Abstract
Background
Targeted next-generation sequencing (NGS) is a cost-effective approach for rapid and accurate detection of genetic mutations in patients with suspected genetic disorders, which can facilitate effective diagnosis.
Methodology/Principal Findings
We designed a capture array to mainly capture all the coding sequence (CDS) of 2,181 genes associated with 561 Mendelian diseases and conducted NGS to detect mutations. The accuracy of NGS was 99.95%, which was obtained by comparing the genotypes of selected loci between our method and SNP Array in four samples from normal human adults. We also tested the stability of the method using a sample from normal human adults. The results showed that an average of 97.79% and 96.72% of single-nucleotide variants (SNVs) in the sample could be detected stably in a batch and different batches respectively. In addition, the method could detect various types of mutations. Some disease-causing mutations were detected in 69 clinical cases, including 62 SNVs, 14 insertions and deletions (Indels), 1 copy number variant (CNV), 1 microdeletion and 2 microduplications of chromosomes, of which 35 mutations were novel. Mutations were confirmed by Sanger sequencing or real-time polymerase chain reaction (PCR).
Conclusions/Significance
Results of the evaluation showed that targeted NGS enabled to detect disease-causing mutations with high accuracy, stability, speed and throughput. Thus, the technology can be used for the clinical diagnosis of 561 Mendelian diseases.
Introduction
At present, about 7000 Mendelian diseases have been recognized according to the Online Mendelian Inheritance in Man (OMIM) database. Although most of these diseases are individually rare, the total number of population affected by Mendelian diseases is vast. According to the data released by the World Health Organization (WHO), the global prevalence of all Mendelian diseases at birth is approximately 1:10000. The diagnosis is difficult for many Mendelian diseases and many are not onset at birth [1]. Except for some Mendelian diseases can be corrected by surgery, most are responsible for a heavy loss of life, disability or malformation. There was little relevant medicine to rare monogenic disorders [2]. It also lacks other effective treatment. Diagnostic testing can not only accurately determine whether a person carries the disease-causing gene or not, providing the effective basis for the diagnosis of the disease, but also estimate the risk of such a genetic disease in offspring, which plays an important role in genetic diagnosis and fertility guidance. So effective and exact diagnostic testing is very necessary.
Sanger sequencing is a common method of first generation of sequencing technology. Shortcomings such as narrow screening spectrum and poor stability of experiment limit the application in wide range screening of gene mutations. The primary advantage of NGS over Sanger method is the inexpensive production of large amounts of sequencing data [3]. With unprecedented high throughput, highly parallel and base-pair resolution data of NGS, whole-exome sequencing (WES) and whole-genome sequencing (WGS) are all applied in the diagnostic testing or carrier screening of Mendelian genetic disorders [4–7]. WGS and WES yield myriad of genetic variations which have an advantage in carrier screening. However, identifying the few disease-causing mutations among the vast variants present in human genomes remains a major challenge [8]. Targeted NGS with lower cost and high throughput can efficiently target disease-associated regions of the genome and detect variants with high sensitivity, which is widely used in the clinical screening of mutations and effective diagnostic testing of genetic diseases [9–12], including some cancers [13–15]. Therefore, we designed a capture array which could mainly capture the CDS of 2,181 genes associated with 561 Mendelian diseases and developed a pipeline including experiment, sequencing, bioinformatic analysis, mutation identification and confirmation.
Materials and Methods
Information of Samples
We totally collected 94 samples for this study. To evaluate the stability of our method, S1 was sequenced three times in a batch (S1-1, S1-2 and S1-3) and two times in different batches (S1-4 and S1-5). Four samples (S1-1, S2, S3 and S4) from normal human adults were selected to evaluate the accuracy of our method. In addition, we selected 84 patient samples and 6 samples (P44, P45, P77, P78, P81 and P82) of suspected carriers from clinical cases (S1 Table). Among them, 75 samples were diagnosed and 15 samples were undiagnosed (Table 1). Written informed consent was obtained from all of the adult participants or the parents of the minors enrolled in our study. This project and the protocols for the investigation involving human tissues were approved by the ethics committee of BGI-Shenzhen (BGI-IRB 14061).
Table 1. Summary of disease-causing mutations in 90 clinical cases.
| ID | Sex, age a | Clinical diagnosis b | Candidate causative genes | Mutations c , status | Consistency d |
|---|---|---|---|---|---|
| P1 | F, 4 years | AxD | GFAP | GFAP NM_001242376.1:c.236G>A (p.R79H) | Yes |
| P2 | M, 23 days | AS | COL4A4, COL4A5, et al. | COL4A5 NM_033380.2:c.1769A>C (p.K590T) (Hem), novel | Yes |
| P3 | F, 6 years | AS | COL4A4, COL4A5, et al. | COL4A5 NM_033380.2:c.2414G>T (p.G805V) | Yes |
| P4 | F, 13 years | AS | COL4A4, COL4A5, et al. | COL4A5 NM_033380.2:c.187G>T (p.G63*), novel | Yes |
| P5 | M, 27 years | APS-1? | AIRE | negative result | No |
| P6 | M, 37 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.9914_9915delCT (p.A3305Pfs*10), novel | Yes |
| P7 | F, 32 years | ADPKD | PKD1, PKD2 | negative result | No |
| P8 | F, 22 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.7210-1G>A, novel | Yes |
| P9 | M, 29 years | ADPKD | PKD1, PKD2 | negative result | No |
| P10 | M, 42 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.1589G>A (p.C530Y) | Yes |
| P11 | M, 32 years | ADPKD | PKD1, PKD2 | negative result | No |
| P12 | M, 74 years | ADPKD | PKD1, PKD2 | negative result | No |
| P13 | F, 39 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.7210-2A>G | Yes |
| P14 | F, 30 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.8698C>T (p.Q2900*) | Yes |
| P15 | M, - | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.7204C>T (p.R2402*) | Yes |
| P16 | M, 28 years | ADPKD | PKD1, PKD2 | negative result | No |
| P17 | M, 51years | ADPKD | PKD1, PKD2 | negative result | No |
| P18 | M, 30 years | ADPKD | PKD1, PKD2 | negative result | No |
| P19 | F, 28 years | ADPKD | PKD1, PKD2 | negative result | No |
| P20 | M, 33 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.7566 C>A (p.C2522*), novel | Yes |
| P21 | M, 67 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.10618G>T (p.G3540*), novel | Yes |
| P22 | M, 30years | ADPKD | PKD1, PKD2 | negative result | No |
