DEFINITION AND BACKGROUND
Horizontal gene transfer (HGT) is the movement of genetic information between organisms, a process that includes the spread of antibiotic resistance genes among bacteria (except for those from parent to offspring), fueling pathogen evolution.
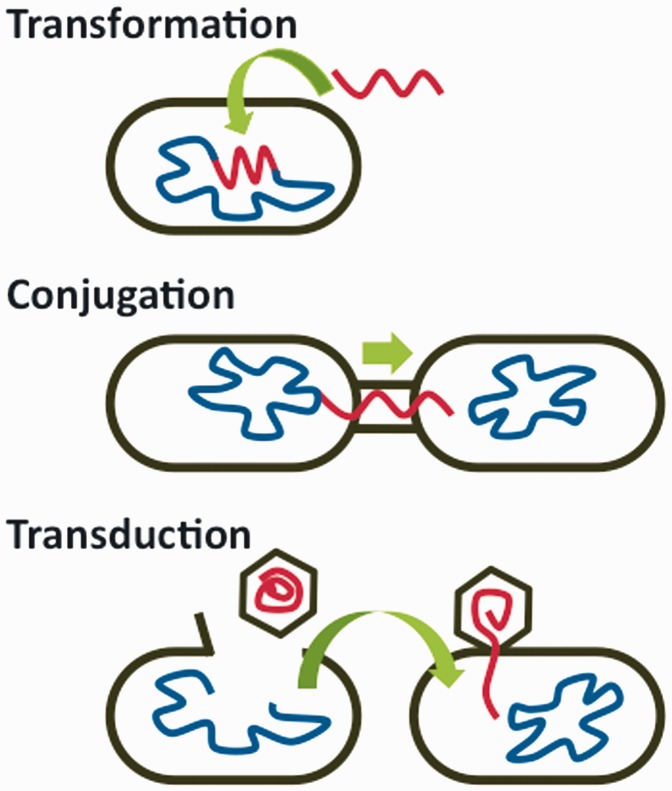
Many resistance genes evolved long ago in natural environments with no anthropogenic influence but these genes are now rapidly spreading to and among human pathogens. HGT occurs by three well-understood genetic mechanisms (Fig. 1):
Figure 1.
Mechanisms of bacterial horizontal gene transfer
Transformation: Bacteria take up DNA from their environment
Conjugation: Bacteria directly transfer genes to another cell
Transduction: Bacteriophages (bacterial viruses) move genes from one cell to another
Once transferred, the genes and pathogens continue to evolve, often resulting in bacteria with greater resistance [1, 2, 3, 4]. All genes—not just those causing drug resistance—may be horizontally transferred and proliferate by natural selection, including virulence determinants [5].
EXAMPLES IN HUMAN BIOLOGY AND PUBLIC HEALTH
Antibiotic use in human medicine and agriculture continually selects for resistant bacteria [2, 6]. For example, tetracycline and β-lactams commonly fed to animals provide a selective environment for tetracycline and methicillin resistance. Genes conferring resistance to these antibiotics have horizontally transferred into a sensitive human-associated Staphylococcus aureus strain, resulting in methicillin-resistant strain CC398 [7]. After a strain gains resistance by HGT, the bacteria proliferate and continue to evolve as they move among patients and hospitals [1]. This process occurs in many bacterial lineages, resulting in diverse populations of a variety of strains, such as USA300 [5].
EXAMPLES IN CLINICAL MEDICINE
Ongoing HGT poses a problem for clinical surveillance and treatment. Bacterial populations evolve rapidly, resulting in diversity that necessitates individual screening to determine effective treatments and to detect new strains, such as methicillin and high level vancomycin resistant S.aureus (MRSA and VRSA) [2]. Even when new drugs and diagnostic tools become available, the persistence of HGT will require ongoing surveillance for newly resistant pathogens, leaving practitioners and researchers racing with evolution.
FUNDING
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1424871.
REFERENCES
- 1. Harris SR, Feil EJ, Holden MTG, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 2010;327:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindsay J. A. Hospital-associated MRSA and antibiotic resistance-what have we learned from genomics? Int J Med Microbiol 2013;303:318–23. [DOI] [PubMed] [Google Scholar]
- 3. McCarthy AJ, Loeffler A, Witney AA, et al. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol 2014;6:2697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stanczak-Mrozek KI, Manne A, Knight GM, et al. Within-host diversity of MRSA antimicrobial resistances. J Antimicrob Chemother 2015;70:2191–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McDougal LK, Steward CD, Killgore GE, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 2003;41:5113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. US Department of Health and Human Services. Antibiotic Resistance Threats in the United States. Atlanta: CDC, 2013; p, 11–14. [Google Scholar]
- 7. Price LB, Stegger M, Hasman H, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 2012;3 [DOI] [PMC free article] [PubMed] [Google Scholar]