Abstract
The many advantages of 13C NMR are often overshadowed by its intrinsically low sensitivity. Given that carbon makes up the backbone of most biologically relevant molecules, 13C NMR offers a straightforward measurement of these compounds. Two-dimensional 13C-13C correlation experiments like INADEQUATE (incredible natural abundance double quantum transfer experiment) are ideal for the structural elucidation of natural products and have great but untapped potential for metabolomics analysis. We demonstrate a new and semi-automated approach called INETA (INADEQUATE network analysis) for the untargeted analysis of INADEQUATE datasets using an in silico INADEQUATE database. We demonstrate this approach using isotopically labeled Caenorhabditis elegans mixtures.
Introduction
Though many techniques exist to study the metabolome, the identification of metabolites is a universal challenge. The primary analytical techniques are liquid or gas chromatography coupled with mass spectrometry (LC-MS or GC-MS) and nuclear magnetic resonance (NMR). With LC-MS or GC-MS, chemical separation provides an extra dimension that can aid in feature recognition and in database matching, but it is difficult or impossible to confidently identify compounds that are not in databases from just masses or even fragmentation patterns using tandem MS. NMR most often does not utilize an initial chromatographic step but rather analyzes complex mixtures directly. Thus, a goal of most NMR metabolomics approaches is the deconvolution of complex spectra into resonances that correspond to individual metabolites, which then makes compound identification simpler.
NMR, however, suffers from relatively low sensitivity, especially in comparison with MS techniques. The low sensitivity has led to the nearly ubiquitous use of 1H NMR, because 1H is almost 100% abundant and has the highest NMR frequency—and thus the highest sensitivity—of any commonly analyzed isotope. Several methods have been developed to directly analyze 1D 1H NMR spectra of metabolomic mixtures including multivariate methods such as principal component analysis (PCA) to identify resonances that differ between groups,1 statistical total correlation spectroscopy (STOCSY) to obtain correlations between nuclei in the same molecule or pathway,2 and direct fitting of spectra using library databases.3
In contrast to 1H, 13C has only 1.1% natural abundance and has a frequency that is ¼ that of 1H, translating to lower intrinsic NMR sensitivity. However, 13C NMR has several advantages over 1H, namely larger chemical shift dispersion, detection of quaternary carbons, and a direct measure of the backbone structure of most metabolites.4 Isotopic labeling of metabolites significantly enhances the feasibility of 13C detection and allows for more efficient utilization of this important isotope. Other 13C detected NMR methods have been developed and proposed for metabolomics.4,5 One-dimensional 13C spectral analysis has been shown to enhance the ability to identify compounds in mixtures when compared to 1D 1H analysis alone.4 In addition, 2D methods such as 13C-13C constant time TOCSY in conjunction with the 13C-13C total correlation spectroscopy (TOCCATA) database was used to identify metabolites in a complex mixture.5
In this study we present an alternate approach to NMR untargeted (a.k.a. global) metabolomics using INDADEQUATE (incredible natural abundance double quantum transfer experiment).6–8 The 2D INADEQUATE experiment records 13C chemical shifts in the acquisition dimension and 13C-13C double quantum correlations in the indirect dimension, leading to directly bonded 13C-13C correlation networks. Because INADEQUATE provides complete carbon correlated networks, it is the most direct NMR experiment for metabolite and natural product identification. However, it is also extremely challenging at natural abundance 13C, because in addition to the intrinsically low 13C sensitivity discussed above, INADEQUATE correlations require two adjacent 13C nuclei, which at natural abundance has a probability of 1 in 8264 (0.0112).
Markley’s laboratory showed that INADEQUATE-like experiments could be used for 13C-13C correlations using 13C labeled proteins,9 and more recently Wagner and coworkers have used directly detected 13C for assignments in perdeuterated proteins with fast relaxation times.10 Sumner and collaborators described the use of 13C NMR experiments to characterize and track the metabolism of selectively 13C labeled small molecules in rats, mice, and humans.11–14 INADEQUATE spectroscopy was used to determine the 13C-13C connectivity of signals arising from these metabolites. These studies using 13C enriched samples demonstrate that INADEQUATE is a feasible approach to obtain direct 13C-13C correlations in complex spectra.
Here we present a new approach to NMR untargeted metabolomics using uniformly 13C-enriched Caenorhabditis elegans. We utilized a custom 1.5-mm 13C-optimized high temperature superconducting (HTS) probe,15 which allowed us to efficiently analyze sample volumes of just 40 µL. To demonstrate the approach, we compared unfractionated INADEQUATE spectra from both the endometabolome (i.e. metabolites within the worm) and exometabolome (i.e. metabolites released into the environment) of worms that had been heat shocked and control animals that were maintained at room temperature. These spectra were compared using HATS-PR,16 a method previously developed in our lab for 2D NMR multivariate analysis. We wrote a new software package that we are calling INETA (INADEQUATE network analysis) to automatically identify correlated networks. Finally, we created a database of INADEQUATE spectra from assigned metabolites with 13C 1D data obtained from the BMRB.17,18 Using INETA, we were able to identify 53 and 49 networks from the endo- and exometabolomes, respectively, which could be semi-automatically assigned to 31 compounds from the BMRB.
Experimental Methods
Sample preparation
Worms were isotopically labeled with 99% uniformly labeled 13C glucose (Cambridge Isotope Labs) as the primary carbon source and heat shocked as described in a previous publication.19 One million worms were used for each NMR sample, and endo- and exometabolomes were collected as previously described.19 Additional Information on sample preparation is available in the supplement (text and Figure S1).
NMR Data Collection and Processing
All spectra were collected on an Agilent VNMRS-600 spectrometer using a custom 1.5 mm 13C high temperature superconducting (HTS) probe with a 40 µL sample volume.15 The total time for each INADEQUATE experiment was ~14 hours. A spectral width of 202 ppm (30487.8 Hz) along F2 and a 404 ppm (60975.6 Hz) along F1 was used. Spectra were collected using a 90° pulse (pw=15.6) with a 180° adiabatic pulse at 57 dB, τ corresponding to 13C-13C coupling of 55 Hz was used with 2048 t1 increments with 4 scans, a 3 sec relaxation delay with WALTZ-16 1H decoupling at 599.68 MHz with a power of 37 dB.