| P23 | F, 30 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.2085_2086insC (p.A696Rfs*18) | Yes |
| P24 | F, 37 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.9377C>T (p.T3126I) | Yes |
| P25 | M, 32 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.8350 C>T (p.Q2784*) | Yes |
| P26 | M, 60 years | ADPKD | PKD1, PKD2 | negative result | No |
| P27 | M, 29 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.9569G>T (p.G3190V), novel | Yes |
| P28 | M, - | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.11721_11722insCT (p.L3909Cfs*37), novel | Yes |
| P29 | M, - | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.11343C>A (p.Y3781*), novel | Yes |
| P30 | F, 58 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.6574_6580delACCGCCA (p.T2192Afs*18) | Yes |
| P31 | M, - | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.12608_12635delGGCTGGGGACAAGGTGTGAGCCTGAGCC (p.R4203Pfs*93) | Yes |
| P32 | F, 27 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.8578C>T (p.Q2860*), novel | Yes |
| P33 | M, 31 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.12003+2T>C | Yes |
| P34 | F, 31 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.8590G>T (p.E2864*), novel | Yes |
| P35 | F, 28 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.12373C>T (p.Q4125*) | Yes |
| P36 | F, 26 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.12373C>T (p.Q4125*) | Yes |
| P37 | M, 26 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.5014_5015delAG (p.R1672Gfs*98) | Yes |
| P38 | M, 26 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.8590G>T (p.E2864*), novel | Yes |
| P39 | F, 27 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.7915C>T (p.R2639*) | Yes |
| P40 | M, 42 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.4306C>T (p.R1436*) | Yes |
| P41 | F,56 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.7546C>T (p.R2516C) | Yes |
| P42 | M, 32 years | ADPKD | PKD1, PKD2 | PKD1 NM_001009944.2:c.7546C>T (p.R2516C) | Yes |
| P43 | -, 11 years | ARCI | ALOX12B, ALOXE3, et al. | ALOXE3 NM_001165960.1:c.814C>T (p.R272*) (Hom), novel | Yes |
| P44 | M, - | ARCI, carrier | ALOX12B, ALOXE3, et al. | ALOXE3 NM_001165960.1:c.814C>T (p.R272*), novel | Yes |
| P45 | F, - | ARCI, carrier | ALOX12B, ALOXE3, et al. | ALOXE3 NM_001165960.1:c.814C>T (p.R272*), novel | Yes |
| P46 | F, 27 years | BS | SLC12A1, CLCNKB, et al. | negative result | No |
| P47 | F, 7 months | BS? | BSND, KCNJ1, et al. | CFTR NM_000492.3:c.1116+1G>A c.3062C>T (p.P1021L), novel | CF |
| P48 | M, 6 months | BS? | BSND, KCNJ1, et al. | CFTR NM_000492.3:c.2909G>A (p.G970D) (Hom) | CF |
| P49 | F, 33 years | CMT | AARS, EGR2, et al. | AARS NM_001605.2:c.2042G>T (p.G681V), novel | Yes |
| P50 | F, 35 years | CMT | AARS, EGR2, et al. | EGR2 NM_001136177.1:c.928ins>GCC (p.Y310delinsCH), novel | Yes |
| P51 | F, 3 years | CMT | AARS, EGR2, et al. | negative result | No |
| P52 | F, 24 years | CMT | AARS, EGR2, et al. | PMP22 NM_153322.1:c.215C>T (p.S72L) | Yes |
| P53 | M, 36 years | CMT | AARS, EGR2, et al. | MFN2 NM_014874.3:c.1039-2A>G, novel; GJB1 NM_000166.5:c.265C>G (p.L89V) (Hem), novel | Yes |
| P54 | M, 28 years | CMT | AARS, EGR2, et al. | GJB1 NM_000166.5:c.223C>T (p.R75W) (Hem) | Yes |
| P55 | F, 12 years | CMT | AARS, EGR2, et al. | GDAP1 NM_018972.2:c.767A>G (p.H256R) | Yes |
| P56 | M, 10 years | CMT | AARS, EGR2, et al. | PMP22 NM_153322.1:CDS1-4 dup | Yes |
| P57 | M, 37 years | CMT | AARS, EGR2, et al. | GJB1 NM_000166.5:c.269T>C (p.L90P) (Hem), novel | Yes |
| P58 | F, 3.5 years | Congenital Afibrinogenemia | FGA, FGB, FGG | FGA NM_000508.3:c.1368delC (p.T457Rfs*27) (Hom), novel | Yes |
| P59 | F, 5 years | CDA? | CDAN1, SEC23B, KLF1 | PKLR NM_000298.5:c.1528C>T (p.R510*) c.661G>A (p.D221N) | PK Deficiency |
| P60 | -, 3 years | CdLS | NIPBL, SMC1A, et al. | NIPBL NM_133433.3:c.2207_2211delCAAAG (p.Q738Rfs*2), novel | Yes |
| P61 | M, 8 years | Dent Disease | CLCN5, OCRL | OCRL NM_000276.3:c.215_216delTT (p.L73Dfs*2) (Hem) | Yes |
| P62 | M, 1 year | Dent Disease | CLCN5, OCRL | CLCN5 NM_001127899.1:c.778_798delACTCTGGTTATCAAAACCATC (p.T260_I266del) (Hem), novel | Yes |
| P63 | F, 18 months | FHL | PRF1, STX11, et al. | PRF1 NM_005041.4:c.1620A>G (p.Q540Q) | Yes |
| P64 | F, 34 years | GS | SLC12A3, CLCNKB | SLC12A3 NM_000339.2:c.1202C>T (p.A401V) (Hom), novel | Yes |
| P65 | -, - | GSD? | AGL, ALDOA, et al. | GALT NM_000155.3:c.1043A>G (p.D348G) (Hom), novel | Galactosemia |
| P66 | M, - | Hereditary Muscular Disease? | ACTA1, ATP1A3, et al. | negative result | No |
| P67 | M, 3 years | HSAN | IKBKAP, NTRK1, et al. | NTRK1 NM_002529.3:c.963delG (p.L322Sfs*148), novel c.851-33T>A | Yes |
| P68 | -, - | Hypochondroplasia? | FGFR3 | COL2A1 NM_001844.4:c.863G>C (p.G288A), novel; LBR NM_194442.2:c.1640A>G (p.N547S), novel c.1757G>A (p.R586H), novel | Yes |
| P69 | M, 4 years | Hypophosphatasia | ALPL | ALPL NM_000478.4:c.98C>T(p.A33V)(Hom) | Yes |
| P70 | F, 2 years | IP | IKBKG | negative result | No |
| P71 | M, 75 days | IBD deficiency? EE? SCAD Deficiency? | ACAD8, ETHE1, ACADS | ACADS NM_000017.2:c.989G>A (p.R330H) c.1031A>G (p.E344G) | Yes |
| P72 | M, 55 days | IBD deficiency? EE? SCAD Deficiency? | ACAD8, ETHE1, ACADS | ACADS NM_000017.2:c.164C>T(p.P55L) c.1031A>G(p.E344G) | Yes |
| P73 | F, 41 years | LPG | APOE | APOE NM_000041.2:c.127C>T (p.R43C) | Yes |
| P74 | M, - | MMA, carrier | MUT, MMAA, et al. | MMACHC NM_015506.2:c.656_658delAGA (p.K220del) | Yes |
| P75 | F, - | MMA, carrier | MUT, MMAA, et al. | MMACHC NM_015506.2:c.609G>A (p.W203*) | Yes |
| P76 | M, - | Microphthalmia | SOX2, OTX2, et al | OTX2 NM_021728.3:c.538C>T (p.Q180*), novel | Yes |
| P77 | F, 6 years | Mucopolysaccharidosis | ARSB, GALNS, et al. | GNPTAB NM_024312.4:c.3565C>T (p.R1189*) c.2590_2591insG (p.E864Gfs*4) | Mucolipidosis |
| P78 | F, 28 years | Mucopolysaccharidosis, carrier? | ARSB, GALNS, et al. | negative result | No |
| P79 | M, 31 years | Mucopolysaccharidosis, carrier? | ARSB, GALNS, et al. | negative result | No |
| P80 | F, 33 years | OCA | GPR143, TYR, et al. | TYR NM_000372.4:c.455C>A (p.P152H), novel c.832C>T (p.R278*) | Yes |
| P81 | F, 6 years | MCPH | MCPH1, WDR62, et al. | negative result | No |
| P82 | M, 5 years | PCD? | DNAI1, DNAH5, et al. | negative result | No |
| P83 | M, 8 years | SLSN | CEP290, IQCB1, et al. | IQCB1 NM_001023570.2:c.1225C>T (p.Q409*), novel c.1090C>T (p.R364*) | Yes |
| P84 | F, 9 years | SRNS? | ACTN4, ALG1, et al | negative result | No |
| P85 | F, 12 years | SRNS? | ACTN4, ALG1, et al | negative result | No |
| P86 | F, 11 months | WAS? X-SCID? | WAS, IL2RG | RAG1 NM_000448.2:c.874T>C (p.S292P), novel c.1328G>A (p.R443K), novel | OS |
| P87 | M, 14 years | XLA | BTK | BTK NM_000061.2:c.1684C>T (p.R562W) (Hem) | Yes |
| P88 | -, - | Trisomy 10 | - | 47.XX,+10 | Yes |
| P89 | -, - | Trisomy 9 | - | 47.XX,+9 | Yes |
| P90 | -, 8 years | SMS | - | 46,XN,del(17)(p11.2) | Yes |
aSex, age: ‘-’ shows that the sex or age of a sample is not available.
