Abstract
Introduction:
The use of clinically derived data from electronic health records (EHRs) and other electronic clinical systems can greatly facilitate clinical research as well as operational and quality initiatives. One approach for making these data available is to incorporate data from different sources into a joint data warehouse. When using such a data warehouse, it is important to understand the quality of the data. The primary objective of this study was to determine the completeness and concordance of common types of clinical data available in the Knowledge Program (KP) joint data warehouse, which contains feeds from several electronic systems including the EHR.
Methods:
A manual review was performed of specific data elements for 250 patients from an EHR, and these were compared with corresponding elements in the KP data warehouse. Completeness and concordance were calculated for five categories of data including demographics, vital signs, laboratory results, diagnoses, and medications.
Results:
In general, data elements for demographics, vital signs, diagnoses, and laboratory results were present in more cases in the source EHR compared to the KP. When data elements were available in both sources, there was a high concordance. In contrast, the KP data warehouse documented a higher prevalence of deaths and medications compared to the EHR.
Discussion:
Several factors contributed to the discrepancies between data in the KP and the EHR—including the start date and frequency of data feeds updates into the KP, inability to transfer data located in nonstructured formats (e.g., free text or scanned documents), as well as incomplete and missing data variables in the source EHR.
Conclusion:
When evaluating the quality of a data warehouse with multiple data sources, assessing completeness and concordance between data set and source data may be better than designating one to be a gold standard. This will allow the user to optimize the method and timing of data transfer in order to capture data with better accuracy.
Keywords: Data use and quality, health information technology, quality
Introduction
The use of electronic health records (EHRs) and other electronic clinical systems that capture data about clinical encounters and patient’s care continue to grow rapidly.1,2 Linking clinically derived data from these different systems can greatly facilitate clinical research and operational and quality initiatives.1 There are several possible approaches to make these data available for such uses, including (1) maintaining separate repositories for different data sources and linking data after extraction, and (2) incorporating data from different sources into a new data warehouse. Each approach presents unique benefits and challenges, with the institutional choice of approach depending on a variety of conceptual and pragmatic factors. With any approach, evaluation of data quality is necessary to ensure the validity and generalizability of findings from clinical research and operational and quality initiatives. However, data quality evaluation of data linked from different sources faces a number of unique challenges given the complexities of the underlying information systems.
The paper presents a data quality evaluation of a new joint data warehouse formed using the second approach, the Knowledge Program (KP) data warehouse at the Cleveland Clinic. After designing and implementing the approach to form the KP, we focused on evaluating and adjusting our strategy by collecting and assessing data quality using two commonly used metrics of quality: completeness and concordance. “Completeness” refers to the proportion of cases where data are recorded in the system,3 and “concordance” is a measure of value agreement;4 neither relies on the presence of a gold standard. The primary objective of our analysis was to determine the completeness and concordance of common clinical data variables available in the joint warehouse when compared to a patient’s medical record, in preparation for its use in quality and research activities. A novel aspect of this project was that a “gold standard” was not used. Selecting a gold standard may have introduced errors, due to the fact that the EHR was populated with data from multiple sources at variable intervals. To disseminate our findings to others, we discuss the challenges associated with evaluating the data quality of the joint warehouse in this comparative fashion and relate the lessons learned.
Measuring Data Quality
Most commonly, validation studies of electronic data employ conventional statistical tools such as sensitivity, specificity, and positive and negative predictive values, typically comparing the electronic data with a manual medical record review or direct patient report.5 Additional measures that have been recommended when validating data include the following: the ratio of true positive cases to false negative cases (TPFN ratio)5 and the ratio of false negatives cases to the total number of patients in a database multiplied by 10,000 (DBFind10,000).5 These methods require the existence of a reference- or gold standard. Other methods, such as the kappa value, are prevalence dependent with limited generalizability to other settings where prevalence differs.6 Although a manual review of data stored in the EHR seems an obvious choice for such a reference standard when validating data housed in a joint database, several issues complicate this decision. The complexity of data feeds received by a joint database like the KP results in data being obtained from several different sources, such that there is no single complete reference. Data tied to a patient, rather than an encounter, varies over time depending on the date at which it is assessed. In a large, multisite institution, Health-Level 7 (HL7) feeds from different locations rarely “turn on” at a single time point, making it difficult to assess a firm start date for evaluation. The complexity of available locations for entering data into the EHR presents perhaps the greatest challenge, sometimes resulting in less information being available upon review of a single encounter in the EHR, as compared to the joint warehouse. These factors motivate the evaluation of data quality using criteria that do not assume either source to be complete; namely, descriptive statistics in terms of (1) the presence or absence of a given value in each source (completeness), and (2) concordance between the two values in cases where they are both present.