All spectra were processed in NMRPipe.20 INADEQUATE spectra were processed along both dimensions using a Lorentz-to-Gaussian window function, zero-filled 2x, Fourier transformed, and baseline corrected. Spectra were referenced to the most isolated and prominent peaks. Exometabolome spectra were referenced using the anomeric carbon of glucose at 98.6 ppm and endometabolome spectra were referenced to the gamma carbon of isoleucine at 17.5 ppm in the exometabolome.
Statistical Analysis
INADEQUATE spectra were imported into our laboratory’s MATLAB Metabolomics Toolbox and aligned using hierarchical clustering as previously described.16 All spectra were normalized using probabilistic quotient normalization21(PQN) and log scaled. Principal component analysis (PCA) was used to compare heat shock and control C. elegans populations in both the endo- and the exometabolome. In addition, the endo- and exometabolomes were directly compared using PCA across all samples. One outlier in the control exometabolome worms was identified using PCA and removed. Loadings plots were annotated using the compounds found by INETA, as described below. All NMR raw data, processing scripts, and code are deposited in the Metabolomics Workbench database (http://www.metabolomicsworkbench.org/) supported by the NIH Common Fund.
Data analysis using INADEQUATE Network Analysis (INETA)
NMR spectra were converted in MATLAB from NMRPipe (*.ft) files to a matrix of peak intensities that was subsequently imported into Mathematica 922 for INETA analysis. INETA has several user-adjustable parameters, which are defined in Table 1 and discussed for each step below.
Table 1.
Peak picking and Network Parameters
| Parameters | Endometabolome | Exometabolome | |
|---|---|---|---|
| Peak Picking | |||
| Minimum threshold | PPmin | 2×10^7 | 3×10^7 |
| Maximum threshold | PPmax | 8×10^7 | 12×10^7 |
| Chemical shift threshold | PPCS | 1 | 1 |
| Double quantum threshold | PPDQ | 1 | 2 |
| Network Finding | |||
| Double quantum tolerance | DQT | 0.3 ppm | 0.5 ppm |
| Symmetrical/Diagonal tolerance | SDT | 0.3 ppm | 0.3 ppm |
| Chemical shift tolerance | CST | 0.03 ppm | 0.07 ppm |
| Database Match | |||
| Ambiguity allowance | AA | 0.5 | 0.5 |
| Chemical shift match tolerance | CSMT | 0.5 ppm | 0.5 ppm |
| Matches tolerance | NCMT | 2 | 2 |
| Double quantum threshold | DQMT | 4 | 4 |
| Total Networks Found | 53 | 49 | |
| Networks Matched | 20 | 25 | |
1. Peak Picking
INADEQUATE spectra were imported into Mathematica 9.22 In this initial development of INETA, we chose to use the absolute values of the antiphase INADEQUATE peaks to simplify the algorithm. We first tried single values of peak intensity thresholds to define peaks but found that INETA was not able to resolve peaks that were too close together. Therefore, we implemented multiple thresholds, starting with the highest value (least noise), defined in INETA as PPmax. All parameters are defined in Table 1. We then defined the lowest value (most noise) as PPmin and allowed for a user-defined value of n number of steps between PPmax and PPmin. At every threshold INETA finds the local maximum for each peak and then defines an area for that peak where the intensity reaches the value of the current threshold.
Next, peaks with their defined areas from each threshold are compared using the parameters PPCS and PPDQ (Table 1). If the centers of mass for any cluster of peaks on any threshold are within PPCS along the chemical shift axis and PPDQ along the double quantum axis, they are considered one peak. If they exceed either of these values, they are considered separate peaks. This resulted in a single peak picked INADEQUATE spectrum.
2. Finding Networks
A network is a set of spectroscopically correlated peaks. The double quantum frequency in an INADEQUATE experiment is defined as the sum of the single quantum chemical shift values for each pair of directly bonded 13C nuclei. This relationship leads to a double quantum diagonal, which has a slope of 2. Pairs of coupled 13C nuclei are symmetric with respect to the diagonal at a given double quantum frequency. INETA groups peaks if the differences in their DQ values are less than the DQ tolerance (DQT; Table 1). Next, INETA determines that pairs of peaks follow INADEQUATE rules by taking the absolute value of the difference between the sum of their chemical shifts and the mean of their double quantum values. If this difference is smaller than the user defined parameter SDT, then these are considered correlated (i.e. |(CS1 + CS2)-Mean [DQ1 + DQ2]| < SDT). Peaks without DQ pairs are eliminated.
The set of peaks that are found to obey INADEQUATE double quantum rules are then checked for vertical chemical shift correlations by comparing the difference of their chemical shifts to the user adjusted parameter CST (Table 1). The final step is to connect all spins that passed both horizontal and vertical correlations into networks and mapped onto the peak picked spectra.
3. Database Construction
We constructed an INETA database using experimental 1D 13C spectra from the biological magnetic resonance data bank (BMRB).18 The BMRB data fall into different categories. Some metabolites have multiple 13C datasets collected under different conditions such as solvent or metabolite concentration. Some spectra are fully assigned, while others have regions of ambiguity. In cases of incomplete assignments, the BMRB “assigns” all possible resonances to all atoms that were not specifically assigned. For example, in glucose there are 36 different “assignments” of the 12 13C resonances (which include both α and β anomers) to the 6 carbons. In INETA, we define an ambiguity score as CA/CT where CA is the number of carbons with more than one chemical shift assignment and CT is the total number carbons in the molecule. Thus, the ambiguity score ranges from 0 (no ambiguous assignments) to 1 (all assignments are ambiguous). A red-blue scale visually indicates the level of ambiguity for each 13C: red being high ambiguity and dark blue being no ambiguity. An example using Adenosine (completely unambiguous) and NADP+ (with some ambiguity) is given in Figure S2. In this study, we only included library compounds with ambiguity scores less than 0.5 (Table 1). The current INETA database contains 1186 metabolites with 1957 13C spectra. Of these 1186 metabolites, 1085 had at least one spectrum with an ambiguity score less than or equal to 0.5, and 948 had at least one spectrum with an ambiguity score of 0.