bClinical diagnosis: The result of this column indicates the diagnosis of clinical cases in the form of abbreviations (S2 Table). The question mark indicates the case is undiagnosed and the result is noted with ‘carrier’ if the case is a suspected carrier.
cmutations: The results of this column is the summary of disease-causing mutations. Homozygous mutations are marked with (Hom) and hemizygous mutations are marked with (Hem), the other unmarked mutations are heterozygous. Mutation is noted with ‘novel’ if it is novel. ‘*’ indicates a termination codon.
dConsistency: Consistency is defined as ‘Yes’ if our result is consistent with the clinical diagnosis. Consistency is defined as ‘No’ if the result is negative. Consistency is noted with the name of the disease based on our analysis if we could not detect mutations in candidate causative genes but could detect mutations in other genes and our result is inconsistent with the clinical diagnosis.
Chip Design, Experiment and Sequencing
A capture array (NimbleGen, Roche) was designed to mainly capture the CDS of 2,181 known pathogenic genes associated with 561 Mendelian diseases (S3 Table) based on GeneReviews (NCBI) and Genetics Home Reference.
Genomic DNA from peripheral blood or tissues of abortion was fragmented ranging from 200 bp to 250bp. The primers, adapters and indexes were then ligated to the DNA fragments to construct libraries. The DNA fragments were pooled and hybridized to the capture array (Roche NimbleGen, Inc.). After hybridization and enrichment, the DNA sample was sequenced on Illumina HiSeq2500 Analyzers to generate paired-end reads (90 bps). The detailed procedure is roughly the same as used in a previous paper [9].
Bioinformatic Analysis and Mutation Identification
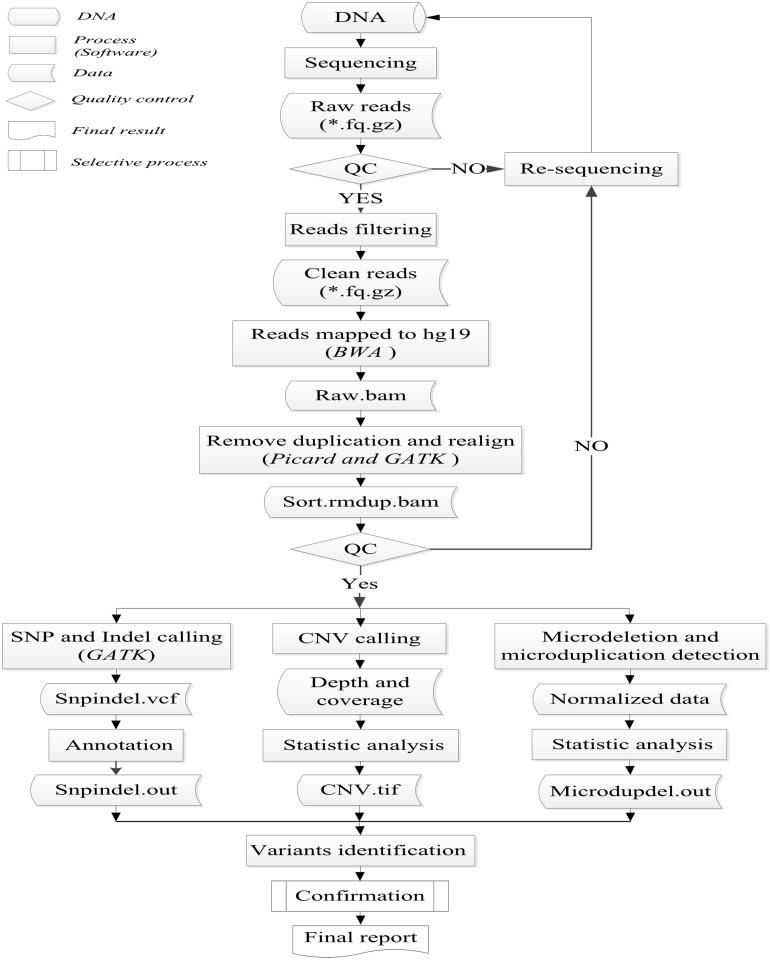
The pipeline of bioinformatic analysis included data filteration, data alignment, variants detection and results annotation (Fig 1). The pipeline started from the sequencing data (Raw reads) which generated from the HiSeq2500. First, after a preliminary quality assessment of sequencing data using the Illumina Pipeline (version 1.3.4), reads containing adapters and with low-quality values which defined as that the average quality value was less than ten were filtered out. Second, the clean reads were mapped to the human reference genome from the NCBI database (Build37) using Burrows Wheeler Aligner (BWA). Third, after removing duplications caused by PCR using Picard and realigning by Genome Analysis Toolkit (GATK), the bam results were used to do variants detection. Single-nucleotide polymorphisms (SNPs) and Indels were detected using GATK. CNVs were identified by a method based on comparison of average depth between patients and normal human samples in the same batch, which was introduced in detail in a previous paper [9]. Normalization analysis was a method to detect microdeletions and microduplications of chromosome using read count-based approach [16]. The microdeletions or microduplications of chromosomes related to our target regions were selected from the database of DECIPHER [17]. Finally, SNPs and Indels were annotated by databases such as dblocal (Mutation frequency database of 100 normal human samples) [9], dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), HapMap (http://hapmap.ncbi.nlm.nih.gov/), dbNSFP (http://varianttools.sourceforge.net/Annotation/DbNSFP), HGMD (http://www.hgmd.org/) and the 1000 Genome (http://www.1000genomes.org/).
