived from fragmented group II intron50. We placed the catalytic core of Group II intron RNA in the tri-snRNP structure by superimposing its exon binding stem-loop48 onto stem I of U5 snRNA. The Group II intron catalytic core fits neatly into the active site cavity after removal of Dib1, which is absent from activated spliceosomes (Fig. 6b), and with small rearrangement it can make contacts with the Thumb/linker region of Prp8, which cross-links with the catalytic RNA core in the spliceosome 9,10. The structure of tri-snRNP illustrates beautifully how the Prp8 domains and other spliceosomal proteins come together to assemble snRNAs and insert their functional segments into the active site cavity of the spliceosome10.
Methods
Brr2-TAPS tagging for yeast U4/U6.U5 tri-snRNP purification
Primers specific for 55 nucleotides of the C-terminus and 3′ UTR of BRR2 were used to PCR-amplify the TAPS-tag cassette together with the KanMX6 gene from pFA6a-TAPS-kanMX6, a modified version of pFA6a-TAP-kanMX6 in which the Calmodulin-binding peptide tag is replaced by 2 tandem copies of the StrepII tag51. The PCR product was used to transform yeast strain BCY123 [MATa pep4::HIS3 prb1::LEU2 bar1::HIS6 lys2::GAL1/10-GAL4 can1 ade2 trp1 ura3 his3 leu2-3, 112] by homologous recombination, selecting for G418-resistance. C-terminal TAPS-tagging of Brr2 was confirmed by PCR analysis of genomic DNA and DNA sequencing.
Sample preparation
The Brr2-TAPS-tagged yeast cells (72 Liters) were grown in YEPD medium to OD600 of 3.5, harvested and resuspended in lysis buffer (100 mM HEPES KOH pH 8.0, 200 mM KCl, 2 mM Mg(OAc)2 and 10% w/v glycerol). The cells were frozen and lysed by a Freezer Mill 6870 (SPEX CertiPrep). The crude lysate was centrifuged at 45krpm for 1 hour. The resulting supernatants were incubated with IgG sepharose overnight at 4 °C. The resin was washed with TAPS wash buffer (20 mM HEPES KOH pH 7.9, 150 mM KCl, 1 mM Mg(OAc)2 and 0.1% NP40) and incubated with TAPS wash buffer in the presence of TEV protease at 4 °C overnight. The flow-through was collected and incubated with Streptactin resin (GE Healthcare) for 3 hours. The resin was washed with TAPS wash buffer and particles were eluted with Strep elution buffer (20 mM HEPES KOH pH 7.9, 150 mM KCl, 1 mM Mg(OAc)2, 0.1% NP40, 5 mM desthiobiotin). The eluate was subsequently applied to a 10-30% v/v glycerol gradient centrifuged at 210,000g at 4 °C in a SWTi60 rotor. The fractions from the gradient were analysed by SDS-PAGE for protein composition. Glycerol was removed from the peak fractions containing tri-snRNP by dialysis against B150 buffer (20 mM HEPES KOH pH 7.9, 150 mM KCl, 1 mM Mg(OAc)2) prior to EM sample preparation (Extended Data Fig. 1a and 1b).
Electron microscopy
For cryo-EM analysis, 3.5 μl of the tri-snRNP sample was applied to Quantifoil R2/2 or R1.2/1.3 grids which were previously coated with a 6 nm thick layer of homemade carbon film and glow-discharged (Extended Data Fig. 1c). The grids were blotted for 2 s at 4 °C and plunged into liquid ethane using an FEI Vitrobot MKIII. The grids were loaded onto a Tecnai F30 Polara transmission electron microscope operated at 300 kV. Images were collected manually in low-dose mode at a calibrated magnification of 79,096×. The micrographs were recorded on either a Falcon II or a (ultra back-thinned) Falcon III detector at the same calibrated pixel size of 1.77 Å in movie mode at 17 frames/s. A total dose of 40e/ Å2 over 2.5s, and a defocus range of 2-4 μm were used.