In this study, completeness and concordance were assessed for demographic information, common laboratory results, vital signs, diagnoses, and medications. These data variables represent the major categories of data in the KP data warehouse that are used for secondary analyses. We anticipated that the assessment would identify three major characteristics of data quality in the KP, including the following: (1) completeness and concordance would be highest with data elements that do not require manual entry into the EHR (e.g., laboratory results); (2) when demographics and vital signs were present, there would be a high level of agreement between the two sources; and, (3) the highest levels of disagreement would be with medication and diagnoses data, due to the variety of locations and ways in which they can be stored, in addition to the wide range of options available for data entry into the EHR.
Methods
Study Setting
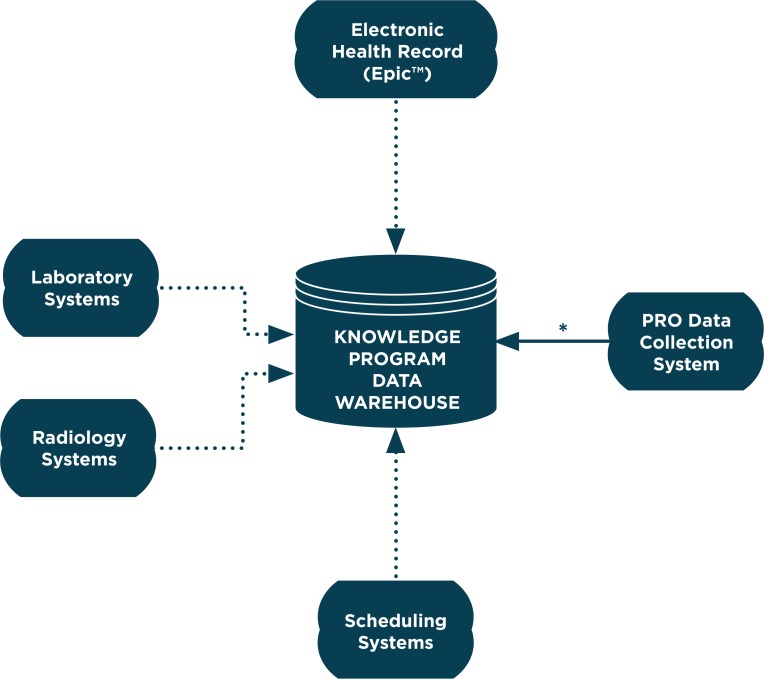
The Cleveland Clinic has developed the KP data warehouse to more effectively use clinical data to inform clinical operations, quality, and research. The KP data warehouse was first used in the Neurological Institute (NI), which consists of four departments: Neurology, Neurosurgery, Psychiatry, and Physical Medicine and Rehabilitation. This warehouse includes direct HL7 feeds from the EHR (Epic, Verona, Wis.), and laboratory, radiology, and scheduling systems (Figure 1). The KP data warehouse also serves as the main repository for patient-reported outcome measures (PROMs) that are collected at the point of care through a Web-based system.7 The availability of PROMs was one of the catalysts to develop this warehouse and include feeds from additional data sources. The focus of this investigation was on the quality of warehouse data compared to the EHR, since that can be also be used as a data source for aggregate analyses.
Figure 1.
Knowledge Program Data Warehouse
Solid Arrow represents a direct data feed. Dotted arrow represents a data feed via HL7 interface.
PRO=patient reported outcomes
*Knowledge Program Date Warehouse is primary data repository for PRO Data Collection System
A manual review of electronic charts was performed by first reviewing an electronic chart based on one specific encounter in a commercially available EHR, followed by a comparison with the same data available in the KP database. Completeness and concordance were determined for the five categories shown in Table 1.
Table 1.
Categories Evaluated for Completeness and Concordance
| 1. DEMOGRAPHIC INFORMATION | 2. LABORATORY RESULTS | |
|
|
|
| 3. VITAL SIGNS | 4. MEDICATIONS | |
|
|
|
| 5. DIAGNOSES | ||
|
|
|
Notes: CO2 = carbon dioxide, BUN = blood urea nitrogen, HgbA1C = hemoglobin A1C, WBC = white blood count, TSH= thyroid stimulating hormone
Sample Size
For this study, 250 charts of patients seen in the NI were reviewed. One encounter was reviewed from each of the 250 patient charts. The NI was chosen as a focus for this review as it represented the largest user of the data warehouse. The sample size was chosen to limit the time-consuming process of manual review, but still provide adequate power to estimate the sensitivity of a chosen diagnosis, with a reasonable margin of error (e.g., coronary artery disease with an expected prevalence of 18 percent). In order to represent adequately all 15 centers in the NI, a random sample of charts were proportionally stratified by center, based on the patient volumes of each center. We excluded patients under 18 years old at the time of the clinical encounter. The analysis included patient encounters occurring between January 2010 and September 2012.