Fig 1. Flowchart for the process of bioinformatic analysis.
According to the annotation of the variants, we could obtain the frequency of occurrence and pathogenicity of the variants in different selected databases above. Based on the patient's clinical working diagnosis, candidate disease-causing genes could be found in our own database. According to the variants detected, we first focused on the variants in the candidate genes. If we could not find disease-causing mutations in the candidate genes, we would look for mutations in the other genes.
Confirmation
To verify some SNVs and Indels detected by our method, Sanger sequencing (ABI 3730 DNA Analyzer; Applied Biosystems) was performed for three inconsistent genotypes in two samples (S1-1 and S4) and 5 pedigrees. To quantify duplications in the PMP22 gene, real-time PCR (7500 Real-time PCR system, Applied Biosystems) was performed for patient P56.
Results
Analysis of Sequencing Data
The total size of target regions of the capture array is 6.19Mb. Except all the CDS of 2,181 genes associated with 561 Mendelian diseases, the target regions also included some disease-related introns and UTRs of the 2,181 genes and the immediately adjacent region of 10bps of each region. The DNA probes in the array can hybridize with all of the targeted regions and flanking sequences of 100 bps on each side of the target region.
To evaluate the technology and detect mutations in patients and suspected carriers, we obtained at least 6, 925, 841 reads (P72) mapped to target regions per sample. The reads length was 90bps, so we got at least 623.33M bases mapped to target region per sample. The average sequencing depth of each sample was above 77.43-fold (P72) and the highest sequencing depth was 277.43-fold (P32). At least 90.34% of target regions with more than 20-fold were successfully captured and the coverage of target region was at least 99.4% (S1 Table).
Evaluation of Accuracy
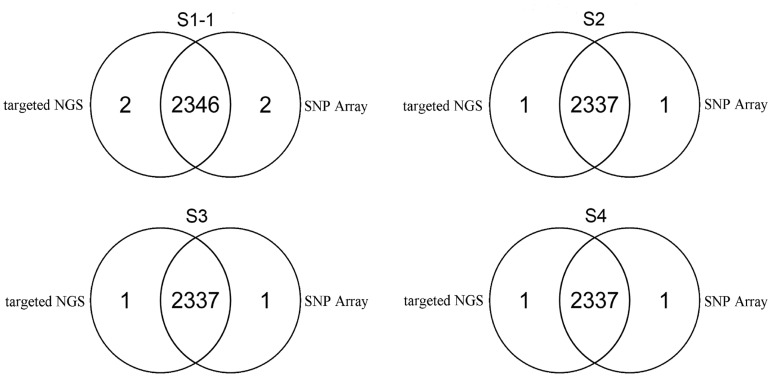
To assess the accuracy of the technology, we selected four samples (S-1, S2, S3 and S4) from normal human adults. The average depth of S1-1 was only 79.49-fold and the average depth of other three samples was about 200-fold (S1 Table). Genotypes were detected using targeted NGS and SNP Array (Illumina's Human Zhonghua-8 Bead Chips) respectively. In sample S1-1, 99.91% (2346/2348) genotypes of selected loci detected by targeted NGS were accordant to the genotypes detected by SNP Array. In each of the other three samples (S2, S3 and S4), 99.96% (2337/2338) genotypes of selected loci detected by targeted NGS were accordant to the genotypes detected by SNP Array (Fig 2). So the average consistency was 99.95% and the inconsistent genotypes (Table 2) were validated by Sanger sequencing. So, our method had a high accuracy even in the case of low average depth which was only about 80-fold.
Fig 2. Evaluation of the accuracy of our method.
Venn diagrams of the number of genotypes detected by targeted NGS and SNP Array in four samples (S1-1, S2, S3 and S4).
Table 2. Inconsistent genotypes detected by our method and SNP Array.
| Sample | Sex | Chr | Position | Rs number | Reference | Alleles of NGS | Alleles of SNP Array |
|---|---|---|---|---|---|---|---|
| S1-1 | male | chr4 | 106158216 | rs3796927 | G | A/G | A |
| S1-1 | male | chrX | 8503641 | rs809446 | C | T | T/C |
| S2 | female | chrY | 14851554 | rs2032599 | - | - | T |
| S3 | female | chrY | 14851554 | rs2032599 | - | - | T |
| S4 | male | chrX | 8503641 | rs809446 | C | T | T/C |
Evaluation of Stability
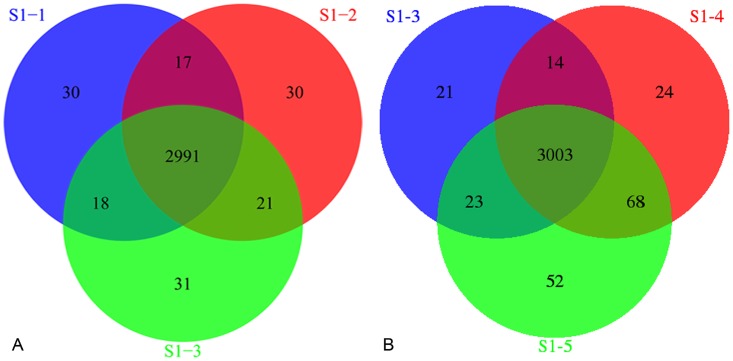
To assess the stability of the technology, S1 were sequenced three times (S1-1, S1-2 and S1-3) in a batch and the average depth of each test was about 80-fold. S1 were sequenced two times (S1-4 and S1-5) in two different batches and the average depth of the two tests was 98.99-fold and 180.71-fold respectively (S1 Table). Due to most of target regions were CDS, SNVs in noncoding regions were incredible, so we only compared the SNVs in CDS. The total numbers of SNVs of three tests (S1-1, S1-2 and S1-3) in a batch were 3056, 3059 and 3061 respectively, the same SNVs of three tests were 2991, the proportion of same SNVs in each test were 97.87%, 97.78% and 97.71% respectively, with an average of 97.79%. The total number of SNVs of three tests (S1-3, S1-4 and S1-5) in three different batches were 3061, 3109 and 3146 respectively, the same SNVs of three tests were 3003, the proportion of same SNVs in each test were 98.11%, 96.59% and 95.45% respectively, with an average of 96.72% (Fig 3). So an average of 97.79% and 96.72% of SNVs can be stably detected in a same batch and different batches respectively, which indicated good stability of our method.
Fig 3. Evaluation of the stability of our method.
(A) Venn diagram of S1 sequenced three times in a same batch. (B) Venn diagram of S1 sequenced three times in different batches.
In addition, we also compared genotypes of S1-2, S1-3, S1-4 and S1-5 with that of SNP Array respectively. We found that there were only two inconsistent loci in each test which were the same as in S1–1 (Table 2). So, the accordance ratio of genotypes was all 99.91% (2346/2348) whether in a test from the same batch or not, showing high consistency, which also indicated good stability of our method.
