Abstract

Ebola virus (EBOV), a member of the family Filoviridae, is a nonsegmented negative-sense RNA virus that causes severe, often lethal, disease in humans. EBOV RNA synthesis is carried out by a complex that includes several viral proteins. The function of this machinery is essential for viral gene expression and viral replication and is therefore a potential target for antivirals. We developed and optimized a high-throughput screening (HTS) assay based on an EBOV minigenome assay, which assesses the function of the polymerase complex. The assay is robust in 384-well format and displays a large signal to background ratio and high Z-factor values. We performed a pilot screen of 2080 bioactive compounds, identifying 31 hits (1.5% of the library) with >70% inhibition of EBOV minigenome activity. We further identified eight compounds with 50% inhibitory concentrations below their 50% cytotoxic concentrations, five of which had selectivity index (SI) values >10, suggesting specificity against the EBOV polymerase complex. These included an inhibitor of inosine monophosphate dehydrogenase, a target known to modulate the EBOV replication complex. They also included novel classes of inhibitors, including inhibitors of protein synthesis and hypoxia inducible factor-1. Five compounds were tested for their ability to inhibit replication of a recombinant EBOV that expresses GFP (EBOV-GFP), and four inhibited EBOV-GFP growth at sub-cytotoxic concentrations. These data demonstrate the utility of the HTS minigenome assay for drug discovery and suggest potential directions for antifiloviral drug development.
Keywords: antiviral, Ebola virus, filovirus, high-throughput screen, RNA polymerase, translation
INTRODUCTION
Ebola virus (EBOV), a member of the family Filoviridae, is associated with outbreaks of highly lethal hemorrhagic fever.1 Currently, no therapies are approved to specifically treat these infections. The need for approved antivirals and/or therapeutics has been emphasized by the recent EBOV outbreak in West Africa, which is of unprecedented scope, having caused >10000 deaths and resulted in cases that were exported to the United States and Europe.2
Several approaches have been taken to screen for inhibitors of EBOV replication, including high-content screening to detect viral antigen and the use of recombinant EBOVs expressing GFP or luciferase.3–7 However, any use of live EBOV requires biosafety level 4 (BSL4) containment, limiting opportunities for antiviral drug development. An alternative approach is to develop high-throughput screening assays that are based on critical viral functions, but do not involve infectious material.
EBOV transcription and genome replication require four viral proteins, nucleoprotein (NP), VP35, VP30, and the enzymatic component of the complex, the large (L) protein. Because this RNA polymerase complex is essential for viral propagation, it is an attractive target for inhibitor development. A functional EBOV RNA polymerase complex can be reconstituted by transfection of the four components into mammalian cells. Its activity can be measured through the coexpression of a model viral RNA that encodes a reporter gene flanked by the appropriate virus-derived cis-acting regulatory sequences.8 Such systems can be used to identify inhibitors of EBOV RNA synthesis.5,9,10 Chemical compound libraries can be screened in biosafety level 1 (BSL1) conditions, identifying hit compounds that can then be confirmed using infectious virus. Previously, optimization of an EBOV minigenome system in 96-well format has been described; however, this is not optimal for screening large numbers of compounds.5,10
In this study, we optimized an EBOV minigenome-based HTS assay in 384-well format to identify small-molecule inhibitors of the EBOV RNA synthesis machinery. A pilot screen of a bioactive library that includes 2080 compounds identified hit compounds that inhibit EBOV minigenome activity. The inhibitory activity of these compounds was further assessed against recombinant EBOV expressing GFP (EBOV-GFP), showing inhibition of infectious EBOV replication at sub-cytotoxic concentrations. Together, these data illustrate the utility of the HTS EBOV minigenome assay in 384-well format to identify inhibitors of infectious EBOV.
MATERIALS AND METHODS
Cell Culture
HEK293T cells were maintained in Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and cultured at 37 °C and 5% CO2. Vero E6 cells were maintained in minimum essential media (MEM) supplemented with 2% FBS, 0.1% gentamicin, 1% nonessential amino acids and 1% sodium pyruvate and cultured at 37 °C and 5% CO2.
Plasmids
The EBOV minigenome luciferase (pTM1-eMGLuc) reporter was previously described.11 pCAGGS expression plasmids for L, VP30, VP35, and NP were kind gifts from Thomas Hoenen and Heinz Feldmann (Rocky Mountain Laboratories, NIAID).
Chemicals
The chemical library used for the primary screen at the Mount Sinai Integrated Screening Core was purchased from Microsource Discovery (Gaylordsville, CT, USA) and contains 2080 bioactive compounds. 6-Azauridine and guanosine were purchased from Sigma-Aldrich. Mycophenolic acid, emetine, cycloheximide, and azacitidine used in infection assays were purchased from Sigma-Aldrich. Gedunin used in infection assays was purchased from Santa Cruz Biotechnology. All chemicals, except guanosine, were diluted to a concentration of 50 mM in DMSO before use. Guanosine was diluted in DMEM to a final concentration of 50 μM.
