Abstract
Early life nutritional intervention causes adult onset insulin resistance and obesity in rats. Thyroid hormone receptor (TR) in turn transcriptionally enhances skeletal muscle Glut4 expression. We tested the hypothesis that reduced circulating TSH and T4 concentrations encountered in postnatal (PN4-PN24) high carbohydrate (HC) milk formula-fed versus the mother-fed controls (MF) would epigenetically interfere with TR induction of adult (100d) male rat skeletal muscle Glut4 expression, thereby providing a molecular mechanism mediating insulin resistance. We observed increased DNA methylation of the CpG island with enhanced recruitment of Dnmt3a, Dnmt3b and MeCP2 in the glut4 promoter region along with reduced acetylation of histone (H)2A.Z and H4 particularly at the H4.lysine (K)16 residue, which was predominantly mediated by histone deacetylase 4 (HDAC4). This was followed by enhanced recruitment of heterochromatin protein 1β to the glut4 promoter with increased Suv39H1 methylase concentrations. These changes reduced TR binding of the T3 response element of the glut4 gene (TREs; −473 to −450 bp) detected qualitatively in-vivo (EMSA) and quantified ex-vivo (ChIP). In addition, the recruitment of steroid receptor co-activator and CREB-binding protein to the glut4 promoter-protein complex was reduced. Co-immunoprecipitation experiments confirmed interaction between TR and CBP to be reduced and HDAC4 to be enhanced in HC versus MF groups. These molecular changes were associated with diminished skeletal muscle Glut4 mRNA and protein concentrations. We conclude that early postnatal exposure to HC diet epigenetically reduced TR induction of adult male skeletal muscle Glut4 expression, uncovering novel molecular mechanisms contributing to adult insulin resistance and obesity.
Keywords: Overnutrition programming, DNA methylation, histone code modifications, histone deacetylase, DNA methylase, heterochromatin, insulin resistance, obesity
Introduction
Early life nutritional perturbations cause long term consequences contributing towards the world-wide epidemic of insulin resistance, diabetes mellitus and more recently obesity with concomitant complications [1–4]. Our group has previously investigated the effect of reduced calories during prenatal and postnatal period of rat development and observed adult onset consequences such as glucose intolerance [1] and obesity [2] particularly in the male offspring. At the other end of the spectrum, early prenatal and postnatal exposure to a high fat diet caused insulin resistance and adult-onset obesity [3]. Similarly, early postnatal exposure to a high carbohydrate milk formula was also observed to cause adult onset insulin resistance and obesity in rat males [4]. Thus, overnutriton during early life sets the stage for predetermining the adult phenotype of insulin resistance with obesity.
Skeletal muscle is the main tissue that is responsible for the majority of insulin responsive glucose uptake and metabolism [5]. Perturbations in skeletal muscle metabolic pathways play a major role in producing total body insulin resistance [6]. One such mechanism is the insulin responsive glucose transporter Glut4, which can be perturbed transcriptionally and post-translationally, i.e. defective insulin responsive translocation to the plasma membrane [7, 8]. Genetically manipulated mouse models where skeletal muscle Glut4 was specifically targeted demonstrated that its absence gave rise to insulin resistance and obesity [13].
Thyroid hormones (T4/T3) which are perturbed in conditions of obesity and diabetes mellitus [9] stimulates basal and insulin mediated rat skeletal muscle glucose uptake [15,16]. This stimulatory effect is predominantly due to its action on skeletal muscle Glut4 expression [10, 11]. Such an action is mediated by the binding of the thyroid hormone receptor (TR), a member of the nuclear steroid hormone receptor superfamily, to the thyroid responsive element (TRE) found in the 5′-flanking region of the glut4 gene [12]. TRs function as monomers, homodimers or heterodimers with retinoid X receptor (RXR) and modulate transcription activity (repression or activation) by interacting with co-repressors and co-activators, which associate with TR in the absence or presence of T3, respectively. Such an interaction is modified by heterodimerization with RXR or in the presence of the CREB-binding protein (CBP), a histone acetyl-transferase (HAT), and/or steroid co-activator (SRC1) that interact with the thyroid hormone receptor nuclear protein-DNA complex.
While prior studies have shown the in-vitro interaction between thyroid hormone receptor and the glut4 gene, the role of this interaction under in-vivo conditions of overnutrition has remained controversial and thereby elusive. Furthermore this thyroid hormone receptor-glut4 gene interaction in early exposure to a high carbohydrate diet that is only limited to the suckling phase of development, with long term consequences of insulin resistance and obesity in the adult male [4,13], has never been investigated. The molecular mechanisms whereby long term phenotypic effects result as a consequence of early life nutritional manipulation have been assigned to epigenetic influences. More specifically DNA methylation of CpG islands within promoter regions of genes and concomitant histone code modifications have been observed to play a role in mediating such changes in gene expression that ultimately affect the adult phenotype [3]. Similar epigenetic modifications have been previously reported by us in the case of skeletal muscle glut4 gene expression regulated by MyoD and MEF2 nuclear protein-DNA binding when the offspring was exposed to prenatal and postnatal caloric restriction [7].
Based on this accumulated information, we hypothesized that early postnatal exposure to a high carbohydrate diet would modify DNA methylation and the histone code thereby interfering with thyroid hormone receptor induction of the adult male rat skeletal muscle glut4 gene expression. To test this hypothesis, we engaged the previously described gastrostomy fed rat model where a high carbohydrate diet exposure occurred only during the suckling period extending from PN4 to PN24 after which the offspring was weaned to a regular chow diet until adult life [4,13]. At 100 days of age, we observed novel perturbations in both DNA methylation and histone acetylation that interfered with the induction of skeletal muscle glut4 gene expression by thyroid hormones. The circulating thyroxine (T4) concentrations were reduced only during early life at 12 days of age, displaying normalcy in the adult. These findings support high carbohydrate diet induced T4 - thyroid hormone receptor programming of skeletal muscle glut4 gene expression.
Methods and Materials
Materials
Oligonucleotides - Synthetic oligonucleotides (Retrogen Inc., Carlsbad, CA; Integrated DNA Tech. Inc., San Diego, CA) were used in these experiments. Double-stranded oligonucleotides were generated by annealing synthetic oligonucleotides with respective complementary sequence. Antibodies - Rabbit polyclonal anti-HP1β (D-15): sc-10217, anti-HDAC4, anti-HDAC5, anti-Dnmt1, anti-Dnmt3a, anti-Dnmt3b, anti-dimethyl-histone H3 (Lys9), anti-thyroid hormone receptor (alpha 1+2) (ab1131) and monoclonal anti-thyroid hormone receptor (alpha 1 and beta 1) (ab2743), anti-H2A.Z and anti-SRC1 antibodies were purchased from Abcam Inc. (Cambridge, MA) and anti-CBP was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-acetyl-histone H3, anti-acetyl-histone H4, anti-acetyl-histone H4 (Lys5), anti-acetyl-histone H4 (Lys8), anti-acetyl-histone H4 (Lys16), anti-acetyl-histone H4 (Lys12) and anti-MeCP2 were purchased from Upstate Biotechnology (Lake Placid, NY). Anti-polymerase II antibody was from Active Motif (Carlsbad, CA). Horseradish peroxidase linked anti-rabbit and anti-mouse IgGs were from Amersham Biosciences (Piscataway, NJ).
Methods
In-vitro C2C12 cell experiments
Murine skeletal myotubular C2C12 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM glutamine, sodium pyruvate (110 mg/ml), penicillin (100 units/ml), streptomycin (100 units/ml), 4.5% glucose media with 10% FBS. After achieving 90% confluence, these cells were washed with PBS and re-plated in the same DMEM described above but with charcoal-stripped 4% FBS for 24h. Then the cells were treated with 10−7M concentrations of TSH, T3 or T4 (Sigma, St. Louis, MO) in the same charcoal-stripped 4% FBS-DMEM for 6h at 37°C.
In-vivo studies: Animal Model
All animal protocols employed in this study were approved by the Institutional Animal Care and Use Committee of the University at Buffalo, State University of New York (PI: MSP). Pregnant Sprague-Dawley rats were obtained from Zivic Miller Laboratories (Zellenople, PA), maintained under 12:12 light:dark cycles and had access to a standard rodent laboratory diet (Harlan-Teklad, Madison, WI) and water ad libitum. Newborn pups that were born to these rats were pooled and randomly assigned to each nursing dam (10 pups/dam) and left so until postnatal day 4. On postnatal day 4, pups were assigned randomly to control and experimental groups. In the mother-fed (MF) control group, pups were reared by their nursing mothers receiving rat milk (caloric distribution: 8% carbohydrate, 68% fat and 24% protein) whereas pups in the high carbohydrate (HC) experimental group were artificially reared on a HC milk formula (caloric distribution: 56% carbohydrate, 24% protein, and 20% fat) [13]. The artificial rearing technique employed in this study has previously been described in detail [14] and the need for artificial rearing on rat milk as an additional group was eliminated, since this group was no different from the pups reared by nursing dams (MF) [4, 15, 16]. Animals in both groups (HC and MF) were weaned on postnatal day 24 and given ad libitum access to standard rodent laboratory diet (caloric distribution: 61% carbohydrate, 4% fat, 16% protein) and water. Each rat was housed individually and kept at a constant temperature (23°C) on fixed (12:12-h) artificial dark:light cycles until postnatal 100d.