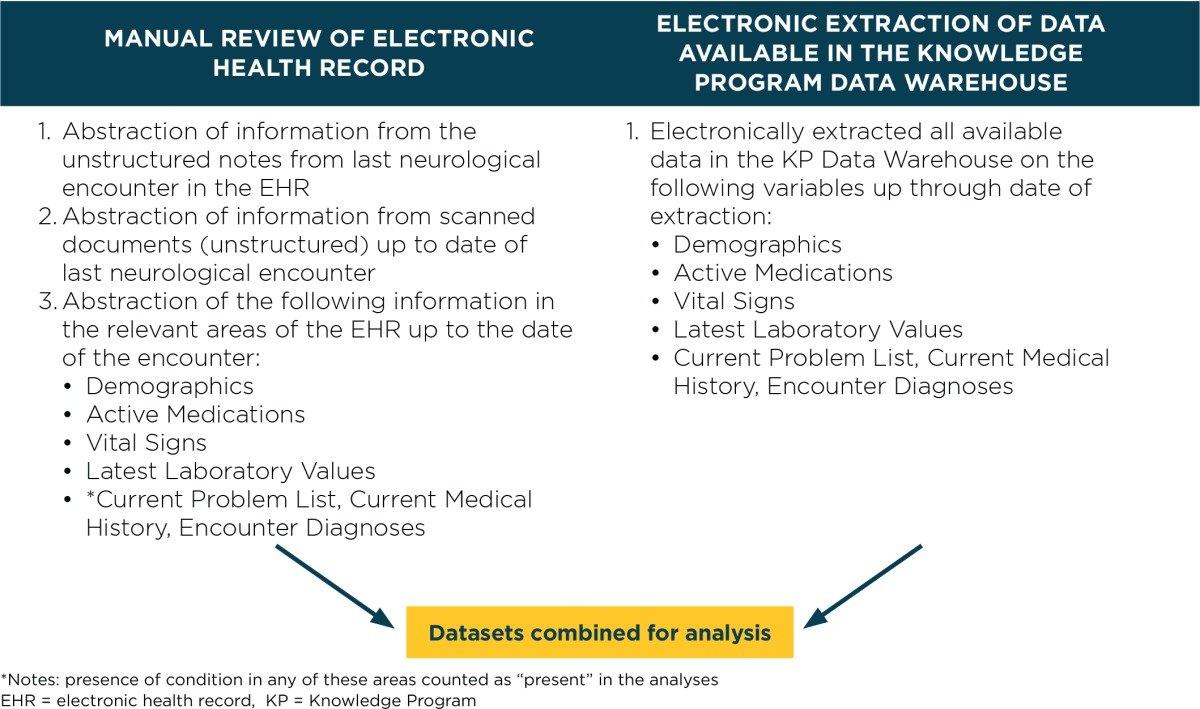
Manual Chart Abstraction of the Electronic Health Record (EHR)
Manual chart abstractions of the EHR were performed by a physician who reviewed the most recent NI-based encounter during the study period. Clinical information in the reviewed encounter note was abstracted, including vital signs, diagnoses, and medications, regardless of whether it was entered as free-text or through autopopulation of clinical data from other areas in the EHR. Scanned documents were reviewed only if they were recorded at or before the date of the encounter. Information available in the other areas of EHR at the time of the review was abstracted for demographic information, laboratory results, vital signs, active medications, and diagnoses. Level of agreement of data elements between the EHR and KP was determined for the following diagnoses: cancer, coronary artery disease, diabetes, stroke, chronic renal insufficiency, depression, hypertension, atrial fibrillation, hypothyroidism, obesity, and sleep apnea. Three areas of the electronic chart were considered when evaluating diagnoses: past medical history, problem list, and the encounter diagnosis section. Notes from nursing, home care, physical and occupational therapy, speech therapy, pastoral care, social work, telephone encounters, and financial counselors were excluded from the review, as was additional documentation unrelated to a specific encounter.
Knowledge Program (KP) Data Warehouse Extraction
Data from the KP data warehouse on the 250 subjects were electronically extracted and placed into a study data set, which included all laboratory values, vital signs, diagnoses, and medications in the KP data warehouse up until the time of extraction.
A data registry registered nurse reformatted the extracted data set and merged this with the manually abstracted EHR data on these subjects. The manual EHR abstraction occurred over a period of approximately one year. The KP data extraction into the study data set occurred at a single time point. The difference in these time points is an additional source of potential disagreement.
Data Management and Statistical Analysis
The data sets for this study (Table 2) were housed in the Research Electronic Data Capture (REDCap) tool hosted at Cleveland Clinic.8 REDCap is a secure, Web-based application designed to support data capture for research studies, providing the following: (1) an intuitive interface for validated data entry; (2) audit trails for tracking data manipulation and export procedures; (3) automated export procedures for seamless data downloads to common statistical packages; and (4) procedures for importing data from external sources.
Table 2.
