Abstract
The crystal structure of the four-stranded DNA Holliday junction has now been determined in the presence and absence of junction binding proteins, with the extended open-X form of the junction seen in all protein complexes, but the more compact stacked-X structure observed in free DNA. The structures of the stacked-X junction were crystallized because of an unexpected sequence dependence on the stability of this structure. Inverted repeat sequences that contain the general motif NCC or ANC favor formation of stacked-X junctions, with the junction cross-over occurring between the first two positions of the trinucleotides. This review focuses on the sequence dependent structure of the stacked-X junction and how it may play a role in structural recognition by a class of dimeric junction resolving enzymes that themselves show no direct sequence recognition.
Keywords: recombination, Holliday junction, resolvases, protein recongnition DNA
INTRODUCTION
Homologous recombination is involved in a variety of cellular processes. Originally described as a means to generate genetic diversity by creating new gene combinations (Holliday, 1964,1974), homologous recombination is now recognized to be important for viral integration (Subramaniam et al., 2003), for maintaining genome stability (Flores-Rozas and Kolodner, 2000) through recombination dependent repair of DNA lesions (Kreuzer, 2004; Smith, 2004) and restart of stalled replication forks (Cox et al., 2000; Cox, 2001), and for proper segregation of homologous chromosomes during meiosis (McKim et al., 2002; Morrison et al., 2003; Kreuzer, 2004; McKee, 2004; Sherratt et al., 2004). Loss of recombination functions results in increased mutagenesis, mitotic and meiotic aneuploidy (MacDonald et al., 1994; Kamstra et al., 1999; Kwan et al., 2003), and DNA instability, which has been related to various diseases, including fragile-X syndrome (Bowater and Wells, 2001; Fleming et al., 2003), colon cancer (Grady, 2004), and aging (Lombard et al., 2005; Rodier et al., 2005). In addition, methods are currently being developed to apply recombination strategies to promote genetic therapy (see, for example, Urnov et al., 2005). The central intermediate in homologous recombination is the four-stranded DNA complex known as the Holliday junction. Thus, it is important to characterize the Holliday junction intermediate and how this DNA structure is recognized by recombinases, repair enzymes, and other junction binding proteins to fully understand the basic mechanism of recombination. In this review, we will focus primarily on the detailed structure and structural determinants of the Holliday junction in free DNA, and speculate on how the DNA structure itself may be involved in how proteins recognize and bind to such junctions.
STRUCTURE OF THE DNA HOLLIDAY JUNCTION
Early Models of junctions
The structure and conformation of the DNA Holliday junction (Fig. 1) have been of interest since it was first proposed by R. Holliday in 1964 (Holliday, 1964). The basic structural features of DNA junctions in solution were elucidated in the 1980's by several groups using asymmetric sequence constructs that prevent migration and resolution of the junction off the ends (Kallenbach et al., 1983; Seeman and Kallenbach, 1983; Seeman et al., 1985; Cooper and Hagerman, 1987, 1989; Duckett et al., 1988; Lilley, 1999, 2000). The general structure was found to be dependent on both the type and concentration of cations present in the solution, with the DNA junction adopting either a low salt extended-X form (Fig. 1b) or a high-salt compact stacked-X form of the junction (Fig. 1c, d) (reviewed in Lilley, 1999, 2000). At low salt, the negatively charged phosphates remain largely unshielded and, thus, the arms are extended away from each other in an approximate 4-fold symmetric structure. At higher salt concentrations, condensation of cations around these phosphates allow formation of a more compact structure in which the four arms pair and coaxially stack into two nearly continuous double-helices that are interrupted only by the crossing of strands. The model for the stacked-X junction relates the stacked duplexes by a positive (right-handed) rotation of ~60° (Duckett et al., 1988). The strands of this latter stacked-X junction were proposed to be aligned antiparallel to each other, thereby forcing the two cross-over strands that link the stacked duplex arms to form a sharp U-turn. Notably, this antiparallel form of the junction would be topologically incapable of migrating along the DNA strands, while both the parallel stacked-X (as initially proposed by Holliday, Fig. 1a, Holliday, 1964) and the extended open-X forms would be free to migrate along the duplex arms of homologous sequences.
Figure 1.
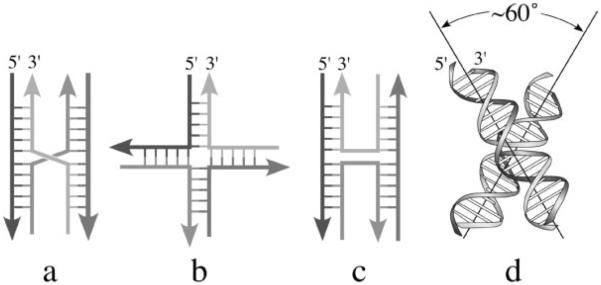
Structural forms of DNA Holliday junctions. a. The parallel stacked-X junction initially proposed by Holliday as the recombination intermediate (Holliday, 1964). b. The extended open-X form of a DNA junction. c. The antiparallel stacked-X junction does not allow for migration of the junction along the DNA strands. d. Model of the antiparallel stacked-X junction proposed from solution studies (Duckett et al., 1988).