Serum TSH, T4 and T3 measurements
Blood was collected from the 12d and 100d old rats and the serum separated and stored at −70°C. Thyroid stimulating hormone (TSH), triiodothyronine (T3) and thyroxine (T4) concentrations were quantified by the rat thyroid hormone panel kit using rat standards, species specific antibodies (Milliplex Map Rat Thyroid Hormone Immunoassay, EMD Millipore Co., Billerica, MA) and employing the Bioplex 200 system (Bio-Rad Laboratories Hercules, CA). The values were calculated based upon generated standard curves.
Skeletal muscle preparation
Hind limb skeletal muscle was rapidly separated from surrounding tissues, quickly snap frozen in liquid nitrogen, and stored at −70°C as previously described [8]. Skeletal muscle was powdered under liquid nitrogen prior to use for varying extractions and assays.
Reverse-transcription and quantitative real time PCR
RNA isolation and real-time PCR - RNA was isolated using RNeasy (Qiagen, cat #74106). cDNAs were generated by reverse transcription using oligo (dT) and SuperScript III reverse transcriptase (Invitrogen). Real-time RT-PCR was performed with SYBR Green Master Mix (Life technologies, Carlsbad, CA). RT-PCR reactions were performed in triplicate with glyceraldehyde-3-phosphate dehydrogenase (gapdh) serving as the internal control on a StepOnePlus real-time PCR System (Applied Biosystems, Foster City, CA), using forward primer 5′ccgaaagagtctaaaagcgcct3′ and reverse primer 5′cgtttctcatccttcagctca3′ that encompass exons 5–6 of the rat glut4 gene [8]. Amplification conditions consisted of initial 12 min activation at 95°C followed by 40 cycles of denaturation at 95°C for 30s, annealing at 58°C for 30s, and extension at 72°C for 30s. Relative quantification of amplified DNA was performed using the comparative critical threshold (CT) method.
Electromobility Shift Assay (EMSA)
The primers encompassing a TRE binding site were employed to amplify a ~100 bp sized DNA fragment that was then end-labeled with [γ32P]-ATP (Perkin Elmer Life and Analytical Sciences Inc., Boston, MA) and T4 polynucleotide kinase. To amplify the TRE probe the TRE-sense 5′-ccatccgggttacttcggggcattgttttc-3′ and TRE-anti-sense 5′-gaaaacaatgccccgaagtaacccggatgg-3′ primers were used. Approximately 6 fmole (specific activity=3000 Ci/mmol) of the labeled DNA probe was added to 5 μg of skeletal muscle nuclear extract in a final volume of 20 μl containing 1 μg of poly (dI-dC), 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.5 mM EDTA, 1 mM MgCl2, 4% glycerol, 1 mM dithiothreitol and incubated for 15 min at room temperature. Competition occurred in the presence of 50 to 500-fold excess of unlabeled DNA probe. Subsequently the DNA-protein complex was separated from unbound DNA by electrophoresis through a 5% non-denaturing polyacrylamide gel in a 90 mM Tris borate, 2 mM EDTA buffer. The gels were dried and subjected to autoradiography [17]. Super shift experiments consisted of additional incubation with the antibodies targeting TR (α1+2 and α1+β1) (Abcam, Cambridge, MA) for 2 hrs at room temperature and then left over night at 4°C.
DNA methylation assay
Five μg of skeletal muscle genomic DNA in a 40 μl reaction mix was extracted and subjected to MspI or HpaII digestion. Southern blot analysis was performed with the digested DNA after electrophoresing in a 1.5% agarose-gel and transferring by capillary blotting to nylon membranes (Hybond-N; Amersham, Piscataway, NJ). The blots containing DNA were UV-cross-linked, prehybridized for 2h at 42°C, and subsequently hybridized to a 32P-labeled rat glut4 DNA fragment as the probe in 0.5 M sodium phosphate (pH 7.2), 1 mM EDTA, 1% bovine serum albumin (fraction V), 7% sodium dodecyl sulfate (SDS) and 15% formamide. Inter-lane loading variability of Southern blots was standardized by using 32P-labeled rat β-actin probe. The β-actin probe primers were as follows: forward - 5′-acctgacagactacctcatg-3′ and reverse - 5′-taacagtccgcctagaagca-3′. The glut4 probe primers were: forward - 5′-aggtcgtgccctctcaact-3′ and reverse - 5′-cgaagtaacccggaatggct-3′. The blots were then washed in 40 mM sodium phosphate (pH 7.2) solution containing 1% SDS and 1 mM EDTA at 42°C twice for 30 min each. These washed blots were exposed to Phosphorimager screens. The results were expressed as a ratio between glut4 and β-actin Phosphorimager captured values (Fig. 1D).
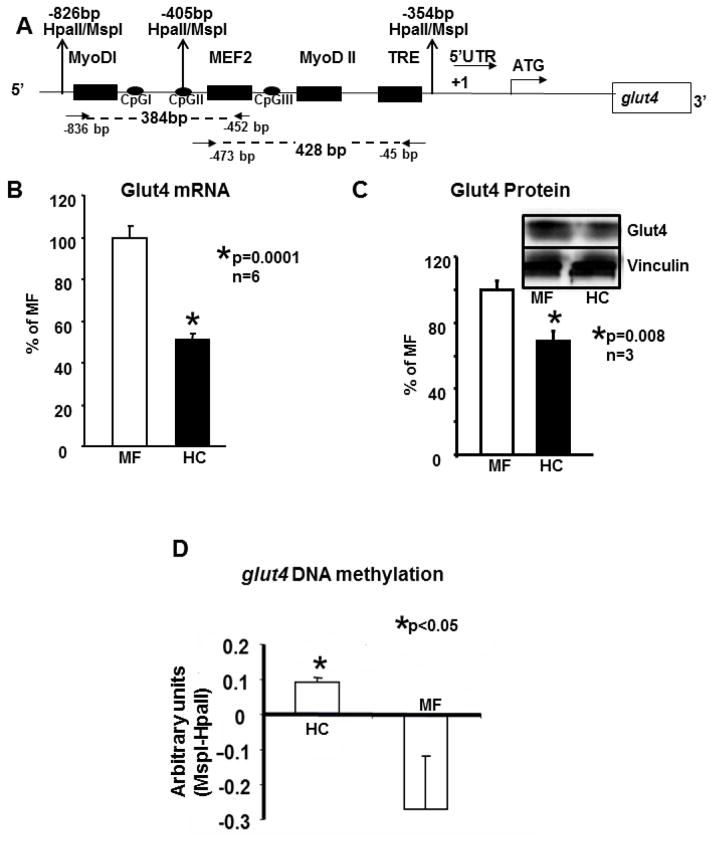
Figure 1.
A. Schematic representation of the one kb upstream region of the rat glut4 gene cloned in a pGL3 vector.
One kb of the rat glut4 promoter region is depicted. This region contains the MyoD-I, MyoD-II, MEF2 and the TRE DNA binding consensus elements 5′- to the transcriptional (+1) and translational (ATG) start sites. In addition the three CpG sites, I, II and III and the MspI/HpaII restriction enzyme sites are shown. Arrows show the location of forward and reverse primers employed to generate the 384 bp and 428 bp amplification DNA fragments (shown as dotted lines) for chromatin immunoprecipitation assays. B. Skeletal muscle Glut4 mRNA. In 100 day old skeletal muscle of early postnatal mother fed (MF) and high carbohydrate (HC) fed male rats, RT-qPCR assessment of Glut4 mRNA employing β-actin as the control gene. The relative expression of Glut4 is shown as a percent of MF value depicted as 100%. Data are shown as mean ± SEM of triplicate determinations in an n=6 samples for each group, *p<0.0001 between the two groups. C. Skeletal muscle Glut4 protein. The inset demonstrates representative Western blots showing Glut4 (above) and vinculin (internal control, below) protein bands in MF and HC groups. Quantification of 100d old skeletal muscle Glut4 protein concentrations as a ratio to vinculin is depicted as a percent of MF. Mean±SEM values are shown in an n=3 for each group, *p=0.008. D. Global methylation of rat skeletal muscle glut4 promoter around MyoDI-TRE region and difference in quantification of the glut4 bands. Southern blot analysis of MspI or HpaII digested genomic DNA obtained from the 100 day old skeletal muscle of MF and HC groups was quantified (after expression as a ratio to β-actin loading control) as a difference between corresponding MspI and HpaII digestions. Data is shown as Mean±SEM for an n=6 each, *p<0.05.