EBOV Minigenome High-Throughput Screening Assay
HEK293T cells (7.5 × 106) were transfected in a T75 flask using Lipofectamine 2000 (Invitrogen), at a 2:1 ratio of Lipofectamine 2000 to micrograms of DNA, with pCAGGS-L (7.5 μg), pCAGGS-NP (3.7 μg), pCAGGS-VP30 (1.5 μg), pTM1-eMGLuc (3.0 μg), pCAGGS-T7 (3.0 μg), and either pCAGGS (1.9 μg) (empty vector) for the negative control or pCAGGS-VP35 (1.9 μg). Multiple transfections of individual T75 flasks were required to produce enough cells for the screening assay. Twenty-three hours post-transfection cells were trypsinized and resuspended in DMEM without phenol red supplemented with 10% FBS. The cells from separately transfected flasks were pooled prior to distribution in 384-well plates (white opaque CulturPlate, PerkinElmer) at 20000 cells/30 μL/well using a Multidrop Combi Reagent dispenser (Thermo Scientific). Cells were allowed to rest for 1 h, after which library compounds were transferred by pin tool (V&P Scientific) in duplicate (22 nL, final concentration = 7–7.7 μM). 6-Azauridine was added as a positive control for inhibition using a HP D300 digital dispenser (Tecan) (final concentration = 7 μM).5 Twenty-four hours post-treatment Renilla-Glo (Promega) substrate was added (5 μL), and the luciferase signal was read using an EnVision plate reader (PerkinElmer). Z-Factor values for each plate were calculated using the equation Z-factor = 1 − [(3σc+ + 3σc−)/(|μc+ − μc−|)]; the positive control contained all components of the polymerase complex, and the negative control lacked VP35. Z-Scores for each compound were calculated using the equation Z-score = (Xcompound − μplate)/σplate; the plate mean (μ) and plate standard deviation (σ) were determined for individual plates from all wells treated with library compounds.
Determining Half-Maximal Inhibitory Concentration (IC50)
HEK293T cells were transfected and plated as under EBOV Minigenome High-Throughput Screening Assay. After plating, cells were allowed to rest for 1 h, and compounds were added to reach the indicated final concentrations (0–50 μM, 2-fold dilution series) in triplicate using an HP D300 digital dispenser (Tecan). Twenty-four hours post-treatment, Renilla-Glo (Promega) substrate was added, and the luciferase signal was read using an EnVision plate reader (PerkinElmer). The 50% inhibitory concentration (IC50) values were calculated with Prism using a four-parameter, nonlinear regression analysis.
Determining Half-Maximal Cell Cytotoxicity Concentration (CC50)
HEK293T cells (1 × 103 cells/30 μL/well) were plated in 384-well plates (white opaque CulturPlate, PerkinElmer). One hour after plating, compounds were added as under Determining Half-Maximal Inhibitory Concentration (IC50) to reach the indicated final concentrations (0–50 μM, 2-fold dilution series) in triplicate. Twenty-four hours post-treatment CellTiter-Glo (Promega) was added, and ATP content was determined by reading luminescence using an EnVision plate reader. CC50 values were calculated with Prism using a four-parameter, nonlinear regression analysis.
Assessing the Inhibitory Mechanism of Mycophenolic Acid
HEK293T cells were transfected with the minigenome system as before. Twenty-four hours post-transfection, 1 × 105 cells/100 μL/well were plated in a 96-well plate in the absence or presence of 50 μM guanosine. One-hour post-plating mycophenolic acid was added in triplicate to reach final concentrations (0–50 μM, 3-fold dilution series). Twenty-four hours post-treatment, Renilla-Glo substrate was added, and luciferase activity was read using a Glomax Multi+Microplate reader (Promega).
EBOV-GFP Confirmation Assay
All experiments using infectious EBOV were performed under biosafety level 4 (BSL-4) conditions at the Galveston National Laboratory. Vero E6 cells (1 × 104 cells/well) were plated in 96-well plates (black clear bottom, Costar) overnight, and the next day compounds (azacitidine, cycloheximide, emetine, gedunin, or mycophenolic acid) were added at 0.4, 2, 10, and 50 μM using an epMotion 5075 robot (Eppendorf). One-hour post-treatment, EBOV-GFP3 was added at an MOI of 0.3 (40 μL) to media containing compound (60 μL) and left on the cells for the course of the experiment. Three days post-infection the mean fluorescence intensity (MFI) was measured using an EnVision plate reader. To assess cell viability, 1 × 104 cells/well were plated in 96-well plates (white polystyrene, Costar) overnight and treated with compound as above. Cell viability was assessed 5 days after compound treatment using Viral ToxGlo (Promega), and ATP content was determined by reading luminescence using a BioTek Synergy HT plate reader.
