Key Points
Our study delineates a signaling pathway and molecular mechanism that band-stage neutrophils acquire to gain monocytic characteristics.
Band-stage neutrophils from G-CSF–mobilized blood differentiate into monocytic cells upon recruitment to inflammatory sites.
Abstract
During inflammation, neutrophils are rapidly mobilized from the bone marrow storage pool into peripheral blood (PB) to enter lesional sites, where most rapidly undergo apoptosis. Monocytes constitute a second wave of inflammatory immigrates, giving rise to long-lived macrophages and dendritic cell subsets. According to descriptive immunophenotypic and cell culture studies, neutrophils may directly “transdifferentiate” into monocytes/macrophages. We provide mechanistic data in human and murine models supporting the existence of this cellular pathway. First, the inflammatory signal–induced MKK6-p38MAPK cascade activates a monocyte differentiation program in human granulocyte colony-stimulating factor–dependent neutrophils. Second, adoptively transferred neutrophils isolated from G-CSF–pretreated mice rapidly acquired monocyte characteristics in response to inflammatory signals in vivo. Consistently, inflammatory signals led to the recruitment of osteoclast progenitor cell potential from ex vivo–isolated G-CSF–mobilized human blood neutrophils. Monocytic cell differentiation potential was retained in left-shifted band-stage neutrophils but lost in neutrophils from steady-state PB. MKK6-p38MAPK signaling in HL60 model cells led to diminishment of the transcription factor C/EBPα, which enabled the induction of a monocytic cell differentiation program. Gene profiling confirmed lineage conversion from band-stage neutrophils to monocytic cells. Therefore, inflammatory signals relayed by the MKK6-p38MAPK cascade induce monocytic cell differentiation from band-stage neutrophils.
Introduction
Gain- and loss-of-function studies of particular transcription factors showed that leukocyte lineage identity can be plastic (eg, B cells can be converted into macrophages).1 In addition, leukocytes may lose lineage identity in response to specific microenvironmental signals as shown for CD4+ helper T-cell subpopulations2 and myelomonocytic cells.3 For example, macrophages may develop into M1 or M2 phenotypes,4 or into myeloid-derived suppressor cells (MDSCs), depending on microenvironmental signals.5 In addition, in vitro studies demonstrated that murine6,7 or human8,9 differentiated postmitotic neutrophils can acquire a monocytic/macrophage/dendritic cell (DC) phenotype. This latter finding was surprising, because granulocyte/monocyte lineage separation was believed to occur at the clonogenic progenitor cell stage, and monocyte committed progenitor cells have recently been isolated.10,11 Moreover, certain transcription factors (eg, C/EBPα, Gfi-1, Egr-1, Klf4, Pu.1) are differentially involved in granulocyte vs monocyte development from shared progenitors.12,13 It is interesting to speculate that “transdifferentiation” of neutrophils into monocytic cells only occurs in response to inflammatory signals. Left-shifted granulopoiesis is defined as the appearance in the peripheral blood (PB) of band-stage neutrophils. Upon egressing from blood vessels, bone marrow–derived band-stage neutrophils can enter disease-specific microenvironments.
Activation states and expression levels of lineage-determining transcription factors are tightly controlled by upstream kinase pathways whose activity in turn is dependent on extracellular cues. p38MAPK is strongly activated by environmental stress and inflammatory cytokines but not by growth factors.14 Four different isoforms (α, β, γ, and δ) of p38 have been described with overlapping substrate specificity and are activated by upstream mitogen-activated protein kinase kinases 3 and 6 (MKK3 and MKK6). MKK6 was shown to phosphorylate all isoforms of p38, whereas MKK3 was shown to be more restricted (activation of p38α, p38γ, p38δ).15 Notably, phosphorylated MKK6 was identified as a strong immunohistology marker for lesional monocytic cells and synoviocytes in rheumatoid arthritis.16 p38 signaling has also been implicated in neutropenia17 and is activated by toll-like receptor (TLR) signaling in hematopoietic progenitor cells concomitant with TLR-induced monocyte/DC differentiation.18
The present study was prompted by our initial observations that conditional MKK6-p38MAPK activation in granulocyte colony-stimulating factor (G-CSF)–dependent neutrophils leads to the activation of a monocyte differentiation program. Because thus far the existence of this cellular pathway was supported only by in vitro cell culture data, we considered mechanistic experiments to be of substantial relevance. Here, data from mouse and human support a model of monocytic cell differentiation from G-CSF–mobilized band-stage neutrophils under inflammatory conditions via MKK6 activation.
Materials and methods
Cell lines and primary cell culture
Control vector- or dominant active (d.a.)MKK6-expressing HL60 cells were generated by cotransducing TA-mCD8α either together with HR-NGFR or HR-d.a.MKK6-NGFR followed by single-cell cloning for obtaining optimal DOX-inducible clones. Cell lines were maintained in RPMI medium, 10% fetal calf serum, penicillin/streptomycin (P/S; 125 U/mL each), and l-glutamine (complete RPMI). Cord-blood samples were collected during healthy full-term deliveries with both approval from the Medical University of Vienna institutional review board and informed consent according to the Declaration of Helsinki. CD34+ cells, neutrophils, and monocytes were isolated as previously described.19 Neutrophils and monocytes from CD34+ cells were generated as previously described.19 To generate neutrophils, CD34+ cells (2-5 × 104/mL per well) were plated in X-VIVO15 medium supplemented with G-CSF (100 ng/mL) and stem cell factor (20 ng/mL) for 12 to 14 days. The medium was partially exchanged for fresh medium containing cytokines every 3 days. A detailed list of cytokines is found in the supplemental data, available on the Blood Web site. Osteoclasts derived from neutrophil-derived monocytic cells were generated with macrophage colony-stimulating factor (M-CSF) (25 ng/mL) and receptor activator of nuclear factor κB ligand (RANKL) (100 ng/mL) as described.20 Cytokines and reagents are listed in the supplemental methods. Neutrophils of G-CSF–mobilized (10 µg/kg body weight Neupogen [Amgen Europe] on 4 consecutive days) were isolated by density-gradient centrifugation using Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) according to the manufacturer’s protocol.
Retroviral vectors
Gene transduction and the tetracycline-inducible gene expression system was previously described.21 This enables inducible expression of d.a.MKK6 followed by either IRES-GFP (HR-GFP) or IRES-Nerve growth factor receptor (HR-NGFR). cDNA encoding dominant-negative c-Jun (d.n. c-Jun; S63A, S73A, T91A, and T93A; kindly provided by G. Chen, Medical College of Wisconsin)22 was subcloned into the HR vectors. To induce gene expression, 1 to 2 µg/mL DOX was added.
Flow cytometry
Flow cytometry analysis was performed as previously described.19 For a detailed list of antibodies, see the supplemental methods. Sorting and fluorescence-activated cell sorting (FACS) analyses were carried out on BD FACSAria and LSRII cytometers. Data were analyzed with FlowJo software (BD Biosciences).
Reverse-transcriptase polymerase chain reaction (RT-PCR) and microarray analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. For real-time RT-PCR analysis, the SYBR Green detection system was used (Invitrogen). Microarray analysis was performed as previously described.23 The whole-gene datasets have been deposited in the GEO database (accession no. GSE58920). A detailed protocol and list of primers are described in supplemental Methods.
Western blot
Total cell extracts were prepared as described.21 Proteins from equal numbers of cells were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto polyvinylene difluoride membrane (Immobilon-P; Millipore, Billerica, MA). Protein detection was performed with chemiluminescence (SuperSignal WestPico; Pierce Biotechnology). The detailed protocols and a complete list of antibodies used are available in supplemental Methods.
In vivo transdifferentiation
Neutrophils from G-CSF–mobilized lysM-EGFP mice (kindly provided by T. Graf) were obtained from PB. Peritonitis was induced by instillation of 4% ThG into wild-type mice as described.24 The Animal Care and Use Committee of the Medical University of Vienna approved all experiments. Typically, 2 to 4 × 106 GFP+Ly6G+F4/80– neutrophils were injected intraperitoneally 4 hours postinduction of peritonitis. Peritoneal leukocytes were collected from the peritoneal cavity and analyzed by FACS. The detailed protocol of neutrophil isolation and a complete list of antibodies used are available in supplemental Methods.
Statistics
Statistical analysis was performed using the paired and unpaired, 2-tailed Student t test.