4. Database Matching
We used three steps to match experimental networks to the INETA database. First, all single quantum chemical shifts in each network are compared with the INETA database. If the absolute value of the difference between a given network peak and database entry is less than the user defined parameter, CMT, then that database entry is considered in subsequent steps (Figure S3A). In the second step, a user-defined threshold NCMT defines the lowest number of chemical shift matches from the first step that are necessary to constitute a match. The default value for NCMT is 2. The third step involves consideration of the double quantum axis in order to determine 13C connectivity. The user defined DQMT is the threshold that is used to search the subset of the INETA database that passed the first two steps for matches in both the chemical shift and double quantum axes (Figure S3B).
At this point, there may be several candidate database matches to a particular experimental network. These candidates are ranked according to CM/CCB where CM is the number of matched connections (C-C bonds) between an experimental network and INETA candidate and CCB is the number of total C-C bonds in the INETA candidate.
The first 3 steps outlined above are largely automatic, given user-defined parameter values. The output from these first 3 steps is all the networks and possible database matches ranked as described above. These matches can be overlaid onto the experimental peak picked spectrum for validation. This important step allows for partial networks to be identified, as described below in the results. If no matches were found for a given network, the user has the option to visualize any match that passed the first automated chemical shift-matching step described above. These chemical shift matches are ranked according to the number of successful matches.
Results and Discussion
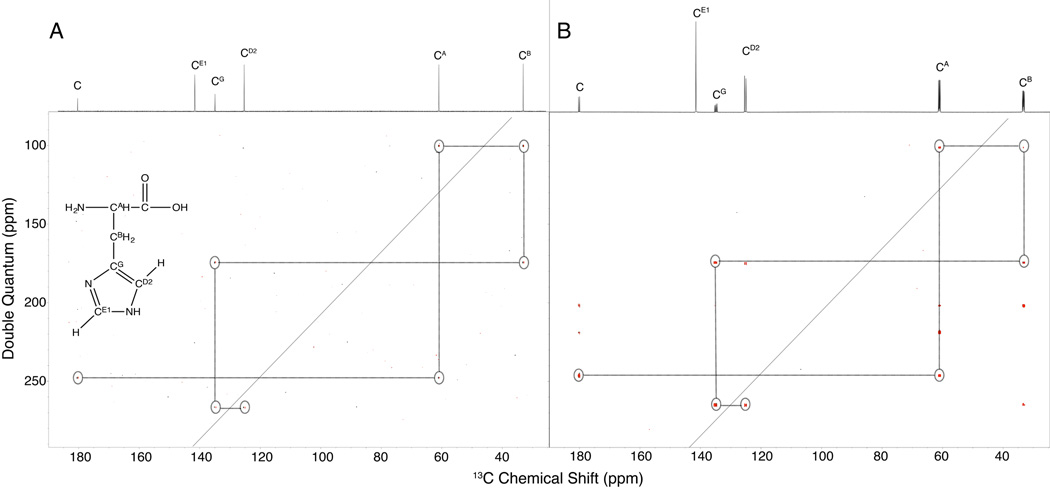
As described above, one of the major challenges with INADEQUATE at natural abundance is the low probability of adjacent 13C nuclei. Isotopic labeling significantly improves the situation, as shown in Figure 1. In Figure 1A we show our previously published results of an INADEQUATE spectrum recorded using 200 mM (1.1 mg) of histidine at natural abundance 13C using our specialized HTS 13C optimized NMR probe.15 This spectrum required about 48 hours and has relatively low signal-to-noise (S/N). Figure 1B shows an INADEQUATE spectrum collected with the same probe and spectrometer with 2 mM (100×lower concentration) uniformly 13C histidine (Cambridge Isotope Labs) in just 4 hours with much greater S/N.23 NMR probe mass sensitivity scales as the inverse of the diameter of the coils, so the highest mass sensitivity probes also have the smallest volumes.24 Larger volume 13C probes could be used in this sort of study, but in general they would provide lower mass sensitivity, and less efficient use of the 13C labeling.
Figure 1.
INADEQUATE of 200 mM natural abundance (A) and 2 mM 99% 13C-labeled histidine (B). This experiment provides direct one bond 13C-13C coupling information that allows the molecular backbone of histidine to be traced (A). Fully isotopically labeled compounds give significantly increased signal to noise but also results in additional coherences (resonances not circled) beyond the typical double quantum that are essentially artifacts of multiple interacting spin systems (B). Peaks in (B) that are not in the drawn network are extra coherences not seen at natural abundance.
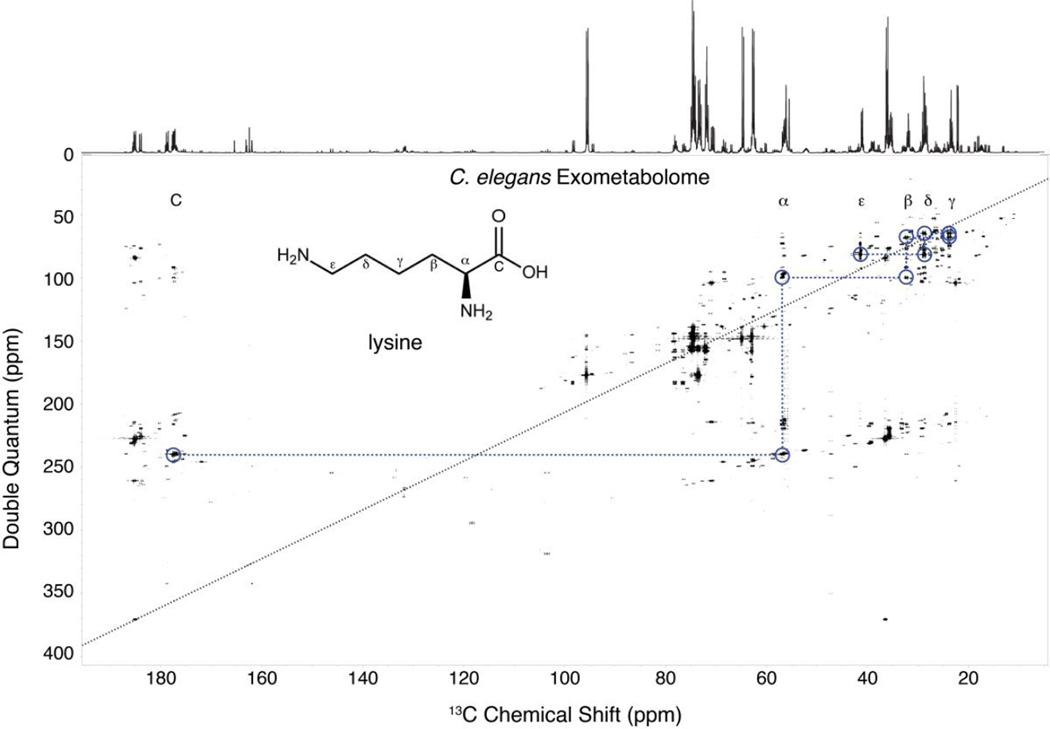
The 99% 13C labeling used in this study provides high overall sensitivity, but that comes at a cost of extra peaks. Markley and coworkers have shown that a 13C enrichment of 26% reduces long-range 13C couplings while maintaining high sensitivity,9 but to maximize this benefit of reduced long-range interactions one would ideally use 26% randomly labeled 13C glucose, as opposed to a 26% mixture of 99% 13C glucose and 74% natural abundance glucose. Figure 1B shows that uniform 13C-labeling results in additional correlations beyond the standard double quantum couplings shown in Figure 1A. Although these extra correlations complicate the spectrum, the rules of INADEQUATE (i.e. the double quantum frequency is the sum of the two correlated resonances) allow easy discrimination between the directly bonded double quantum correlations. Moreover, these other correlations provide additional information that can help in connecting fragments separated by heteroatoms or with weak INADEQUATE correlations. This additional information has not yet been implemented into INETA and has thus far been used to aid in the manual verification of compounds. Figure 2 shows an INADEQUATE spectrum of the C. elegans exometabolome, with the corresponding 1D 13C spectrum above. The double quantum diagonal is drawn in a black dashed black line, and the spin system for lysine is highlighted to illustrate the rich information content of the spectrum. An example of the endometabolome is given in Figure S4.