RESULTS
Optimization of EBOV Minigenome System for 384-Well Format
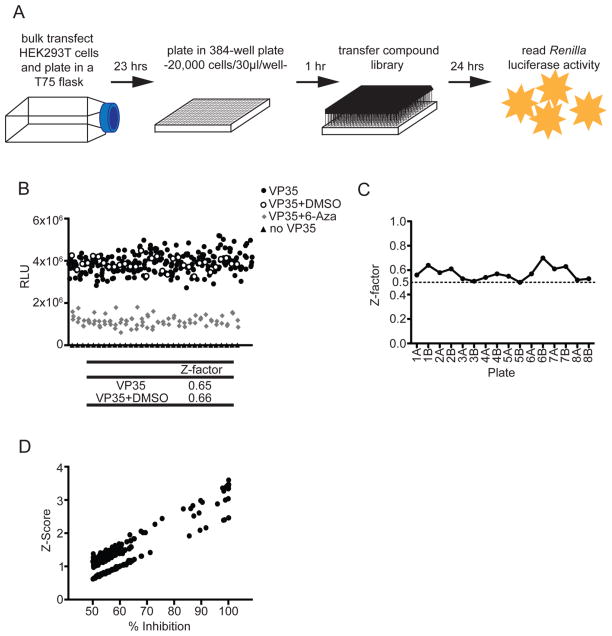
To ensure that our minigenome system could be adapted to a high-throughput format, we first tested the activity of the system in HEK293T cells in 96-well format. The EBOV minigenome Renilla luciferase reporter (eMGLuc) was transfected with plasmids expressing the components of the polymerase complex, L, NP, VP35, and VP30, in a T75 flask to produce a uniform transfection of a large number of cells. As a negative control, to assess background luciferase levels in the absence of a complete polymerase complex, pCAGGS-VP35 was replaced with an empty pCAGGS vector (“no VP35” control). Twenty-four hours post-transfection, cells were trypsinized and dispensed in a 96-well plate for an additional 24 h, after which Renilla luciferase activity was assessed. This produced a robust assay with nearly 900-fold induction over the negative control and a Z-factor of 0.8 (Figure 1A). A Z-factor value, also known as a Z′ value, >0.5 indicates a high-quality screening assay.12
Figure 1.
The EBOV minigenome assay is robust in 96- and 384-well formats. (A) HEK293T cells were transfected with the components of the EBOV minigenome system, and 24 h post-transfection, cells were plated in a 96-well plate. “No VP35” indicates samples in which the plasmid expressing VP35 was replaced with pCAGGS empty vector. “VP35” indicates samples in which a complete polymerase complex, including VP35, was transfected. Forty-eight hours post-transfection luciferase activity was assessed. The data represent the mean and standard error of the mean (SEM) of 48 wells for each condition and are reported in relative light units (RLU). (B) HEK293T cells were transfected as in (A) and plated in a 384-well plate. At each time point post-transfection (hours post-transfection, hpt) luciferase activity was assessed. The data represent the mean and SEM of 16 wells each, reported in RLU; No VP35, black triangles; VP35, black circles. Z-Factor values were calculated in both (A) and (B) using the formula Z-factor = 1 − [(3σc+ + 3σc−)/(|μc+ − μc−|)].
We next evaluated the capacity of the EBOV minigenome system to be further miniaturized to a 384-well format. Transfection of HEK293T cells was carried out as before. Twenty-four hours post-transfection cells were trypsinized and plated in a 384-well plate. Luciferase activity was assessed at several time points post-transfection (Figure 1B). The luciferase signal and Z-factor increased over the course of the assay (Figure 1B). There was little activity and a negative Z-factor at 24 h post-transfection (fold induction of 21 and Z-factor = −0.7), which increased to >2000-fold induction and a Z-factor of 0.6 at 48 h post-transfection (Figure 1B). We therefore chose 48 h post-transfection as the end-point of our EBOV minigenome assay due to the robust activity and Z-factor observed. The robust activity and Z-factors are consistent between experiments and from day to day (data not shown). These data show that the EBOV minigenome assay can be successfully adapted for HTS in 384-well format.