Ex-vivo studies - Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described previously [7] with some modifications. Powdered skeletal muscle was fixed in 1% formaldehyde for 45 min at room temperature. The tissue pellet was re-suspended in cell lysis buffer (5 mM Pipes [KOH] pH 8.0, 85 mM KCl, 0.5% NP-40) containing protease inhibitors (100 mM PMSF at 1:100 dilution in ethanol, aprotinin at 10 mg/ml concentration diluted to 1:1000 in 0.01M HEPES, pH 8.0 and leupeptin at 10 mg/ml diluted to 1:1000 in water) and homogenized with an Omni tissue homogenizer, 115V (Omni International, Inc., Marietta, GA). The separated nuclei were lysed in nuclear lysis buffer (50 mM Tris pH 8.1, 10 mM EDTA, 1% SDS) containing protease inhibitors. The resultant chromatin was sonicated (Fisher Scientific model 100 Sonic Dismembrator, Pittsburgh, PA) on ice with 20 pulses for 15 sec each at setting 4 with a 1 min rest interval in between pulses. The average length of sonicated chromatin was determined by resolving on a 1.5% agarose gel and found to be ~500 bp. The sample was then centrifuged at 4°C (10 min at 14,000 rpm) to remove cell debris from the crude chromatin lysate.
Ten percent of the lysate was used as the input control for PCR. Fifty μg (at OD260) of sheared chromatin was added to a final volume of 500 μl of the immunoprecipitation (IP) dilution buffer and pre-cleared with 25 μl of salmon sperm DNA/protein A agarose slurry (Upstate Biotechnology, Lake Placid, NY) at 4°C for 30 min. The pre-cleared chromatin was subsequently incubated overnight on a nutator with 2 μg of a primary polyclonal antibody at 4°C. Fifty μl of pre- blocked Protein A-agarose beads were then added to the pre-cleared chromatin for 2 hrs, followed by centrifugation at 14,000 rpm for 3 min at room temperature. The pellet was washed twice with 0.4 ml of a wash buffer A (50 mM HEPES pH 8.0, 0.1% SDS, 1% Triton X-100, 0.1% deoxycholate, 1 mM EDTA, 140 mM NaCl) followed by two washes with 0.4 ml each of wash buffer B (50 mM HEPES pH 8.0, 0.1% SDS, 1% Triton X-100, 0.5% deoxycholate, 1 mM EDTA, 500 mM NaCl), then LiCl containing buffer (20 mM Tris-HCl pH 8.0, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA, 250 mM LiCl and TE [10 mM Tris-Cl, pH 8.0, 1 mM EDTA]). Antibody/protein/DNA complexes were eluted from Protein A agarose beads by adding 100 μl of the elution buffer (50 mM Tris-HCl pH 8.0, 1 mM EDTA and 1% SDS) followed by 150 μl TE buffer with 0.67% SDS at 65°C for 10 min each. The combined supernatants were incubated overnight at 65°C after addition of 1 μl of RNAse A (10 mg/ml) to reverse the formaldehyde cross-links. The complex was then treated with proteinase K at 55°C for 2 hours, extracted once with phenol/chloroform/isoamyl alcohol and once with chloroform/isoamyl alcohol and subsequently precipitated overnight at −20°C with ethanol in the presence of 5 μg of tRNA and 5 μg of glycogen. The DNA concentration in the complex was determined by a Dip Stick kit (Invitrogen Inc., Carlsbad, CA), and ~2–4 ng of immunoprecipitated DNA was used as a template in each PCR reaction.
PCR consisted of 25 μl of the reaction mix containing 8 μl of the DNA template, 0.75 μM of forward primer, 0.25 μM of reverse primer, 10 X PCR buffer with 1.5 mM MgCl2, 200 μM dNTPs, and 0.25 μl of Taq polymerase (5 units/μl). The PCR amplification in a T3 thermocycler (Biometra, Goettingen, Germany) for the glut4 promoter regions from −836 bp to −452 bp (corresponding to 384 bp PCR product) and from −473 bp to −45 bp (corresponding to the 428 bp PCR product) was performed initially at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30s, annealing at 55°C for 30s and extension at 72°C for 4 min. PCR employed for gapdh (amplification product targeted at the translational start site 1 to 231 bp spans exons 1 to 4) consisted of 25 μl of the reaction mix containing 2 μl of the DNA template, 0.5 μM of forward primer, 0.5 μM of reverse primer, 10 X PCR buffer with 1.5 mM MgCl2, 200 μM dNTPs and 0.25 μl of Taq polymerase (5 units/μl), which was subjected to amplification in a T3-thermocycler. PCR was performed as described above except for an annealing temperature of 60°C over 60s [7, 17]. qPCR as described above was also performed for validation and accurate quantification of ChIP. The primers used in these PCRs are listed in Table 1. Other controls consisted of a 248 bp amplification product ~1 kb upstream from MyoD-MEF2 binding region of rat glut4 (forward primer spanning −2172 bp to −2149 bp: 5′-accaagtgtaatcccaggcctcat-3′ and reverse primer spanning −1947 bp to −1924 bp: 5′-ttgttgccctgtggtcaagtttgg-3′) in the presence or absence of anti-RNA polymerase II IgG (+) and a non-specific IgG (−).
Table 1.
Primers used in ChIP assays
| Primers | DNA Sequence | Position |
|---|---|---|
| glut4-384 bp -For | 5−-ccaaaacaggagctgactctg-3− | −836 bp |
| glut4-384 bp–Rev | 5−-aatggctatttttagctccca-3− | −452 bp |
| glut4-428 bp-For | 5−-gtgggagctaaaaatagccatt-3− | −473 bp |
| Glut4-428 bp) Rev | 5−-cacctcctcactcccgcccc -3− | −45 bp |
| gapdh-For | 5−-ccggaattcgaaggtcggtgtcaacggatttgg -3− | +1 bp |
| gapdh-Rev | 5−-cacacctgcagcctggaagatggtgatgggtttcc -3− | +230 bp |
Western blot analysis
Fifty μg of the extracted nuclear protein fraction (per the nuclear extraction kit manufacturer’s instructions; Active Motif, Carlsbad, CA) was solubilized in 50 mM Tris pH 6.8, containing 2% SDS and the protein concentration determined by the Bio-Rad dye binding assay [18] prior to undertaking Western blots as previously described [17].
Co-immunoprecipitation
Hundred μg of nuclear protein was pre-cleared with 40μl of 50% Protein A-agarose slurry (1:1 dilution) (Pierce Biotechnology, Rockford, IL) in a lysis buffer consisting of 25 mM Tris (pH 8.0), 100 mM NaCl, 10% glycerol, Nonidet P-40, 1.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 20 μg/ml leupeptin and 1 μg/ml pepstatin A at 4°C (all procedures were carried out in the cold unless stated otherwise) by gentle agitation for 30 min. The samples were centrifuged (2000 rpm, 2 min) and 10 μl of anti-TR (α1+2) antibody was added and incubation carried out overnight with gentle agitation. Fifty μl of the protein A-agarose slurry (1:1 dilution) was added and incubation was continued for 2h. Protein A-agarose beads were washed twice for 5 min each in 500 μl of the lysis buffer and then boiled for 5 min in SDS sample buffer with 100 mM DTT. The proteins were separated on 4–15% SDS/PAGE and then processed for Western blotting [17]. The membranes were probed with 1:100 dilution of the anti-CBP, 1 μg/ml of anti-HDAC4, or 1:200 dilution of anti- HDAC5 antibodies.
Data Analysis
All data were calculated as a percent of control with the MF group being normalized to a mean of 100%. Data are depicted as mean ± S.E. Differences between the two groups were detected by the Student’s t test after ensuring normality of data distribution. Significance was assigned when the p value was ≤0.05.
Results
Table 2 demonstrates the serum concentrations of TSH, T4 and T3 concentrations in 12 day old and 100 day old male rats from HC and MF groups. A significant reduction in both TSH and T4 with no change in T3 concentrations was evident early in life at 12 days of age during the high carbohydrate challenge. Subsequently at 100d of age, in the absence of the high carbohydrate challenge but the presence of insulin resistance and obesity [16], while the reduction in serum TSH persisted, no change in both circulating T3 and T4 was seen.
Table 2.
Serum TSH, T3 and T4 concentrations in 12 day and 100 day old male rats
| 12 day | 100 day | |||
|---|---|---|---|---|
| MF | HC | MF | HC | |
| Serum TSH ng/ml (n) | 4.62 ± 0.572 (4) | 2.52 ± 0.652* (5) | 10.45 ± 1.97 (10) | 3.954 ± 1.001** (10) |
| Serum T3 ng/ml (n) | 4.806 ±0.453 (4) | 6.2 ± 0.515 (5) | 6.3 ±0.217 (8) | 5.84 ± 0.186 (9) |
| Serum T4 ng/ml (n) | 125.39 ±3.83 (4) | 91.96 ± 4.1*** (4) | 302.5 ± 6.332 (10) | 292.534 ± 4.011 (10) |
p =0.05,
p =0.009,
p = 0.0009
Figure 1A demonstrates a cartoon of the 5′-flanking region of the rat glut4 gene which contains the well characterized MEF and MyoD binding regions, the thyroid response element (TRE), the three CpG containing regions, three HpaII/MspI restriction sites and the transcription start site. The forward and reverse primers used to generate a 428 and 384 bp amplification products by qPCR are schematically shown. While the 428 bp product contains the MEF2, MyoDII and TRE regions with CpG III, the 384 bp contains the MyoDI and MEF2 regions alone without the TRE region but with CpG I and CpG II regions.