Figure 2.
INADEQUATE of C. elegans exometabolome. Above the INADEQUATE spectrum is the 1D 13C spectrum. Circled in blue is the 13C-13C trace of lysine. This connectivity information allows the simple identification of metabolites.
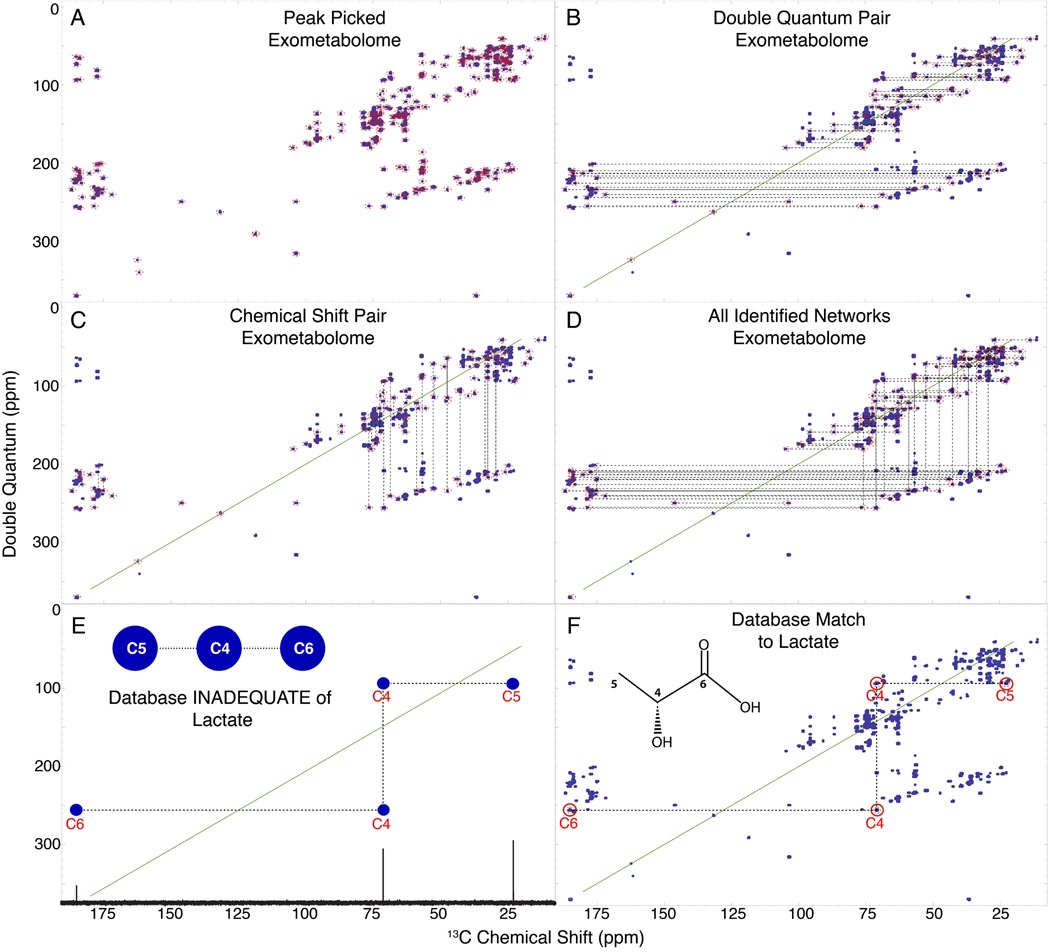
The INETA software was written to identify INADEQUATE networks and match them to compounds in the database. The entire process is shown in Figure 3 using parameters defined in Table 1. The first step is peak picking the experimental spectrum (Figure 3A). After peak picking we then identified all pairs of peaks that follow the rules of INADEQUATE (Figure 3B). Next, for each pair of INADEQUATE peaks, INETA identifies overlapping chemical shifts along the single quantum chemical shift dimension, resulting in vertically connected pairs as shown in Figure 3C. Finally, the vertical and horizontal correlations from Figure 3B and Figure 3C are connected into networks, which are shown for the C. elegans exometabolome in Figure 3D. These networks represent whole or partial carbon topology maps that can be matched to a database or analyzed de novo. Figure S5A-D outlines this process in the endometabolome.
Figure 3.