High-Throughput Screen for Identifying Inhibitors of EBOV RNA Synthesis
Using the optimized conditions for 384-well format, we screened a library of bioactive chemicals (2080 compounds) to identify compounds that inhibit EBOV RNA synthesis (Figure 2A). Briefly, the optimized conditions included bulk transfecting HEK293T cells in a T75 flask overnight. The cells were then replated in a 384-well plate and allowed to rest for 1 h before compound addition. Due to the facts that little luciferase activity was detected at 24 h post-transfection and the signal greatly increased by 48 h (Figure 1B), we reasoned that for library screening, 24 h would be an appropriate time to add compounds and 48 h would be an appropriate time to read the luciferase activity (Figure 2A). The quality control plate showed that DMSO at the concentration used in the screen, 0.07%, had no effect on the activity of the minigenome (Figure 2B). 6-Azauridine, an inhibitor of both EBOV minigenome activity and EBOV, was used as a positive control at 7 μM, a concentration that inhibited activity by approximately 70% (Figure 2B).5 The eight plates in the library were screened in duplicate, with all plates having a Z-factor ≥0.5, indicating a robust assay (Figure 2C). Using the duplicate plates, average percent inhibition and Z-scores were calculated for each compound, and those compounds with ≥50% inhibition were plotted against their Z-score (Figure 2D). We identified 257 compounds (12.4% of the library) that inhibited minigenome activity by >50%, 31 (1.5% of the total library) of which reduced minigenome activity by >70%.
Figure 2.
High-throughput screen for identifying EBOV RNA synthesis inhibitors. (A) Schematic diagram of EBOV minigenome HTS assay. HEK293T cells were transfected in bulk in a T75 flask. Twenty-four hours post-transfection cells were plated in a 384-well plate and allowed to rest for 1 h, after which the compound library was transferred via pin tool (final concentration = 7–7.7 μM). Twenty-four hours following compound addition Renilla luciferase activity was measured. (B) A quality control plate was used to assess the effects of DMSO and the efficiency of pin tool transfer prior to the screen. Twenty-four hours post-transfection cells were plated in 384-well format. DMSO (final concentration = 0.07%) was added via pin tool transfer, and 6-azauridine (6-Aza) (final concentration = 7 μM) was added using an HP D300 digital dispenser. Twenty-four hours post-treatment luciferase activity was assessed, and Z-factor values were calculated using the formula Z-factor = 1 − [(3σc+ + 3σc−)/(|μc+ − μc−|)]. No VP35, black triangles, 32 wells; VP35, black circles, 240 wells; VP35+DMSO, open circles, 32 wells; VP35 + 6-Aza, gray diamonds, 80 wells. (C) Z-Factor values were calculated from the controls on each plate in the screen of known bioactive compounds. Duplicate plates are designated A and B (i.e., duplicates of library plate 1 are 1A and 1B). (D) Z-Scores and percent inhibitions were calculated for each compound (two replicates), and those with inhibition ≥50% were plotted against their Z-score.
Validating Hits Identified in Primary Screen
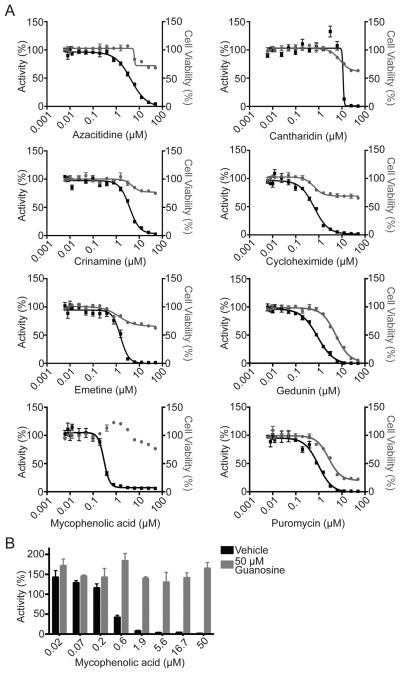
To evaluate the reproducibility of identified hits, 19 compounds that varied in inhibition from 56 to 100% in the screen were chosen for retesting. The 50% inhibitory concentrations (IC50) and 50% cell cytotoxic concentrations (CC50) were assessed for the compounds in parallel to allow for elimination of those that inhibited EBOV minigenome activity by causing cell death (Figure 3A and Table 1). The selectivity index (SI) of each compound was calculated using the CC50 and IC50 values (Table 1). Of the 19 compounds retested, 5 had inhibited minigenome activity by ≤70% in the primary screen. None of these yielded significant inhibition in the retest, as they did not achieve 50% inhibition with any of the concentrations tested (0–50 μM, 2-fold dilution series) (Table 1). Several of the compounds that were highly inhibitory in the screen were also removed following retesting, as they were also highly cytotoxic (Table 1). Of the 19 compounds retested, we identified 8 compounds with IC50 values below their CC50 values, 5 of which have SI values >10 (Figure 3A and Table 1). Three compounds are protein synthesis inhibitors: puromycin, emetine, and cycloheximide.13,14 Azacitidine, a cytidine analogue, is known to inhibit both protein and DNA synthesis.15 Crinamine is an inhibitor of hypoxia-inducible factor 1 (HIF1).16 Cantharidin is an inhibitor of protein phosphatases 1 and 2A (PP1 and PP2A).17 Gedunin inhibits heat shock protein 90 (HSP90).18 Finally, mycophenolic acid is an inhibitor of inosine monophosphate dehydrogenase (IMPDH), an enzyme that catalyzes the rate-limiting step toward the de novo biosynthesis of guanine (GTP) nucleotides.19 Hit compounds emetine, cycloheximide, and mycophenolic acid had SI values of greater than 34, 82, and 158, respectively, suggesting that they inhibit EBOV minigenome activity at concentrations independent of their cytotoxic effects. It should be noted, however, that although these compounds did not reach a CC50 value when assayed at 24 h after addition in our assays, it is possible that some, such as the protein synthesis inhibitors, would have reached a CC50 value if the cytotoxicity assay was extended to 48 or 72 h after compound addition.