No change in skeletal muscle Glut4 mRNA and protein in the 12d old male rat offspring between HC and MF groups was observed (negative data not shown). However in the 100d old adult male rat offspring, HC versus MF group demonstrated a 50% reduction in skeletal muscle Glut4 mRNA (Figure 1B) and protein (Figure 1C) concentrations. Hence the rest of the experiments focused on the 100d skeletal muscle Glut4 changes alone. Examination of methylation status of the CG rich region (will be referred to as the CpG island and contains CpGI, CpGII and CpGIII sites) within the 5′-flanking region of the glut4 gene revealed an increase in percent methylation in HC versus MF as detected by resistance to HpaII digestion translated as the intensity of the DNA band generated when compared to that of MspI digestion (Figure 1D). In the MF group, as expected there was unmethylation of the CpG island as is usual to promote skeletal muscle glut4 gene expression under conventional nutritional state. To determine the role of various Dnmt isoforms in DNA methylation, using ChIP we determined a 2-fold enhanced recruitment of Dnmt3b to the 384 bp fragment of the glut4 gene in HC versus the MF groups (HC:0.89±0.06 versus MF:0.41±0.07 arbitrary units; p=0.047, n=4 each). In contrast, the 428bp glut4 DNA recruited Dnmt3a at a 1.75-fold greater extent in the HC versus the MF group (HC:0.73±0.04 versus MF:0.42±0.02 arbitrary units; p=0.0005, n=4 each). This suggests that CpG I and II interacted with Dnmt3b and CpG III with Dnmt3a.
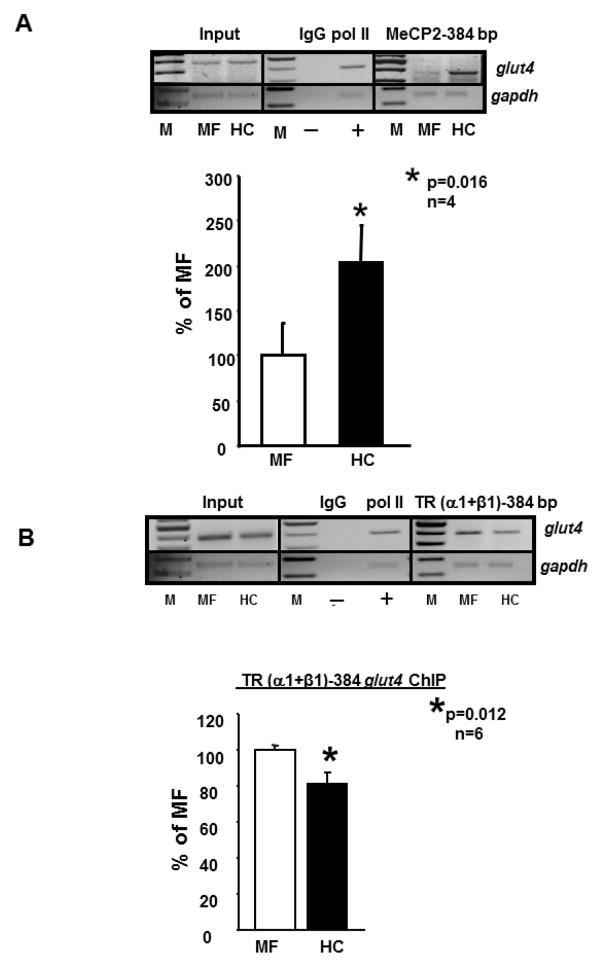
We next examined recruitment of the MeCP2 protein to the CpG island (384 bp) and noted a 1.87-fold enhanced recruitment in HC versus the MF groups (p=0.016, n=4 each; Figure 2A). In keeping with this recruitment, we observed a 20% reduction in the recruitment of TR (α1+2 or α1+β1) (Figure 2B) perhaps related to interference by Dnmt3b/MeCP2 binding the glut4 gene. No differences were observed between the HC and MF groups with respect to recruitment of either MyoD or MEF2A/2D to the glut4 gene (384 bp and 428 bp fragments; negative data not shown).
Figure 2.
A., B. Chromatin immunoprecipitation (ChIP) assay demonstrating MeCP2- and TR-glut4 interaction.
Top panel depicts representative 2% agarose gels demonstrating the input chromatin PCR amplified glut4 and gapdh control without an antibody (left panels), in the presence of nonspecific (IgG, -) and anti-polymerase II (pol II, +) IgGs (middle panels), and ChIP assay demonstrating the PCR amplified 384 bp fragment containing the glut4 DNA (top gels) or 230 bp fragment with the gapdh DNA (internal control) (bottom gels) from 100d old male MF and HC skeletal muscle in the presence of MeCP2 (A) or anti-TR (α1+β1) IgG (B) (respective right panels). Bottom panel depicts quantification (qPCR) of the 384 bp glut4 amplification product from the MeCP2 (A) or TR (α1+β1) (B) ChIP as a ratio to that of the respective gapdh DNA product after correction for the input control and expressed as a percent of MF. M = DNA size markers. Data is shown as mean±SEM, *p=0.016, n=4 (A) or *p=0.012, n=6 (B) samples in each group. TR (α1+2) ChIP demonstrated similar results as TR (α1+β1) (not shown).
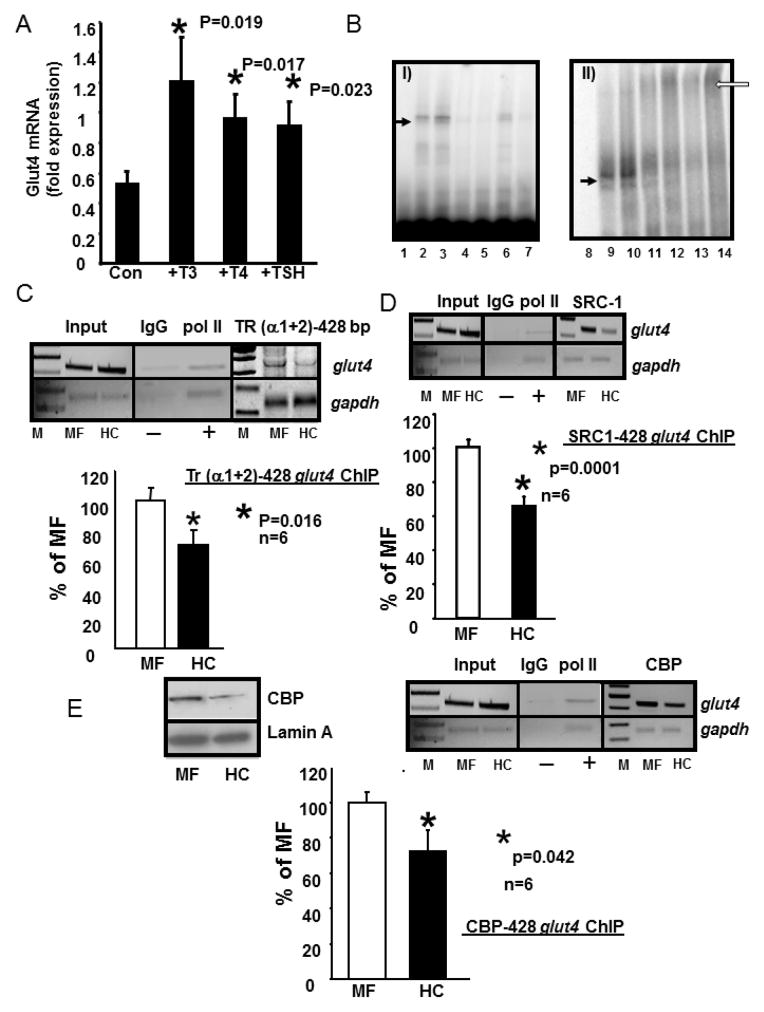
We investigated the effect of T3, T4 and TSH on Glut4 mRNA in-vitro employing C2C12 skeletal myotubular mouse cells. The addition of T3 and T4 but not TSH increased Glut4 mRNA concentrations (n=3 each; p=0.0004 each) as assessed by reverse transcription-qPCR (Figure 3A). We next undertook electromobility gel shift and super-gel shift assays with nuclear extracts obtained from skeletal muscle of 100d old adult male HC and MF groups. In these studies, the nuclear protein binding to the TRE region of the glut4 gene was specifically displaced by increasing amounts of the same unlabeled DNA probe assigning specificity in both the HC and MF groups (Figure 3B, panel I). Furthermore, the anti-TR antibody (both α1+2 and α1+β1) (super-shifted the TR-TRE complexed glut4 DNA region detected as supershift bands (Figure 3B, panel II). To confirm this in-vivo finding, and quantify inter-group changes, we next employed ex-vivo ChIP to determine whether there is a reduction in thyroid hormone receptor (TR) recruitment to the glut4 gene promoter region. Figure 3C demonstrates that while no change is seen with the input DNA, a significant reduction in TR bound glut4 is observed (p=0.016, n=6). This 30–35% reduction was slightly greater when the 428 bp fragment containing the −473 to −450 bp TRE region of the glut4 gene was amplified (Figure 3C) when compared to the 20% reduction with the 384 bp fragment that lacked the TRE containing region (Figure 2B). Since a reduction was seen with both fragments, this observation suggests that TR while binding the TRE region of the 428 bp glut4 DNA fragment was perhaps recruited to the 384 bp fragment by a protein-protein interaction.
Figure 3.