Variants Identification
Using targeted NGS, we obtained high-quality reads of 90 samples. After alignment and variants detection, the variants were obtained. Based on the annotation of the variants, some disease-causing mutations (62 SNVs, 14 Indels, 1 CNV, 1 microdeletion and 2 microduplications of chromosomes) were identified in 65 patients and 4 suspected carriers, 35 of which were novel. Table 1 summarized the total mutations of the 90 samples.
There were three couples of 6 suspected carriers (P44, P45, P74, P75, P78 and P79) from normal human adults. P44 and P45 are parents of patient P43 who was diagnosed with autosomal recessive Congenital Ichthyosis. An ALOXE3 gene nonsense mutation (c.814C>T, p.R272*, #MIM 606545) was detected in the three subjects. The mutation was homozygous in the proband (P43) and heterozygous in his parents, which followed the segregation rules. The incidence of this mutation in human populations was extremely low based on annotated databases. According to database of OMIM, the ALOXE3 gene mutations can cause autosomal recessive Nonbullous Congenital Ichthyosiform Erythroderma, so the mutation was causative for the proband. We detected a heterozygous MMACHC gene mutation (c.656_658delAGA, p.K220del, OMIM# 277400) in P74 and a heterozygous MMACHC gene nonsense mutation (c.609G>A, p.W203*, OMIM# 277400) in P75 who previously bore a child with Methylmalonic Acidemia. It was reported that the mutations c.609G>A could cause Methylmalonic Academia [18]. Although there was no literature reported about the mutation of c.656_658delAGA, the incidence in human populations was very low based on annotated databases. So the two mutations were suspected causative for the child. P78 and P79 had born a child who was dead with suspected Mucopolysaccharidosis VII, but we found no pathogenic mutations in them.
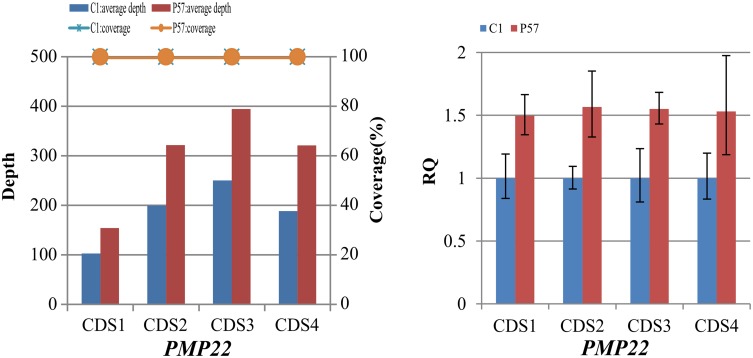
Based on the method of CNV detection above, we identified heterozygous duplications of CDS1-4 in the PMP22 gene in patient P57 who were diagnosed with Charcot-Marie-Tooth (CMT) disease (Fig 4A). Duplication of PMP22 gene could cause over expression of encoded protein and affect the normal function of cells which was the most common factor leading to CMT [19–21]. Moreover, it was reported in GeneReviews that CMT1A (70%-80% of all CMT1) involves duplication of PMP22 gene. So we speculated that the duplications of CDS1-4 in the PMP22 gene were the causative mutations for patient P57 which was confirmed by real-time PCR.
Fig 4. Identification and confirmation of heterozygous duplications involved in CMT in P57.
(A) Diagram of the average sequencing depth and coverage for CDS1-4 in the PMP22 gene. (B) Quantitative real-time PCR analysis. Relative amplification value calculated from the data of quantitative real-time PCR for detecting possible duplications in the PMP22 gene of patient P57.
Two samples (P88, P89) were tissues of spontaneous abortion. After normalization analysis, we identified the genders of the two samples were all female and detected a duplication of chromosome 10 in patient P88, the gender ratio was about 1.42. We also detected a duplication of chromosome 9 in patient P89, the gender ratio was about 1.31 (S4 Table). Based on karyotype analysis, patient P88 was diagnosed with trisomy 10 syndrome and patient P89 was diagnosed with trisomy 9 syndrome. So our results were consistent with the results of karyotype analysis. Clinical features of patient P90 were lordosis of lower lip, crooked teeth, mental retardation and she was talkative but the words fail to convey the idea. Previously, two microdeletions (del 6p21.1; del 17p11.2) were detected using WGS. As is well known, Smith-Magenis Syndrome (SMS) is caused by mutations in the RAI1 gene on chromosome 17p11.2 (SMS, OMIM# 1882290). Due to analyzed regions of the array did not include chromosome 6, we couldn’t detect the mutation of del 6p21.1,but we detected a microdeletion in chromosome 17 (16773072–20222149) and the gender ration was about 0.55 (S4 Table), which was consistent with the result of WGS.
Variants Confirmation
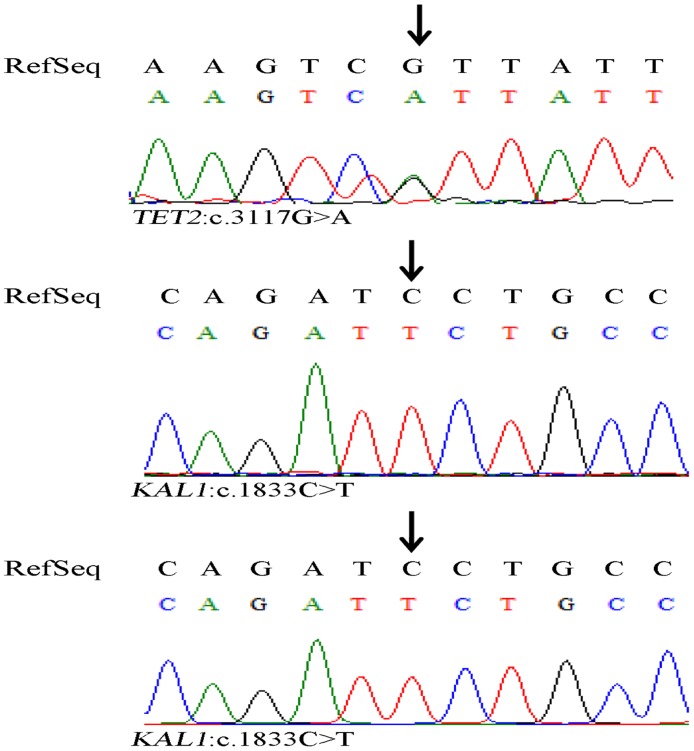
We did validation for the inconsistent genotypes detected by targeted NGS and SNP Array respectively. The inconsistent genotype of T in locus of 14851554 in Y chromosome was detected in both S2 and S3 by SNP Array, but it was not detected by targeted NGS (Table 2). It is obviously wrong to detect a genotype in the Y-chromosome of a normal female. In addition, we designed primers (S5 Table) and try to target the site, but we got no PCR products. Moreover, it turned out to be that all the genotypes of other three inconsistent loci detected by targeted NGS were consistent with that of Sanger sequencing (Fig 5), which suggested that our method provides high accuracy.
Fig 5. Confirmation of inconsistent genotypes detected by targeted NGS and Snp Array using Sanger sequencing.