Results
d.a.MKK6 expression in neutrophils induces a phenotypic shift to monocytes
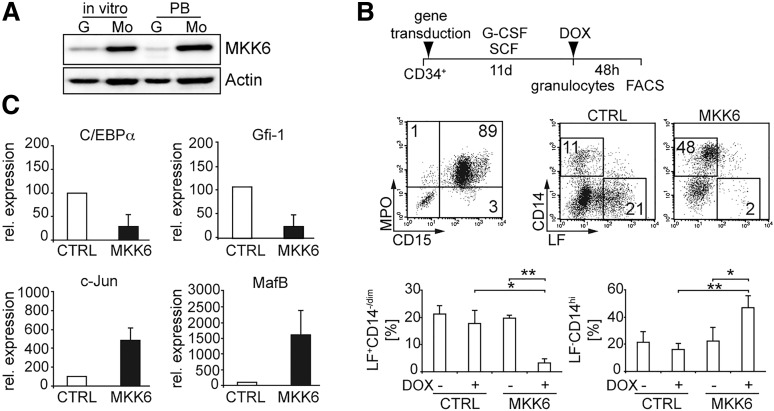
We compared endogenous MKK6 levels in neutrophils with monocytes either generated in vitro or isolated from PB. MKK6 levels were substantially higher in monocytes than in neutrophils (Figure 1A). We generated G-CSF–dependent neutrophils or M-CSF–dependent monocytes from CD34+ cord blood progenitors and conditionally expressed d.a.MKK625 in differentiated cells by using a tetracycline-inducible retroviral system.21 The vast majority of G-CSF–dependent day 11–generated cells represented CD15+MPO+ neutrophils with a subset expressing the neutrophil-associated lysosomal marker molecule lactoferrin (LF26; Figure 1B19). Although CD15 and MPO are induced early during in vitro granulopoiesis of CD34+ cells, a subset of MPOhi cells subsequently begins to express the secondary (specific) neutrophil granule marker LF, after in vitro generated cells had acquired a myelocyte/metamyelocyte phenotype.27,28 In line with their granulopoietic phenotype, these cells exhibited robust oxidative burst activity (supplemental Figure 1A). We analyzed transduced d.a.MKK6-IRES-GFP or empty control vector cells for hallmark phenotypic characteristics of neutrophils vs monocytes (ie, LF vs the monocyte marker CD14). d.a.MKK6 expression in G-CSF–dependent neutrophils resulted within 48 hours in the generation of LF–CD14hi monocytelike cells at the expense of LF+CD14–/dim and double-negative cells (Figure 1B). Conversely, d.a.MKK6 expression in M-CSF–dependent monocytes did not alter percentages of CD14+ cells significantly (supplemental Figure 1B). Thus, MKK6 signaling in G-CSF–dependent human primary neutrophils induces their differentiation toward a CD14hi monocytic phenotype. In line with this, mRNA expression levels of neutrophil-affiliated transcription factors C/EBPα and Gfi-1 were down-regulated, whereas monocyte-associated transcription factors c-Jun and MafB were upregulated in d.a.MKK6 expressing primary neutrophils (Figure 1C). These reciprocal changes of transcription factors were similarly detected at the protein levels (supplemental Figure 1C).
Figure 1.
d.a.MKK6 expression in neutrophils induces a phenotypic shift to monocytes. (A) MKK6 expression, assessed by western blotting, of neutrophils (G) and monocytes (Mo) either generated in vitro from CD34+ cells or isolated from PB. Proteins from equal numbers of cells were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), blotted, and probed with anti-MKK6 antibody. (B) Schematic representation of culture conditions; d.a.MKK6-transduced day 11 neutrophils were analyzed for CD15 vs MPO; 48 hours later NGFR+ cells (d.a.MKK6 expressing) were analyzed for neutrophil (LF) vs monocytic (CD14) marker. Bars represent mean percentages (± standard deviation [SD]) of 3 independent experiments (*P < .05, **P < .01). (C) Expression of neutrophil (C/EBPα, Gfi-1) and monocyte (c-Jun, MafB)-associated transcription factors in FACS-sorted d.a.MKK6-induced or empty-vector control cells in vitro–generated neutrophils was assessed after 24 hours by real-time RT-PCR (means ± SD of 3 independent experiments).
Proteasomal degradation of C/EBPα and subsequent de-repression of c-Jun is critical for monocytic cell differentiation of HL60 cells
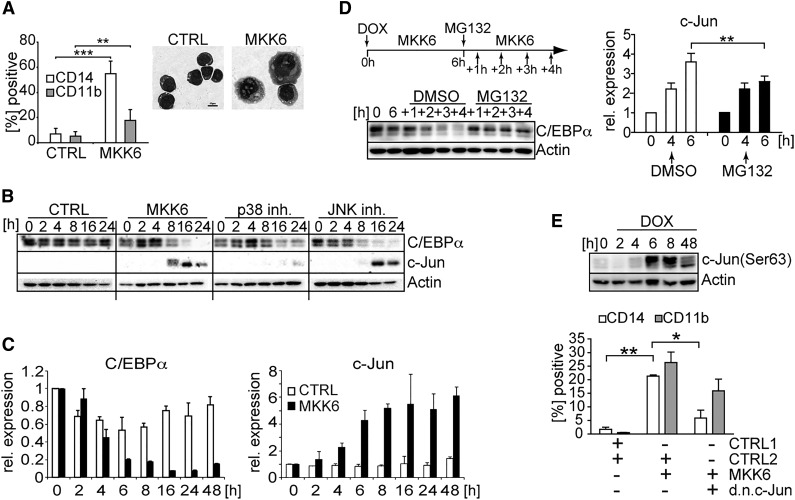
Similarly as observed for primary cells (Figure 1), d.a.MKK6 expression in HL60 cells resulted in the induction of CD14 and CD11b expression and in the acquisition of well-described monocyte/macrophage-associated morphologic changes such as increased cell size, ruffled cell shape, and basophilic cytoplasm (Figure 2A). Moreover, time-course analyses revealed reciprocal regulation of c-Jun vs C/EBPα proteins within 8 hours in d.a.MKK6-induced HL60 cells (Figure 2B). Therefore, HL60 cells mimicked primary cells in phenotypic and transcription factor changes. These developmentally arrested homogeneous myeloid model cells offer the advantage to study initial molecular events in d.a.MKK6-mediated monocytic cell differentiation. c-Jun vs C/EBPα transcription factor expression changes were found to be p38-dependent but JNK-independent (ie, impaired by p38 inhibitor but unaffected by JNK inhibitor; Figure 2B). Short-term (6 hours) d.a.MKK6 expression was sufficient to induce an approximately fivefold reduction in basal C/EBPα levels and a reciprocal fourfold increase in c-Jun mRNA levels (Figure 2C). Because CEBP family members were shown to be regulated via ubiquitin-proteasome–dependent degradation,29 we tested whether MKK6 activation leads to the downregulation of C/EBPα levels via proteasomal degradation. Blocking of the proteasomal pathway by proteasomal inhibitor MG132 indeed led to the stabilization of C/EBPα levels upon MKK6 signaling, whereas under control conditions, C/EBPα was degraded as expected (Figure 2B-D). C/EBPα negatively regulates c-Jun promoter activity by blocking of c-Jun binding to its own promoter.30 Thus, we examined whether stabilization of C/EBPα levels inhibits c-Jun upregulation in response to MKK6. Four hours induction of d.a.MKK6 followed by proteasomal inhibition stabilized C/EBPα levels and significantly interfered with MKK6-dependent upregulation of c-Jun mRNA (Figure 2D, bar diagram). Under control conditions c-Jun upregulation in response to MKK6 occurred at the expected rate (Figure 2D-C). Egr-1/Egr-2 positively regulates monocyte gene expression and counteracts Gfi-1, a granulocyte-associated gene activated by C/EBPα.13 Inhibition of proteasomal C/EBPα degradation correlated with the inhibition of Egr-1 induction; conversely, the downregulation of Gfi-1 was unaffected (supplemental Figure 1D). Because d.a.MKK6-p38 activation led to the rapid induction of c-Jun and triggered c-Jun phosphorylation (Figure 2E), we used a dominant-negative (d.n.)c-Jun22 to analyze whether phosphorylation is functionally required for monocyte differentiation. Hence, HL60 cells were cotransduced with DOX-inducible d.a.MKK6 and d.n.c-Jun. d.a.MKK6-induced upregulation of CD14 and CD11b was impaired in d.n.c-Jun transduced cells relative to control cells (Figure 2E). Moreover, the expression of d.n.c-Jun failed to restore C/EBPα levels upon MKK6-p38 signaling (supplemental Figure 1E). As expected, d.a.MKK6-activation–induced monocyte differentiation was associated with inhibition of cell proliferation (supplemental Figure 1F). In conclusion, MKK6-induced monocyte differentiation of HL60 model cells depends on C/EBPα proteasomal degradation, resulting in the rapid induction of c-Jun; moreover it is critically dependent on c-Jun phosphorylation.