A. Induction of Glut4 mRNA by T3, T4 and TSH treatment in C2C12 murine skeletal muscle cells.
C2C12 cells were treated with nothing (Con) or T3 (10−7M), T4 (10−7M) and TSH (5 mIU/ml) for 6 h. RT-qPCR generated CT values were normalized to that of gapdh, and expressed in arbitrary units. Data are expressed as the mean±SEM of triplicate independent determinations (n=3 each).*denotes statistical difference between the treatment and the Con group, with p values shown in the figure. B. Electromobility shift (EMSA) and supershift (EMSSA) assays. Panel I: Representative polyacrylamide gel demonstrating no gel-shift in the absence of nuclear extracts with only the free 32P-end-labeled DNA probe (lane 1 = free probe, FP) that spans the glut4 gene containing the TRE-binding site, A gel-shifted band (solid arrow) is seen with the labeled glut4 DNA probe in the presence of skeletal muscle nuclear extracts obtained from MF (lane 2) or HC (lane3). This gel-shift (solid arrow) is competed by increasing concentrations (50x in lanes 4 and 6; 500x in lanes 5 and7) of the unlabeled oligoprobe. Panel II: Representative polyacrylamide gel demonstrates supershifted band (open arrow) only in the presence of anti-TR IgG (lanes 11 and 12 are MF and HC incubated for 2hr; lanes 13 and14 are MF and HC when incubated overnight). Free probe alone = lane 8 and free probe with nuclear extracts from MF and HC respectively demonstrate a gel-shift (solid arrow) but no supershift in lanes 9 and 10. C., D., E. Chromatin immunoprecipitation (ChIP) assay demonstrating TR-, SRC-1-, CBP-glut4 interaction. Top panel depicts representative 2% agarose gels demonstrating the input chromatin PCR-amplified glut4 and gapdh control without an antibody (left panels), in the presence of nonspecific (IgG, −) and anti-polymerase II (pol II, +) IgGs (middle panels), and ChIP assay demonstrating the PCR amplified 428 bp fragment containing the glut4 DNA (top gels) or 230 bp fragment with the gapdh DNA (internal control, bottom gels) from 100d old male MF and HC skeletal muscle in the presence of anti-TR (α1+2) (C), SRC-1 (D) or CBP (E) IgGs (respective right panels). In addition, left top panel (E) demonstrates representative Western blots showing nuclear CBP (top gels) and lamin A (nuclear internal loading control; bottom gels) protein concentrations in 100 day old male MF and HC skeletal muscle. Bottom panels depict quantification (qPCR) of the 428 bp glut4 amplification product from the TR (C), SRC-1 (D) or CBP (E) ChIP as a ratio to that of the respective gapdh DNA product after correction for the input control and expressed as a percent of MF. M = DNA size markers, *p=0.016 (C), *p=0.0001 (D), *p=0.042, (E) with n=6 samples for each protein and in each experimental group.
Further, we observed a reduction in the recruitment of the steroid receptor co-activator (SRC1) to the complex containing the glut4 gene (428 bp) (p=0.0001, n=6) (Figure 3D). Similarly, a reduction in the recruitment of CREB-binding protein (CBP) to the glut4 gene (428 bp) complex was also noted in HC versus the MF group (Figure 3E). In fact, by Western blot analysis, a decrease in nuclear amounts of CBP was evident (Figure 3E left inset). Both SRC-1 and CBP serve as co-activators of TR within the nucleus, thus the recruitment of these players that form a protein-protein complex with TR was also diminished at the glut4 428 bp region.
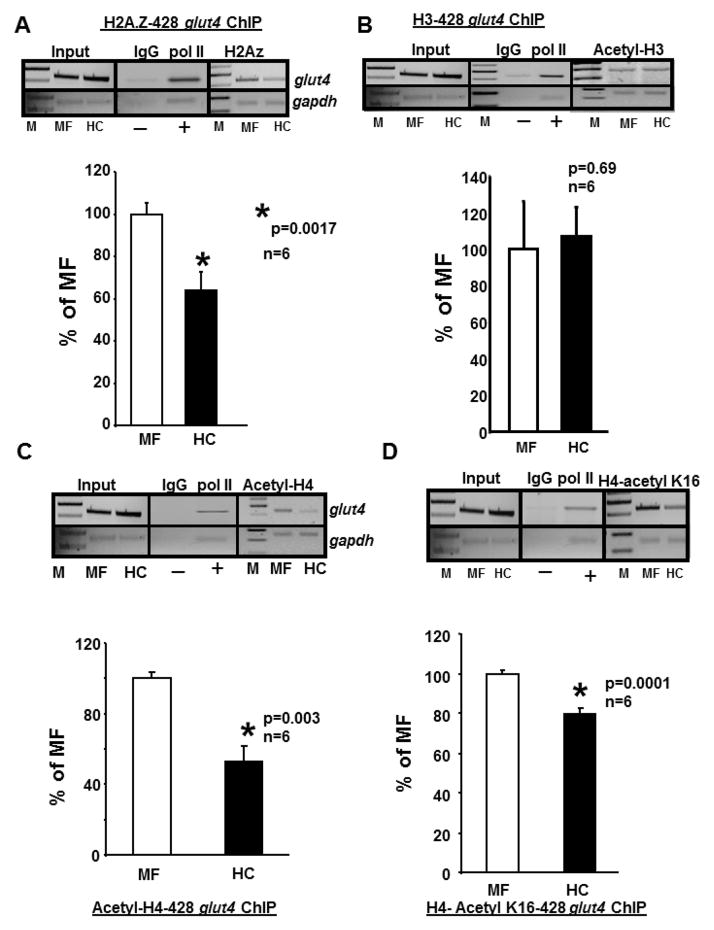
We subsequently analyzed the role of histone acetylation. First we assessed acetylation of the variant form histone 2A.Z known to confer epigenetic memory of the transcriptional state of genes [19]. Next we examined acetylation of both histone 3 and histone 4. We observed that acetylation of both the variant form histone 2A.Z (p=.0017, n=6) (Figure 4A) and the regular histone 4 (p=0.003, n=6) (Figure 4C) were reduced in HC versus MF groups however no similar change in histone 3 was observed (Figure 4B). Specifically acetylation of lysine 16 residue of histone 4 was reduced (p=0.0001, n=6) (Figure 4D) while lysine 5, 8 and 12 were not affected (negative data not shown).
Figure 4. Chromatin immunoprecipitation (ChIP) assays demonstrating histones (H) associated with the glut4 gene.
A–D. Top panels - Representative 2% agarose gels demonstrate the input chromatin PCR amplified glut4 (top gels) and gapdh control (bottom gels) without an antibody (left panels), in the presence of nonspecific (IgG, −) and anti-polymerase II (pol II, +) IgGs (middle panels), and ChIP assay demonstrating the PCR amplification products of the 428 bp glut4 DNA or 230 bp gapdh DNA (internal control) from 100 day old male MF and HC skeletal muscle chromatin in the presence of anti-H2A.Z (A), anti-H3 (B), anti-H4 (C) or anti-H4, lysine (K)16 (D) IgGs. Bottom panels depict the quantification of the 428 bp glut4 amplification product in the H2A.Z (A), H3 (B), H4 (C) or H4.K16 (D) ChIP as a ratio to that of gapdh DNA after correction for the input control and expressed as a percent of MF. M = DNA size markers, *p=0.0017 (A), p=0.69 (B), *p=0.003 (C), *p=0.0001 (D), n=6 samples for each histone and in each experimental group.
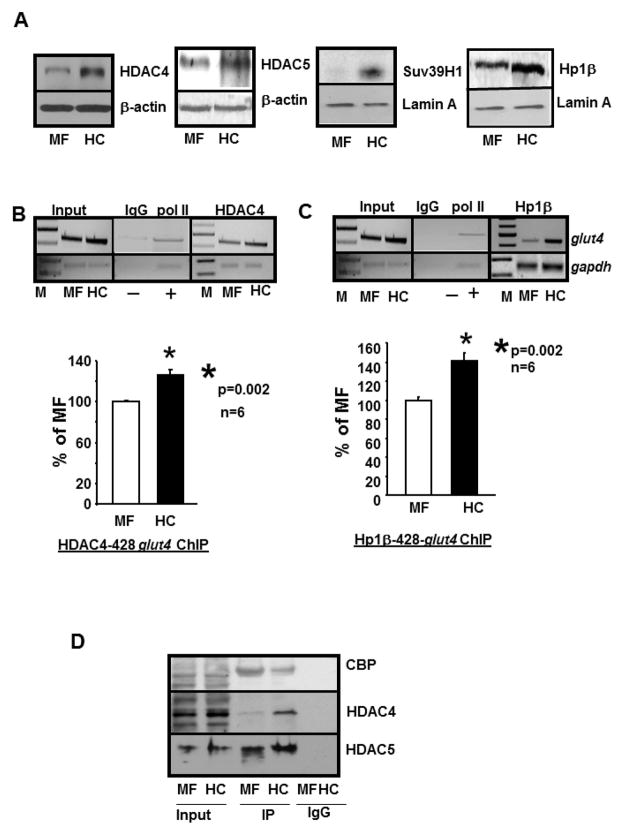
To determine the involvement of HDACs, we observed that there were increased nuclear amounts of HDAC4 (left panel) and HDAC5 (second panel) (Figure 5A inset), however enhanced recruitment of HDAC4 and not HDAC5 (negative data not shown) to the complex containing the glut4 gene (428 bp) (Figure 5B) was evident in HC versus MF groups. In addition, nuclear amounts of Suv39H1 methylase and HP1 proteins were also enhanced in HC versus MF group (Figure 5A, third and right panels). Further, enhanced recruitment of HP1β to this 428 bp glut4 DNA containing complex was also seen (p=0.002, n=6) (Figure 5C), but not with the 384 bp amplified DNA fragment (negative data not shown).