Data processing
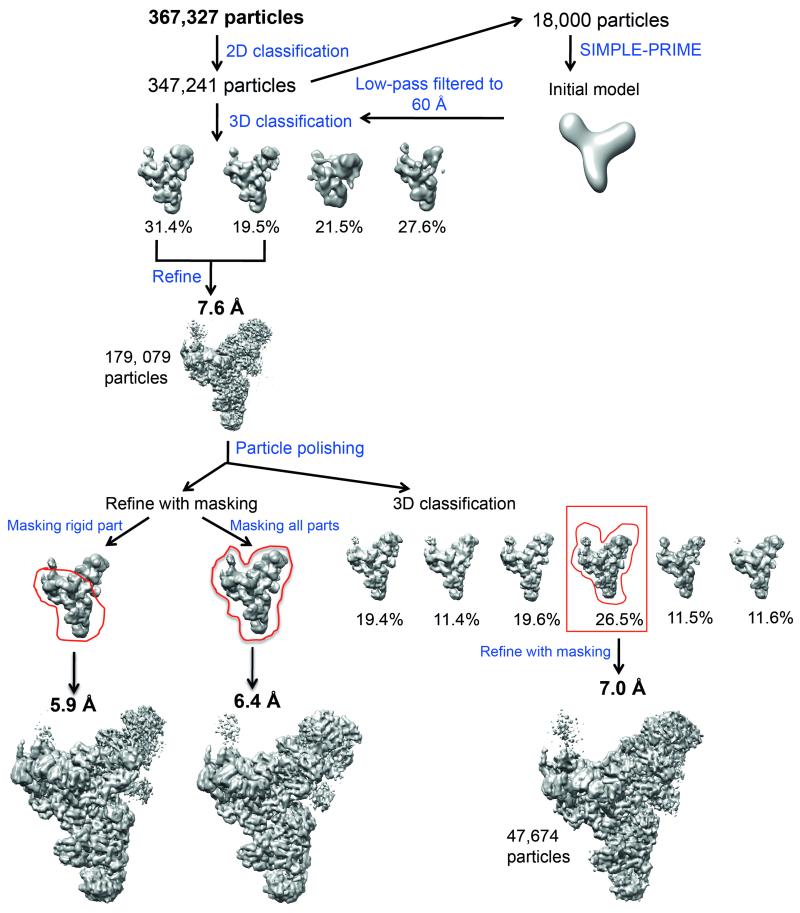
Most steps of data processing were performed in RELION22 unless otherwise stated. The 42 movie frames for each micrograph were corrected for whole-image drift using MOTIONCORR21, and contrast transfer function (CTF) parameters were estimated from the resulting micrographs using CTFFIND3 and CTFFIND4 (ref. 52). A subset of 5000 particles was picked manually and extracted with a 2802 pixel box, followed by reference-free 2D classification to obtain initial 2D class averages, which were then used as references for automatic particle picking. Particles resulting from the first round of automatic picking were extracted with a 2802 pixel box for reference-free 2D classification to obtain better references for the next round of automatic particle picking. All particles from the second round of automatic particle picking were manually checked before extracting them for reference-free 2D classification (Extended Data Fig. 1d, e). Prior to both autopicking runs, the templates were low-pass filtered to 20 Å to prevent high-resolution noise bias. A total of 347,241 particles from 2035 micrographs were selected from good 2D classes, and these particles were used for subsequent 3D processing (Extended Data Fig. 2).
A subset of 18,000 particles from only the best 2D classes of one of the datasets was used for ab initio 3D reconstruction by SIMPLE-PRIME53 to obtain an initial model of the complex, which was low-pass filtered to 60 Å for 3D classification. 3D classification with four classes was run for 25 iterations, using an angular sampling of 7.5° and a regularization parameter T of 4. This resulted in two classes with much better reconstructed features than the others. These classes were combined into a subset of 179,079 particles. Auto-refinement of these particles resulted in a 7.6 Å reconstruction. These particles were subsequently used for particle based beam-induced movement correction. For these calculations, we only used the first 30 frames of each micrograph with running averages of seven frames and a standard deviation of 1 pixel for translational alignments. We used the new “particle-polishing” approach, which fits linear tracks through the optimal translations for all running averages and takes into account the movements of neighbouring particles on the micrographs, to further improve the accuracy of the particle-based movement corrections24. The B-factors for the resolution and dose-dependent model for radiation damage were estimated using reconstructions from running averages of 3 frames.
Auto-refinement of the movement-corrected particles with a soft mask (with 12-pixel fall-off) around the entire map resulted in a map at 6.4 Å resolution while refinement with a similar soft mask around the more rigid part resulted in a map at 5.9 Å resolution (Extended Data Figure 2 and 3a). The 5.9 Å map was used for interpretation. Our map was also validated by a tilt-pair test (Extended Data Fig. 3d). Local resolution analysis showed a wide range of resolution from 5.0 Å to 20 Å (Extended Data Fig. 4a), indicating flexibility within some parts of the structure. Further 3D classification with a finer angular sampling interval of 1.8° and local angular search range of 10° revealed conformational heterogeneity of the head, body and foot domains of the structure and did not improve the overall resolution of the map (Extended Data Fig. 2).
We used a modified refinement approach in RELION, which we term “multi-body refinement”, to improve the density for the flexible arm domain (Extended Data Fig. 3b and c). In this approach, we used masks to divide the reference map into four “bodies”, approximately corresponding to the body, head, foot and arm domains. In each iteration of the auto-refine procedure, we independently aligned every experimental particle image against projections from the four distinct bodies. To minimize errors in these alignments, prior to the alignment against a given body we subtracted projections from the other three bodies from the experimental particle. Because we assume that the four bodies may adopt different relative orientations in each particle, we kept track of the most likely orientation for each of the four bodies for every particle during the course of the refinement. Thereby, subtraction of the other bodies should become ever more accurate, and this resulted in four sets of relative orientations for each particle. To express our expectation that the relative movements between the different bodies were limited, we used only local orientation searches (±22.5°) in the multi-body refinement, and centered the local searches around the orientations determined for the unmasked auto-refinement mentioned above. Details of this methodology will be described elsewhere (S.H.W.S. unpublished results), and its implementation will be made available through incorporation into RELION.
All refinements used gold standard Fourier shell correlation (FSC) calculations25 and reported resolutions are based on the FSC = 0.143 criterion. The FSC curves are calculated using a soft spherical mask (Extended Data Fig. 4d). Prior to visualization, all maps were corrected for the modulation transfer function of the detector and sharpened by applying a negative B-factor.
