Abstract
Protein GPI anchorage to the cell surface is important for various biological processes, but GPI-anchored proteins are difficult to study. This paper developed an effective strategy for metabolic engineering of cell surface GPIs and GPI-anchored proteins by using inositol derivatives carrying an azido group. The azide-labeled GPIs and GPI-anchored proteins on live cells were then tagged with biotin via click reaction and with a fluorescent molecule. The strategy can be used to label GPI-anchored proteins with various tags for biological studies.
Keywords: GPI, glycolipid, carbohydrate, metabolic engineering, inositol
Glycosylphosphatidylinositol (GPI) attachment to the protein C-terminus is one of the most common posttranslational protein modifications in eukaryotic species, which helps anchor proteins onto the cell membrane.[1] Many GPIs and GPI-anchored proteins have been identified[2] and shown to play a critical role in various biological processes,[3] thus GPI-anchored proteins need GPI anchor to function properly[2a, 4] and losing GPI anchoring ability is lethal for mammals and conditionally lethal for yeasts.[3k] Moreover, elevation in GPI-anchored proteins was observed on human cancer cells, indicating their potential as biomarkers for cancer detection and therapy.[3n]
No matter of their origin, all GPI-anchored proteins share a conserved construct, having the protein C-termini linked to the phosphoethanolamine [(P)-EtNH2] moiety at the mannose-III 6-O-position of the GPI core (Figure 1).[1] This suggests a conserved biosynthetic pathway for GPIs and GPI-anchored proteins in eukaryotic cells. GPIs and GPI-anchored proteins are biosynthesized in the endoplasmic reticulum via a series of membrane-bound enzymatic transformations involving >20 gene products.[2a, 3a, 5] First, intact GPI anchors are synthesized, which were then attached to target proteins bearing a GPI attachment signal at the C-termini by GPI transamidase.[5c, 6] After lipid remodeling and other subtle modifications, GPI-linked proteins are transported and anchored to the cell membrane (Figure 1).[2a]
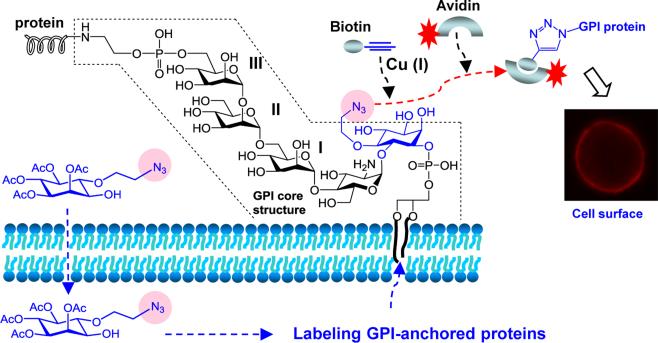
Figure 1.
The designed strategy for GPI-anchored protein labeling on the cell surface through metabolic engineering of the GPI core structure. Cells are incubated with an inositol derivative bearing a chemically reactive, bio-orthogonal functionality such as the azido group. The unnatural inositol is taken up by cells and incorporated into GPIs and GPI-anchored proteins. The modified GPI-anchored proteins on the cell surface can be subsequently elaborated by a specific, bio-orthogonal reaction for attachment of various molecules, such as fluorescent tags to make cell surface GPI-anchored proteins visible, affinity tags to facilitate GPI-linked protein isolation, and so on.