(A) A heterozygous substitution of G with A was confirmed in the TET2 gene in sample S1-1. (B) A Homozygous substitution of C with T was confirmed in the KAL1 gene in sample S1-1. (C) A Homozygous substitution of C with T was confirmed in the KAL1 gene in sample S4.
For 5 patients (P2, P49, P53, P58 and P76), in order to verify the results of our method and determine whether the disease-causing mutations were inherited from father or mother of the proband, 6 primer pairs were designed (S5 Table) to target the mutated loci and the 6 mutations of pedigrees were confirmed by Sanger sequencing. A hemizygous COL4A5 gene missense mutation (c.1769A->C, p.K590T, MIM# 301050) was detected in patient P2 with Alport Syndrome. Sanger sequencing confirmed this mutation in the proband and showed that his mother was a carrier of the heterozygous mutation, which was accorded with X-linked inheritance pattern (Figure A in S1 Fig). A heterozygous AARS gene missense mutation (c.2042G>T, p.G681V, MIM# 613287) was detected in patient P49 with autosomal dominant CMT disease type 2N. Sanger sequencing confirmed this mutation in the proband and showed that his father did not carry the mutation and although his mother was a carrier of the heterozygous mutation but she was asymptomatic (Figure B in S1 Fig). There were two possible assumptions, the mutation was pathogenic but the inheritance was incomplete in his mother or the mutation was non-pathogenic and the disease of the proband was caused by other unknown reasons. We detected two different mutations in patient P53 with CMT disease. One was a heterozygous gene mutation (c.1039-2A>G, MIM# 609260) in the AG splicing region of MFN2 gene. This mutation could cause autosomal dominant CMT disease type 2A2. Sanger sequencing showed that his mother was not a carrier of this mutation and although his father was a carrier of the heterozygous mutation, he was asymptomatic, so the mutation was not the cause of the disease of patient P53 (Figure C in S1 Fig). The other was a hemizygous GJB1 gene missense mutation (c.265C>G, p.L89V, MIM# 302800). It was reported that mutation c.266T>C in GJB1 gene could cause X-linked CMT disease type 1, indicating that the amino acid of this site was conservative [22]. Sanger sequencing showed that his father was not a carrier and his mother was a carrier of the heterozygous mutation, which was accorded with X-linked inheritance pattern, so this was a suspected pathogenic mutation (Figure D in S1 Fig). A homozygous FGA gene frame shift mutation (c.1368delC, p.T457Rfs*27, MIM# 202400) was detected in patient P58 with autosomal recessive Congenital Afibrinogenemia. Sanger sequencing showed that his parents were all carriers of the heterozygous mutation (Figure E in S1 Fig), so the mutation was the disease-causing mutation. A heterozygous OTX2 gene nonsense mutation (c.538C>T, p.Q180*, MIM# 610125) was detected in patient P76 with autosomal dominant syndromic Microphthalmia type 5. However, his parents did not carry the mutation according to Sanger sequencing (Figure F in S1 Fig). Maybe the mutation of proband was caused by mosaicism of germ cells or spontaneous mutation.
Using quantitative real-time PCR, heterozygous duplications of CDS1-4 in the PMP22 gene of patient P57 were confirmed. A total of 4 primer pairs (S5 Table) were designed to target CDS1-4 of PMP22 gene. The relative quantification (RQ) of CDS1–4 in patient P57 were 1.4965, 1.5674, 1.5513 and 1.5306 respectively, ~150% of the RQ in control sample (C1) (Fig 4B), suggesting that there were heterozygous duplications of CDS1–4 in the PMP22 gene of this patient. The results showed complete consistency of targeted NGS and real-time PCR.
Discussion
Targeted NGS has equal quality to Sanger sequencing in detecting mutations in disease-specific genes, so it can replace Sanger sequencing in diagnostic test [23]. Moreover, the feasibility of using targeted NGS sequencing for genetic diagnosis of inherited diseases has been demonstrated [24]. In this study, we used targeted NGS to mainly capture the CDS of 2,181 genes associated with 561 Mendelian diseases and detected variants for clinical diagnosis. The 561 Mendelian diseases belonged to 17 classifications of diseases, which included genetic bone disease, genodermatosis, hereditary neuropathy, neurocutaneous syndrome, hereditary neuromuscular disease, hereditary muscular disease, hereditary metabolic disease, immune system disorder, endocrine system disease, digestive system disease, respiratory disease, cardiovascular and cerebrovascular disease, hereditary kidney disease, genetic blood disease, hereditary ophthalmopathy, cancer and carcinoid syndrome and multisystemic syndrome. The test of the evaluation showed that the method could detect SNVs with high accuracy and good stability even in the case of low average depth which was only about 80-fold. Moreover, using the pipeline of bioinformatic analysis, we could not only detect SNVs but also detect Indels, CNVs, microdeletions and microduplications of chromosomes. Six candidate mutations in 5 pedigrees and duplications in PMP22 gene were confirmed by Sanger sequencing or real-time PCR, indicating the accuracy and reliability of our method in clinical diagnosis of Mendelian diseases.
For 90 clinical cases, we at first found candidate disease-causing genes in our list of 561 disorders based on the clinical diagnosis and used targeted NGS to find pathogenic mutations in the candidate genes. If the mutation was uncertain based on the results of information analysis and annotated databases we would draw the distribution diagram of reads aligning to reference for the mutated site with a single base resolution. Sanger sequencing or real-time PCR would be performed if the mutation still could not be confirmed. However, the pathogenic mutations of some cases (P47, P48, P59, P65, P77 and P86) were not in the candidate genes. We would find candidate mutations in the other genes in the target regions and backward infer the disease. Clinical features of patient P47 were low sodium and low potassium and the clinical diagnosis was suspected Bartter Syndrome (BS). Two heterozygous CFTR gene mutations (c.1116+1G>A; c.3062C>T, p.P1021L, MIM# 219700) were detected in patient P47. c.1116+1G>A was a splicing mutation which could cause abnormal splice of mRNA and change primary structure of protein. It was included in the database of cystic fibrosis (http://www.genet.sickkids.on.ca/app). c.3062C>T was a missense mutation and Pro1021 was very conservative. Moreover, it was reported that the exchange of proline residue could influence the function of chloride channel [25]. So the compound heterozygous mutations were the causing mutations of cystic fibrosis for this patient. Clinical features of patient P48 were potassium and sodium deficiency, electrolyte instability and mild iodine poisoning. The clinical diagnosis