Local resolution analyses
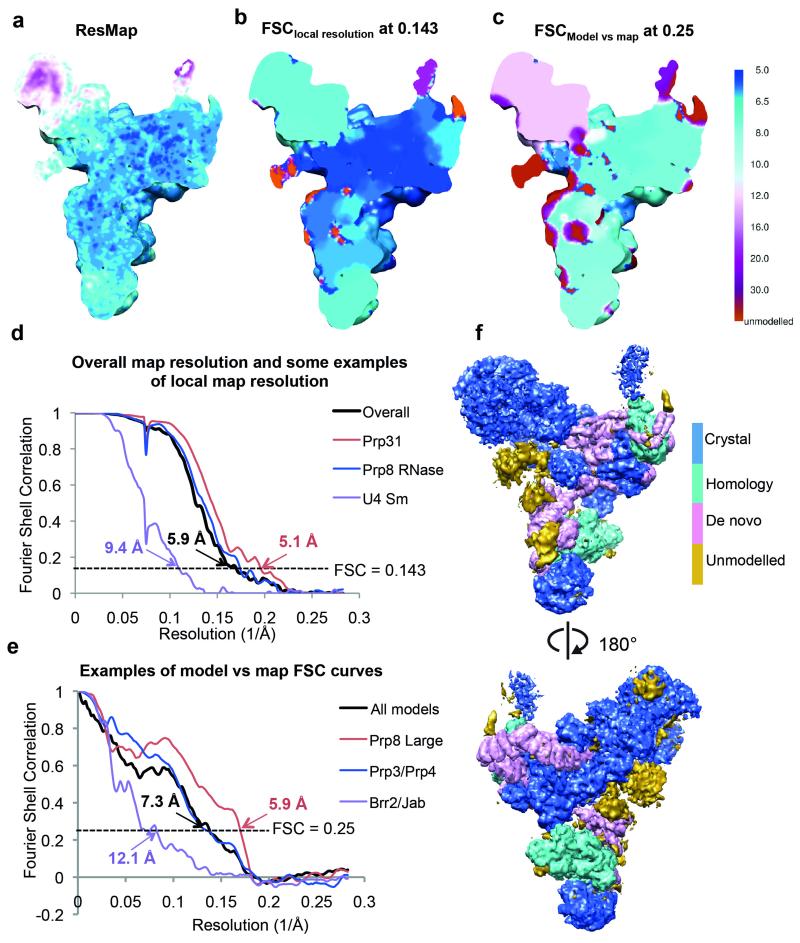
Local resolution analyses were performed by Resmap54 and compared with that calculated by us for each protein/RNA component of the map (Extended Data Fig. 4a-c). For the latter calculations, FSC curves are calculated using a soft spherical mask (with a 30-pixel fall-off) around each protein/RNA component of interest. Convolution effects of the masks on the FSC curves were corrected using high-resolution noise substitution55. Resolution was estimated at FSC=0.143. These calculations were performed for each of the following components: Prp8 Large Domain, Prp8 RNaseH domain, Prp8 Jab domain, Prp8 N-terminal domain, Brr2, U4 Sm with U4 3′ SL, U5 Sm with Sm site, Snu114, Dib1, Prp6, Prp3, Prp4, Snu13, Prp31, LSm, U5 snRNA and U4/U6 snRNA. Extended Data Fig. 4d shows some representative curves from these calculations.
FSC curves of model versus map were calculated using the Xmipp package56 and the reported resolutions were based on the FSC = 0.25 criterion. FSC curves of model versus map were calculated for not only the entire model of all components but also different parts of the maps. The map of each modelled component was extracted from the tri-snRNP map using a soft mask (with a 5-pixel soft-edge) surrounding the component. A map of each model was created by the program pdb2mrc within the EMAN package57. Some proteins/domains that are close together were grouped together for these calculations, including Prp8 N-terminal domain/Dib1, Brr2/Prp8-Jab domain, Prp3/Prp4 and Prp8 N-terminal domain/Dib1. Extended Data Fig. 4e shows some representative curves from these calculations.
Model fitting and building
Locations of available X-ray or homology models were fitted initially by visual inspection of the tri-snRNP map and low-pass filtered maps (to 10 Å) were generated for each model in Chimera followed by fit optimization in Chimera58. For the LSm proteins that are in the flexible arm region, the map resulting from multi-body refinement (Extended Data Fig. 3c and 5l) was used for fitting. Further rigid-body fitting was performed in Coot59. The homology model for Snu114 was prepared by I-TASSER web server60 based on the crystal structure of the yeast Elongation Factor 2 (ref. 26; PDB ID: 1N0V) (Fig. 4, Extended Data Fig. 5c). The model was manually inspected and the disordered regions were removed. The model for the ferredoxin-like domain of Prp3 is available at the Yeast Genome Center (http://www.yeastrc.org), which contains its structure predictions61. The model with the highest Mammoth Confidence Metric (MCM) score was selected for fitting. For Prp4, the protein sequence was input into Robetta Beta Full-chain Protein Structure (http://robetta.bakerlab.org), which yielded a model for the C-terminal part of Prp4 based on the structure of the WDR5 protein62 (PDB ID: 3MXX). Double stranded RNA helices and idealized poly-alanine helices were built into the masked map in Coot when possible. U4 snRNA 5′SL was modeled based on the structure of the human hPrp31-15.5K-U4snRNA complex33 (PDB ID: 2OZB) using ModeRNA modeling tool63. Yeast Snu13 structure64 (PDB ID: 2ALE) was fitted into the map. U4 snRNA 3′SL partial model was adapted from the structure of the human U4 snRNP core domain65 (PDB ID: 4WZJ). The short and long forms of U5 snRNA66 are present in our sample (Extended data Fig. 1b) but no density for 3′ stem-loop (3′SL) was observed, presumably because 3′SL attached to the Sm site with a long single stranded stretch is disordered or the particle population with the long U5 snRNA is classified out during classification. U5 snRNP Sm core with only the Sm site was also adapted from the human U4 snRNP core domain. The LSm proteins67 (PDB ID: 4M77) were placed in the low-resolution arm region of the map with the flat surface of the LSm complex facing the entrance side of U6 snRNA. The register of the LSm proteins cannot be accurately determined. Human Dib1 structure29 (PDB ID: 1QGV) was used for fitting. Extended Data Table 1 and Extended Data Fig. 4f summarise all the details of tri-snRNP components and modeling in our study. The active site cavity of Prp8 was described previously10 and defined by cross-links with crucial elements of U5 snRNA, U6 snRNA and pre-mRNA9 and suppressors of defective splice site mutations68-74. U4-cs1 mutants have been described46,75. Extended Data Table 2 summarises all the U4-cs1 mutants and their locations in tri-snRNP.