Methods for Electronic Health Record Abstraction and Knowledge Program Data Warehouse Extraction
Descriptive statistics were computed for demographics (mean and standard deviation for age, frequency counts, and percentages for gender, race, etc.). Completeness of demographic, lab, diagnoses, medical use, and vital signs data was determined by calculating the percent of cases in which each was present in the EHR and KP, respectively, and computing the discrepancy (number and percent) between the two sources. Concordance was determined for categorical variables by calculating the number and percent of cases in which the data values were identical (including missing) in both data sets. Comparisons between the EHR and KP were made both excluding and including free text EHR fields (scanned documents and progress notes). Completeness and concordance of common clinical data values in the joint warehouse was performed without identifying a gold standard. Having a gold standard is a potential source of errors, since the EHR contains data from a variety of sources that are entered at different intervals.
Results
The mean age of the patients in this study was 51.4 (SD = 16.1), consisting of more women than men (59.6 percent vs. 40.4 percent). The group was primarily white (79.2 percent), and a minority was black (14.0 percent). The majority of patients were married (62.0 percent), followed by single (19.6 percent), divorced (9.2 percent), and widowed (5.2 percent).
The results comparing completeness and concordance between the two sources are displayed in Tables 3–5. Data elements for demographic data (Table 3) were present in more cases in the EHR (100 percent) compared to the KP data warehouse (98.4–99.6 percent) in all instances except for date of death, for which there were more data elements in the KP data warehouse than the EHR (3.2 percent vs. 1.2 percent). In cases where the demographic data elements were present in both the EHR and the KP data warehouse, values between the two sources were concordant over 98 percent of the time. Select labs (Table 4) were recorded in the KP data warehouse in 1.6 percent (fasting blood sugar) to 52.4 percent (creatinine, blood urea nitrogen, and glucose) of cases. The EHR review was more complete relative to the KP data warehouse with a 0.8–6.4 percent difference in the presence of specific lab values between the two. Vital signs were recorded in 25.6 percent (height) to 69.6 percent (blood pressure) of cases. For all vital signs (Table 5), the EHR was 3.2–9.2 percent more complete, relative to the KP. There was complete agreement in values for laboratory results and vitals when they were present in both the EHR and KP data set.
Table 3.
Completeness and Concordance of Demographic Data Elements between the Electronic Health Record and the Knowledge Program
| TOTAL N1= 250 | COMPLETENESS | CONCORDANCE | ||
|---|---|---|---|---|
| EHR | KP | % EHR ONLY, (#EHR – #KP) | % MATCH (Nm2), INCLUDE BOTH MISSING CASE | |
| % OF TOTAL RECORDS (# PRESENT) | % OF TOTAL RECORDS (# PRESENT) | |||
| Date of Birth | 100 (250) | 99.6 (249) | 0.4 (1) | 99.6 (249) |
| Date of Death | 1.2 (3) | 3.2 (8) | −2 (−5) | 98.0 (245) |
| Sex | 100 (250) | 99.6 (249) | 0.4 (1) | 99.6 (249) |
| Race | 100 (250) | 99.2 (248) | 0.8 (2) | 98.8 (247) |
| Marital Status | 100 (250) | 98.4 (246) | 1.6 (4) | 98.4 (246) |
| City | 100 (250) | 99.6 (249) | 0.4 (1) | 98.8 (247) |
| State | 100 (250) | 99.6 (249) | 0.4 (1) | 99.6 (249) |
| ZIP Code | 100 (250) | 99.6 (249) | 0.4 (1) | 98.4 (246) |
Notes: Includes EHR data extracted from nonstructured fields
EHR= electronic health record, KP= Knowledge Program
N, total number of observations
Nm, the number of matches between EHR and KP data, including cases where both are missing
Table 4.