INETA uses the rules of INADEQUATE to connect peaks. Peaks are picked (A) and their double quantum pair is found (B). Peaks lacking a partner are not considered in the analysis. Vertical pairs are matched based on chemical shifts (C). Vertical and horizontal pairs are connected to form a network. All networks are shown in D. 1D 13C spectra and carbon connectivity information is downloaded from the BMRB and a 2D INADEQUATE in silico database is generated by adding the chemical shifts of two directly bonded carbons (E). The INADEQUATE of database matches or candidates can then be plotted onto the peakpicked experimental spectra (denoted by red circles above carbon numbers) to give positive identification (F).
Although reference INADEQUATE spectra are not readily available, we can easily create a computed INADEQUATE spectrum from an assigned 13C 1D spectrum. This is illustrated in Figure 3E for lactate. In the study presented here we only used compounds with an ambiguity score less than or equal to 0.5. Algorithms to utilize ambiguous data could expand the database matching and overall utility of INETA. Figure 3F shows lactate automatically mapped onto the peak picked spectrum by INETA. A red circle above the carbon numbers in the matched network indicates the area in which the peak for lactate should be found, based on BMRB values. This is particularly useful if not all the peaks in the network are correlated, either due to low S/N, overlap, threshold cutoffs, or carbons separated by heteroatoms.
Table 2 summarizes the networks and matched compounds that were found by INETA for both the endo- and the exometabolomes using parameters given in Table 1. Compounds with greater than 0.5 ambiguity in the database, such as sugars and some fatty acids, were not included in the analysis and thus not considered here. There were 53 networks found for the endometabolome and 49 compounds found for the exometabolome. Of the 53 networks that were found in the endometabolome, 20 were matched to the database resulting in 16 identified compounds. These included amino acids, carbohydrates, organic acids, and some fatty acid species. Of the 49 networks found in the exometabolome, 25 networks were matched to the database resulting in 20 identified compounds. As have been shown previously,25 the worm exudates are dominated by amino acids.
Table 2.
Number of networks found by software for the endo- and exometabolome.
| C. elegans Endometabolome (53 Networks*) | C. elegans Exometabolome (49 Networks*) | ||
|---|---|---|---|
| Alanine (1) | Lactic Acid (1) | Creatine (1) | Isoleucine (2) |
| Proline (1) | Valine (2) | Glutathione Oxidized (2) | Valine (1) |
| Arabitol (1) | Threonine (1) | Lysine (2) | Alanine (1) |
| Glucose 1,6-Bisphosphate (1) | 3-Hydroxybutyric Acid (1) | Glucoronate (1) | Allantoin (2) |
| Succinic Acid (2) | Stearic Acid (1) | Threonine (1) | Putrescine (1) |
| Propionic Acid (1) | Valeric Acid (3) | Glycine (1) | Succinic Acid (1) |
| Isoleucine (1) | Palmitic Acid (1) | Glutamine (1) | Lactic Acid (1) |
| N-Acetylglycine (1) | Histidine (1) | Proline (2) | Acetate (1) |
| Ornithine (1) | Uracil (1) | ||
| Serine (1) | Methionine (1) | ||
Number of networks found by INETA
Number of networks associated with each metabolite
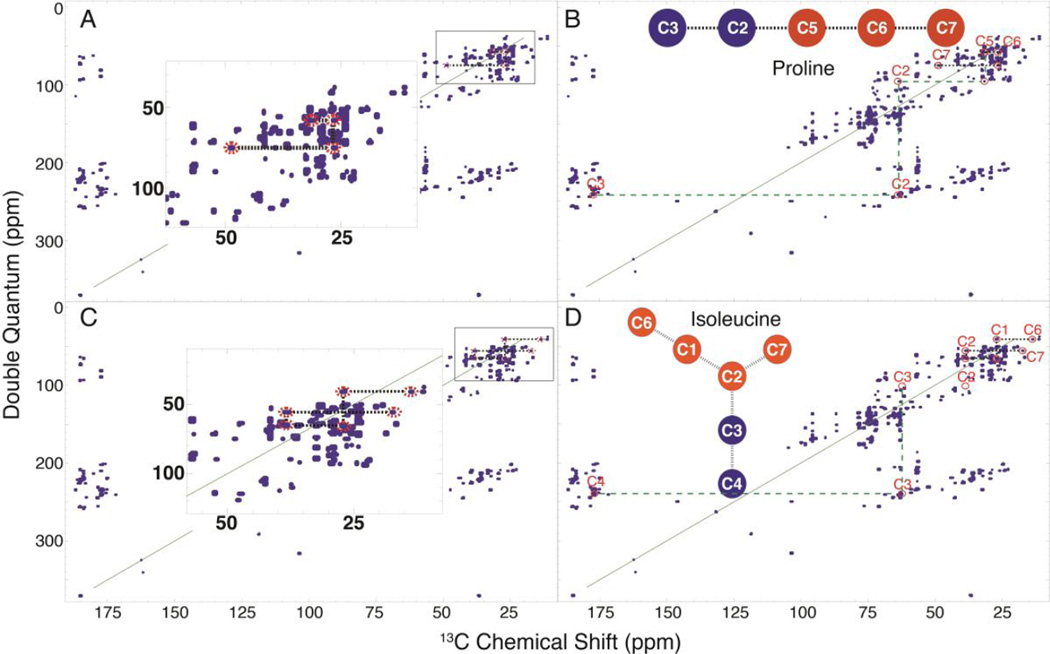
For a given set of INETA parameters, complete networks of some metabolites could not always be made (Figure 4A-D). Given these restrictions, INETA was still able to identify many of these compounds. Figure 4A shows a partial network of proline found by INETA. Though only part of the proline network was initially found (orange circles in backbone molecule), INETA still identified the network to be proline. The database-generated proline INADEQUATE spectrum (red circles below carbon numbers) was mapped by INETA onto the peak picked INADEQUATE spectrum and displayed where other proline peaks should be found (Figure 4B). To verify the identity, we simply traced the rest of the molecule (green lines).
Figure 4.
Partial INADEQUATE networks. For the INETA parameters used in this study, the network in (A) was incomplete. INETA was able to identify this network as proline. The in silico INADEQUATE database network of proline was mapped onto the experimental peak picked spectrum, and red circles below the carbon numbers indicate where other proline peaks should should be. We can thus manually validate the rest of the compound (denoted by green lines) (B). In other cases, due to low S/N of a specific molecule or incomplete labeling, not all crosspeaks will show up in the INADEQUATE, as shown in (C and D). INETA was able to match these partial networks with a missing correlation as isoleucine (D).