Natural GPIs are structurally diverse. Moreover, GPI-anchored proteins are amphiphilic and associated with the cell membrane, making their isolation from nature difficult. Most importantly, GPI-anchored proteins are present in low quantity on cells. Therefore, biological studies of GPI-anchored proteins at the molecular level and on live cells are difficult. Attempts to deal with this issue included chemical and chemoenzymatic syntheses of GPI-anchored proteins,[7] which have resulted in only unnatural products yet and are not applicable to live cells. Attempts to study GPI-anchored proteins via engineering GPIs on live cells have gained limited success.[4]
This work aimed at developing a practical method for GPI-anchored protein labeling on live cells to facilitate their identification, purification, and biological study. We planned to introduce a molecular handle to the GPI common core via cell metabolic engineering, namely, giving cells a modified GPI precursor for biosynthetic incorporation (Figure 1). The precursor must be accepted by involved enzymes and the handle should be flexible to allow for further elaboration. In this regard, we were interested in modified inositol, due to its unique presence in GPIs, with the azido group as a molecular handle, as it is small and can be easily elaborated through chemoselective and bio-orthogonal click reaction.[8] Although a similar strategy has been used for glycobiological studies based on cell surface sialic acid, fucose, and N-acetylgalactosamine engineering,[9] it has not been applied to GPI and GPI-anchored protein engineering yet.
To find a proper precursor for cell surface GPI engineering, we prepared inositol derivatives 1-3 (Figure 2 and Supporting Information) that had an azidoethyl group linked to the inositol 3−, 4−, and 5-O-positions, respectively. We were interested in 1-3, not those with modifications at the 1−, 6− and 2-O-positions, as these positions are either linked to a phospholipid and glycan in GPIs or required in GPI biosynthesis. Moreover, instead of directly replacing the hydroxyl groups with azides, an azidoethyl group was introduced to improve the molecular space and flexibility, which might help click reaction later on. Saccharomyces cerevisiae, a fast-growing yeast that expresses abundant GPI-anchored proteins, was used to examine the efficiency of 1-3 incorporation in GPI-anchored proteins. Our experimental protocols were to incubate yeast cell with 1-3 and then subject the cell to click reaction with alkynylated biotin 4. Several bio-orthogonal click reactions are available, but we chose the Cu (I)-catalyzed azide-alkyne cycloaddition since it is convenient and fast.[10] Finally, the treated cell was stained with the streptavidin-allophycocyanin (APC) conjugate and used to analyze 1-3 incorporation in GPIs and GPI-anchored proteins.
Figure 2.
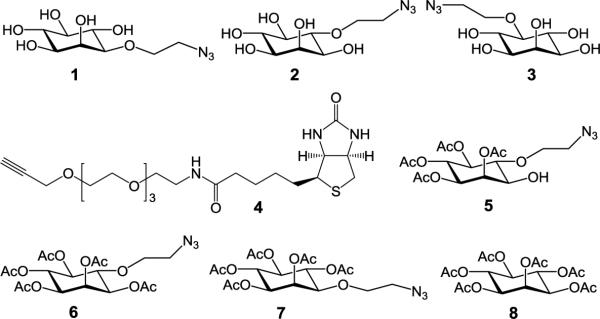
Structures of the synthesized inositol derivatives 1-3 and 5-7 used for metabolic engineering of GPIs and GPI-anchored proteins on cells and the biotin derivative 4 used to tag modified GPIs on cells via click reaction.
Flow cytometry results of the stained cells shown in Figure 3a and Figure S1 (Supporting Information) revealed that the mean fluorescent intensities (MFI) of cells treated with 1-3 (3 mM) were similar to that of the control, suggesting that 1-3 were not incorporated by yeast cell. Among various potential reasons to cause problem, it was possible that 1-3 could not effectively penetrate the cell membrane due to high polarity. Inspired by the finding that acylation of free sugars could improve their efficacy to engineer cell,[11] we prepared and studied acetylated inositol derivatives 5-7 (Figure 2). As shown in Figure 3b, yeast cell was effectively labeled upon treatment with 5-7 (3 mM). The MFI of cells treated with 5 was >1000 folds higher than that of the control (Figure S1), confirming that 5 could be used by yeast cell to label GPIs and GPI-anchored proteins.