Completeness and Concordance of Numeric Data Elements between the Electronic Health Record and the Knowledge Program
| TOTAL N1= 250 | COMPLETENESS | CONCORDANCE | ||
|---|---|---|---|---|
| EHR REVIEW | KP | % EHR ONLY, (#EHR – #KP) | % MATCH (Nm2), INCLUDE BOTH MISSING CASE | |
| % OF TOTAL RECORDS (# PRESENT) | % OF TOTAL RECORDS (# PRESENT) | |||
| LABORATORY VALUES | ||||
| Sodium | 57.6 (144) | 51.6 (129) | 6 (15) | 94 (235) |
| Potassium | 58 (145) | 52 (130) | 6 (15) | 94 (235) |
| CO2 | 57.6 (144) | 52 (130) | 5.6 (14) | 94.4 (236) |
| Chloride | 58 (145) | 52 (130) | 15, 6 (15) | 94 (235) |
| BUN | 58 (145) | 52.4 (131) | 5.6 (14) | 94.4 (236) |
| Creatinine | 58.8 (147) | 52.4 (131) | 6.4 (16) | 93.6 (234) |
| Glucose | 58.4 (146) | 52.4 (131) | 6 (15) | 94 (235) |
| Calcium | 56.8 (142) | 50.8 (127) | 6 (15) | 94 (235) |
| Albumin | 50 (125) | 45.6 (114) | 4.4 (11) | 95.6 (239) |
| Total Protein | 48 (120) | 43.6 (109) | 4.4 (11) | 95.6 (239) |
| HgbA1C | 14.8 (37) | 13.6 (34) | 1.2 (3) | 98.8 (247) |
| Fasting Blood Sugar | 2.4 (6) | 1.6 (4) | 0.8 (2) | 99.2 (248) |
| WBC | 54.8 (137) | 48.4 (121) | 6.4 (16) | 93.6 (234) |
| Hemoglobin | 54.8 (137) | 48.4 (121) | 6.4 (16) | 93.6 (234) |
| Hematocrit | 54.8 (137) | 48.4 (121) | 6.4 (16) | 93.6 (234) |
| TSH | 36.4 (91) | 30 (75) | 6.4 (16) | 93.6 (234) |
| VITAL SIGNS | ||||
| Blood Pressure | 78.8 (197) | 69.6 (174) | 9.2 (23) | 90.8 (227) |
| Heart Rate | 70.8 (177) | 62.8 (157) | 8 (20) | 92 (230) |
| Height | 28.8 (72) | 25.6 (64) | 3.2 (8) | 96.8 (242) |
| Weight | 51.2 (128) | 46.8 (117) | 4.4 (11) | 95.6 (239) |
Notes: Includes EHR data extracted from nonstructured fields
EHR= electronic health record, KP= Knowledge Program
N, total number of observations
Nm, the number of matches between EHR and KP data, including cases where both are missing
BUN= blood urea nitrogen, CO2 = carbon dioxide, WBC = white blood cells, HgbA1c = hemoglobin A1c, TSH = thyroid stimulating hormone
Table 5.
Completeness of Binary Data Elements between the Electronic Health Record and Knowledge Program Data Repositories
| TOTAL N1= 250 | COMPLETENESS | ||
|---|---|---|---|
| EHR REVIEW | KP | % EHR ONLY, (#EHR – #KP) | |
| % OF TOTAL RECORDS (# PRESENT) | % OF TOTAL RECORDS (# PRESENT) | ||
| DIAGNOSES | |||
| Cancer | 16.4 (41) | 13.6 (34) | 2.8 (7) |
| Coronary Artery Disease | 10.4 (26) | 13.6 (18) | 3.2 (8) |
| Diabetes | 12.4 (31) | 9.2 (23) | 3.2 (8) |
| Stroke | 7.6 (19) | 8 (20) | −0.04 (−1) |
| Chronic Renal Insufficiency | 3.2 (8) | 2.8 (7) | 0.4 (1) |
| Depression | 29.6 (74) | 18.4 (46) | 11.2 (28) |
| Hypertension | 36.4 (91) | 25.2 (63) | 11.2 (28) |
| Atrial Fibrillation | 4.4 (11) | 3.2 (8) | 1.2 (3) |
| Sleep Apnea | 13.6 (36) | 13.6 (34) | 0.8 (2) |
| Hypothyroidism | 8.4 (21) | 6.4 (16) | 2 (5) |
| Obesity | 10 (25) | 9.6 (24) | 0.4 (1) |
| Hypertension | 42.4 (106) | 47.2 (118) | −4.8 (−12) |
| Diabetes | 10.4 (26) | 14 (35) | −3.6 (−9) |
| MEDICATIONS | |||
| Aspirin | 24.4 (60) | 30.8 (77) | −6.8 (−17) |
Notes: Includes EHR data extracted from nonstructured fields
EHR= electronic health record, KP= Knowledge Program
N, total number of observations
The prevalence for the selected diagnoses varied widely in both the EHR and KP data warehouse (3.2–29.6 percent vs. 2.8–25.2 percent; Table 5). Hypertension and depression were the most frequently documented diagnoses, with rates of 18.4 and 25.2 percent, respectively, in the KP data warehouse. For all diagnoses except stroke, a higher prevalence was identified using EHR data than KP data, with the discrepancies ranging from 0.4 to 11.2 percent.
The documented use of medication also varied widely. According to the KP data warehouse, 14 percent were on diabetic medications, 47.2 percent were on antihypertensives, and 30.8 percent were on aspirin. In contrast to the other data variables assessed in this study, there was higher prevalence of medication use in the KP records than in the EHR review ranging from 3.6 to 6.8 percent higher rates than the EHR.