Figure 5. HDAC4, Suv39H1 methylase and HP1β are associated with the glut4 gene.
A. Representative Western blots demonstrating nuclear concentrations of HDAC4 (left most panel), HDAC5 (second panel), Suv39H1 methylase (third panel) and Hp1β protein (right most panel) concentrations in MF and HC skeletal muscle with either β-actin in the case of HDACs and lamin A in the case of Suv39H1 and Hp1β serving as an internal loading control. B.,C. Chromatin immunoprecipitation (ChIP) assay demonstrating HDAC4- and Hp1β-glut4 interaction. Top panels: Representative 2% agarose gels demonstrate the input chromatin PCR amplified glut4 (top gels) and gapdh (bottom gels) control without an antibody (left panel), in the presence of nonspecific (IgG, −) and anti-polymerase II (pol II, +) IgGs (middle panel), and ChIP assay demonstrating the PCR amplification products of the 428 bp glut4 DNA or 230 bp gapdh DNA (internal control) from 100 day old male MF and HC skeletal muscle chromatin in the presence of anti-HDAC4 (B) or anti-Hp1β (C) IgGs. Bottom panels: demonstrate quantification of the 428 bp glut4 amplification product in the HDAC4 (B) or Hp1β (C) ChIP as a ratio to that of gapdh DNA product corrected for the input control and expressed as a percent of MF. M = DNA size markers, *p=0.002 (B, C), n=6 samples for HDAC4 and Hp1β and in each experimental group. D. Co-immunoprecipitation demonstrates direct interaction of CBP, HDAC4 and HDAC5 with TR in MF and HC skeletal muscle. Representative Western blots demonstrate CBP, HDAC4 and HDAC5 protein band in MF and HC nuclear protein extract in the absence (input) or presence of nuclear anti-TR (α1+2) immunoprecipitate (IP) or in the presence of a nonspecific IgG (IgG). While the anti-TR antibody detected CBP in both MF and HC input nuclear fraction, a significantly reduced interaction was evident in HC versus MF. On the other hand, a significantly higher interaction of TR with HDAC4 was evident in the IP obtained from HC versus MF, with no such difference observed with HDAC5.
Employing co-immunoprecipitation assays using the anti-TR (α1+2) antibody for immunoprecipitation and immunoblotting with various antibodies, we observed that the interaction between TR and CBP was diminished, TR and HDAC4 increased and TR and HDAC5 remained unchanged in HC group when compared to that of the MF group (Figure 5D).
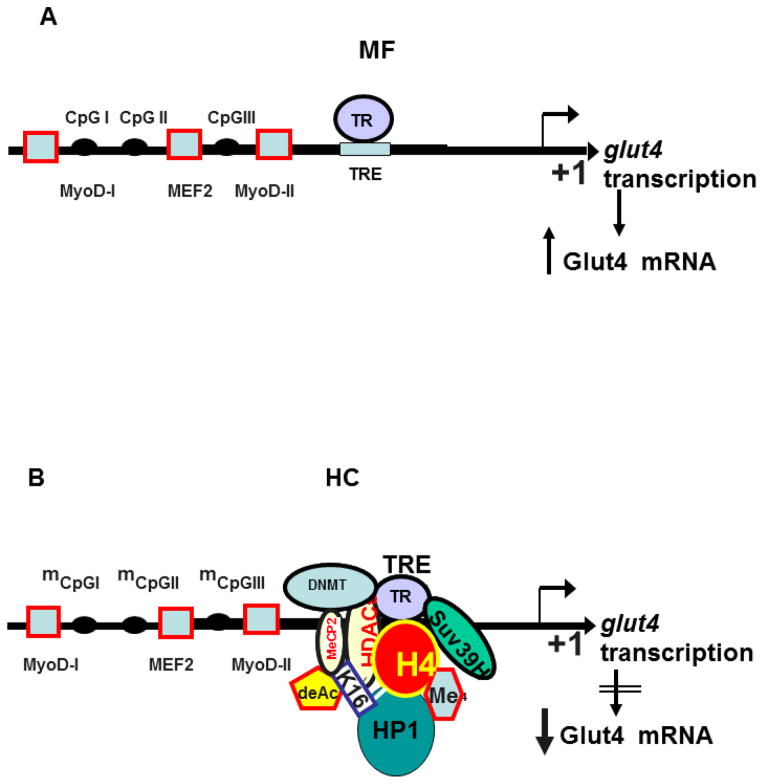
Schematic representation of the baseline under MF dietary conditions (Figure 6A) and the proposed molecular changes encountered in the adult skeletal muscle responsible for Glut4 expression changes in response to postnatal HC diet exposure (Figure 6B) is depicted.
Figure 6.
Schematic representation of the molecular machinery involved in A. mother fed group (MF) where TR binds the TRE element of the glut4 gene transcription to result in Glut4 mRNA, and B. early postnatal high carbohydrate diet (HC) induced recruitment of nuclear proteins responsible for epigenetic control of the glut4 gene in rat skeletal muscle to interact with the TR-TRE complex, which results in reduced Glut4 mRNA that translates into decreased Glut4 protein.
Discussion
We have shown that postnatal high carbohydrate diet exposure limited to the suckling phase of life is associated with a reduction in adult male skeletal muscle Glut4 expression. This reduction which occurs during adult life contributes to the adult onset obesity and insulin resistance that was previously described as the emerging phenotype [16]. This phenotype mimics that observed in the adult offspring born to rat dams reared on a high fat diet from weaning through pregnancy and lactation [3]. In this case of early postnatal HC milk feeding as well, adult onset obesity with glucose intolerance, dyslipidemia and insulin resistance is evident in males [4]. Thus overnutrition during early life molds the ultimate adult phenotype of the male offspring predisposing towards the development of insulin resistance and obesity with associated type 2 diabetes mellitus. Various investigations have defined the central basis for adult onset insulin resistance to involve key changes in skeletal muscle Glut4 mRNA and protein in both rats [8, 20, 21] and human [20] along with aberrant post-translational modifications [8] that dictate translocation to the sarcolemma.
Our current artificially reared nutritional intervention consisting of exposure to a high carbohydrate diet only during the suckling period results in the immediate onset of hyperinsulinemia which persists into adulthood without any further nutritional stimuli [15]. The growth rate of HC rats increases around postnatal day 55 and these animals are distinctly obese by day 100 [4,13]. In 100-day old HC rats, hypertrophy of the β-cells and an altered glucose-stimulated insulin secretory pattern was observed. The timing of the dietary modification in this rat model coincides with the neonatal endocrine pancreatic development and it appears that the HC milk formula potentiates certain changes in the pancreatic islets and their function resulting in hyperinsulinemia which eventually leads to the development of obesity later in adult life [16]. Significant increase in the preproinsulin mRNA and insulin biosynthesis in 12-day HC islets compared to that from MF control rats has been reported [16]. This is particularly important, since at 12 days of age during exposure to the HC diet, while hyperinsulinemia was observed, we did not see any changes in skeletal muscle Glut4 concentrations (negative data not shown). It is only at 100d of age that a reduction in skeletal muscle total Glut4 concentrations was noted in our present study. This suggests that the concomitant hyperinsulinemia that develops first and persists perhaps over time chronically downregulates skeletal muscle Glut4 expression that is observed subsequently.
However, changes in skeletal muscle Glut4 expression related to aberrant glucose and insulin metabolism have previously been observed to affect myocyte enhancer factor 2 (MEF2) mediated transcriptional control of the skeletal muscle glut4 gene. Transgenic investigations established the crucial in-vivo transcriptional role for the conserved glut4 promoter consisting of the −502 to −420 bp fragment that encompasses the MyoD, MEF2 and TRα1 binding cis-elements that regulate skeletal muscle glut4 gene expression [22]. Disruption of the MEF2-binding site (−473 to −464 bp) ablated tissue-specific Glut4 expression in transgenic mice [23, 24]. This is why we initially examined the effect of postnatal high carbohydrate diet exposure on the adult MEF2 and MyoD induced skeletal muscle Glut4 expression [7]. However, no changes were observed between the high carbohydrate and the relatively high fat rat mother’s milk fed adult rats with regard to either MEF2A/D or MyoD recruitment to the glut4 gene.