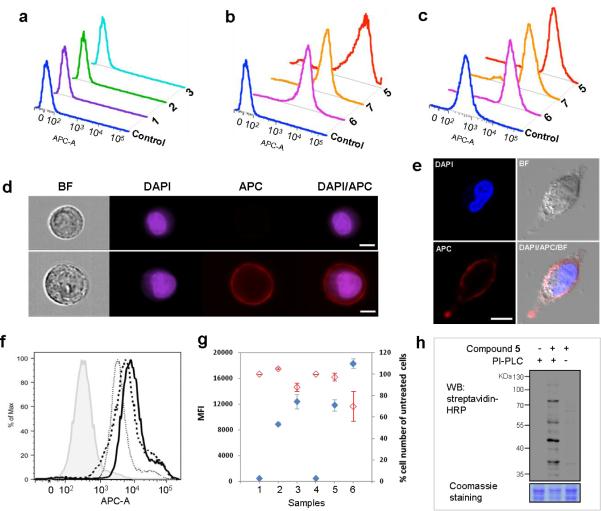
Figure 3.
Detection of GPIs and GPI-anchored proteins on cells treated with or without modified inositol derivatives after click reaction with 4 and staining with streptavidin-APC (for cell analysis) or streptavidin-peroxidase (for protein detection). Flow cytometry results of yeast cells incubated with (a) 1-3 (3 mM) and (b) 5-7 (3 mM); and (c) of A549 cells treated with 5-7 (200 μM). (d) Imaging cytometry results of 5-treated A549 cell suspended in PBS buffer. BF: bright field image; DAPI: the nuclear stain. The upper and lower images are untreated and treated cells, respectively (white scale bar: 7 μm). (e) Confocal microscopy images of 5-treated A549 cell cultured in the dish (white scale bar: 10 μm). (f) Flow cytometry results of A549 cell incubated with 5 (100 μM) in the presence of 0 (solid line), 100 (dashed line), and 200 μM (dotted line) of 8. The filled histogram shows the result of cell not treated with 5 and 8. (g) Influences of the concentration of 5 and incubation time on metabolic engineering of GPIs and GPI-anchored proteins on A549 cell. The concentrations of 5 were 0 (samples 1 & 4, negative controls), 100 (samples 2 & 5), and 200 μM (samples 3 & 6). The cell was incubated for 1 day (samples 1-3) or 2 days (samples 4-6). Blue and red rhombuses show cell labeling and growth, respectively. (h) Western blot of GPI-anchored proteins expressed on 5-treated A549 cell. The supernatants of cells treated with PI-PLC only (left), with 5 and PI-PLC (middle), and with 5 only (right) were subjected to click reaction with 4 and then probed with streptavidin-peroxidase (HRP). Bottom: Coomassie staining to show the protein loading in each lane.
Acetylated 5-7 were clearly more efficient than free 1-3 for yeast cell GPI engineering. As suggested in the literature,[11] lipophilic 5-7 may be more effectively taken up by cells via passive diffusion. Inside the cell, the acetyl groups could be removed by esterases[11] to give free inositol derivatives that were incorporated in GPIs. Moreover, the enzymes involved in GPI biosynthesis must be able to tolerate modified inositols, especially 3/4-O-modified ones. It is also noteworthy that partially acetylated 5 was more efficient for GPI engineering than fully acetylated 6, maybe because 5 had more proper hydrophilic/hydrophobic balance to favor its entry into cell.
This strategy was also tested using a human lung cancer cell A549 to probe the application scope. After treatment with 5-7 (200 μM), the cell was subjected to click reaction with 4, staining with streptavidin-APC, and flow cytometry. As shown in Figures 3c and S2, cells treated with 5-7 were effectively labeled with fluorescent tags, proving the incorporation of 5-7. The MFIs of cells treated with 5 and 6 or 7 were ca. 30 and 16 folds higher than that of the control. Again, partially acetylated 5 was better than peracetylated 6. Flow distribution analysis revealed that >98% of 5-treated cells were stained with APC (Figure S3), suggesting extensive cell metabolic engineering. The stained cells were also analyzed by imaging flow cytometry and confocal microscopy. As depicted in Figures 3d and 3e, bright red fluorescence was concentrated on the cell surface. In addition, strong fluorescence on the cell surface also indicated the abundant labeling of GPIs and GPI-anchored proteins and the high efficiency of GPI metabolic engineering.