Analysis of completeness and concordance of the data was repeated, comparing the KP to the EHR review limited to variables available in structured fields (Appendices 1 and 2), excluding EHR data present in scanned documents or free-text fields. This provides a comparison of the discretely available electronic data in the two data sets. As expected, there were more cases of matching data between the structured fields of the EHR and the KP data warehouse. Still, vital signs, laboratory values, and diagnoses were more complete in the EHR than in the KP data warehouse; there were 2.8–6.8 percent more cases with vitals, 1–4.4 percent more cases with laboratory values, and 0–9.6 percent more cases with diagnoses in the EHR than in the KP. In our comparison (N = 250), completeness of demographic data (excluding date of death) was 100 percent in the EHR, and 98.4–99.6 percent in the KP data warehouse. The KP data warehouse registered more active medications than in the EHR. Overall, most data abstracted in the EHR review came from structured fields. Data from EHR were available only in a nonstructured format for up to 2.4 percent of laboratory values, 2.4 percent of vitals, and 1.6 percent of diagnoses.
Discussion
In this evaluation comparing completeness and level of agreement of the KP data warehouse and the EHR, there was a mismatch in available data in most of the categories assessed—up to 6.4 percent of laboratory values, 9.2 percent of vital signs, and 11.2 percent of diagnoses, which could potentially have an impact on analyses using these data. The EHR generally had more complete data than the KP data warehouse except for information on deaths and medications, for which the KP data warehouse had more. When data were available in both sources, there was a high, but imperfect, concordance in values. The reasons for these discrepancies were explored in depth, and our findings highlight many factors that need to be considered when constructing, evaluating, and utilizing an external data set derived from an EHR and other clinical data sources.
Data Completeness
The biggest factor that has an impact on the completeness of data was the start date of data feeds into the KP data warehouse. Data were deposited into the KP data warehouse from the EHR only after specific data feeds were turned on, and any data recorded in the EHR prior to this were not available in the KP data warehouse. This lack of historical data resulted in the absence of up to 9.6 percent of evaluated diagnoses in the KP data warehouse compared to the EHR (Appendix 2). Similarly, there were separate KP data feeds for different laboratory test categories and locations within the health system, which were turned on in a staggered fashion. Laboratory test results were not available in the KP data warehouse if the tests were performed and reported before the data feeds to the warehouse were turned on. This staggered implementation of laboratory feeds was responsible for the majority of the mismatches in laboratory values in this current study. Because of the uneven nature of missing values, with laboratory values available for some but not all patients, this issue may not be apparent to an end user who might falsely conclude that certain laboratory tests were not performed. The systematic missingness of data has the potential to confound analyses if it is not identified and adequately addressed in the study design or analysis. Thus, when constructing and utilizing an external data source, it is necessary to have a clear understanding of when data variables for different collection locations are accessible.
An unexpected finding of this analysis was that, in contrast to other data variables, there was a higher prevalence of medications in the KP data warehouse compared to the data from the EHR review. After investigation, this was found to be due to a systematic issue with medication data feeds—the KP data warehouse occasionally did not receive updates on medication discontinuations. This resulted in an incorrect inflation of medications designated as active. Finding this significant unexpected error is an example of why it is critical to systematically assess the data quality of all data variables.
Another relevant factor contributing to the completeness of data in the KP and other data warehouses that obtain data from EHRs was the inability to incorporate provider-entered nonstructured clinical data, such as free-text or scanned documents. This issue was evaluated in our study with a manual chart review. We found that outside results (e.g., laboratory tests) were sometimes entered into the EHR in the form of a scanned document. Other data including vital signs, medications, or medical history were occasionally entered only as free-text in an encounter note, even though discrete fields for data entry were available and recommended as part of standard documentation practices. Data that are in nonstructured parts of the EHR were not sent to the KP data warehouse, which resulted in less complete data in the KP in these areas compared to the manual EHR review. Our analysis indicates that up to 2.4 percent of external laboratory information and vitals were not recorded in the KP due to this issue (Table 4 and Appendix 1). Although these percentages are fairly low, they reflect a limitation of performing automated surveys of electronic data sources. These findings may differ for databases from other institutions due to differences in documentation practices across specialties and institutions.
Concordance
The imperfect concordance between the KP and EHR in this study when variables were present in both data sets was due to the dynamic nature of some of the clinical data elements in the EHR. Patient-level data, such as medications, diagnoses, or demographics (except for death) can change over time, sometimes outside of a clinic visit. Encounter-based data, such as laboratory values or an encounter note, are typically static and do not change once entered unless a correction or amendment is made. The KP data warehouse is designed to only receive updates to a patient’s data when a clinical visit is completed, and only if the visit is in a clinical area utilizing the KP. These factors, along with an occasional difference in timing of data reviews of the EHR and KP data, resulted in imperfect agreement for some demographic data between the two databases. Given the dynamic nature of some data variables in an EHR and other clinically derived data sources, it is critical to be aware of the frequency and extent of data updates to the external warehouse. Ideally, there would be real-time feeds that immediately upload any changes in the original data set to the joint database. Scheduled updates are an alternative, however, these types of processes may be resource intensive to set up.