Previous investigations involving a high fat diet during the fetal stages of development disrupted the thyroid hormone axis in both sheep [25] and non-human primates [26]. However similar investigations have not been carried out during exposure to modified nutrition during the early postnatal period of development, our present study being the first. We observed in our HC group there was a reduction in serum T4 and TSH concentrations evident during the postnatal stage of development at 12 days of age, consistent with hypothyroidism. However, these changes were not associated with any changes in skeletal muscle Glut4 expression. During the adult stage while reduced serum TSH concentrations persisted, the reduction in serum T4 concentrations encountered at 12 days of age did not persist in the 100 day old male adult offspring. Despite normal circulating T4 concentrations at 100 days of age, the presence of low TSH concentrations may suggest a response to low TRH (not measured in the present study) or a functionally hyperthyroid state if examined in isolation as an adult alone. However in light of low TSH associated with low T4 concentrations at 12 days of age, this may demonstrate persistence of low TSH in the adult secondary to early life hypothyroidism as a sign of hormonal programming, encountered in response to postnatal introduction of a high carbohydrate diet. Regardless of the circulating T4 concentrations as an adult, the early onset of reduced TSH/T4 was associated with a subsequent reduction in skeletal muscle Glut4 expression observed at 100 days of age. Based on this finding we examined the effect on thyroid hormone receptor (TR) binding of the skeletal muscle glut4 gene.
Previous studies in diabetic and obese humans have provided controversial results related to thyroid hormones. It is known that thyroxine deficiency is associated with obesity related to myxedema [23, 27, 28], while obese individuals have demonstrated mixed results with respect to circulating thyroxine concentrations. Thyroid hormone (T3/T4) stimulates basal and insulin-mediated glucose uptake in rat skeletal muscle [10, 11], primarily by induction of skeletal muscle Glut4 protein concentrations. In our present study, we too observed T4/T3 induction of enhanced C2C12 skeletal myotubular Glut4 expression. This increase in Glut4 expression despite no change in the number of C2C12 myotubes encountered in response to T3 as previously reported [29], supports a direct T4/T3 effect on Glut4 gene expression independent of the cellular state.
Thyroid hormones (TH) regulate gene expression via thyroid hormone receptors (TR) and members of the steroid-thyroid superfamily of nuclear transcription factors [30, 31]. TR binds to specific DNA sequences, T3 response elements (TREs), contained primarily in the 5′-flanking region of T3-regulated genes. The core TRE sequence consists of a hexameric sequence (A/GGGTC/AA) and can include a single binding domain or two binding domains arranged as direct repeats, a palindrome, or an inverted palindrome [30, 32, 33]. The major constituent of nuclear proteins that heterodimerize with TR, is the retinoid-X receptor (RXR) [34–37], other interactions including CBP or SRC1.
A 281 bp region containing the T3 responsive element was identified in the upstream sequences of the glut4 gene. This TRE site was demonstrated in the rat glut4 gene promoter region spanning −450 and −434 bp region and oriented as in the classical DR+4 motif [38]. These observations support a direct effect of thyroid hormones on glut4 gene transcription. In our present investigation, we observed a reduction in TR binding of TRE in adult male skeletal muscle glut4 gene when exposed to early postnatal high carbohydrate diet.
The insulin resistant phenotype of the adult males exposed to nutritional perturbations during early life has been trans-generationally propagated [39–41]. These observations support an epigenetic basis for the transmission of insulin resistance. Changes in chromatin structure and locus accessibility predetermine the epigenetic imprint on gene expression [42]. DNA methylation and concerted changes in the combinatorial histone code regulate tissue-specific gene expression [42–46]. Exposure postnatally to a high carbohydrate diet increased the relative DNA methylation of the CpG island contained within the skeletal muscle glut4 promoter region which was evident in adult males in the present study. This change contributes to reduction in skeletal muscle Glut4 expression during adult life.
Many recent studies suggest that histone code modifications also have central roles in regulating transcription. Multiple lysine or arginine residues in core histones, particularly H3 and H4, are subject to posttranslational modifications including methylation and acetylation, and many of these modifications are associated with distinct transcriptional states [47–50]. Several studies confirmed not only that hyperacetylation of histone H3 and H4 is associated with the euchromatin region of the genome, but also that it induces transcription by recruiting key transcriptional complexes to target genes [51, 52]. Our present observations due to exposure to early postnatal high carbohydrate diet consist of reduced acetylation of H2A.Z and H4 in association with the glut4 promoter in adult skeletal muscle further mediating diminished Glut4 expression. This association of perturbed acetylation of H2A.Z and H4 together bring about a concerted effect on transcriptional regulation of downstream genes [53]. Particularly, H2A.Z-mediated localization of altered genes occurs at the nuclear periphery and is known to represent an epigenetic state that confers memory of previously altered transcriptional state [19].
These nutritionally induced transcriptional effects may merely diminish rather than silence gene expression. An example being the association of MEF2 with class II histone de-acetylating enzymes suppressing MEF2 mediated downstream gene expression [54]. Similarly, postnatal high carbohydrate dietary introduction reduced acetylation of H4.K16 due to increased recruitment of HDAC4 and enhanced histone methylation by increased amounts and perhaps recruitment of Suv39H1 methylase. Both de-acetylation and methylation of histones complement the heightened DNA methylation of the CpG island, all collectively contributing towards heterochromatin formation attracting HP1β and thereby reducing the adult skeletal muscle glut4 gene expression.
Thus introduction of a high carbohydrate diet early during the suckling period has long lasting implications on the adult metabolic phenotype. This observation has significant translatability to the human condition, where fruit juices and other high carbohydrate containing foods and carbonated drinks are introduced early during infancy. Such a practice sets the stage for subsequent development of childhood and adult onset insulin resistance and obesity. Our uncovering of epigenetic regulation of the molecular machinery that controls skeletal muscle glut4 gene expression forms the basis of adult-onset insulin resistance, a forerunner of diabetes mellitus and obesity, and may explain the trans-generational persistence of this phenotype. Our present molecular findings although not intuitive in the face of normal circulating T3/T4 concentrations in the adult obese offspring but perhaps related to postnatal T4 perturbations, provide novel targets for developing future therapeutic strategies aimed at reversing morbidity and mortality due to early exposure to high carbohydrate consumption.
Acknowledgments
This work was supported by grants from the NIH-HD41230 and HD25024 (to SUD) and DK61518 (to MSP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garg M, Thamotharan M, Dai Y, Lagishetty V, Matveyenko AV, Lee WN, et al. Glucose intolerance and lipid metabolic adaptations in response to intrauterine and postnatal calorie restriction in male adult rats. Endocrinology. 2013;154:102–13. doi: 10.1210/en.2012-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garg M, Thamotharan M, Dai Y, Thamotharan S, Shin BC, Stout D, et al. Early postnatal caloric restriction protects adult male intrauterine growth-restricted offspring from obesity. Diabetes. 2012;61:1391–8. doi: 10.2337/db11-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srinivasan M, Katewa SD, Palaniyappan A, Pandya JD, Patel MS. Maternal high-fat diet consumption results in fetal malprogramming predisposing to the onset of metabolic syndrome-like phenotype in adulthood. Am J Physiol Endocrinol Metab. 2006;291:E792–9. doi: 10.1152/ajpendo.00078.2006. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan M, Mitrani P, Sadhanandan G, Dodds C, Shbeir-ElDika S, Thamotharan S, et al. A high-carbohydrate diet in the immediate postnatal life of rats induces adaptations predisposing to adult-onset obesity. J Endocrinol. 2008;197:565–74. doi: 10.1677/JOE-08-0021. [DOI] [PubMed] [Google Scholar]
- 5.O’Doherty RM, Halseth AE, Granner DK, Bracy DP, Wasserman DH. Analysis of insulin-stimulated skeletal muscle glucose uptake in conscious rat using isotopic glucose analogs. Am J Physiol. 1998;274:E287–96. doi: 10.1152/ajpendo.1998.274.2.E287. [DOI] [PubMed] [Google Scholar]