To further verify the incorporation of 5 in GPIs, we did a competitive inhibition analysis, in which A549 cell was treated with 5 and peracetylated inositol 8. After staining, the cell showed decreased MFI with increased concentration of 8 (Figures 3f and S4). Clearly, 8 inhibited cell incorporation of 5 and GPI engineering, thus 5 and 8 were involved in the same biosynthetic pathway. The impact of precursor concentration and incubation time on the engineering of GPIs and GPI-anchored proteins was also studied (Figures 3g and S5). A549 cell treated with 200 μM of 5 had significantly stronger fluorescence than the cell treated with 100 μM of 5, confirming that the intensity of cell labeling depended on the concentration of 5. In addition, cells incubated with 100 and 200 μM of 5 had increased MFIs after elongated incubation, and the cell treated with 200 μM of 5 had the highest MFI after 2 days of incubation. However, at 200 μM, 5 inhibited cell growth by 30%, while it did not show obvious inhibition on cell growth at 100 μM.
Phosphatidylinositol-specific phospholipase C (PI-PLC) can be used to detach GPI-anchored proteins from the cell membrane,[3n] which was proved by the detection of a GPI-anchored cancer marker, placental alkaline phosphatase, in the supernatant of PI-PLC-treated A549 cell (Figure S6). To verify that GPI-anchored proteins on engineered cell were labeled with azides, after treatment with 5, A549 cell was incubated with PI-PLC, and the released GPI-anchored proteins in the cell supernatant were subjected to click reaction with 4 and then probed with streptavidin-peroxidase for Western blot. As shown in Figure 3h, the cell treated with both 5 and PI-PLC gave a series of protein bands that were absent or very faint when 5 or PI-PLC was missing. This result confirmed the expression of azide-labeled GPI-anchored proteins on 5-treated A549 cell. We are currently pursuing further verification and characterization of these proteins.
Above studies using different cells indicated that the new strategy for GPI and GPI-anchored protein engineering may be widely useful. To verify this, four other human cancer cell lines, MCF-7, Hela, K562, and SKM28, were treated with 5, click reaction and fluorescent staining, and then analyzed. Flow cytometry showed that all four cell lines were labeled with the fluorescent tag, and imaging cytometry showed that the fluorescence was focused on the cell surface (Figure S7-S10). These results ultimately proved the efficiency, flexibility, and broad applicability of 5 for GPI and GPI-anchored protein engineering. It is also noteworthy that, except for MCF-7, 5 did not show a significant impact on cell growth at concentrations up to 100 μM.
In summary, we have developed a facile and effective method for cell surface GPI and GPI-anchored protein labeling, which was realized by giving cell an azide-modified inositol derivative and allowing cell to incorporate it in GPIs. The azide-labeled GPIs and GPI-anchored proteins on the cell surface were then elaborated by bio-orthogonal click reaction to enable their visualization and potentially the study of GPI and GPI-anchored protein trafficking and organization on cells by quantitative fluorescence microscopy,[12] and so on. The new GPI labeling strategy was effective and applicable to different cells. Moreover, we envision that this technique can be used for GPI-anchored protein capture, GPI-anchored proteomics analysis, and discovery of new GPI-anchored protein markers. It can also be used for targeted drug delivery and for the diagnosis and therapy of diseases via monitoring cell surface GPI-anchored proteins. Therefore, this technique can be a powerful tool to have a significant impact on GPI biology.
Supplementary Material
Footnotes
This work was supported in part by NIH (R01 GM090270), National Major Scientific and Technological Special Program for “New Drug Development” (No. 2012ZX09502001-005), National Basic Research (973) Program (2012CB822102) and National Science Foundation (No. 21472114) of China, and a Chinese Scholarship to L. Liu (No. 201206225032). The Microscopy, Imaging and Cytometry Resources Core is supported in part by an NIH Center grant (P30CA22453) to the Karmanos Cancer Institute, Wayne State University, and the Perinatology Research Branch of the National Institutes of Child Health and Development.