Figure 2.
d.a.MKK6 expression leads to the modulation of expression levels of lineage-associated transcription factors. (A) HL60 cells transduced with d.a.MKK6 (MKK6) or control (CTRL) were induced with DOX (2 µg/mL). Cell morphology and monocytic marker (CD11b and CD14) expression of HL60 cells after 48 hours of d.a.MKK6 induction are shown (bar = 10 µm). Mean values ± SD were calculated from 3 independent experiments (**P < .01, ***P < .001). (B) d.a.MKK6 (MKK6) or control (CTRL) expressing HL60 cells were induced with DOX (2 µg/mL), and whole-cell extracts from an equal number of cells were analyzed at indicated time points. Blocking of the p38 signaling pathway by 10 µM SB203580 (p38 inh.) stabilized C/EBPα and inhibited c-Jun expression. JNK signaling pathway inhibition by 10 µM SP600125 (JNK inh.) had no effect. One representative blot of 3 experiments is shown. (C) Real-time RT-PCR assessment of C/EBPα and c-Jun expression in response to d.a.MKK6 expression. (D) Blocking proteasomal degradation by MG132 (10 µM) stabilizes C/EBPα. D.a.MKK6 was induced by DOX (2 µg/mL); after 6 hours, a proteasomal block was induced by adding MG132, and subsequently stabilization of C/EBPα levels was monitored for up to 4 hours (+1 to +4 hours). Stabilization of C/EBPα by MG132 interferes with c-Jun mRNA induction in d.a.MKK6-expressing HL60 cells (**P < .01). (E) c-Jun phosphorylation, assessed by western blotting, in response to d.a.MKK6 expression. Simultaneous d.a.MKK6 and d.n.c-Jun expression (defined as NGFR+GFP+) impairs the upregulation of CD14 and CD11b. Bars represent mean values ± SD of 3 independent experiments (*P < .05, **P < .005).
Proinflammatory cytokines induce monocytic cell characteristics of band-stage neutrophils from G-CSF–mobilized blood
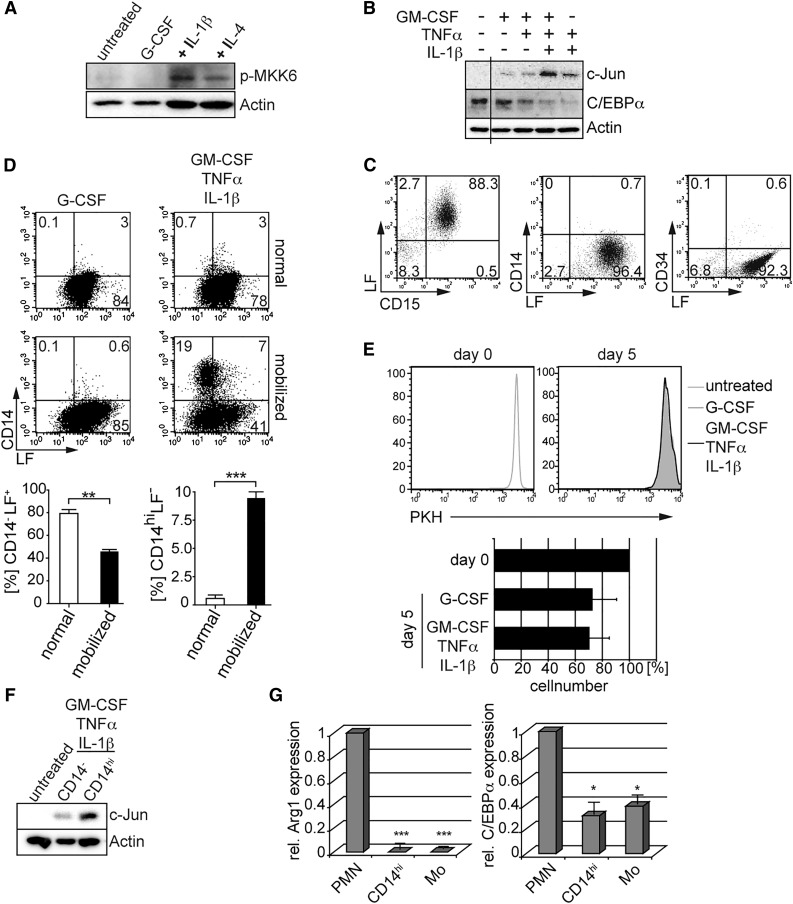
GM-CSF, tumor necrosis factor-α (TNFα), and IL-1β belong to well-documented proinflammatory mediators involved in the pathogenesis of many inflammatory disorders and were shown to trigger MKK6-p38 signaling in myeloid cells.31 Expectedly, GM-CSF/TNFα/IL-1β triggered MKK6 phosphorylation in primary G-CSF–induced neutrophils (Figure 3A). Furthermore, this cytokine combination, but not individual cytokines, potently triggered C/EBPα degradation and c-Jun induction (Figure 3B), associated with the repression of LF+ neutrophils in favor of CD14hiLF– cells (supplemental Figure 2A). These lineage-affiliated transcription factor changes are in line with data obtained by experimental induction of d.a.MKK6 in primary human neutrophils and HL60 cells (Figures 1 and 2).
Figure 3.
Proinflammatory cytokines induce neutrophil-to-monocyte reprogramming. (A) Primary G-CSF–induced neutrophilic granulocytes (untreated) were stimulated with G-CSF (100 ng/mL, lane 2) or proinflammatory cytokines (GM-CSF 10 ng/mL, TNFα 25 ng/mL) plus IL-1β (10 ng/mL; +IL-1β, lane 3) or IL-4 (25 ng/mL; +IL-4, lane 4). After 72 hours, whole-cell lysates were prepared for western blot analysis. (B) Neutrophils generated as in Figure 1 were stimulated with GM-CSF (10 ng/mL), TNFα (25 ng/mL), and IL-1β (10 ng/mL) as indicated, and whole-cell extracts were analyzed for c-Jun and C/EBPα protein levels. (C) Neutrophils isolated from G-CSF–mobilized blood display band-stage neutrophil morphology and are devoid of detectable CD14+ and CD34+ cells. Freshly isolated neutrophils from G-CSF–mobilized donors were analyzed by FACS for neutrophil markers (CD15 and LF), the hematopoietic stem/progenitor cell marker CD34, and the monocyte marker CD14. Quadrants were set according to isotype control stainings. (D) Neutrophils from G-CSF–mobilized, but not normal, donors show a monocytic phenotype in response to proinflammatory cytokines. After 5 days of stimulation with proinflammatory cytokines (GM-CSF 10 ng/mL, TNFα 25 ng/mL, IL-1β 10 ng/mL) or G-CSF (100 ng/mL), CD15+ cells were analyzed for CD14 vs LF expression by FACS. Bars represent mean ± SD (percentages of phenotypically defined cells; n = 4 normal and n = 4 G-CSF mobilized donors; ***P < 0.001, **P < 0.005). (E) Assessment of cell proliferation of neutrophils in culture. Neutrophils from G-CSF–mobilized donors were labeled with PKH26 (day 0) and further cultivated in the presence of G-CSF (100 ng/mL) or proinflammatory cytokines (GM-CSF 10 ng/mL, TNFα 25 ng/mL, IL-1β 10 ng/mL) for 5 days. Data are representative of 3 independent experiments. Bars represent mean percent cell recovery at day 5 after culture of neutrophils in the presence of the indicated cytokines (n = 4). (F) Day 5–stimulated CD14hi cells were separated by magnetic-activated cell sorting (MACS) and analyzed for c-Jun protein expression. (G) Neutrophils from G-CSF–mobilized blood (PMN), MACS-sorted day 5–stimulated CD14hi cells (CD14hi), and MACS-sorted monocytes from normal blood (Mo) were analyzed for arginase-1 and C/EBPα expression by RT-PCR. Mean values ± SD were calculated from 3 independent experiments and donors (*P < .05, ***P < .0001).