Map and model visualization
Maps were visualized in Chimera58. Map segmentation was performed in Chimera using each of the fitted models and the “zone-masking” function (Fig. 1, Extended Data Fig. 5, 6b and 6d). The LSm protein density was obtained from multi-body refinement and low-pass filtered to 20 Å. For all the remaining components, the sharpened tri-snRNP map (B= −214 Å2) low-pass filtered to 5.9 Å was used. Figures were generated using either Chimera58 or Pymol (www.pymol.org) and the video was made in Chimera58.
ATP assays
Purified tri-snRNP from glycerol gradient (~25 nM) was incubated at 30 °C for 30 min in the presence of either no nucleotide or with each of the following nucleotide combinations: ATP, ATP/GTP, ATP/GDP, ADP and AMPPNP (1 mM each). The samples (10 μl) were loaded onto a native agarose gel (0.5% in TB buffer supplemented with 1 mM MgCl2) and run at 75 V at 4 °C for 2.5 hours. The gel was stained with ethidium bromide for 1 hour before being imaged by a Syngene UV imager (Extended Data Fig. 8a). For negative staining, the sample was also treated similarly and stained with 2% uranyl acetate prior to imaging on a Tecnai T12 transmission electron microscope operated at 120 kV (Extended Data Fig. 8b and c).
Extended Data
Extended Data Figure 1. U4/U6.U5 tri-snRNP sample used for this study.
a, Coomassie-blue stained SDS-PAGE gel showing protein composition of the purified tri-snRNP. U5-, U4/U6- and tri-snRNP-specific proteins are labeled in blue, red and teal, respectively. Sm proteins present in both U5 and U4/U6 are in black. b, Toluidine-blue stained denaturing acrylamide (9%) gel showing RNA compositions. c, Electron cryo-micrograph of tri-snRNP where the carbon coated grid was discharged in N-amylamine. d, and e, Reference-free two-dimensional class averages of a data set collected on a grid discharged in air and N-amylamine, respectively.
Extended Data Figure 2. Classification and refinement procedures used in this study.
A total of 367,327 particles were subjected to reference-free 2D classification. A subset of 347,241 particles from good 2D classes was selected for 3D classification using an initial model obtained from SIMPLE-PRIME53, which was low-pass filtered to 60 Å. The data was divided into four 3D classes, two of which (a total of 179,079 particles) showed better features and were combined for refinement. This resulted in a 7.6 Å reconstruction. To further improve the reconstruction, these particles were subjected to beam-induced motion correction (particle polishing)24. Refinement of these polished particles with a soft mask around the rigid part of the map (as indicated by the red envelope) yielded a 5.9 Å reconstruction while refinement with a mask around the whole map yielded a 6.4 Å reconstruction. The polished particles were also subject to further 3D classification with a finer angular sampling of 1.8°. The most populated class (47,674 particles), which also has the best rotational accuracy, was refined with a soft mask around the whole density. This resulted in a 7.0 Å reconstruction. In this study, the 5.9 Å reconstruction was used for subsequent biological interpretation. All steps were performed in RELION22 unless otherwise stated.
Extended Data Figure 3. CryoEM maps and tilt-pair validation.
a, CryoEM density of the whole tri-snRNP at 5.9 Å resolution by “gold standard” Fourier Shell Correlation (FSC) of 0.143 criterion at two different contour levels. The high contour map (gold) shows well-resolved densities for protein and RNA helices and flat densities for beta-sheets. The low contour map (silver) shows densities for the more flexible head and arm. The map was sharpened by a B-factor of −214 Å2 and low-pass filtered to 5.9 Å as determined by RELION. b, The unsharpened full map of tri-snRNP. c, The map resulting from multi-body refinement, in which tri-snRNP is divided into four parts: the head, body, arm and foot. This resulted in better density for the arm domain (indicated by red circles), which is at 20 Å resolution. d, Tilt-pair validation plot for tri-snRNP. This was obtained from 1196 particles from 32 micrograph pairs, imaged at 0° and 10° tilt angles. The position of each dot represents the direction and the amount of tilting for a particle pair in polar coordinates. Blue dots correspond to in-plane tilt transformations; red and purple dots correspond to out-of-plane tilt transformations. Blue dots cluster in the same region of the plot at a tilt angle of approximately 10° as indicated by the red circle.