Missing Documentation
Lack of documentation in the EHR led to missing data in both the KP data warehouse and original EHR. For example, blood pressure measurements were available for only 78.8 percent (197 of 250), height measurements for 28.8 percent (72 of 250), and weight measurements for 51.2 percent (128 of 250) of encounters in the original EHR (Table 4). Missing documentation is a significant limitation of data sets derived from EHRs and other clinically derived data sources.9,10 Phenotyping with EHR data is an area of active research to combat the problem of missing or variably recorded data. With EHR-driven phenotyping, raw EHR data is transformed into clinically relevant features using heuristic rules or modeling to increase the sensitivity and specificity of identifying specific diagnoses or patient characteristics.11 A better understanding of data quality and the implications of missingness can provide information on uncertainty in EHR-derived phenotypes. This current analysis could be part of an initial step toward the development of algorithms to better identify these select conditions.
Use of a Gold Standard
In this KP data quality evaluation, completeness and concordance between the KP data warehouse and data from a manual review of the EHR were compared, rather than designating the EHR as the gold standard. The KP receives direct feeds from the laboratory, radiology, and scheduling systems, in addition to feeds from the EHR, and receives regular updates from the Social Security Death Index on deaths. Given the different direct data sources that feed into the KP, a comparison with the manual EHR review as the gold standard would have misguided our analysis. Indeed, although the KP data warehouse did have less complete information in most data variables compared to the EHR, the KP had more data on deaths. This supports the practice of avoiding the designation of any single data set as the gold standard when evaluating data quality of a joint data warehouse, especially one that has multiple data sources.
Conclusion
This study identifies several factors that should be evaluated when developing a joint data warehouse and subsequently assessing the quality of its data. These include the frequency and method for updating the data warehouse, start date, and maintenance of data feeds, and the dynamic nature of data in the source EHR. The additional limitations of using clinically derived data sources should also be considered, such as an inability to capture information recorded in nonstructured formats and incomplete documentation with data feeds. In addition, when evaluating the quality of an external data set with multiple and complex data sources, assessing completeness and concordance between data set and source data may be more appropriate than designating one to be a gold standard. This study provides an innovative example of how the quality of a data set with multiple sources can be evaluated without designating a gold standard for comparison.
Acknowledgments
The authors thank Srividya Ramachandran PhD who provided editorial assistance with this manuscript. This submission is based on work presented at the 2014 EDM Forum Symposium.
Appendix 1. Completeness and Concordance of Numeric Data Elements between Electronic Health Record Structured Data and Knowledge Program Data
| TOTAL N1= 250 | COMPLETENESS | CONCORDANCE | ||
|---|---|---|---|---|
| EHR (EXCLUDING FREE-TEXT) | KP | #EHR – #KP, % OUT OF TOTAL (N=250) | % MATCH (Nm2), INCLUDE BOTH MISSING CASE | |
| % OF TOTAL RECORDS (# PRESENT) | % OF TOTAL RECORDS (# PRESENT) | |||
| LABORATORY VALUES | ||||
| Sodium | 55.2 (138) | 51.6 (129) | 3.6 (9) | 96.4 (241) |
| Potassium | 55.6 (139) | 52 (130) | 3.6 (9) | 96.4 (241) |
| CO2 | 55.2 (138) | 52 (130) | 3.2 (8) | 96.8 (242) |
| Chloride | 55.6 (139) | 52 (130) | 3.6 (9) | 96.4 (241) |
| BUN | 55.6 (139) | 52.4 (131) | 3.2 (8) | 96.8 (242) |
| Creatinine | 56.4 (141) | 52.4 (131) | 4.0 (10) | 96 (240) |
| Glucose | 56 (140) | 52.4 (131) | 3.6 (9) | 96.4 (241) |
| Calcium | 54.4 (136) | 50.8 (127) | 3.6 (9) | 96.4 (241) |
| Albumin | 48.4 (121) | 45.6 (114) | 2.8 (7) | 97.2 (243) |
| Total Protein | 46.4 (116) | 43.6 (109) | 2.8 (7) | 97.2 (243) |
| HgbA1C | 14 (35) | 13.6 (34) | 0.4 (1) | 99.6 (249) |
| Fasting Blood Sugar | 1.6 (4) | 1.6 (4) | 0 (1) | 100 (250) |
| WBC | 52 (130) | 48.4 (121) | 3.6 (9) | 96.4 (241) |
| Hemoglobin | 52 (130) | 48.4 (121) | 3.6 (9) | 96.4 (241) |