Supporting information for this article is available on the WWW under http://dx.doi.org/----
Supporting Information. Experimental procedures for the synthesis of compounds 1-3 and 5-8; NMR and MS data and spectra of all compounds; protocols for cell metabolic engineering and related analysis, and additional data, figures, and images of flow cytometry, fluorescence, and Western blot studies.
Contributor Information
Dr. Lili Lu, National Glycoengineering Research Center, Shandong University, 29 Shanda Nan Lu, Jinan 250010 (China) Department of Chemistry, Wayne State University, 5101 Cass Avenue, Detroit, Michigan 48202 (the United States)..
Dr. Jian Gao, Department of Chemistry, Wayne State University, 5101 Cass Avenue, Detroit, Michigan 48202 (the United States).
Prof. Dr. Zhongwu Guo, National Glycoengineering Research Center, Shandong University, 29 Shanda Nan Lu, Jinan 250010 (China); Department of Chemistry, Wayne State University, 5101 Cass Avenue, Detroit, Michigan 48202 (the United States)..
References
- 1.Ferguson MAJ, Williams AF. Ann. Rev. Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 2.a Orlean P, Menon AK. J. Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]; b Paulick MG, Bertozzi CR. Biochemistry. 2008;47:6991–7000. doi: 10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Tsai Y, Liu X, Seeberger PH. Angew. Chem. Int. Ed. 2012;51:11438–11456. doi: 10.1002/anie.201203912. [DOI] [PubMed] [Google Scholar]
- 3.a Thomas JR, Dwek RA, Rademacher TW. Biochemistry. 1990;29:5413–5422. doi: 10.1021/bi00475a001. [DOI] [PubMed] [Google Scholar]; b Robinson PJ. Cell Biol. Intern. Rep. 1991;15:761–767. doi: 10.1016/0309-1651(91)90031-d. [DOI] [PubMed] [Google Scholar]; c McConville MJ, Ferguson MAJ. Biochem. J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ferguson MAJ. Parasitology Today. 1994;10:48–52. doi: 10.1016/0169-4758(94)90392-1. [DOI] [PubMed] [Google Scholar]; e Gaulton GN, Pratt JC. Seminars Immunol. 1994;6:97–104. doi: 10.1006/smim.1994.1014. [DOI] [PubMed] [Google Scholar]; f Ferguson MAJ. J. Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]; g Kasahara K, Sanai Y. Glycoconjugate J. 2000;17:153–162. doi: 10.1023/a:1026576804247. [DOI] [PubMed] [Google Scholar]; h Marmor MD, Julius M, Biol J. Regul. Homeostatic Agents. 2000;14:99–115. [PubMed] [Google Scholar]; i Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang S-H, Yang X, Zhang M-Y, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]; j Chan C, Stanners C. Curr. Oncol. 2007;14:70–73. doi: 10.3747/co.2007.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; k Varma Y, Hendrickson T. ChemBioChem. 2010;11:623–636. doi: 10.1002/cbic.200900704. [DOI] [PubMed] [Google Scholar]; l Bradley JE, Chan JM, Hagood JS. Lab Invest. 2013;93:365–374. doi: 10.1038/labinvest.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]; m Park S, Lee C, Sabharwal P, Zhang M, Meyers CLF, Sockanathan S. Science. 2013;339:324–328. doi: 10.1126/science.1231921. [DOI] [PMC free article] [PubMed] [Google Scholar]; n Dolezal S, Hester S, Kirby PS, Nairn A, Pierce M, Abbott KL. Cancer Biomark. 2014;44:55–62. doi: 10.3233/CBM-130377. [DOI] [PMC free article] [PubMed] [Google Scholar]; o Kinoshita T. Proc. Jpn. Acad. Ser. B. 2014;90:130–143. doi: 10.2183/pjab.90.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor DR, Hooper NM. Post-Translational Modifications in Health and Disease. Springer; New York: 2011. pp. 39–55. [Google Scholar]