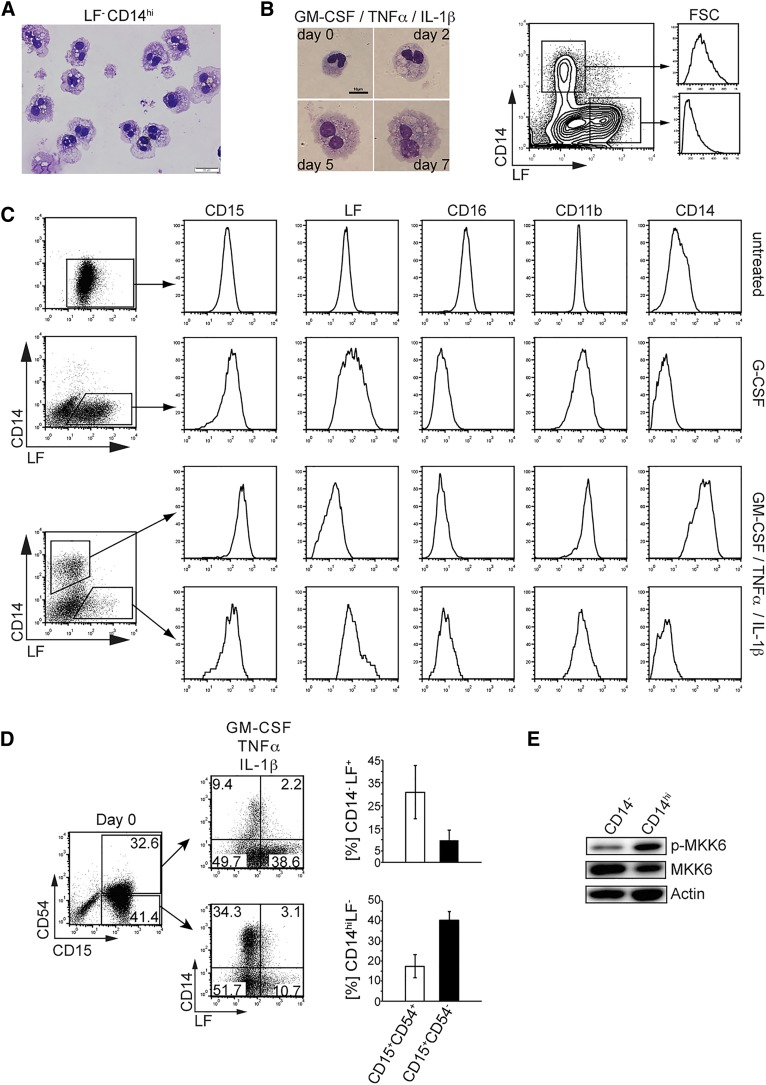
Next, we analyzed whether in vivo occurring neutrophils are similarly capable of acquiring monocytic characteristics in response to GM-CSF/TNFα/IL-1β stimulation. Administration of G-CSF to healthy donors mimics left-shifted granulopoiesis. Isolated neutrophils from G-CSF–mobilized blood mostly show a band-stage neutrophil phenotype and uniformly expressed CD15 and LF (Figure 3C and supplemental Figure 3). A significant portion of neutrophils from mobilized blood but not from normal blood acquired a CD14hiLFlo/– monocyte phenotype within 5 days upon stimulation with GM-CSF/TNFα/IL-1β (Figure 3D). CD14hi cell differentiation from neutrophils occurred without cell division (Figure 3E), with approximately 70% cell recovery rates at day 5 (Figure 3E). Moreover, CD14hi cells generated from neutrophils showed strong c-Jun expression, whereas expression was low in CD14– cell fractions (Figure 3F). In comparison, freshly isolated LF+ neutrophils lacked detectable c-Jun expression (Figure 3F, untreated). C/EBPα expression was also down-regulated in CD14hi cells when compared with freshly isolated neutrophils and was comparable with expression levels of freshly isolated monocytes (Figure 3G). In human, arginase-1 is constitutively expressed by neutrophils but not by monocytes.32 CD14hi cells lacked detectable arginase-1, similarly as observed for freshly isolated monocytes (Figure 3G). LF–CD14hi cells exhibited ruffled cell shapes, with a considerable portion of these cells showing a bilobed nucleus (Figure 4A). In addition, cell size/forward scatter characteristics progressively increased over time for CD14hi cells (Figure 4B), and CD14hi cells showed higher expression levels of CD15 and CD11b and much lower/undetectable levels of CD16 and LF than did freshly isolated neutrophils (Figure 4C). Moreover, day 5–generated CD14hi cells possessed phagocytic activity similar to freshly isolated PB monocytes and neutrophils (supplemental Figure 4A).
Figure 4.
Analysis of CD14hi cells generated from band-stage neutrophils. (A) Neutrophils from G-CSF–mobilized donors were stimulated with proinflammatory cytokines (GM-CSF 10 ng/mL, TNFα 25 ng/mL, IL-1β 10 ng/mL). At day 5, CD14hi cells were FACS-sorted and cell morphology was assessed (bar = 20 µm). Data are representative of 4 independent experiments. (B) Cell morphology and cell size were analyzed by May-Grünwald-Giemsa staining (days 2-7; bar = 10 µm) or FACS (day 5; CD14+ or LF+ cell subsets were gated and analyzed for forward scatter intensity). (C) Day 5–stimulated neutrophils were gated as indicated and analyzed for the expression of informative marker molecules. Data are representative of 3 experiments. (D) CD15+CD54– neutrophils exhibit increased CD14hi cell differentiation potential. Freshly isolated neutrophils from G-CSF–mobilized donors were FACS-sorted for CD15 and CD54. CD15+CD54+ and CD15+CD54– neutrophils were stimulated with the cytokines GM-CSF (10 ng/mL), TNFα (25 ng/mL), and IL-1β (10 ng/mL) for 5 days and analyzed by FACS for CD14 and LF. Bars represent mean ± SD (percentages of phenotypically defined cells; n = 3). (E) Neutrophils from G-CSF–mobilized donors were stimulated as in (A) for 5 days. Phospho-MKK6 and MKK6 expression levels were assessed by western blot comparing proteins from equal number of cells. FACS-sorted CD14hi vs CD14– cells were separately analyzed.
CD54 is expressed by CFU-GM and is gradually down-regulated during granulopoiesis, with band-stage and segmented neutrophils exhibiting low/undetectable CD54; conversely, monocytes are CD54+.33 Neutrophils from G-CSF–mobilized blood can be FACS-sorted into CD15+CD54+ and CD15+CD54– fractions, the latter being enriched for CD14hi cell differentiation potential (Figure 4D and supplemental Figure 4B). Expectedly, phospho-MKK6 can be detected at higher levels in sorted CD14+ monocytelike cells compared with CD14– cells from the same cultures (day 5; Figure 4E).