Figure 3.
Validation of hits identified in primary screen. (A) Twenty-four hours post-transfection, HEK293T cells were plated in a 384-well plate and treated with increasing concentrations of the indicated compounds (0–50 μM) in triplicate. Twenty-four hours post-treatment Renilla luciferase activity was assessed (left axis, black line, solid squares). In parallel, HEK293T cells were plated in a 384-well plate and treated in triplicate with increasing concentrations of compounds (0–50 μM). Twenty-four hours post-treatment, ATP content was assessed to determine cell viability (right axis, gray line, solid circles). Error bars represent the standard error of the mean. (B) Mycophenolic acid inhibits minigenome activity through GTP depletion. Twenty-four hours post-transfection HEK293T cells were plated in a 96-well plate. Guanosine was resuspended in DMEM, and either DMEM (vehicle) (black columns) or 50 μM guanosine (gray columns) was added to the wells. Cells were treated with increasing concentrations of mycophenolic acid, and 24 h post-treatment Renilla luciferase was read. Data represents the mean and standard error of the mean in triplicate, normalized to nontreated transfected cells.
Table 1.
Retest of Hit Compounds from Bioactive Library
| compound | % inhibition in screen | CC50 (μM) | IC50 (μM) | SI (CC50/IC50) | |
|---|---|---|---|---|---|
| 1 | lanatoside C | 100.0 | 0.177 | 0.255 | 0.694 |
| 2 | gambogic amide | 100.0 | 0.454 | 1.01 | 0.45 |
| 3 | strophanthidin | 100.0 | 0.491 | 1.161 | 0.423 |
| 4 | acetyl isogambogic acid | 99.0 | 2.885 | 2.935 | 0.983 |
| 5 | puromycin hydrochloride | 99.7 | 3.594 | 0.889 | 4.043 |
| 6 | emetine | 98.3 | >50 | 1.474 | >33.921 |
| 7 | plumbagin | 98.3 | 0.756 | 0.919 | 0.823 |
| 8 | cycloheximide | 97.9 | >50 | 0.608 | >82.237 |
| 9 | digitoxin | 92.5 | 0.031 | 0.05 | 0.62 |
| 10 | crinamine | 91.5 | >50 | 3.789 | >13.196 |
| 11 | mycophenolic acid | 90.5 | >50 | 0.316 | >158.228 |
| 12 | cantharidin | 87.3 | >50 | 11.325 | >4.415 |
| 13 | azacitidine | 76.9 | >50 | 4.011 | >12.466 |
| 14 | gedunin | 72.9 | 5.484 | 0.803 | 6.829 |
| 15 | fluorouracil | 70.6 | >50 | >50 | |
| 16 | methotrexate | 67.1 | >50 | >50 | |
| 17 | gemfobrozil | 62.1 | >50 | >50 | |
| 18 | dramamine | 57.8 | >50 | >50 | |
| 19 | vidarabine | 56.1 | >50 | >50 |
Previously, mycophenolic acid was demonstrated to inhibit EBOV replication through the depletion of the GTP pool such that the inhibition can be reversed by the addition of exogenous guanosine.20 To assess whether the inhibition of minigenome activity by mycophenolic acid is also due to depletion of the GTP pool, we added mycophenolic acid to the minigenome assay and either did or did not supplement the media with exogenous guanosine. Guanosine addition rescued minigenome activity at all concentrations of mycophenolic acid tested, recapitulating what is seen with infectious virus (Figure 3B). This suggests that the minigenome assay and infectious EBOV are similarly sensitive to cellular nucleotide pools.