Extended Data Figure 4. Resolution estimation of tri-snRNP map.
a, Local resolution of the tri-snRNP map estimated by ResMap using the color scheme shown in panel c. b, Local resolution of the tri-snRNP map calculated by “gold-standard” FSC. For each component of the map that we modeled protein/RNA components, a soft mask (with a 30-pixel soft-edge) surrounding the region of interest was prepared and used for FSC calculations. Convolution effects of the masks on the FSC curves were corrected using high-resolution noise substitution55. Resolution was estimated at FSC=0.143. Local resolution for the unmodeled region of the map (in red) was not estimated. c, Local resolution of model versus map. The map of each modeled component was extracted from the map using a soft mask (with a 5-pixel soft-edge) surrounding the component. The model was converted into density by EMAN57. FSC of model versus map was calculated using Xmipp56. The map is colored according to resolution estimates based on a FSC threshold of 0.25. The lower resolution estimates from the FSC of model versus map compared to the estimates from ResMap and the gold-standard FSCs are explained by the nature of our models. Because of the limited resolution of our map, we did not perform full atomic refinement, but placed known crystal structures and homology models as rigid bodies in the map. d, Gold-standard FSC curves for the whole tri-snRNP map and some of its components calculated as described in b. e, FSC curves of model versus map for the whole model and some of the components. f, The full tri-snRNP map in which portions of the structure produced from crystal structures, homology modeling and de-novo building or unmodeled are colored as indicated.
Extended Data Figure 5. Fitting of protein components into tri-snRNP map.
a, Prp8885-2413 crystal structure10 (PDB ID: 4I43, green) and additional helices built de novo assigned to the N-terminus of Prp8 (blue). b, Brr2-Jab1/MPN complex31 (PDB ID: 4BGD). c, Snu114 homology model based on EF2 (ref. 26). d, The Prp6 TPR motifs built into the tri-snRNP map. e, U5 Sm proteins (grey) with Sm site (blue) based on the human U4 Sm structure (PDB ID: 4WZJ). f, Dib1 (ref. 29) (PDB ID: 1QGV). g, i, Prp31. ii, Comparison between the crystal structure of human Prp3178-333 (ref. 33) (PDB ID: 2OZB, grey) and that in tri-snRNP (yellow and blue). The coiled-coil domain (yellow) rotates by 60° in tri-snRNP with respect to the Nop domain (grey). Additional helices (blue) that extend from the N- and C-termini were built. h, U4 Sm proteins with part of U4 snRNA (blue) based on the human U4 Sm structure. i, Prp3 model. The ferredoxin-like domain was obtained from homology modeling while the extra helices were built de novo. j, Prp4 WD40 homology model with the extra helices built de novo. k, Snu13 (ref. 64) (PDB ID: 2ALE). l, U6 LSm proteins67 (PDB ID: 4M77).
Extended Data Figure 6. Fitting of the RNA components in tri-snRNP map.
a, and c, The sequences and predicted secondary structures of U4/U6 snRNA and the long version of U5 snRNA, respectively. b, and d, The maps of the fitted parts of U4/U6 snRNA and U5 snRNA, respectively. Unmodeled density assigned to U5 snRNA was also shown in d.
Extended Data Figure 7. Sequence alignment of yeast and human Snu114 with yeast and human elongation factor 2 (EF-2).
The secondary structures of our homology model for yeast Snu114 and the yeast EF-2 (ref. 26) (PDB ID: 1N0V) are shown on the top and bottom of the alignment, respectively. Important sequence elements are also shown.
Extended Data Figure 8. The effect of ATP on Brr2-TAPS purified tri-snRNP.
a, Ethidium-bromide stained native agarose gel (0.5%) showing the effects of ATP addition to Brr2-TAPS purified tri-snRNP used in this study. Upon ATP addition either without or with GTP/GDP, tri-snRNP fell apart (lanes 1-4). Under the same conditions, the addition of ADP or the non-hydrolysable ATP-analogue, AMPPNP had no effects on the complex (lanes 5-6). b, and c, The effect of ATP addition observed by negative stain microscopy. When ATP was not present, tri-snRNP particles could be observed. When ATP was added to the sample prior to grid preparations, tri-snRNP particles fell apart as observed by many small components on the micrograph rather than tri-snRNP particles. d, Tri-snRNP model where U4/U6 snRNP proteins are not shown. In tri-snRNP, Brr2/Prp8Jab complex is loosely associated to the remaining of U5 snRNP components including Prp8large, Prp8RNaseH, Prp8Nterm, Snu114, Dib1, U5 Sm proteins and U5 snRNA. After U4/U6 snRNA unwinding by Brr2, Brr2/Prp8Jab could be repositioned within the spliceosome. e, A schematic showing the arrangement of tri-snRNP protein and RNA components.