| Hematocrit | 52 (130) | 48.4 (121) | 3.6 (9) | 96.4 (241) |
| TSH | 34.4 (86) | 30 (75) | 4.4 (11) | 95.6 (239) |
| VITAL SIGNS | ||||
| Blood Pressure | 76.4 (191) | 69.6 (174) | 6.8 (17) | 93.2 (233) |
| Heart Rate | 69.2 (173) | 62.8 (157) | 6.4 (16) | 93.6 (234) |
| Height | 28.4 (71) | 25.6 (64) | 2.8 (7) | 97.2 (243) |
| Weight | 50.8 (127) | 46.8 (117) | 4 (10) | 96 (240) |
Notes: Excludes information in free-text of notes and scanned documents
EHR= electronic health record, KP= Knowledge Program
N, total number of observations
Nm, the number of matches between EHR and KP data, including cases where both are missing
EHR= electronic health record, KP= Knowledge Program
BUN= blood urea nitrogen, CO2 = carbon dioxide, WBC = white blood cells, HgbA1c = hemoglobin A1c, TSH = thyroid stimulating hormone
Appendix 2. Completeness of Binary Data Elements between Electronic Health Record Structured Data and Knowledge Program Data
| TOTAL N1= 250 | EHR (EXCLUDING FREE-TEXT) | KP | #EHR – #KP, % OUT OF TOTAL (N=250) |
|---|---|---|---|
| % OF TOTAL RECORDS (# PRESENT) | % OF TOTAL RECORDS (# PRESENT) | ||
| DIAGNOSES | |||
| Cancer | 16.4 (41) | 13.6 (34) | 2.8 (7) |
| Coronary Artery Disease | 9.6 (24) | 13.6 (18) | 2.4 (6) |
| Diabetes | 12.4 (31) | 9.2 (23) | 3.2 (8) |
| Stroke | 7.6 (19) | 8 (20) | −0.4 (−1) |
| Chronic Renal Insufficiency | 3.2 (8) | 2.8 (7) | 0.4 (1) |
| Depression | 28 (70) | 18.4 (46) | 9.6 (24) |
| Hypertension | 34.4 (86) | 25.2 (63) | 9.2 (23) |
| Atrial Fibrillation | 3.6 (9) | 3.2 (8) | 0.4 (1) |
| Sleep Apnea | 13.6 (34) | 13.6 (34) | 0 (0) |
| Hypothyroidism | 6.8 (17) | 6.4 (16) | 0.4 (1) |
| Obesity | 10 (25) | 9.6 (24) | 0.4 (1) |
| Hypertension | 42.4 (106) | 47.2 (118) | −4.8 (−12) |
| Diabetes | 10.4 (26) | 14 (35) | −3.6 (−9) |
| MEDICATION | |||
| Aspirin | 23.2 (58) | 30.8 (77) | −7.6 (−19) |
Notes: EHR= electronic health record, KP= Knowledge Program
N, total number of observations
Footnotes
Disciplines
Health Information Technology | Health Services Research
References
- 1.Bloomrosen M, Detmer DE. Informatics, evidence-based care, and research; implications for national policy: A report of an american medical informatics association health policy conference. J Am Med Inform Assoc. 2010;17:115–123. doi: 10.1136/jamia.2009.001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jha AK, Burke MF, DesRoches C, Joshi MS, Kralovec PD, Campbell EG, Buntin MB. Progress toward meaningful use: Hospitals’ adoption of electronic health records. Am J Manag Care. 2011;17:SP117–124. [PubMed] [Google Scholar]
- 3.Dixon BE, Siegel JA, Oemig TV, Grannis SJ. Electronic health information quality challenges and interventions to improve public health surveillance data and practice. Public Health Rep. 2013;128:546–553. doi: 10.1177/003335491312800614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikkelsen G, Aasly J. Concordance of information in parallel electronic and paper based patient records. Int J Med Inform. 2001;63:123–131. doi: 10.1016/s1386-5056(01)00152-6. [DOI] [PubMed] [Google Scholar]
- 5.Hassey A, Gerrett D, Wilson A. A survey of validity and utility of electronic patient records in a general practice. BMJ. 2001;322:1401–1405. doi: 10.1136/bmj.322.7299.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Faris P, Hemmelgarn B, Walker RL, Quan H. Measuring agreement of administrative data with chart data using prevalence unadjusted and adjusted kappa. BMC Med Res Methodol. 2009;9:5. doi: 10.1186/1471-2288-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzan I, Speck M, Dopler C, Urchek J, Bielawski K, Dunphy C, Jehi L, Bae C, Parchman A. The knowledge program: An innovative, comprehensive electronic data capture system and warehouse. AMIA Annu Symp Proc. 2011;2011:683–692. [PMC free article] [PubMed] [Google Scholar]
- 8.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (redcap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan KS, Fowles JB, Weiner JP. Review: Electronic health records and the reliability and validity of quality measures: A review of the literature. Med Care Res Rev. 2010;67:503–527. doi: 10.1177/1077558709359007. [DOI] [PubMed] [Google Scholar]
- 10.Roth CP, Lim YW, Pevnick JM, Asch SM, McGlynn EA. The challenge of measuring quality of care from the electronic health record. Am J Med Qual. 2009;24:385–394. doi: 10.1177/1062860609336627. [DOI] [PubMed] [Google Scholar]
- 11.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]