We have found that not all peaks of a particular compound will have the same intensity and may thus lie below thresholds or within the noise. This may be due to low concentrations, unusual values of 13C-13C J couplings, or incomplete (non uniform) labeling of metabolites. Generally, 13C-13C correlations in INADEQUATE spectra are symmetrical in their intensities; however, in some cases, pairs of correlations will not have the same intensity.23 For example, the INADEQUATE spectrum for isoleucine in Figure 4D is missing a correlation between C2 and C3, but INETA was still able to identify isoleucine based on the matches to other carbons (Figure 4D).
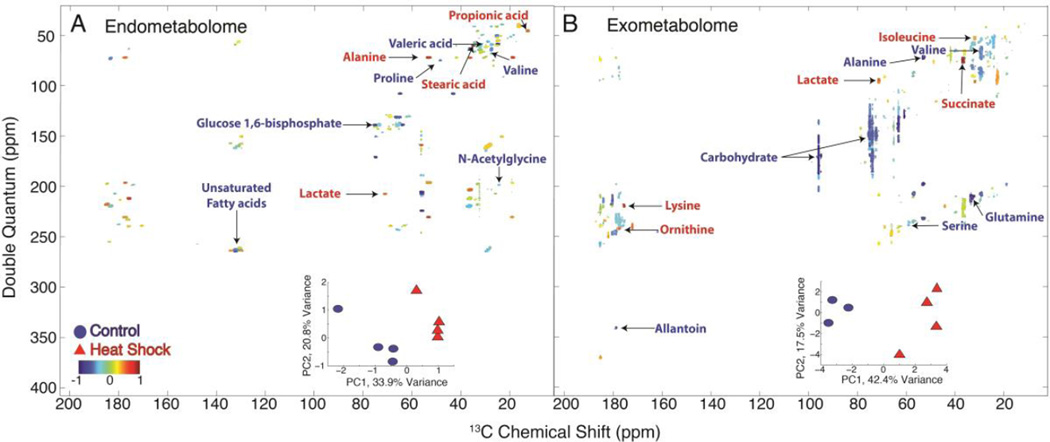
The INETA analysis described above can be done on a single INADEQUATE spectrum, but to obtain useful biological knowledge it is necessary to analyze multiple replicates and different conditions. Our laboratory previously published a method called HATS-PR to perform multivariate statistical analysis on 2D NMR data.16 In this study we obtained 4 replicates each of worms that had been heat-shocked vs. room temperature controls, and each replicate yields an endo- and exometabolome. PCA was performed on both to determine the metabolic differences between heat shock and control (Figure 5A and B and Table 3). In both cases we observe good separation along PC1 and determined the loadings along that axis, which were annotated using results from INETA. In the endometabolome (Figure 5A), lactate, alanine, and propionate were enhanced in the heat shocked worms, while proline, valine, N-acetyl glycine, other N-acetyl amino acids, as well as the unsaturated fatty acids species were enhanced in controls. Fatty acid melting points decrease with increasing degree of unsaturation, suggesting that worms regulate their fatty acid membranes with temperature. This phenomenon of decreased unsaturated fatty acids with increased temperatures has been observed in E. coli as well as other bacterial species.26,27
Figure 5.
PCA comparing control and heat shock endo- and the exometabolome. Scores plots are given as an inset within the loadings plots from PC1. Blue circles indicate control populations and red triangles indicate heat-shocked populations. Blue loadings denote resonances that loaded with the control and red loadings denote those that loaded with the heat shock populations. Compounds such as lactate and propionic acid loaded with the heat shock in the endometabolome where as compounds such as valine and proline loaded with the control worms (A). Overall, in the exometabolome (B), carbohydrates loaded more with the control while compounds such as lactate and succinate loaded with the heat shock.
Table 3.
Changes in metabolites found in the endo- and exometabolome under heat shock.
| Endometabolome Metabolites | Exometabolome Metabolites | ||
|---|---|---|---|
| Alanine | ↑ | Alanine | ↓ |
| Proline | ↓ | Proline | - |
| Arabitol | - | Lysine | ↑ |
| Glucose 1,6-Bisphosphate | ↓ | Glucoronate | - |
| Succinic Acid | - | Succinic Acid | ↑ |
| Propionic Acid | ↑ | Glycine | - |
| Isoleucine | - | Isoleucine | ↑ |
| N-Acetylglycine | ↓ | Glutathione Oxidized | - |
| Lactic Acid | ↑ | Lactic Acid | ↑ |
| Valine | ↓ | Valine | ↓ |
| Threonine | - | Threonine | - |
| 3-Hydroxybutyric Acid | - | Serine | ↓ |
| Stearic Acid | ↑ | Creatine | - |
| Valeric Acid | ↓ | Allantoin | ↓ |
| Palmitic Acid | - | Putrescine | - |
| Histidine | - | Glutamine | ↓ |
| Unsaturated Fatty Acids | ↓ | Ornithine | ↑ |
| Acetate | - | ||
| Uracil | - | ||
| Methionine | - | ||
| Carbohydrates | ↓ | ||
(↑) Metabolite increase under heat shock conditions
(↓) Metabolite decrease under heat shock conditions
(-) No Change
In the exometabolome, compounds such as succinate, isoleucine, lysine, ornithine, and lactate were enhanced in heat shock animals relative to control. Succinate is an intermediate in the TCA cycle and can donate electrons to the electron transport chain. It has been shown to be a signal of inflammation and stress.28,29 In addition lactate production has been shown to be a possible marker of stress in cells.30 Carbohydrates, alanine, valine, glutamine, serine, and allantoin were enhanced in controls. There is an overall increase in lactate, but a decrease in carbohydrates that may indicate high-energy production in the heat shock animals because the carbohydrates are being catabolized resulting in an increase in lactate both in the endo- and the exometabolome. In addition, we used PCA to determine the overall differences between the endo- and exometabolome (Figures S6 and S7). We observed an overall increase in carbohydrates, several amino acids, and metabolic products such as lactate and succinate in the exometabolome and an overall increase in fatty acids in the endometabolome. In a previous study of the exudates of C. elegans,25 NMR, HPLC, and GC-MS were used to identify metabolites in the worm exometabolome and also found that amino acids made up the majority of metabolites present in the exometabolome. The presence of these metabolites in the worm exudates are hypothesized to play a vital role in C. elegans chemical ecology and may even serve to interact with microbes in the environment.25
Conclusion
INADEQUATE is an ideal experiment for the direct structural elucidation of compounds by NMR. We show here that it is also useful in 13C-based metabolomics by providing information about the carbon skeleton of metabolites in mixtures. Mixture analysis is further simplified with the use of INETA that allows us to semi-automate the analysis of complex labeled mixtures by matching to a database library. The overall performance of INETA will improve as assigned 13C spectra are added to the BMRB or other databases.