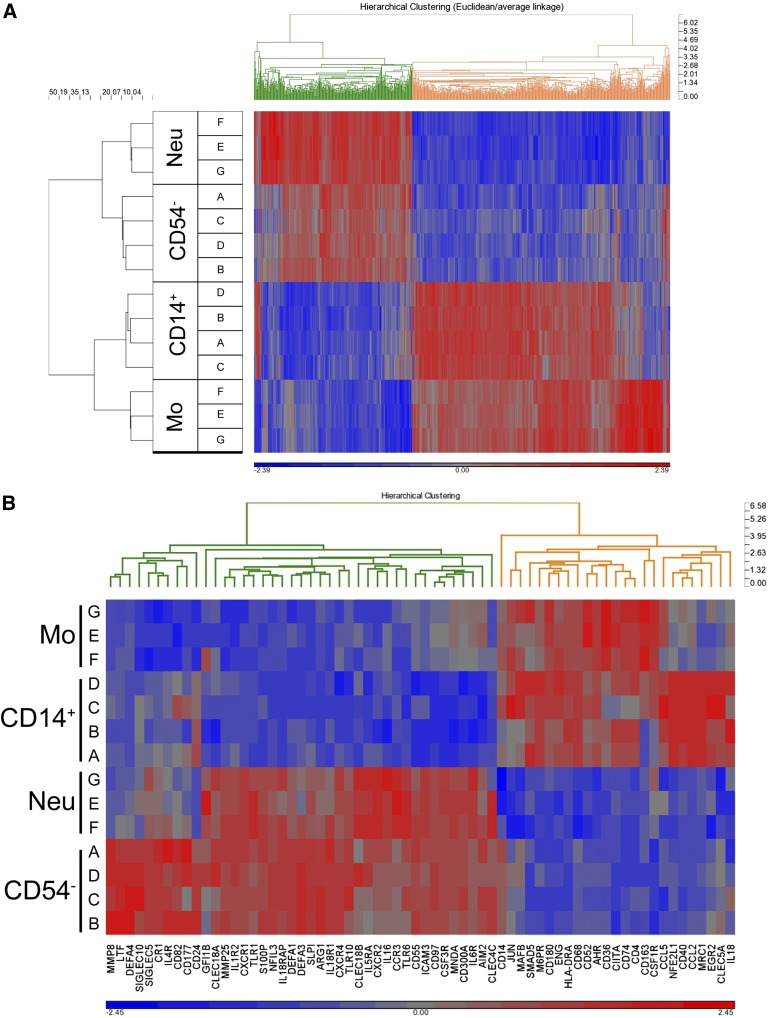
In subsequent experiments, we analyzed the lineage relationship of in vitro–generated CD14hi monocytic cells to blood monocytes vs neutrophils using cDNA microarray gene profiling. CD14hi cells generated from G-CSF–mobilized CD15+CD54– neutrophils exhibited a close resemblance to blood monocytes and are clearly separated from blood neutrophils as well as from their precursors (ie, CD15+CD54– cells) (Figure 5A). For example, well-known granulocytic markers CSF3R (G-CSF receptor), DEFA1, DEFA3, ARG1, and GFI-1 were down-regulated in CD14hi progeny compared with neutrophils. Inversely, monocyte-associated genes CSF1R (M-CSF receptor), CD68, JUN, and MAFB were upregulated in CD14hi cells with similar expression level as observed in PB monocytes (Figure 5B).
Figure 5.
Transcriptional profiling of neutrophil-derived CD14+ monocytic cells. FACS-sorted CD15+CD54– neutrophils from G-CSF–mobilized blood were stimulated with GM-CSF (10 ng/mL)/TNFα (25 ng/mL)/IL-1β (10 ng/mL) for 5 days; subsequently, CD14+ cells were FACS-sorted (donors A-D). In addition, normal blood monocytes and neutrophils (donors G-F) were FACS-sorted from PB. RNA from CD15+CD54– neutrophils (day 0; starting population; CD54–), CD14+ monocytic cells (day 5; CD14+), monocytes (day 0; Mo), and neutrophils (day 0; Neu) were isolated and processed for hybridization on GeneChip Human 2.0 ST arrays (Affymetrix, Santa Clara, CA). (A) Hierarchical clustering (euclidean dissimilarity/average linkage; standard normalization) of differential expressed genes (P < .005) is shown. (B) Heat map and hierarchical clustering of 75 granulocyte- and monocyte-associated genes showing gene expression levels from low (blue) to high (red).
G-CSF–mobilized human blood neutrophils give rise to osteoclasts
Because osteoclasts were previously shown to arise from CD14hi monocytes/macrophages,34 we analyzed whether neutrophil-derived monocytic cells possess osteoclast potential. Hence, purified neutrophils were induced to acquire monocyte characteristics in response to 5-day stimulation with GM-CSF/TNFα/IL-1β (Figure 3D), followed by subculture in osteoclast differentiation–promoting media (see scheme, Figure 6). At day 21, TRAP+ giant multinucleated cells were visible in these cultures (Figure 6, arrowheads). Moreover, ultrastructural analysis of bovine bone discs after 21 days of culture revealed numerous pits formed on the surface of the bone (Figure 6). M-CSF/RANKL stimulation of freshly isolated or in vitro G-CSF pretreated neutrophils (day 5) resulted in massive cell death with no detectable cells after 21 days. Therefore, osteoclast differentiation strictly required prestimulation of neutrophils with GM-CSF/TNFα/IL-1β. Thus, osteoclast differentiation potential of neutrophils correlates positively with the induction of CD14hi monocytic cells, suggesting that they arise from monocytic intermediates.
Figure 6.
Neutrophils from G-CSF–mobilized blood can be induced to acquire osteoclast phenotype. Neutrophils from G-CSF–mobilized donors were stimulated with GM-CSF (10 ng/mL)/TNFα (25 ng/mL)/IL-1β (10 ng/mL) for 5 days, followed by a culture period in osteoclast-promoting conditions (M-CSF, 25 ng/mL; RANKL, 100 ng/mL). TRAP (lower left; original magnification ×10) and immunofluorescence (upper right; nuclei, blue; actin, red; original magnification ×40) at day 21 of osteoclast culture identify cells with osteoclast phenotype (arrowheads). Lower right: Secondary cultures were initiated on bovine bone discs, and lacunae formation was assessed by scanning electron microscopy (bar = 100 µm). The insert shows a single osteoclast (bar = 10 µm) with several lacunae in its surrounding area. Data are representative of 3 experiments.
Neutrophils from G-CSF–pretreated mice acquire a monocytic phenotype in vivo
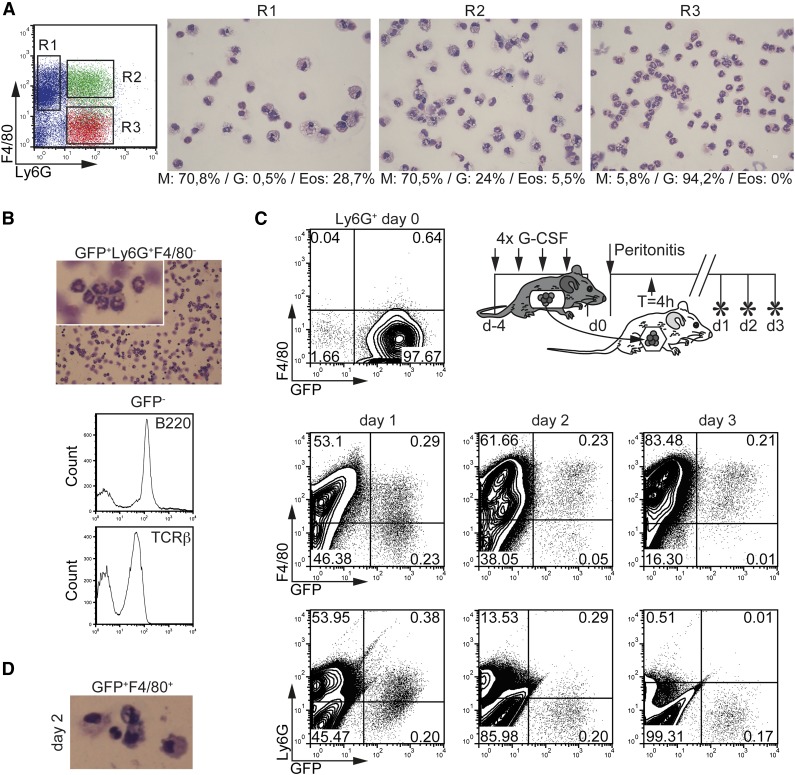
Cell transfer experiments are required to rigorously test whether neutrophils can acquire monocytic cell characteristics in vivo. Thioglycollate (ThG) peritonitis induces a rapid influx of neutrophils to the peritoneal cavity within a short time. ThG-induced peritoneal neutrophils can be driven in vitro to develop into F4/80+ monocytes/macrophages in response to M-CSF.6 Whether these cells acquire monocytic cell characteristics in vivo was not analyzed. F4/80 is a specific monocyte/macrophage marker, whereas Ly6G specifically identifies neutrophils.6 Expectedly, we observed high numbers of neutrophils (F4/80–Ly6G+) and moderate numbers of monocytes/macrophages (F4/80+L6G–) at day 1 after the administration of ThG. Frequencies of monocytes/macrophages increased at days 2 and 3 as expected.35 In addition, F4/80+Ly6G+ cells appeared at day 1 and persisted to later time points. Cell sorting revealed that F4/80+Ly6G+ cells were mostly comprised of monocytes/macrophages with a low percentage of neutrophils and very few eosinophils (Figure 7A, gate R2). We speculated that they may arise from Ly6G+ neutrophils and thus we performed cell transfer experiments. GFP+Ly6G+F4/80– neutrophils were isolated from G-CSF–mobilized lys-EGFP mice.36 Detailed characterization of GFP+Ly6G+F4/80– neutrophils confirmed their neutrophilic phenotype (ie, characteristic neutrophilic morphology; F4/80–) (Figure 7B and supplemental Figure 6). The small portion of GFP– cells among the neutrophil fraction mainly represented lymphocytes (B220+ or T-cell receptor-β+; Figure 7B); importantly, no F4/80+ monocytic cells were detectable. GFP+Ly6G+F4/80– neutrophils were transferred to wild-type mice 4 hours postinduction of ThG-peritonitis by intraperitoneal injection (Figure 7C). Twenty-four hours later, these cells expressed the monocytic marker F4/80, thus phenotypically resembling the Ly6G+F4/80+ population in Figure 7A. Inversely, the initially high expression levels of Ly6G were reduced on GFP+ cells at this time point. Time kinetics analyses revealed a successive upregulation of F4/80 and inverse downregulation of Ly6G expression levels by GFP+ neutrophils within 72 hours after ThG administration (Figure 7C). A monocytic phenotype of FACS-sorted GFP+F4/80+ cells in peritonitis was also confirmed by microscopic examination (Figure 7D). Taken together, GFP+Ly6G+F4/80– neutrophils acquire a monocytic phenotype in vivo in ThG-induced peritonitis. Side-by-side staining revealed similar populations of F4/80+CD11b+ cells among gated Ly6G+ cells in ThG peritonitis, a mouse model of bacterial infection, and in the K/BxN mouse model of rheumatoid arthritis (supplemental Figure 5).