was also suspected BS. A homozygous CFTR gene missense mutation (c.2909G>A, p.G970D, MIM# 219700) was detected in this patient. The incidence of this mutation was very low and it was harmful annotated by SIFT (http://sift.jcvi.org/) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/). It was reported that the mutation could cause Cystic Fibrosis [26] and it was recessive heredity. So c.2909G>A was the pathogenic mutation of patient P48. Clinical features of patient P59 were hemolytic anemia and poor body immunity after birth. She was suspected to suffer from congenital Dyserythropoietic Anemia. Two heterozygous PKLR gene mutations (c.661G>A, p.D221N; c.1528C>T, p.R510*, MIM# 266200) were detected in patient P59. c.661G>A was a missense mutation and the other was a nonsense mutation. They were both harmful annotated by SIFT and PolyPhen-2. It was reported that the two mutations could cause Pyruvate Kinase Deficiency [27–28]. So the two mutations were causative mutations for the case. We did not get the clinical feature of P65 and the clinical diagnosis was suspected Glycogen Storage Disease. A homozygous GALT gene missense mutation (c.1043A>G, p.D348G, MIM# 230400) was detected in this patient. The incidence of this mutation was very low and it was harmful annotated by SIFT and PolyPhen-2. Based on annotated databases, mutations in the GALT gene could cause Galactosemia with an autosomal recessive mode of inheritance, so c.1043A>G was the suspected pathogenic mutation for this case. Clinical features of patient P77 were slow retarded physical development, mental retardation, flattened nose, hepatosplenomegaly, sparse teeth, large head, thick valgus lips, thick eyebrows, claw hand and feet, stubby fingers, kyphosis and short stature, which were very similar to the phenotypes of mucopolysaccharidosis. Two heterozygous GNPTAB gene mutations (c.3565C>T, p.R1189*; c.2590_2591insG, p.E864Gfs*4, MIM# 252500, MIM# 252600) were detected in patient P77. It was reported that c.3565C>T could lead to Mucolipidosis II Alpha/Beta and Mucolipidosis III Alpha/Beta [29]. Although there was no literature reported about the mutation of c.2590_2591insG, the incidence was very low according to annotated databases. So the compound heterozygous mutations were pathogenic mutations of this patient. Clinical features of patient P86 were recurrent multiple pathogen infection (respiratory, digestive, blood), eczema history, idiopathic thrombocytopenic purpura, enlargement of cervical lymph node and submandibular lymphatic nodes. The disease was onset after 3 months old. She was suspected to get Wiskott-Aldrich Syndrome or X-linked Severe Combined Immunodeficiency. Two heterozygous RAG1 gene mutations (c.874T>C, p.S292P; c.1328G>A, p.R443K, MIM# 603554) were detected in patient P86. The two mutations were both harmful annotated by database of SIFT and PolyPhen-2 and the incidence was very low. Mutations in the RAG1 gene could cause autosomal recessive Omenn Syndrome based on annotated databases, therefore, the compound heterozygous mutations were suspected causative mutations of this patient. So our method could not only find pathogenic mutations and confirm the clinical diagnose but also find misdiagnosis of clinical cases. There were 15 samples with negative results. We thought that the reason might be clinical misdiagnosis or the disease-causing gene was not included in the list of 2,181 genes. Cases that remain unsolved by our method can be considered for more time-intensive WES or WGS studies.
Having a newborn with a genetic disease is a disaster for a family. Carrier screening can estimate the risk of offspring with a genetic disease, which plays an important role in fertility guidance and may induce the birth rate of newborns with genetic diseases. Our method can be used as a promising tool for screening asymptomatic couples especially who once have newborns with one of the 561 Mendelian diseases such as the three couples of suspected carriers.
Due to the target regions mainly include CDS of 2,181 genes, there are some limitations of our method. First, deep intronic mutations and complex rearrangements may not be detected. For example, we detected a heterozygous PRF1 gene synonymous mutation (c.1620A>G, p.Q540Q, MIM# 603553) in patient P63 who was diagnosed with Familial Hemophagocytic Lymphohistiocytosis (FHL). It has been reported that the mutation could cause FHL2 with an autosomal recessive inheritance [30]. However, maybe due to the limitation, there were no other disease-causing mutations were detected in the PRF1 gene of this patient. The mutation of c.1620A>G was a suspected disease-causing mutations, which may constitute compound heterozygous mutations combining with other unknown mutations and cause the disease. Second, we may only detect chromosomal microdeletions and microduplications in the list of detection related to the target regions or part of a microdeletion or microduplication in the list. For example, we only detect one mutation and a part (16773072–20222149) of the mutated region of del 17p11.2 in patient P90, partly reflecting SMS.
In summary, we have presented a tool for diagnostic testing that combines capture array and NGS of a panel of 2,181 genes known to be associated with 561 Mendelian diseases. Results of the evaluation showed that our method had high accuracy and stability in detecting disease-causing mutations. The high throughput and speed of our method have been proved in a previous study [9]. So the technology can be used for diagnostic testing, providing effective basis for the clinical diagnosis or genetic counseling of 561 Mendelian diseases.
Supporting Information
A hemizygous COL4A5 gene missense mutation (c.1769A>C) in patient P2 was confirmed (Figure A). A heterozygous AARS gene missense mutation (c.2042G>T) in patient P49 was confirmed (Figure B). A heterozygous gene mutation (c.1039-2A>G) in the AG splicing region of MFN2 gene in patient P53 was confirmed (Figure C). A hemizygous GJB1 gene missense mutation (c.265C>G) in patient P53 was confirmed (Figure D). A homozygous FGA gene frame shift mutation (c.1368delC) in patient P58 was confirmed (Figure E). A heterozygous OTX2 gene nonsense mutation (c.538C>T) in patient P76 was confirmed (Figure F).
(TIF)
(DOC)
(DOC)
(DOC)
We detected a microduplication of chromosome 10 in patient P88, the gender ratio was about 1.42. We detected a microduplication of chromosome 9 in patient P89, the gender ratio was about 1.31. We detected a microdeletion in chromosome 17 (16773072–20222149) in patient P90, the gender ration was about 0.55.
(DOC)
(DOC)
Acknowledgments
We gratefully thank all blood donors for their invaluable contribution to this study.
Data Availability
Data have been uploaded to the NCBI Sequence Read Archive (SRA) and the accession number is SRA274848.
Funding Statement
The authors have no support or funding to report.