Validation of Selected Hit Compounds versus EBOV-GFP Replication
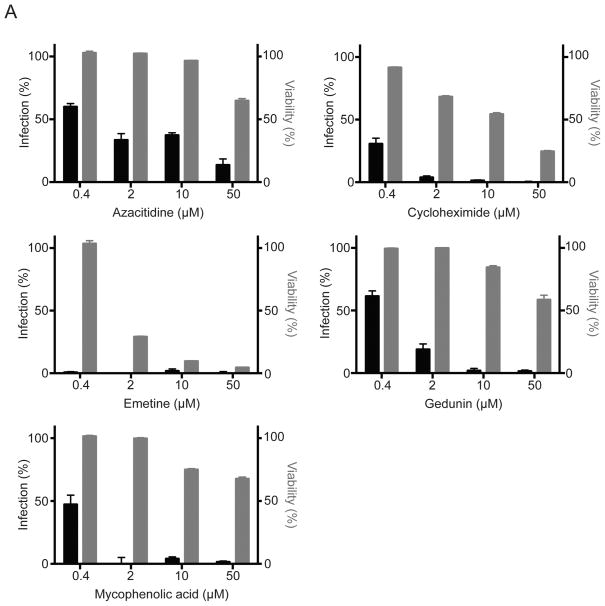
Next, we tested whether five hit compounds, azacitidine, cycloheximide, emetine, gedunin, and mycophenolic acid, also inhibit replication of an infectious EBOV expressing GFP (EBOV-GFP).3 We were unable to find crinamine for purchase and were therefore unable to test its activity against EBOV. Additionally, cantharidin was not further investigated as it is a potent toxin, producing blisters upon skin contact and resulting in severe irritation and ulceration following oral ingestion, and had a high IC50 value against minigenome activity (11.3 μM (Figure 3A)).17 A 96-well assay that assesses GFP expression from a recombinant EBOV that expresses GFP (EBOV-GFP) was used.
Following drug pretreatment, virus was added at an MOI of 0.3 in the presence of compound for the duration of the 3 day experiment, after which virus replication, detected by GFP expression, was measured (Figure 4). Cell cytotoxicity was assessed in parallel on uninfected cells by measuring ATP content. Mycophenolic acid and gedunin significantly reduced virus replication by 96 and 98% at 10 μM (black bars), respectively, while causing little cell death (gray bars) (Figure 4). Emetine and cycloheximide are cytotoxic at higher concentrations. However, at concentrations of 0.4 and 2 μM, cytotoxicity was substantially lower and EBOV replication was still reduced by 99 and 96%, respectively (Figure 4). Inhibition of EBOV replication by azacitidine was 86% at the highest concentration tested, 50 μM, although inhibition titrates out quickly (Figure 4). Taken together, these data demonstrate that the high-throughput minigenome assay is able to identify inhibitors of EBOV replication.
Figure 4.
Antiviral activity of hit compounds. To measure the antiviral activity of the compounds, Vero E6 cells were plated in a 96-well plate overnight and then pretreated with increasing concentrations of compound for 1 h, after which they were infected with EBOV-GFP at an MOI of 0.3. Three days post-infection the mean fluorescence intensity (MFI) was read to assess GFP expression and normalized to untreated controls to determine percent infection (left axis, black bars). In parallel, Vero E6 cells in a 96-well plate were treated with increasing concentrations of compounds, and ATP content was assessed 5 days post-treatment to determine cell viability (right-axis, gray bars). Data represent the mean and standard error of the mean (SEM) of triplicates.
DISCUSSION
The viral polymerase complex is essential for EBOV replication and as such is a potential target for antifilovirus therapeutics. The utility of the EBOV minigenome assay for screening large compound libraries has been limited by the constraints of miniaturization while maintaining assay robustness. Previous optimization of the EBOV minigenome assay has miniaturized the assay to 96-well format.5,10 To further optimize the assay for 384-well format, we increased the ratio of Lipofectamine 2000 transfection reagent to DNA concentration and also established a transfection in bulk to decrease variability. These steps allowed us to miniaturize the minigenome assay for HTS to 384-well format while preserving the robustness of the assay (as assessed by Z-factor and fold induction). Using this HTS minigenome assay, we identified several compounds that inhibit both EBOV minigenome activity and infectious virus. These compounds likely target host factors important for EBOV replication and EBOV polymerase activity.
Interestingly, the HTS identified the compound cantharidin, a potent inhibitor of the phosphatases PP1 and PP2A.17 These phosphatases regulate EBOV polymerase activity by affecting the phosphorylation status of VP30, which, in turn, determines whether viral transcription or replication occurs.21–23 Dephosphorylated VP30 promotes viral transcription, and inhibition of the protein phosphatases 1 and 2A (PP1 and PP2A) prevents VP30 dephosphorylation, impairing transcription and leading to a loss of reporter gene expression in minigenome assays.21–23 Previously identified inhibitors of PP1 and PP2A, including the PP1-specific small-molecule inhibitor 1E7-03 and the PP1/PP2A inhibitor okadaic acid, have been shown to inhibit EBOV transcription and EBOV minigenome activity.22,23
The EBOV minigenome screen also identified an inhibitor of HSP90, gedunin.18 Inhibition of HSP90 results in significant reduction in EBOV replication.24 This may reflect the ability of HSP90 inhibitors to destabilize the L proteins of nonsegmented, negative-sense RNA viruses, because antiviral activity and RNA polymerase degradation following HSP90 inhibition has previously been identified in a range of negative-strand RNA viruses.25 Additionally, mycophenolic acid, a previously identified inhibitor of infectious EBOV replication, was identified by the minigenome assay. Our results show that mycophenolic acid inhibits EBOV minigenome activity through the inhibition of IMPDH, the same mechanism that leads to the inhibition of infectious EBOV.20 Taken together, the identification of inhibitors of PP1/PP2A, HSP90, and IMPDH by the EBOV minigenome HTS illustrates the ability of the minigenome assay to act as a surrogate for infectious virus, requiring similar host factors for efficient polymerase activity.