Figure 7.
Emergence of neutrophils-derived monocytic cells in vivo. (A) Morphology of peritoneal lavage cells at day 1 after peritonitis induction. Leukocyte subpopulations from peritoneal lavage fluid (PLF) were FACS-sorted for Ly6G–F4/80+ (R1), Ly6G+F4/80+ (R2), and Ly6G+F4/80– (R3) and analyzed by May-Grünwald-Giemsa staining (bar = 10 µm). Percentages of cells were calculated (M, monocytes/macrophages; G, neutrophilic granulocytes; Eos, eosinophilic granulocytes). Representative cell morphology is shown. (B) Histomorphologic examination of GFP+Ly6G+F4/80– neutrophils before transfer into ThG-induced mice (bar = 10 µm). FACS histograms: GFP-nonexpressing cells were gated and analyzed for the expression of TCRβ and B220. (C) Time-dependent conversion of GFP+Ly6G+F4/80– neutrophils from G-CSF–mobilized mice to monocytelike cells in vivo. PB GFP+Ly6G+F4/80– neutrophils were isolated from lys-EGFP mice after treatment with G-CSF (upper diagram). Peritonitis in C57BL/6J wild-type mice was induced by intraperitoneal injection of 4% ThG. GFP+Ly6G+F4/80– neutrophils were transferred by intraperitoneal injection 4 hours postinduction of peritonitis. The phenotype of GFP+ cells in PLF was analyzed at indicated time points by FACS for Ly6G and F4/80 expression. Representative from 2 independent experiments are shown. (D) Histomorphologic analysis of sorted GFP+F4/80+ monocytelike cells from PLF at day 2 of peritonitis (original magnification ×20).
Discussion
We here demonstrated that human in vitro–generated G-CSF–dependent neutrophilic granulocytes acquire monocytic cell characteristics in response to MKK6-p38 activation. Neutrophil-derived monocytic cell generation modeled in HL60 cells required proteasomal degradation of C/EBPα and was at least in part mediated by c-Jun induction and c-Jun phosphorylation (see hypothetical model in supplemental Figure 7). Furthermore, we demonstrated to our knowledge for the first time that adoptively transferred GFP+ neutrophils from G-CSF–treated mice are capable of rapidly acquiring a monocytic phenotype in response to inflammatory signals in vivo. Accordingly, human G-CSF–mobilized band-stage neutrophils possessed monocytic cell and osteoclast differentiation potential in response to proinflammatory cytokines in vitro. Together our data indicate that upon immigration into inflammatory sites, left-shifted (band-stage) neutrophils derived from the bone marrow storage pool acquire a monocytic phenotype in response to inflammatory cytokine-mediated MKK6-p38 pathway activation.
We showed that GFP-marked murine neutrophils develop into monocytic cells via F4/80+Ly6G+ intermediates in vivo when injected intraperitoneally in a peritonitis model 4 hours after ThG administration. Percentages of peritoneal GFP+ cells dropped to half between day 1 vs day 3, most likely because of a relative numeric increase of endogenous neutrophils. Nevertheless, given the high frequency and transient presence of endogenous F4/80+Ly6G+ cells (10% at day 1, gradual disappearance at days 2-3), it is likely that large percentages of immigrated neutrophils convert into monocytic cells in this model. The persistence of GFP+ neutrophils until day 3 in the inflammatory lesion is in line with previous data showing that although the half-life of circulating neutrophils is rather short (∼1 day) their tissue half-life is shown to be 6 to 15 times longer than that of circulating neutrophils.37
We found that large percentages of human CD15+CD54– neutrophils from G-CSF– mobilized blood possess monocytic cell differentiation potential. Because CD54 is known to be lost during terminal differentiation of human neutrophils,33,38 our data indicate that the most differentiated neutrophil fraction in G-CSF–mobilized blood possesses enriched monocytic cell differentiation potential. These cells mainly exhibit band-stage morphology. Our observation that neutrophils from steady-state PB lack similar in vitro monocyte differentiation potential indicates that mobilized neutrophils possess higher differentiation plasticity. In severe and persistent inflammation or after mobilization after G-CSF treatment, mobilized neutrophils have been proposed to be functionally different compared with steady-state neutrophils.39-42 In addition, G-CSF–mobilized band-stage neutrophils were shown to exhibit enhanced survival in vitro and in vivo.43,44
The use of a myeloid model cell line was instrumental for studying the initial intracellular signaling events in response to DOX-induced d.a.MKK6 expression. HL-60 cells showed qualitatively the same changes of several granulocyte/monocyte-affiliated transcription factors as did primary granulopoietic cells. Our data are congruent with previous strong C/EBPα expression by neutrophils and C/EBPα expression level–dependent regulation of neutrophil vs monocytes/macrophage differentiation.45 Moreover, previous studies revealed that modulation of c-Jun levels inversely regulated neutrophil vs monocyte lineage choice decisions of primary murine hematopoietic progenitor cells.46 Using this HL60 differentiation model, we additionally found that d.a.MKK6-induced neutrophil-to-monocyte reprogramming is p38-dependent but JNK-independent. In line with this, p38 can function as the upstream kinase for c-Jun.47 Although previous studies showed JNK to be the key kinase phosphorylating c-Jun at S63/S73,22 c-Jun function was shown to be independent of JNK-mediated phosphorylation in monocytopoiesis.48 C/EBPα, MafB, Gfi-1, and Egr-1 participate together with c-Jun in a transcription factor network during inflammatory cytokine-mediated monocytic cell differentiation from neutrophils. Because these mechanistic data mainly relied on HL60 cells, more extensive future studies need to be performed in primary cells.
We here showed that G-CSF–mobilized blood neutrophils differentiate into CD14hic-Jun+ cells in response to GM-CSF/TNFα/IL-1β stimulation and that these cells exhibit enhanced levels of phospo-MKK6 expression. Global gene expression analysis confirmed a close resemblance of these cells with blood monocytes as well as a clear separation from blood neutrophils or CD15+CD54– starting cells. Substitution of IL-1β by IL-4 (ie, GM-CSF/TNFα/IL-4) resulted in weak MKK6 activation by neutrophils (Figure 3A). Therefore, IL-4 may induce alternatively activated macrophage (M2) or DC differentiation from neutrophils, as opposed to the aforementioned description of p38-driven CD14hi inflammatory monocytes/macrophages. In support of this notion, MKK6 activation resulted in the rapid acquisition of the T-cell costimulatory molecule CD86, along with enhancement of T-cell stimulatory capacity in myeloid cells (supplemental Figure 2B-C).