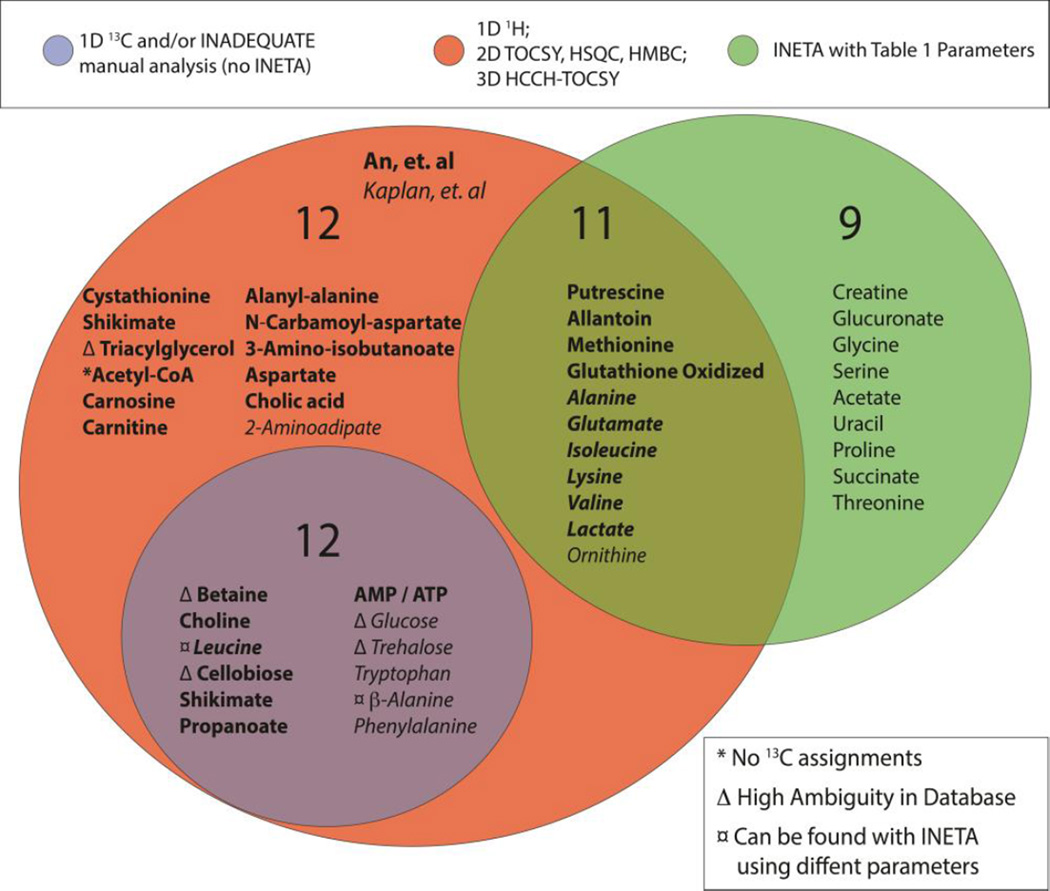
Figure 6 provides a comparison of INETA with other NMR based studies of the C. elegans exometabolome.25,31 Two previous studies used 1H-detected 1D, 2D TOCSY,25 2D 13C-HSQC, and 3D 13C-HSQC-TOCSY31 to identify a total of 35 metabolites in the exometabolome.25,31 Using INETA with the parameters defined in Table 1, we found 11 of the 35 metabolites from previous studies. What about the other 24? Six of the 24 missed compounds either do not have 13C assignments or have high ambiguity and were not used in the INETA library here. We then manually curated the INADEQUATE spectra and found 12 of the 24. Two of those (leucine and β-alanine) could be found by INETA after adjusting search parameters. Finally, INETA was able to identify 9 other metabolites not found in the previous studies (Figure 6).
Figure 6.
Venn diagram of metabolites found in C. elegans exometabolome using various techniques as previously described25,31 and using INETA. Using 1D, 2D, and 3D 1H NMR alone, Kaplan et. al25 and An et. al31 reported 35 compounds, 11 of which were also found using INETA with the parameters set in Table 1. Targeted manual analysis of the 1D 13C and INADEQAUTE data yielded an additional 12 metabolites.
Several networks were found but not matched to a database entry using INETA. Several strategies can be developed to identify metabolites from these networks. First, algorithms could be developed to match the INADEQUATE single quantum chemical shifts of unassigned networks to database entries. There is a good chance that the INADEQUATE data could then be used to assign the unassigned spectra. Second, algorithms could be developed to automatically adjust INETA thresholds to increase the number of full networks for database matching. This may be especially useful in the detection of peaks in heavily overlapped regions. For example, manual adjustment of parameters allowed us to add the two metabolites, leucine and β-alanine discussed above.
Perhaps the most powerful advantage of our 13C INADEQUATE approach will be a combination of traditional natural products analysis, computational chemistry, and metabolomics.32 In this study, only about half of the INETA networks were matched to database entries. The unassigned networks have precise 13C chemical shift and carbon topology information, so one could envision approaches in which calculations of 13C chemical shifts are used in combination with database matching to obtain starting fragments. Quantum mechanical calculations of 13C chemical shifts are quite robust,33 and with computer clusters can be done on a large scale. Using this approach, INADEQUATE data on metabolomics mixtures would be an outstanding way to discover unknown molecules.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Bill Brey, Jerris Hooker, Vijay Ramaswamy, and colleagues at Agilent Technologies for the HTS NMR probe. Jim Rocca provided support in the NMR data collection. Tom Mareci provided helpful information on INADEQUATE experiments. The data obtained in this study (supported by the NIH Common Fund Grant U24 DK097209) were deposited into the NIH Common Fund’s Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) website, http://www.metabolomicsworkbench.org. Additional funding for this study was from the NIH R01EB009772 to A.S.E. NMR data were collected at the National High Magnetic Field Laboratory’s AMRIS Facility, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490 and the State of Florida.