Extended Data Table 1. Components and modeling of yeast U4/U6.U5 tri-snRNP.
| protein | total residues | M.W. | Domain | PDB code | ||
|---|---|---|---|---|---|---|
| US snRNP | Prp8 | 2413 | 279,299 | N-terminal domain | 1-884 | α-helices modeled |
| RT-like | 885-1251 | |||||
| Thumb/X | 1257-1375 | |||||
| Linker | 1376-1649 | 4143 | ||||
| Endonuclease | 1653-1824 | |||||
| RNaseH-like | 1839-1029 | |||||
| Jab1/MPN | 2150-2396 | 4BGD | ||||
|
| ||||||
| Brr2 | 2163 | 246,125 | N-terminal domain | 1-441 | not modeled | |
| 442-478 | ||||||
| N-termnal helicase cassette | 478-1309 | 4BGD | ||||
| C-terminal helicase cassette | 1330-2163 | |||||
|
| ||||||
| Snu114 | 1008 | 114,025 | N-terminal domain | 1-114 | not modeled | |
| G domain | 120-443 | |||||
| domain II | 446-580 | |||||
| domain III | 603-671 | homology model (1NOV) | ||||
| domain IV | 675-853 | |||||
| domain V | 856-990 | |||||
|
| ||||||
| Dib1 | 143 | 16,774 | thioredoxin-like | not modelled | ||
|
| ||||||
| SmB | 196 | 22,403 | Sm fold | |||
| SmD3 | 110 | 11,229 | Sm fold | |||
| SmD1 | 146 | 16,288 | Sm fold | |||
| SmD2 | 110 | 12,856 | Sm fold | 4WJZ(human model) | ||
| SmE | 94 | 10,373 | Sm fold | |||
| SmF | 96 | 9,659 | Sm fold | |||
| SmG | 77 | 8,479 | Sm fold | |||
|
| ||||||
| U5 snRNA-L | 214 | 68,847 | Loop 1 | 92-102 | not modeled | |
| Stem 1 | 84-91;103-110 | A-form double helix | ||||
| IL 1 | 75-83;111-113 | not modeled | ||||
| VSL | 41-74 | A-form double helix | ||||
| Stem 2 | 28-40;114-125 | A-form double helix | ||||
| IL2 | 13-27;126-135 | not modeled | ||||
| Stem 3 | 4-12;136-144 | A-form double helix | ||||
| 3′ SL | 185-212 | not modeled | ||||
|
| ||||||
| U4/U6 snRNP | Snu 13 | 126 | 13,570 | 2ALE | ||
|
| ||||||
| Prp31 | 494 | 56,305 | N-terminal domain | α-helices modeled | ||
| coiled-coil domain | 2OZB (human model) | |||||
| Nop domain | ||||||
| C-termal domain | α-helices modeled | |||||
|
| ||||||
| prp3 | 469 | 55,877 | N-terminal domain | α-helices modeled | ||
| Ferredoxin-like domain | Model obtained from Robetta prediction | |||||
|
| ||||||
| prp4 | 465 | 52,425 | N-terminal domain | α-helices modeled | ||
| β-propeller domain | 166-465 | 3MXX from Robetta prediction | ||||
|
| ||||||
| SmB | 196 | 22,403 | Sm fold | |||
| SmD3 | 110 | 11,229 | Sm fold | |||
| SmD1 | 146 | 16,288 | Sm fold | |||
| SmD2 | 110 | 12,856 | Sm fold | 4WJZ (human model) | ||
| SmE | 94 | 10,373 | Sm fold | |||
| SmF | 96 | 9,659 | Sm fold | |||
| SmG | 77 | 8,479 | Sm fold | |||
|
| ||||||
| Lsm2 | 95 | 11,164 | Sm fold | |||
| Lsm3 | 89 | 10,020 | Sm fold | |||
| Lsm4 | 172 | 20,304 | Sm fold | |||
| Lsm5 | 93 | 10,415 | Sm fold | 4M77 | ||
| Lsm6 | 86 | 9,396 | Sm fold | |||
| Lsm7 | 115 | 13,010 | Sm fold | |||
| Lsm8 | 109 | 12,385 | Sm fold | |||
|
| ||||||
| U4 snRNA | 160 | 51,390 | Stem I | 57-64 | A-form double helix | |
| Stem II | 1-17 | A-form double helix | ||||
| 5′ SL | 20-53 | homology model (2OZB) | ||||
| central domain | 65-80 | partial homology model (2P6R) | ||||
| 3′SL | 91-142 | partial modei 4WJZ (human model) | ||||
|
| ||||||
| U6 snRNA | 112 | 36,088 | 5′ SL | 1-25 | not modelled | |
| Stem I | 55-62 | A-form double helix | ||||
| Stem II | 64-80 | A-form double helix | ||||
|
| ||||||
| tri-snRNP specific | Prp6 | 899 | 104,234 | 1-191 | not modelled | |
| TPR domain | 192-899 | α-helices modeled | ||||
|
| ||||||
| Snu66 | 587 | 66,426 | not modeled | |||
|
| ||||||
| Prp38 | 242 | 27,957 | not modeled | |||
|
| ||||||
| Snu23 | 194 | 22,682 | C2H2 zinc finger-like | not modeled | ||
|
| ||||||
| Spp381 | 291 | 33,764 | not modeled | |||
See Methods for details.