We chose to stimulate neutrophils with GM-CSF, TNFα + IL-1β because this cytokine combination was previously reported to trigger MKK6-p38 signaling in myeloid cells.31 GM-CSF, TNFα + IL-1β abundantly occur in the synovial fluid of patients with rheumatoid arthritis, and these cytokines were previously used as an equivalent of synovial fluid in experimental in vitro studies of RA pathogenesis.49,50 TLR triggering may substitute for IL-1β given shared downstream signaling together, with the demonstration that TLR stimulation induces monocyte differentiation from murine progenitor cells.18 Future studies should compare TLR ligands vs IL-1β in the presence or absence of specific cosignals to better define the signals capable of monocyte differentiation from G-CSF–mobilized band-stage neutrophils.
We demonstrated here that neutrophils from G-CSF–mobilized blood can give rise to osteoclasts. Several studies previously showed that G-CSF promotes osteopenia,51,52 and transgenic mice encoding human G-CSF developed osteoporosis with increased osteoclast activity.53 Consistently, mononuclear cells from G-CSF–mobilized blood showed higher osteoclast differentiation potential than those isolated from healthy controls,54 and G-CSF augments osteoclast formation in vitro.51 Moreover, G-CSF treatment can exacerbate arthritis, and rheumatoid arthritis is associated with elevated serum G-CSF levels,55 whereas G-CSF receptor–deficient mice are refractory to collagen-induced arthritis.56 Of note, arthritis patients contain increased percentages of activated neutrophils, and these cells may represent a major reservoir of osteoclasts in psoriatic arthritis patients.57 In support for a role of MKK6 in osteoclastogenesis, d.a.MKK6 was shown to enhance the survival of osteoclasts,58 and osteoclast differentiation as well as arthritis activity were diminished in MKK6 knockout mice.59
Although we show here that MKK6-p38 activation is sufficient to induce neutrophil-to-monocytic cell differentiation, we did not address whether it is also required. Because of instability, small-molecule p38 inhibitors could not be added to prolonged differentiation cultures; moreover, mice double-deficient for MKK3/6 (redundant upstream activators of p38) die of embryonic lethality,60 leaving this question open.
Several pathways of macrophage differentiation have been described thus far: monocyte-derived macrophages61; hematopoietic progenitor-cell–derived macrophages62; and bone marrow–independent macrophages that proliferate in situ.63,64 Monocyte/macrophage differentiation from G-CSF–mobilized band-stage neutrophils in response to inflammatory signals appears to constitute a distinct pathway of monocyte/macrophage differentiation.
Acknowledgments
The authors thank Thomas Bauer for graphical illustrations; Stefan Blüml and Peter Pietschmann for help with osteoclast experiments; Leonhard Müllauer and Albert Wöfler for cell morphology assessment; and Gerhard Zlabinger, Diana Mechtcheriakova, Georg Schett, Wilfried Ellmeier, Johannes Stöckl, and laboratory members for critical discussion.
This work was supported by grants P23215-B11 (R.K.) and P19425 and P22058-B20 (H.S.) from the Austrian Science Fund FWF.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.K. designed research, performed most experiments, analyzed data, and wrote the paper; A.M., J.W., A. Hennig, K.W., A.J., D.G., D.M., A. Hladik, and B.G. performed experiments; W.v.d.B. and M.K. contributed reagents; U.H. analyzed data; M.B.F., C.S., and S.K. contributed reagents and provided key advice in research design; and H.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Herbert Strobl, Institute of Pathophysiology and Immunology, Center of Molecular Medicine, Medical University Graz, Heinrichstrasse 31A, A-8010 Graz, Austria; e-mail: herbert.strobl@medunigraz.at.
References
- 1.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 2.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Curr Opin Immunol. 2012;24(3):297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat Immunol. 2011;12(11):1035–1044. doi: 10.1038/ni.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasmono RT, Ehrnsperger A, Cronau SL, et al. Mouse neutrophilic granulocytes express mRNA encoding the macrophage colony-stimulating factor receptor (CSF-1R) as well as many other macrophage-specific transcripts and can transdifferentiate into macrophages in vitro in response to CSF-1. J Leukoc Biol. 2007;82(1):111–123. doi: 10.1189/jlb.1206713. [DOI] [PubMed] [Google Scholar]
- 7.Matsushima H, Geng S, Lu R, et al. Neutrophil differentiation into a unique hybrid population exhibiting dual phenotype and functionality of neutrophils and dendritic cells. Blood. 2013;121(10):1677–1689. doi: 10.1182/blood-2012-07-445189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araki H, Katayama N, Yamashita Y, et al. Reprogramming of human postmitotic neutrophils into macrophages by growth factors. Blood. 2004;103(8):2973–2980. doi: 10.1182/blood-2003-08-2742. [DOI] [PubMed] [Google Scholar]
- 9.Oehler L, Majdic O, Pickl WF, et al. Neutrophil granulocyte-committed cells can be driven to acquire dendritic cell characteristics. J Exp Med. 1998;187(7):1019–1028. doi: 10.1084/jem.187.7.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sathe P, Metcalf D, Vremec D, et al. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage-dendritic cell-restricted progenitor. Immunity. 2014;41(1):104–115. doi: 10.1016/j.immuni.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Hettinger J, Richards DM, Hansson J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14(8):821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- 12.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 13.Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 14.Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta. 2007;1773(8):1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Enslen H, Raingeaud J, Davis RJ. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273(3):1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 16.Chabaud-Riou M, Firestein GS. Expression and activation of mitogen-activated protein kinase kinases-3 and -6 in rheumatoid arthritis. Am J Pathol. 2004;164(1):177–184. doi: 10.1016/S0002-9440(10)63108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navas TA, Mohindru M, Estes M, et al. Inhibition of overactivated p38 MAPK can restore hematopoiesis in myelodysplastic syndrome progenitors. Blood. 2006;108(13):4170–4177. doi: 10.1182/blood-2006-05-023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai Y, Garrett KP, Ohta S, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24(6):801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taschner S, Koesters C, Platzer B, et al. Down-regulation of RXRalpha expression is essential for neutrophil development from granulocyte/monocyte progenitors. Blood. 2007;109(3):971–979. doi: 10.1182/blood-2006-04-020552. [DOI] [PubMed] [Google Scholar]
- 20.Quinn JM, Elliott J, Gillespie MT, Martin TJ. A combination of osteoclast differentiation factor and macrophage-colony stimulating factor is sufficient for both human and mouse osteoclast formation in vitro. Endocrinology. 1998;139(10):4424–4427. doi: 10.1210/endo.139.10.6331. [DOI] [PubMed] [Google Scholar]
- 21.Jörgl A, Platzer B, Taschner S, et al. Human Langerhans-cell activation triggered in vitro by conditionally expressed MKK6 is counterregulated by the downstream effector RelB. Blood. 2007;109(1):185–193. doi: 10.1182/blood-2006-05-022954. [DOI] [PubMed] [Google Scholar]
- 22.Qi X, Borowicz S, Pramanik R, Schultz RM, Han J, Chen G. Estrogen receptor inhibits c-Jun-dependent stress-induced cell death by binding and modifying c-Jun activity in human breast cancer cells. J Biol Chem. 2004;279(8):6769–6777. doi: 10.1074/jbc.M311492200. [DOI] [PubMed] [Google Scholar]
- 23.Yasmin N, Bauer T, Modak M, et al. Identification of bone morphogenetic protein 7 (BMP7) as an instructive factor for human epidermal Langerhans cell differentiation. J Exp Med. 2013;210(12):2597–2610. doi: 10.1084/jem.20130275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boring L, Gosling J, Chensue SW, et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J Clin Invest. 1997;100(10):2552–2561. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J, Lee JD, Jiang Y, Li Z, Feng L, Ulevitch RJ. Characterization of the structure and function of a novel MAP kinase kinase (MKK6). J Biol Chem. 1996;271(6):2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 26.Rado TA, Bollekens J, St Laurent G, Parker L, Benz EJ., Jr Lactoferrin biosynthesis during granulocytopoiesis. Blood. 1984;64(5):1103–1109. [PubMed] [Google Scholar]