- 5.a Englund PT. Ann. Rev. Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]; b Takeda J, Kinoshita T. Trends Biochem. Sci. 1995;20:367–371. doi: 10.1016/s0968-0004(00)89078-7. [DOI] [PubMed] [Google Scholar]; c Udenfriend S, Kodukula K. Ann. Rev. Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 6.a Bailey CA, Gerber L, Howard AD, Udenfriend S. PNAS. 1989;86:22–26. doi: 10.1073/pnas.86.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Maxwell SE, Ramalingam S, Gerber LD, Brink L, Udenfriend S. J. Biol. Chem. 1995;270:19576–19582. doi: 10.1074/jbc.270.33.19576. [DOI] [PubMed] [Google Scholar]
- 7.a Guo Z. Curr. Org. Synth. 2013;10:366–383. doi: 10.2174/1570179411310030003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yu S, Guo Z, Johnson C, Gu G, Wu Q. Curr. Opin. Chem. Biol. 2013;17:1006–1013. doi: 10.1016/j.cbpa.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wehle M, Vilotijevic I, Lipowsky R, Seeberger PH, Varon Silva D, Santer M. J. Am. Chem. Soc. 2012;134:18964–18972. doi: 10.1021/ja302803r. [DOI] [PubMed] [Google Scholar]; d Wu Z, Guo X, Wang Q, Swarts BM, Guo Z. J. Am. Chem. Soc. 2010;132:1567–1571. doi: 10.1021/ja906611x. [DOI] [PubMed] [Google Scholar]; e Ferguson MA. Nat. Chem. Biol. 2008;4:223–224. doi: 10.1038/nchembio0408-223. [DOI] [PubMed] [Google Scholar]; f Becker CFW, Liu X, Olschewski D, Castelli R, Seidel R, Seeberger PH. Angew. Chem., Int. Ed. 2008;47:8215–8219. doi: 10.1002/anie.200802161. [DOI] [PubMed] [Google Scholar]; g Paulick MG, Wise AR, Forstner MB, Groves JT, Bertozzi CR. J. Am. Chem. Soc. 2007;129:11543–11550. doi: 10.1021/ja073271j. [DOI] [PubMed] [Google Scholar]
- 8.a Kolb HC, Finn MG, Sharpless KB. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; b Prescher JA, Bertozzi CR. Cell. 2006;126:851–854. doi: 10.1016/j.cell.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 9.a Campbell CT, Sampathkumar SG, Yarema KJ. Mol. BioSyst. 2007;3:187–194. doi: 10.1039/b614939c. [DOI] [PubMed] [Google Scholar]; b Laughlin ST, Agard NJ, Baskin JM, Carrico IS, Chang PV, Ganguli AS, Hangauer MJ, Lo A, Prescher JA, Bertozzi CR. Methods Enzym. 2006;415:230–250. doi: 10.1016/S0076-6879(06)15015-6. [DOI] [PubMed] [Google Scholar]; c Lemieux GA, Bertozzi CR. Chem. Biol. 2001;8:265–275. doi: 10.1016/s1074-5521(01)00008-4. [DOI] [PubMed] [Google Scholar]; d Bertozzi CR, Kiessling LL. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 10.Jiang H, Zheng T, Lopez-Aguilar A, Feng L, Kopp F, Marlow FL, Wu P. Bioconjug. Chem. 2014;25:698–706. doi: 10.1021/bc400502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Prescher JA, Dube DH, Bertozzi CR. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]; b Lemieux GA, Yarema KJ, Jacobs CL, Bertozzi CR. J. Am. Chem. Soc. 1999;121:4278–4279. [Google Scholar]; c Jacobs CL, Goon S, Yarema KJ, Hinderlich S, Hang HC, Chai DH, Bertozzi CR. Biochemistry. 2001;40:12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- 12.a Esposito A, Schlachter S, Schierle GSK, Elder AD, Diaspro A, Wouters FS, Kaminski CF, Iliev AI. Cytoskeleton Methods and Protocols. Springer; 2010. pp. 117–142. [DOI] [PubMed] [Google Scholar]; b Mayor S, Riezman H. Nat. Rev. Mol. Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.