Extended Data Table 2. U4−CS1 suppressors.
| region | mutations | domains | locations | contact |
|---|---|---|---|---|
| region a | R236G | N-terminal domain | unknown | |
| L261P | ||||
| L280P | ||||
|
| ||||
| region b | K611R | N-terminal domain | unknown | |
| E624G | ||||
| N643S | ||||
| V644A | ||||
| D651G or N | ||||
| H659P | ||||
| K684E | ||||
|
| ||||
| region c | E788G or V | N-terminal domain | unknown | |
| N796S | ||||
| W856R | ||||
| E860K | ||||
| Q861R | ||||
|
| ||||
| cluster 1 | D1094A or N or V |
RT domain |
loop in 4 stranded β-sheet | Interface with Prp31 |
| M1095T | loop in 4 stranded β-sheet | |||
| V1098D | near in 4 stranded β-sheet | |||
| N1099K | near in 4 stranded β-sheet | |||
| I1104M | near in 4 stranded β-sheet | |||
| R1105L | near in 4 stranded β-sheet | |||
|
| ||||
| cluster 2 | P1191L or 5 or T | within loop following α12 | interface with Snu114 domain III | |
| D1192Y | within loop following α12 | |||
| N1194D | within loop following α12 | |||
|
| ||||
| cluster 3 | L1624M | endonuclease | top surface | |
| L1634F | top surface | |||
| L1641F | top surface | |||
| T16851 | top surface | |||
| P1688L or R | top surface | |||
| A1754V | side surface | |||
| N1809D | side surface | |||
|
| ||||
| region f | F1851L | RNaseH | on inner surface | |
| V1860D or N | β-finger | |||
| T1861P | β-finger | |||
| V1862A or D or Y | β-finger | |||
| I1875T | β-finger | |||
Supplementary Material
The video sequences showing the cryoEM density at two different contour levels; tri-snRNP map with all modeled components; fitting of available crystal structures into the cryoEM density: Brr2-Jab1/MPN (Prp8) complex31, Prp8 RNase H and large domains10, U4 and U5 Sm core domains65, Lsm core domain67 fitted into the multi-body map, Snu13 (ref. 64), human Prp31 (ref. 33) with remodeling, human Dib1 (ref. 29); fitting of homology models: Snu114 based translation factor EF2 (ref. 26), WD40 domain of Prp4, ferredoxin-like domain of Prp3 (ref. 36), TPR domain of Prp6; fitting of double helical RNA of the U4/U6 snRNA duplex and U5 snRNA; fitting of α-helices attributed to the N-terminal domain Prp8, Prp3 and Prp4; near complete pseudo-atomic structure of the yeast U4/U6.U5 tri-snRNP.
Acknowledgements
We thank S Chen, G. McMullan, J. Grimmett and T. Darling for smooth running of the EM and computing facilities, P. da Fonseca, N. Unwin, I. Sanchez Fernandez, A. Amunts, P. Emsley, G. Murshudov and A. Brown for advice, L. Passmore for reagents, M. Skehel for mass spectrometry, J. Li, Y. Kondo and the members of the spliceosome group for help and advice throughout the project. We are grateful to R. Henderson, D. Barford, S. Fica, P.-C. Lin and L. Strittmatter for critical reading of the manuscript. We thank V. Ramakrishnan, J. Löwe and R. Henderson for their continuing support and encouragements. T.H.D.N. was supported in part by a Herchel Smith Research Studentship. X.-C.B. was supported by a European Union Marie Curie Fellowship. The project was supported by the Medical Research Council (MC_U105184330 to K.N. and MC_UP_A025_1013 to S.H.W.S.).
Footnotes
Author Information
3D cryo-EM density maps of yeast U4/U6.U5 tri-snRNP have been deposited with the Electron Microscopy Database under accession number EMD-2966.
The authors declare no competing financial interests.
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:pii: a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan SP, Cheng SC. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- 3.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 4.Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 6.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 7.Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Biochemical and genetic analyses of the U5, U6, and U4/U6.U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- 8.Gottschalk A, Neubauer G, Banroques J, Mann M, Lührmann R, Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner IA, Norman CM, Churcher MJ, Newman AJ. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA. 2006;12:375–386. doi: 10.1261/rna.2229706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartels C, Urlaub H, Luhrmann R, Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 2003;278:28324–2834. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- 14.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 15.Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXHbox protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Rauhut R, Vornlocher HP, Lührmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Lührmann R, Stark H. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol. Cell. 2006;24:267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Häcker I, Sander B, Golas MM, Wolf E, Karagöz E, Kastner B, Stark H, Fabrizio P, Lührmann R. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nature Struct. Mol. Biol. 2008;15:1206–1212. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- 20.McMullan G, Faruqi AR, Henderson R, Guerrini N, Turchetta R, Jacobs A, van Hoften G. Experimental observation of the improvement in MTF from backthinning a CMOS direct electron detector. Ultramicroscopy. 2013;109:1144–1147. doi: 10.1016/j.ultramic.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheres SH. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nature Struct. Biol. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- 27.Grainger RJ, Barrass JD, Jacquier A, Rain JC, Beggs JD. Physical and genetic interactions of yeast Cwc21p, an ortholog of human SRm300/SRRM2, suggest a role at the catalytic center of the spliceosome. RNA. 2009;15:2161–273. doi: 10.1261/rna.1908309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dix I, Russell CS, O’Keefe RT, Newman AJ, Beggs JD. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1675–1686. doi: 10.1017/s1355838298412998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuter K, Nottrott S, Fabrizio P, Lührmann R, Ficner R. Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein. J. Mol. Biol. 1999;294:515–25. doi: 10.1006/jmbi.1999.3258. [DOI] [PubMed] [Google Scholar]
- 30.Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nature Struct. Mol. Biol. 2009;16:42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen THD, Li J, Galej WP, Oshikane H, Newman AJ, Nagai K. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure. 2013;21:910–919. doi: 10.1016/j.str.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science. 2013;341:80–84. doi: 10.1126/science.1237515. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Lührmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 34.Schultz A, Nottrott S, Hartmuth K, Lührmann R. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J. Biol. Chem. 2006;281:28278–2886. doi: 10.1074/jbc.M603350200. [DOI] [PubMed] [Google Scholar]