References
- 1. Carter C. Monogenic disorders. J Med Genet. 1977; 14 (5): 316–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinkman RR, Dube MP, Rouleau GA, Orr AC, Samuels ME. Human monogenic disorders—a source of novel drug targets. Nature reviews Genetics. 2006; 7(4): 249–60. [DOI] [PubMed] [Google Scholar]
- 3. Metzker ML. Sequencing technologies—the next generation. Nature reviews Genetics. 2010; 11(1): 31–46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- 4. Qing J, Yan D, Zhou Y, Liu Q, Wu W, Xiao Z, et al. Whole-exome sequencing to decipher the genetic heterogeneity of hearing loss in a Chinese family with deaf by deaf mating. PloS one. 2014; 9(10): e109178 10.1371/journal.pone.0109178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England journal of medicine. 2013; 369(16): 1502–11. 10.1056/NEJMoa1306555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews Genetics. 2011; 12(11): 745–55. 10.1038/nrg3031 [DOI] [PubMed] [Google Scholar]
- 7. Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Science translational medicine. 2011; 3(65): 65ra4 10.1126/scitranslmed.3001756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nature reviews Genetics. 2011; 12(9): 628–40. 10.1038/nrg3046 [DOI] [PubMed] [Google Scholar]
- 9. Wei X, Ju X, Yi X, Zhu Q, Qu N, Liu T, et al. Identification of sequence variants in genetic disease-causing genes using targeted next-generation sequencing. PloS one. 2011; 6(12): e29500 10.1371/journal.pone.0029500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alkorta-Aranburu G, Carmody D, Cheng YW, Nelakuditi V, Ma L, Dickens JT, et al. Phenotypic heterogeneity in monogenic diabetes: the clinical and diagnostic utility of a gene panel-based next-generation sequencing approach. Molecular genetics and metabolism. 2014; 113(4): 315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weaver JM, Edwards PA. Targeted next-generation sequencing for routine clinical screening of mutations. Genome medicine. 2011; 3(9): 58 10.1186/gm274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meder B, Haas J, Keller A, Heid C, Just S, Borries A, et al. Targeted next-generation sequencing for the molecular genetic diagnostics of cardiomyopathies. Circulation Cardiovascular genetics. 2011; 4(2): 110–22. 10.1161/CIRCGENETICS.110.958322 [DOI] [PubMed] [Google Scholar]
- 13. Choudhary A, Mambo E, Sanford T, Boedigheimer M, Twomey B, Califano J, et al. Evaluation of an integrated clinical workflow for targeted next-generation sequencing of low-quality tumor DNA using a 51-gene enrichment panel. BMC medical genomics. 2014; 7(1): 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guan Y, Hu H, Peng Y, Gong Y, Yi Y, Shao L, et al. Detection of inherited mutations for hereditary cancer using target enrichment and next generation sequencing. Familial cancer. 2015; 14(1): 9–18. 10.1007/s10689-014-9749-9 [DOI] [PubMed] [Google Scholar]
- 15. Johnson DB, Dahlman KH, Knol J, Gilbert J, Puzanov I, Means-Powell J, et al. Enabling a genetically informed approach to cancer medicine: a retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. The oncologist. 2014; 19(6): 616–22. 10.1634/theoncologist.2014-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Magi A, Tattini L, Pippucci T, Torricelli F, Benelli M. Read count approach for DNA copy number variants detection. Bioinformatics. 2012; 28(4): 470–8. 10.1093/bioinformatics/btr707 [DOI] [PubMed] [Google Scholar]
- 17. Bragin E, Chatzimichali EA, Wright CF, Hurles ME, Firth HV, Bevan AP, et al. DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic acids research. 2014; 42(Database issue): D993–D1000. 10.1093/nar/gkt937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lerner-Ellis JP, Tirone JC, Pawelek PD, Dore C, Atkinson JL, Watkins D, et al. Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nature genetics. 2006; 38(1): 93–100. [DOI] [PubMed] [Google Scholar]
- 19. Hodapp JA, Carter GT, Lipe HP, Michelson SJ, Kraft GH, Bird TD. Double trouble in hereditary neuropathy: concomitant mutations in the PMP-22 gene and another gene produce novel phenotypes. Archives of neurology. 2006; 63(1): 112–7. [DOI] [PubMed] [Google Scholar]
- 20. Huang J, Wu X, Montenegro G, Price J, Wang G, Vance JM, et al. Copy number variations are a rare cause of non-CMT1A Charcot-Marie-Tooth disease. Journal of neurology. 2010; 257(5): 735–41. 10.1007/s00415-009-5401-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang F, Seeman P, Liu P, Weterman MA, Gonzaga-Jauregui C, Towne CF, et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. American journal of human genetics. 2010; 86(6): 892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Janssen EA, Kemp S, Hensels GW, Sie OG, de Die-Smulders CE, Hoogendijk JE, et al. Connexin32 gene mutations in X-linked dominant Charcot-Marie-Tooth disease (CMTX1). Human genetics. 1997; 99(4): 501–5. [DOI] [PubMed] [Google Scholar]
- 23. Sikkema-Raddatz B, Johansson LF, de Boer EN, Almomani R, Boven LG, van den Berg MP, et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Human mutation. 2013; 34(7): 1035–42. 10.1002/humu.22332 [DOI] [PubMed] [Google Scholar]
- 24. Yohe S, Hauge A, Bunjer K, Kemmer T, Bower M, Schomaker M, et al. Clinical validation of targeted next-generation sequencing for inherited disorders. Archives of pathology & laboratory medicine. 2015; 139(2): 204–10. [DOI] [PubMed] [Google Scholar]
- 25. Sheppard DN, Travis SM, Ishihara H, Welsh MJ. Contribution of proline residues in the membrane-spanning domains of cystic fibrosis transmembrane conductance regulator to chloride channel function. The Journal of biological chemistry. 1996; 271(25): 14995–5001. [DOI] [PubMed] [Google Scholar]
- 26. Wagner JA, Vassilakis A, Yee K, Li M, Hurlock G, Krouse ME, et al. Two novel mutations in a cystic fibrosis patient of Chinese origin. Human genetics. 1999; 104(6): 511–5. [DOI] [PubMed] [Google Scholar]
- 27. Fermo E, Bianchi P, Chiarelli LR, Cotton F, Vercellati C, Writzl K, et al. Red cell pyruvate kinase deficiency: 17 new mutations of the PK-LR gene. British journal of haematology. 2005; 129(6): 839–46. [DOI] [PubMed] [Google Scholar]
- 28. Demina A, Varughese KI, Barbot J, Forman L, Beutler E. Six previously undescribed pyruvate kinase mutations causing enzyme deficiency. Blood. 1998; 92(2): 647–52. [PubMed] [Google Scholar]
- 29. Otomo T, Muramatsu T, Yorifuji T, Okuyama T, Nakabayashi H, Fukao T, et al. Mucolipidosis II and III alpha/beta: mutation analysis of 40 Japanese patients showed genotype-phenotype correlation. Journal of human genetics. 2009; 54(3): 145–51. 10.1038/jhg.2009.3 [DOI] [PubMed] [Google Scholar]
- 30. Cappellano G, Orilieri E, Comi C, Chiocchetti A, Bocca S, Boggio E, et al. Variations of the perforin gene in patients with multiple sclerosis. Genes and immunity. 2008; 9(5): 438–44. 10.1038/gene.2008.35 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A hemizygous COL4A5 gene missense mutation (c.1769A>C) in patient P2 was confirmed (Figure A). A heterozygous AARS gene missense mutation (c.2042G>T) in patient P49 was confirmed (Figure B). A heterozygous gene mutation (c.1039-2A>G) in the AG splicing region of MFN2 gene in patient P53 was confirmed (Figure C). A hemizygous GJB1 gene missense mutation (c.265C>G) in patient P53 was confirmed (Figure D). A homozygous FGA gene frame shift mutation (c.1368delC) in patient P58 was confirmed (Figure E). A heterozygous OTX2 gene nonsense mutation (c.538C>T) in patient P76 was confirmed (Figure F).
(TIF)
(DOC)
(DOC)
(DOC)
We detected a microduplication of chromosome 10 in patient P88, the gender ratio was about 1.42. We detected a microduplication of chromosome 9 in patient P89, the gender ratio was about 1.31. We detected a microdeletion in chromosome 17 (16773072–20222149) in patient P90, the gender ration was about 0.55.
(DOC)
(DOC)
Data Availability Statement
Data have been uploaded to the NCBI Sequence Read Archive (SRA) and the accession number is SRA274848.