Footnotes
ASSOCIATED CONTENT
Supporting Information: Supporting figures and tables are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
C.S.C and A.S.E planned and designed the experiments and workflow. MATHEMATICA scripts are available upon request and were written by C.P. with substantial contributions from C.S.C. C.S.C. and R.A. planned and executed the worm labeling and sample preparation; C.S.C. collected and analyzed the NMR data. C.S.C. and A.S.E. wrote the paper with significant contributions from all authors.
The authors declare no competing financial interests.
REFERENCES
- 1.Choi YH, Kim HK, Hazekamp A, Erkelens C, Lefeber AWM, Verpoorte R. Journal of natural products. 2004;67:953. doi: 10.1021/np049919c. [DOI] [PubMed] [Google Scholar]
- 2.Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J, Blancher C, Gauguier D, Lindon JC, Holmes E, Nicholson J. Anal. Chem. 2005;77:1282. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- 3.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Anal. Chem. 2006;78:4430. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 4.Clendinen CS, Lee-McMullen B, Williams CM, Stupp GS, Vandenborne K, Hahn DA, Walter GA, Edison AS. Anal. Chem. 2014;86:9242. doi: 10.1021/ac502346h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingol K, Zhang F, Bruschweiler-Li L, Bruschweiler R. Anal. Chem. 2012;84:9395. doi: 10.1021/ac302197e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bax A, Freeman R, Frenkiel TA. Journal of the American Chemical Society. 1981;103:2102. [Google Scholar]
- 7.Sørensen OW, Freeman R, Frenkiel T, Mareci TH, Schuck R. Journal of Magnetic Resonance (1969) 1982;46:180. [Google Scholar]
- 8.Buddrus J, Bauer H. Angew Chem Int Edit. 1987 [Google Scholar]
- 9.Oh BH, Westler WM, Darba P, Markley JL. Science. 1988;240:908. doi: 10.1126/science.3129784. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi K, Sun Z-YJ, Wagner G. Journal of the American Chemical Society. 2008;130:17210. doi: 10.1021/ja806956p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sumner SCJ. Toxicological Sciences. 2003;75:260. doi: 10.1093/toxsci/kfg191. [DOI] [PubMed] [Google Scholar]
- 12.Garner CE, Sumner SCJ, Davis JG, Burgess JP, Yueh Y, Demeter J, Zhan Q, Valentine J, Jeffcoat AR, Burka LT. Toxicology and Applied Pharmacology. 2006;215:23. doi: 10.1016/j.taap.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Fennell TR. Toxicological Sciences. 2006;93:256. doi: 10.1093/toxsci/kfl069. [DOI] [PubMed] [Google Scholar]
- 14.Sumner SCJ, Stedman DB, Clarke DO, Welsch F, Fennell TR. Chem. Res. Toxicol. 1992;5:553. doi: 10.1021/tx00028a015. [DOI] [PubMed] [Google Scholar]
- 15.Ramaswamy V, Hooker JW, Withers RS, Nast RE, Brey WW, Edison AS. J Magn Reson. 2013;235C:58. doi: 10.1016/j.jmr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinette SL, Ajredini R, Rasheed H, Zeinomar A, Schroeder FC, Dossey AT, Edison AS. Anal. Chem. 2011;83:1649. doi: 10.1021/ac102724x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. Nucleic acids research. 2008;36:D402. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markley JL, Ulrich EL, Berman HM, Henrick K, Nakamura H, Akutsu H. Journal of biomolecular NMR. 2008;40:153. doi: 10.1007/s10858-008-9221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stupp GS, Clendinen CS, Ajredini R, Szewc MA, Garrett T, Menger RF, Yost RA, Beecher C, Edison AS. Anal. Chem. 2013;85:11858. doi: 10.1021/ac4025413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Journal of biomolecular NMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Dieterle F, Ross A, Schlotterbeck G, Senn H. Anal. Chem. 2006;78:4281. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 22.Wolfram Research, Inc. Mathematica. 2012. [Google Scholar]
- 23.Bain AD, Hughes DW, Anand CK, Nie Z, Robertson VJ. Magnetic resonance in chemistry : MRC. 2010;48:630. doi: 10.1002/mrc.2639. [DOI] [PubMed] [Google Scholar]
- 24.Ramaswamy V, Hooker JW, Withers RS, Nast RE, Edison AS, Brey WW. eMagRes. 2013;2:215. doi: 10.1016/j.jmr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan F, Badri DV, Zachariah C, Ajredini R, Sandoval FJ, Roje S, Levine LH, Zhang F, Robinette SL, Alborn HT, Zhao W, Stadler M, Nimalendran R, Dossey AT, Bruschweiler R, Vivanco JM, Edison AS. Journal of chemical ecology. 2009;35:878. doi: 10.1007/s10886-009-9670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mejía R, Gómez-Eichelmann MC, Fernández M. TBMB. 1999;47:835. doi: 10.1080/15216549900201923. [DOI] [PubMed] [Google Scholar]
- 27.Álvarez-Ordóñez A, Fernández A, López M, Arenas R, Bernardo A. International Journal of Food Microbiology. 2008;123:212. doi: 10.1016/j.ijfoodmicro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Mills E, O'Neill L. Trends in cell biology. 2014 doi: 10.1016/j.tcb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 29.McGettrick AF, O'Neill LAJ. Journal of Biological Chemistry. 2013;288:22893. doi: 10.1074/jbc.R113.486464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limonciel A, Aschauer L, Wilmes A, Prajczer S. Toxicology in Vitro. 2011 doi: 10.1016/j.tiv.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 31.An YJ, Xu WJ, Jin X, Wen H, Kim H, Lee J, Park S. ACS chemical biology. 2012;7:2012. doi: 10.1021/cb3004226. [DOI] [PubMed] [Google Scholar]
- 32.Robinette SL, Bruschweiler R, Schroeder FC, Edison AS. Accounts of chemical research. 2011 [Google Scholar]
- 33.Wang B, Dossey AT, Walse SS, Edison AS, Merz KMJ. Journal of natural products. 2009;72:709. doi: 10.1021/np8005056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.