- 35.Ayadi L, Callebaut I, Saguez C, Villa T, Mornon JP, Banroques J. Functional and structural characterization of the prp3 binding domain of the yeast prp4 splicing factor. J. Mol. Biol. 1998;284:673–687. doi: 10.1006/jmbi.1998.2183. [DOI] [PubMed] [Google Scholar]
- 36.Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res. 2012;40:7046–7065. doi: 10.1093/nar/gks347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nottrott S, Urlaub H, Lührmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galisson F, Legrain P. The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res. 1993;21:1555–1562. doi: 10.1093/nar/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarov EM, Makarova OV, Achsel T, Lührmann R. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J. Mol. Biol. 2000;298:567–575. doi: 10.1006/jmbi.2000.3685. [DOI] [PubMed] [Google Scholar]
- 40.Boon KL, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, Beggs JD. Prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nature Struct. Mol. Biol. 2007;14:1077–1083. doi: 10.1038/nsmb1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mozaffari-Jovin S, Santos KF, Hsiao HH, Will CL, Urlaub H, Wahl MC, Lührmann R. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 2012;26:2422–2434. doi: 10.1101/gad.200949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn D, Kudla G, Tollervey D, Beggs JD. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 2012;26:2408–2421. doi: 10.1101/gad.199307.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Büttner K, Nehring S, Hofner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nature Struct. Mol. Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 44.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation. Cell. 2015;160:219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhn AN, Brow DA. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155:1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Brow DA. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA. 1996;2:879–894. [PMC free article] [PubMed] [Google Scholar]
- 48.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nature Struct. Mol. Biol. 2013;21:464–471. doi: 10.1038/nsmb.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharp PA. Five easy pieces. Science. 1991;254:663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
Additional References
- 51.Schreieck A, et al. RNA polymerase II termination involves C-terminal domain tyrosine dephosphorylation by CPF subunit Glc7. Nat. Struct. Mol. Biol. 2014;21:175–179. doi: 10.1038/nsmb.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 53.Elmlund H, Elmlund D, Bengio S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure. 2013;21:1299–1306. doi: 10.1016/j.str.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheres SHW, Nuñez-Ramirez R, Sorzano COS, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using Xmipp. Nature Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang G, et al. EMAN2: an extensive image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta. Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC. Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malmström L, et al. Superfamily assignments for the yeast proteome through integration of structure prediction with the gene ontology. PLoS Biol. 2007;5:e76. doi: 10.1371/journal.pbio.0050076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu XH, Chen RC, Gao Y, Wu YD. The effect of Asp-His-Ser/Thr-Trp tetrad on the thermostability of WD40-repeat proteins. Biochemistry. 2010;49:10237–10245. doi: 10.1021/bi101321y. [DOI] [PubMed] [Google Scholar]
- 63.Rother M, Milanowska K, Puton T, Jeleniewicz J, Rother K, Bujnicki JM. ModeRNA server: an online tool for modeling RNA 3D structures. Bioinformatics. 2011;27:2441–2442. doi: 10.1093/bioinformatics/btr400. [DOI] [PubMed] [Google Scholar]
- 64.Dobbyn HC, McEwan PA, Krause A, Novak-Frazer L, Bella J, O’Keefe RT. Analysis of pre-mRNA and pre-rRNA processing factor Snu13p structure and mutants. Biochem. Biophys. Res.Commun. 2007;360:857–862. doi: 10.1016/j.bbrc.2007.06.163. [DOI] [PubMed] [Google Scholar]
- 65.Leung AK, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011;473:536–539. doi: 10.1038/nature09956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chanfreau G, Elela SA, Ares M, Jr., Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou L, et al. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature. 2014;506:116–120. doi: 10.1038/nature12803. [DOI] [PubMed] [Google Scholar]
- 68.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 69.Umen JG, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nature Struct. Mol. Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 71.Dagher SF, Fu XD. Evidence for a role of Sky1p-mediated phosphorylation in 3′ splice site recognition involving both Prp8 and Prp17/Slu4. RNA. 2001;7:1284–1297. doi: 10.1017/s1355838201016077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ben-Yehuda S, et al. Extensive genetic interactions between PRP8 and PRP17/CDC40, two yeast genes involved in pre-mRNA splicing and cell cycle progression. Genetics. 2000;154:61–71. doi: 10.1093/genetics/154.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collins CA, Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuhn AN, Li Z, Brow DA. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video sequences showing the cryoEM density at two different contour levels; tri-snRNP map with all modeled components; fitting of available crystal structures into the cryoEM density: Brr2-Jab1/MPN (Prp8) complex31, Prp8 RNase H and large domains10, U4 and U5 Sm core domains65, Lsm core domain67 fitted into the multi-body map, Snu13 (ref. 64), human Prp31 (ref. 33) with remodeling, human Dib1 (ref. 29); fitting of homology models: Snu114 based translation factor EF2 (ref. 26), WD40 domain of Prp4, ferredoxin-like domain of Prp3 (ref. 36), TPR domain of Prp6; fitting of double helical RNA of the U4/U6 snRNA duplex and U5 snRNA; fitting of α-helices attributed to the N-terminal domain Prp8, Prp3 and Prp4; near complete pseudo-atomic structure of the yeast U4/U6.U5 tri-snRNP.