- 27.Strobl H, Takimoto M, Majdic O, et al. Myeloperoxidase expression in CD34+ normal human hematopoietic cells. Blood. 1993;82(7):2069–2078. [PubMed] [Google Scholar]
- 28.Scheinecker C, Strobl H, Fritsch G, et al. Granulomonocyte-associated lysosomal protein expression during in vitro expansion and differentiation of CD34+ hematopoietic progenitor cells. Blood. 1995;86(11):4115–4123. [PubMed] [Google Scholar]
- 29.Hattori T, Ohoka N, Inoue Y, Hayashi H, Onozaki K. C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene. 2003;22(9):1273–1280. doi: 10.1038/sj.onc.1206204. [DOI] [PubMed] [Google Scholar]
- 30.Rangatia J, Vangala RK, Treiber N, et al. Downregulation of c-Jun expression by transcription factor C/EBPalpha is critical for granulocytic lineage commitment. Mol Cell Biol. 2002;22(24):8681–8694. doi: 10.1128/MCB.22.24.8681-8694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki K, Hino M, Hato F, Tatsumi N, Kitagawa S. Cytokine-specific activation of distinct mitogen-activated protein kinase subtype cascades in human neutrophils stimulated by granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, and tumor necrosis factor-alpha. Blood. 1999;93(1):341–349. [PubMed] [Google Scholar]
- 32.Munder M, Mollinedo F, Calafat J, et al. Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood. 2005;105(6):2549–2556. doi: 10.1182/blood-2004-07-2521. [DOI] [PubMed] [Google Scholar]
- 33.Elghetany MT. Surface antigen changes during normal neutrophilic development: a critical review. Blood Cells Mol Dis. 2002;28(2):260–274. doi: 10.1006/bcmd.2002.0513. [DOI] [PubMed] [Google Scholar]
- 34.Udagawa N, Takahashi N, Akatsu T, et al. Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc Natl Acad Sci USA. 1990;87(18):7260–7264. doi: 10.1073/pnas.87.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuziel WA, Morgan SJ, Dawson TC, et al. Severe reduction in leukocyte adhesion and monocyte extravasation in mice deficient in CC chemokine receptor 2. Proc Natl Acad Sci USA. 1997;94(22):12053–12058. doi: 10.1073/pnas.94.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96(2):719–726. [PubMed] [Google Scholar]
- 37.Cheretakis C, Leung R, Sun CX, Dror Y, Glogauer M. Timing of neutrophil tissue repopulation predicts restoration of innate immune protection in a murine bone marrow transplantation model. Blood. 2006;108(8):2821–2826. doi: 10.1182/blood-2006-04-018184. [DOI] [PubMed] [Google Scholar]
- 38.Boyd AW, Dunn SM, Fecondo JV, et al. Regulation of expression of a human intercellular adhesion molecule (ICAM-1) during lymphohematopoietic differentiation. Blood. 1989;73(7):1896–1903. [PubMed] [Google Scholar]
- 39.Kaufmann I, Hoelzl A, Schliephake F, et al. Polymorphonuclear leukocyte dysfunction syndrome in patients with increasing sepsis severity. Shock. 2006;26(3):254–261. doi: 10.1097/01.shk.0000223131.64512.7a. [DOI] [PubMed] [Google Scholar]
- 40.Otten MA, Leusen JH, Rudolph E, et al. FcR gamma-chain dependent signaling in immature neutrophils is mediated by FcalphaRI, but not by FcgammaRI. J Immunol. 2007;179(5):2918–2924. doi: 10.4049/jimmunol.179.5.2918. [DOI] [PubMed] [Google Scholar]
- 41.Pillay J, Ramakers BP, Kamp VM, et al. Functional heterogeneity and differential priming of circulating neutrophils in human experimental endotoxemia. J Leukoc Biol. 2010;88(1):211–220. doi: 10.1189/jlb.1209793. [DOI] [PubMed] [Google Scholar]
- 42.Scapini P, Cassatella MA. Social networking of human neutrophils within the immune system. Blood. 2014;124(5):710–719. doi: 10.1182/blood-2014-03-453217. [DOI] [PubMed] [Google Scholar]
- 43.Adachi S, Kubota M, Lin YW, et al. In vivo administration of granulocyte colony-stimulating factor promotes neutrophil survival in vitro. Eur J Haematol. 1994;53(3):129–134. doi: 10.1111/j.1600-0609.1994.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 44.Colotta F, Re F, Polentarutti N, Sozzani S, Mantovani A. Modulation of granulocyte survival and programmed cell death by cytokines and bacterial products. Blood. 1992;80(8):2012–2020. [PubMed] [Google Scholar]
- 45.Dedhia PH, Keeshan K, Uljon S, et al. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood. 2010;116(8):1321–1328. doi: 10.1182/blood-2009-07-229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schüler A, Schwieger M, Engelmann A, et al. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. Blood. 2008;111(9):4532–4541. doi: 10.1182/blood-2007-10-116343. [DOI] [PubMed] [Google Scholar]
- 47.Qi X, Pramanik R, Wang J, et al. The p38 and JNK pathways cooperate to trans-activate vitamin D receptor via c-Jun/AP-1 and sensitize human breast cancer cells to vitamin D(3)-induced growth inhibition. J Biol Chem. 2002;277(29):25884–25892. doi: 10.1074/jbc.M203039200. [DOI] [PubMed] [Google Scholar]
- 48.Behre G, Whitmarsh AJ, Coghlan MP, et al. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J Biol Chem. 1999;274(8):4939–4946. doi: 10.1074/jbc.274.8.4939. [DOI] [PubMed] [Google Scholar]
- 49.McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 2007;7(6):429–442. doi: 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- 50.MacDonald KP, Pettit AR, Quinn C, Thomas GJ, Thomas R. Resistance of rheumatoid synovial dendritic cells to the immunosuppressive effects of IL-10. J Immunol. 1999;163(10):5599–5607. [PubMed] [Google Scholar]
- 51.Hirbe AC, Uluçkan O, Morgan EA, et al. Granulocyte colony-stimulating factor enhances bone tumor growth in mice in an osteoclast-dependent manner. Blood. 2007;109(8):3424–3431. doi: 10.1182/blood-2006-09-048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yakisan E, Schirg E, Zeidler C, et al. High incidence of significant bone loss in patients with severe congenital neutropenia (Kostmann’s syndrome). J Pediatr. 1997;131(4):592–597. doi: 10.1016/s0022-3476(97)70068-4. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi T, Wada T, Mori M, Kokai Y, Ishii S. Overexpression of the granulocyte colony-stimulating factor gene leads to osteoporosis in mice. Lab Invest. 1996;74(4):827–834. [PubMed] [Google Scholar]
- 54.Purton LE, Lee MY, Torok-Storb B. Normal human peripheral blood mononuclear cells mobilized with granulocyte colony-stimulating factor have increased osteoclastogenic potential compared to nonmobilized blood. Blood. 1996;87(5):1802–1808. [PubMed] [Google Scholar]
- 55.Campbell IK, Rich MJ, Bischof RJ, Hamilton JA. The colony-stimulating factors and collagen-induced arthritis: exacerbation of disease by M-CSF and G-CSF and requirement for endogenous M-CSF. J Leukoc Biol. 2000;68(1):144–150. [PubMed] [Google Scholar]
- 56.Eyles JL, Hickey MJ, Norman MU, et al. A key role for G-CSF-induced neutrophil production and trafficking during inflammatory arthritis. Blood. 2008;112(13):5193–5201. doi: 10.1182/blood-2008-02-139535. [DOI] [PubMed] [Google Scholar]
- 57.Chiu YG, Shao T, Feng C, et al. CD16 (FcRgammaIII) as a potential marker of osteoclast precursors in psoriatic arthritis. Arthritis Res Ther. 2010;12(1):R14. doi: 10.1186/ar2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamashita T, Kobayashi Y, Mizoguchi T, et al. MKK6-p38 MAPK signaling pathway enhances survival but not bone-resorbing activity of osteoclasts. Biochem Biophys Res Commun. 2008;365(2):252–257. doi: 10.1016/j.bbrc.2007.10.169. [DOI] [PubMed] [Google Scholar]
- 59.Yoshizawa T, Hammaker D, Boyle DL, et al. Role of MAPK kinase 6 in arthritis: distinct mechanism of action in inflammation and cytokine expression. J Immunol. 2009;183(2):1360–1367. doi: 10.4049/jimmunol.0900483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brancho D, Tanaka N, Jaeschke A, et al. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 2003;17(16):1969–1978. doi: 10.1101/gad.1107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19(1):71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 62.Massberg S, Schaerli P, Knezevic-Maramica I, et al. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell. 2007;131(5):994–1008. doi: 10.1016/j.cell.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenkins SJ, Ruckerl D, Cook PC, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332(6035):1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schulz C, Gomez Perdiguero E, Chorro L, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336(6077):86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]