Nearly half of the hit compounds with inhibitory concentrations below cell cytotoxic concentrations are protein synthesis inhibitors: puromycin, emetine, and cycloheximide.13–15 The activity of the EBOV minigenome assay is sensitive to the levels of the polymerase complex components; decreasing or increasing amounts of plasmids for L, VP30, VP35, or NP can greatly reduce reporter activity (data not shown). Therefore, protein synthesis inhibitors could result in reduced expression of polymerase complex components, leading to inhibition of minigenome activity. These results were mirrored in the inhibitory activity of the compounds against infectious virus, as emetine and cycloheximide were able to significantly reduce EBOV replication at concentrations that caused little cell death by 24 h after compound addition. Interestingly, emetine and cycloheximide both inhibit protein synthesis at the level of translation elongation, preventing release of the nascent peptide.13 A previous study has shown that EBOV transcribes very low levels of L-encoding mRNA, suggesting that little L protein is expressed and that, perhaps, L expression will be particularly sensitive to protein synthesis inhibitors.26 More specifically, a recombinant EBOV with a mutation in the upstream-untranslated region (UTR) of L that leads to an increase in L expression also results in reduced virus replication. This suggests that modulating the expression of the components in the polymerase complex could be a useful therapeutic strategy. Further study is required to determine the basis for the sensitivity of EBOV to protein synthesis inhibitors and whether this might be exploited to specifically target the expression of the viral polymerase.
In summary, through the use of bulk transfection to limit variability, we have successfully optimized the EBOV minigenome HTS for a 384-well format. The assay was demonstrated to successfully identify compounds that inhibit EBOV minigenome activity and infectious EBOV, including compounds that support previous antifilovirus therapeutic directions, such as HSP90 and protein phosphatase inhibitors. The HTS format will allow for screening of large compound libraries for inhibitors of EBOV RNA synthesis, accelerating the search for filovirus therapeutics.
Acknowledgments
This work was supported by NIH Grants U19AI109664 (Basler) to C.F.B. and A.B. and R01AI101308 (Shaw) to M.L.S. and C.F.B. HTS of the bioactive library took place in the Integrated Screening Core at the Icahn School of Medicine at Mount Sinai. We thank Dr. Sharmila Sivendran and Dr. Dan Felsenfeld for their help with the HTS. We thank Christine Schwall for critically reading the manuscript.
Footnotes
The authors declare no competing financial interest.
References
- 1.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agua-Agum J, Ariyarajah A, Aylward B, Blake IM, Brennan R, Cori A, Donnelly CA, Dorigatti I, Dye C, Eckmanns T, Ferguson NM, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D, Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills HL, Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L, Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van Kerkhove MD, Varsaneux O, Wijekoon Kannangarage N. West African Ebola epidemic after one year - slowing but not yet under control. N Engl J Med. 2015;372:584–587. doi: 10.1056/NEJMc1414992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- 4.Ebihara H, Theriault S, Neumann G, Alimonti JB, Geisbert JB, Hensley LE, Groseth A, Jones SM, Geisbert TW, Kawaoka Y, Feldmann H. In vitro and in vivo characterization of recombinant Ebola viruses expressing enhanced green fluorescent protein. J Infect Dis. 2007;196(Suppl 2):S313–S322. doi: 10.1086/520590. [DOI] [PubMed] [Google Scholar]
- 5.Uebelhoer LS, Albarino CG, McMullan LK, Chakrabarti AK, Vincent JP, Nichol ST, Towner JS. High-throughput, luciferase-based reverse genetics systems for identifying inhibitors of Marburg and Ebola viruses. Antiviral Res. 2014;106:86–94. doi: 10.1016/j.antiviral.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Hoenen T, Groseth A, Callison J, Takada A, Feldmann H. A novel Ebola virus expressing luciferase allows for rapid and quantitative testing of antivirals. Antiviral Res. 2013;99:207–213. doi: 10.1016/j.antiviral.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madrid PB, Chopra S, Manger ID, Gilfillan L, Keepers TR, Shurtleff AC, Green CE, Iyer LV, Dilks HH, Davey RA, Kolokoltsov AA, Carrion R, Jr, Patterson JL, Bavari S, Panchal RG, Warren TK, Wells JB, Moos WH, Burke RL, Tanga MJ. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One. 2013;8:e60579. doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muhlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoenen T, Groseth A, de Kok-Mercado F, Kuhn JH, Wahl-Jensen V. Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 2011;91:195–208. doi: 10.1016/j.antiviral.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasenosky LD, Neumann G, Kawaoka Y. Minigenome-based reporter system suitable for high-throughput screening of compounds able to inhibit Ebolavirus replication and/or transcription. Antimicrob Agents Chemother. 2010;54:3007–3010. doi: 10.1128/AAC.00138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, Amarasinghe GK, Basler CF. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe. 2013;14:74–84. doi: 10.1016/j.chom.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 13.Grollman AP. Structural basis for inhibition of protein synthesis by emetine and cycloheximide based on an analogy between ipecac alkaloids and glutarimide antibiotics. Proc Natl Acad Sci U S A. 1966;56:1867–1874. doi: 10.1073/pnas.56.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathans D. Puromycin Inhibition of Protein Synthesis: Incorporation of Puromycin into Peptide Chains. Proc Natl Acad Sci U S A. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson-Thompson M, Albury D. Azapyrimidine analogues: inhibition of viral DNA synthesis and protein synthesis in SV40 infected BSC-1 cells. In Vitro Cell Dev Biol. 1988;24:1114–1120. doi: 10.1007/BF02620813. [DOI] [PubMed] [Google Scholar]
- 16.Kim YH, Park EJ, Park MH, Badarch U, Woldemichael GM, Beutler JA. Crinamine from Crinum asiaticum var. japonicum inhibits hypoxia inducible factor-1 activity but not activity of hypoxia inducible factor-2. Biol Pharm Bull. 2006;29:2140–2142. doi: 10.1248/bpb.29.2140. [DOI] [PubMed] [Google Scholar]
- 17.Honkanen RE. Cantharidin, another natural toxin that inhibits the activity of serine/threonine protein phosphatases types 1 and 2A. FEBS Lett. 1993;330:283–286. doi: 10.1016/0014-5793(93)80889-3. [DOI] [PubMed] [Google Scholar]
- 18.Patwardhan CA, Fauq A, Peterson LB, Miller C, Blagg BS, Chadli A. Gedunin inactivates the co-chaperone p23 protein causing cancer cell death by apoptosis. J Biol Chem. 2013;288:7313–7325. doi: 10.1074/jbc.M112.427328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Y, Gu J, Makhov AM, Griffith JD, Mitchell BS. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J Biol Chem. 2006;281:206–212. doi: 10.1074/jbc.M507056200. [DOI] [PubMed] [Google Scholar]
- 20.Olschlager S, Neyts J, Gunther S. Depletion of GTP pool is not the predominant mechanism by which ribavirin exerts its antiviral effect on Lassa virus. Antiviral Res. 2011;91:89–93. doi: 10.1016/j.antiviral.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Martinez MJ, Volchkova VA, Raoul H, Alazard-Dany N, Reynard O, Volchkov VE. Role of VP30 phosphorylation in the Ebola virus replication cycle. J Infect Dis. 2011;204(Suppl 3):S934–S940. doi: 10.1093/infdis/jir320. [DOI] [PubMed] [Google Scholar]
- 22.Modrof J, Muhlberger E, Klenk HD, Becker S. Phosphorylation of VP30 impairs ebola virus transcription. J Biol Chem. 2002;277:33099–33104. doi: 10.1074/jbc.M203775200. [DOI] [PubMed] [Google Scholar]
- 23.Ilinykh PA, Tigabu B, Ivanov A, Ammosova T, Obukhov Y, Garron T, Kumari N, Kovalskyy D, Platonov MO, Naumchik VS, Freiberg AN, Nekhai S, Bukreyev A. Role of protein phosphatase 1 in dephosphorylation of Ebola virus VP30 protein and its targeting for the inhibition of viral transcription. J Biol Chem. 2014;289:22723–22738. doi: 10.1074/jbc.M114.575050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DR, McCarthy S, Chrovian A, Olinger G, Stossel A, Geisbert TW, Hensley LE, Connor JH. Inhibition of heat-shock protein 90 reduces Ebola virus replication. Antiviral Res. 2010;87:187–194. doi: 10.1016/j.antiviral.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connor JH, McKenzie MO, Parks GD, Lyles DS. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology. 2007;362:109–119. doi: 10.1016/j.virol.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabman RS, Jabado OJ, Mire CE, Stockwell TB, Edwards M, Mahajan M, Geisbert TW, Basler CF. Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells. mBio. 2014;5:e02011. doi: 10.1128/mBio.02011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]