- 6.Dai Y, Thamotharan S, Garg M, Shin BC, Devaskar SU. Superimposition of postnatal calorie restriction protects the aging male intrauterine growth- restricted offspring from metabolic maladaptations. Endocrinology. 2012;153:4216–26. doi: 10.1210/en.2012-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raychaudhuri N, Raychaudhuri S, Thamotharan M, Devaskar SU. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283:13611–26. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thamotharan M, Shin BC, Suddirikku DT, Thamotharan S, Garg M, Devaskar SU. GLUT4 expression and subcellular localization in the intrauterine growth-restricted adult rat female offspring. Am J Physiol Endocrinol Metab. 2005;288:E935–47. doi: 10.1152/ajpendo.00342.2004. [DOI] [PubMed] [Google Scholar]
- 9.Wang C. The Relationship between Type 2 Diabetes Mellitus and Related Thyroid Diseases. J Diabetes Res. 2013:390534. doi: 10.1155/2013/390534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casla A, Rovira A, Wells JA, Dohm GL. Increased glucose transporter (GLUT4) protein expression in hyperthyroidism. Biochem Biophys Res Commun. 1990;171:182–8. doi: 10.1016/0006-291x(90)91374-2. [DOI] [PubMed] [Google Scholar]
- 11.Weinstein SP, O’Boyle E, Haber RS. Thyroid hormone increases basal and insulin-stimulated glucose transport in skeletal muscle. The role of GLUT4 glucose transporter expression. Diabetes. 1994;43:1185–9. doi: 10.2337/diab.43.10.1185. [DOI] [PubMed] [Google Scholar]
- 12.Richardson JM, Pessin JE. Identification of a skeletal muscle-specific regulatory domain in the rat GLUT4/muscle-fat gene. J Biol Chem. 1993;268:21021–7. [PubMed] [Google Scholar]
- 13.Hiremagalur BK, Vadlamudi S, Johanning GL, Patel MS. Long-term effects of feeding high carbohydrate diet in pre-weaning period by gastrostomy: a new rat model for obesity. Int J Obes Relat Metab Disord. 1993;17:495–502. [PubMed] [Google Scholar]
- 14.Patel MS, Vadlamudi S, Johanning GL. Artificial rearing of rat pups: implications for nutrition research. Annu Rev Nutr. 1994;14:21–40. doi: 10.1146/annurev.nu.14.070194.000321. [DOI] [PubMed] [Google Scholar]
- 15.Mitrani P, Srinivasan M, Dodds C, Patel MS. Role of the autonomic nervous system in the development of hyperinsulinemia by high-carbohydrate formula feeding to neonatal rats. Am J Physiol Endocrinol Metab. 2007;292:E1069–78. doi: 10.1152/ajpendo.00477.2006. [DOI] [PubMed] [Google Scholar]
- 16.Vadlamudi S, Hiremagalur BK, Tao L, Kalhan SC, Kalaria RN, Kaung HL, et al. Long-term effects on pancreatic function of feeding a HC formula to rats during the preweaning period. Am J Physiol. 1993;265:E565–71. doi: 10.1152/ajpendo.1993.265.4.E565. [DOI] [PubMed] [Google Scholar]
- 17.Rajakumar A, Thamotharan S, Raychaudhuri N, Menon RK, Devaskar SU. Trans-activators regulating neuronal glucose transporter isoform-3 gene expression in mammalian neurons. J Biol Chem. 2004;279:26768–79. doi: 10.1074/jbc.M402735200. [DOI] [PubMed] [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 19.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozanne SE, Jensen CB, Tingey KJ, Storgaard H, Madsbad S, Vaag AA. Low birthweight is associated with specific changes in muscle insulin-signalling protein expression. Diabetologia. 2005;48:547–52. doi: 10.1007/s00125-005-1669-7. [DOI] [PubMed] [Google Scholar]
- 21.Agote M, Goya L, Ramos S, Alvarez C, Gavete ML, Pascual-Leone AM, et al. Glucose uptake and glucose transporter proteins in skeletal muscle from undernourished rats. Am J Physiol Endocrinol Metab. 2001;281:E1101–9. doi: 10.1152/ajpendo.2001.281.5.E1101. [DOI] [PubMed] [Google Scholar]
- 22.Santalucia T, Moreno H, Palacin M, Yacoub MH, Brand NJ, Zorzano A. A novel functional co-operation between MyoD, MEF2 and TRalpha1 is sufficient for the induction of GLUT4 gene transcription. J Mol Biol. 2001;314:195–204. doi: 10.1006/jmbi.2001.5091. [DOI] [PubMed] [Google Scholar]
- 23.Liu ML, Olson AL, Edgington NP, Moye-Rowley WS, Pessin JE. Myocyte enhancer factor 2 (MEF2) binding site is essential for C2C12 myotube-specific expression of the rat GLUT4/muscle-adipose facilitative glucose transporter gene. J Biol Chem. 1994;269:28514–21. [PubMed] [Google Scholar]
- 24.Thai MV, Guruswamy S, Cao KT, Pessin JE, Olson AL. Myocyte enhancer factor 2 (MEF2)-binding site is required for GLUT4 gene expression in transgenic mice. Regulation of MEF2 DNA binding activity in insulin-deficient diabetes. J Biol Chem. 1998;273:14285–92. doi: 10.1074/jbc.273.23.14285. [DOI] [PubMed] [Google Scholar]
- 25.Johnsen L, Kongsted AH, Nielsen MO. Prenatal undernutrition and postnatal overnutrition alter thyroid hormone axis function in sheep. J Endocrinol. 2013;216:389–402. doi: 10.1530/JOE-12-0389. [DOI] [PubMed] [Google Scholar]
- 26.Suter MA, Sangi-Haghpeykar H, Showalter L, Shope C, Hu M, Brown K, et al. Maternal high-fat diet modulates the fetal thyroid axis and thyroid gene expression in a nonhuman primate model. Mol Endocrinol. 2012;26:2071–80. doi: 10.1210/me.2012-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno M, de Lange P, Lombardi A, Silvestri E, Lanni A, Goglia F. Metabolic effects of thyroid hormone derivatives. Thyroid. 2008;18:239–53. doi: 10.1089/thy.2007.0248. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt T, Luqman W, McCool C, Lenz F, Ahmad U, Nolan S, et al. Unresponsiveness to exogenous TSH in obesity. Int J Obes. 1977;1:185–90. [PubMed] [Google Scholar]
- 29.Sultan KR, Henkel B, Terlou M, Haagsman HP. Quantification of hormone-induced atrophy of large myotubes from C2C12 and L6 cells: atrophy-inducible and atrophy-resistant C2C12 myotubes. Am J Physiol Cell Physiol. 2006;290(2):C650–9. doi: 10.1152/ajpcell.00163.2005. [DOI] [PubMed] [Google Scholar]
- 30.Brent GA. The molecular basis of thyroid hormone action. N Engl J Med. 1994;331:847–53. doi: 10.1056/NEJM199409293311306. [DOI] [PubMed] [Google Scholar]
- 31.Glass CK, Holloway JM. Regulation of gene expression by the thyroid hormone receptor. Biochim Biophys Acta. 1990;1032:157–76. doi: 10.1016/0304-419x(90)90002-i. [DOI] [PubMed] [Google Scholar]
- 32.Desvergne B. How do thyroid hormone receptors bind to structurally diverse response elements? Mol Cell Endocrinol. 1994;100:125–31. doi: 10.1016/0303-7207(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 33.Williams GR, Bland R, Sheppard MC. Retinoids modify regulation of endogenous gene expression by vitamin D3 and thyroid hormone in three osteosarcoma cell lines. Endocrinology. 1995;136:4304–14. doi: 10.1210/endo.136.10.7664649. [DOI] [PubMed] [Google Scholar]
- 34.Mangelsdorf DJ, Borgmeyer U, Heyman RA, Zhou JY, Ong ES, Oro AE, et al. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–44. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 35.Sugawara A, Yen PM, Darling DS, Chin WW. Characterization and tissue expression of multiple triiodothyronine receptor-auxiliary proteins and their relationship to the retinoid X-receptors. Endocrinology. 1993;133:965–71. doi: 10.1210/endo.133.3.8396023. [DOI] [PubMed] [Google Scholar]
- 36.Berrodin TJ, Marks MS, Ozato K, Linney E, Lazar MA. Heterodimerization among thyroid hormone receptor, retinoic acid receptor, retinoid X receptor, chicken ovalbumin upstream promoter transcription factor, and an endogenous liver protein. Mol Endocrinol. 1992;6:1468–78. doi: 10.1210/mend.6.9.1331778. [DOI] [PubMed] [Google Scholar]
- 37.Leid M, Kastner P, Lyons R, Nakshatri H, Saunders M, Zacharewski T, et al. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell. 1992;68:377–95. doi: 10.1016/0092-8674(92)90478-u. [DOI] [PubMed] [Google Scholar]
- 38.Torrance CJ, Usala SJ, Pessin JE, Dohm GL. Characterization of a low affinity thyroid hormone receptor binding site within the rat GLUT4 gene promoter. Endocrinology. 1997;138:1215–23. doi: 10.1210/endo.138.3.4982. [DOI] [PubMed] [Google Scholar]
- 39.Thamotharan M, Garg M, Oak S, Rogers LM, Pan G, Sangiorgi F, et al. Transgenerational inheritance of the insulin-resistant phenotype in embryo-transferred intrauterine growth-restricted adult female rat offspring. Am J Physiol Endocrinol Metab. 2007;292:E1270–9. doi: 10.1152/ajpendo.00462.2006. [DOI] [PubMed] [Google Scholar]
- 40.Zambrano E, Martinez-Samayoa PM, Bautista CJ, Deas M, Guillen L, Rodriguez-Gonzalez GL, et al. Sex differences in transgenerational alterations of growth and metabolism in progeny (F2) of female offspring (F1) of rats fed a low protein diet during pregnancy and lactation. J Physiol. 2005;566:225–36. doi: 10.1113/jphysiol.2005.086462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boloker J, Gertz SJ, Simmons RA. Gestational diabetes leads to the development of diabetes in adulthood in the rat. Diabetes. 2002;51:1499–506. doi: 10.2337/diabetes.51.5.1499. [DOI] [PubMed] [Google Scholar]
- 42.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 44.Devaskar SU, Raychaudhuri S. Epigenetics--a science of heritable biological adaptation. Pediatr Res. 2007;61:1R–4R. doi: 10.1203/pdr.0b013e31805cdbd8. [DOI] [PubMed] [Google Scholar]
- 45.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–12. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–7R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 47.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–52. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–6. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 49.Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer. 2005;41:2381–402. doi: 10.1016/j.ejca.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–4. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 51.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–7. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 52.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–71. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8:e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–5. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]