How the arms of these asymmetric junctions pair defines different conformational isomer forms of the stacked-X junction. These conformational isomers are determined by the nucleotide sequences immediately around the junction cross-over. The interconversion between isomeric forms has been shown to be, again, cation dependent. Furthermore, recent single-molecule studies (McKinney et al., 2003) show that the interconversion between isomeric forms goes through the extended open-X structure. In homologous sequences, this conversion to the open-X form results in migration and subsequent resolution of the junction into discrete B-DNA duplexes (Lushnikov et al., 2003; McKinney et al., 2003).
First single-crystal structures of junctions
Despite this wealth of physical data in solution, obtaining the single-crystal structure of the Holliday junction had long been considered the `Holy Grail' in DNA crystallography. The detailed molecular structure of the junction was first elucidated in the mid-1990's as complexes of DNA with various repair and recombination proteins, including the RuvA DNA repair protein (Hargreaves et al., 1998; Roe et al., 1998), RuvC (Bennett and West, 1995a), Cre recombinase (Guo et al., 1997), and flp recombinase (Chen et al., 2000). In all of these protein bound structures, the DNA junction adopts some version of the extended open-X form, presumably to allow for migration of the junction cross-over along the DNA strands. The crystal structure of a four-way junction in the absence of protein was first seen in an RNA/DNAzyme complex (Nowakowski et al., 1999, 2000), while the structures of the DNA junction in its native state were finally solved nearly simultaneously by two different laboratories (Fig. 2) (Ortiz-Lombardía et al., 1999; Eichman et al., 2000). Both of these DNA junction structures were obtained serendipitously—the first was from a sequence that was designed to study the structure of adjacent G A mismatched base pairs (Ortiz-Lombardía et al., 1999), while the second was intended to study the structure induced by interstrand thymine-thymine cross-links by the drug psoralen (Eichman et al., 2000, 2001). In the former structure, the sequence was an inverted repeat interrupted by G·A mismatches (5′-CCGGGACCGG-3′), while in the latter case, the junction formed in a true inverted-repeat (IR) sequence with all standard Watson-Crick type base pairs (5′-CCGGTACCGG-3′). The general features of both crystal structures were surprisingly similar to the molecular model proposed from solution work in 1988 (Duckett et al., 1988), with the junctions adopting the antiparallel stacked-X form, and the stacked duplexes related by a right-handed, albeit slightly less twisted (at ~40° rather than the 60° rotation relating the two pairs of stacked helical arms, Fig. 1) (reviewed in Ho and Eichman, 2001; Hays et al., 2003b).
Figure 2.
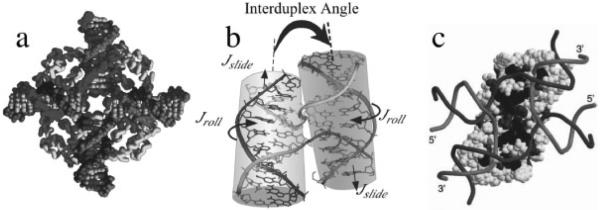
Single-crystal structures of Holliday junctions. a. The open-X junction in complex with the DNA repair protein RuvA (Hargreaves et al., 1998). b. Antiparallel stacked-X junction in the sequence CCGGTACCGG (Eichman et al., 2000). c. Model of the junction-resolving enzyme Hjc from Sulfolobus solfataricus bound to a stacked-X junction (Middleton et al., 2004).
Effect of sequence and sequence dependent interactions on formation and conformation
Comparison of the two junction forming DNA sequences to other similar sequences that had, to that point, been crystallized as standard B-DNA double-helices implicated the ACC trinucleotide at the N6N7N8 nucleotide positions within the sequence motif CCnnnN6N7N8GG as a common motif (a junction core) that stabilized the four-stranded structure in crystals (Eichman et al., 2000; Ho, 2001; Hays et al., 2003b). This hypothesis was supported by the observation that these nucleotides are found at the cross-over of the junction and that the cytosine base at cytosine C8 formed direct hydrogen bonds to the phosphate at the U-turn of the crossing strands. Interestingly, none of the divalent cations present in the crystallization solutions were identified in either crystal structure (Ortiz-Lombardía et al., 1999; Eichman et al., 2000). Thus, contrary to our expectations, there is a strong sequence dependence for the formation and stabilization of DNA junctions in inverted-repeats, which had always been considered to be freely migrating. This raises the question of how sequence, substituent groups, and cations affect the formation and conformation of Holliday junctions.
Crystal structures of Holliday junctions have now been determined from several IR sequences that contain the ACC trinucleotide motif, and show that the junction can form (i) with terminal C·G base pairs replaced by T·A (Thorpe et al., 2003), (ii) with the base at cytosine C8 methylated (Vargason and Ho, 2002), (iii) with the central thymine bases photocross-linked by psoralen (Eichman et al., 2001), (iv) with inosine, 2-aminopurine (Hays et al., 2004) and brominated (Hays et al., 2003a) base analogues at the N6N7N8 positions, and (v) in the presence of various divalent cations including magnesium (Eichman et al., 2000), calcium (Hays et al., 2003a), and strontium (Thorpe et al., 2003). The structures confirm that (i) the ACC core triplet is important for formation of junctions, (ii) the interaction between the N4 amino at the major groove surface of cytosine C8 with the cross-over phosphate is important not only for the formation of the junction but also in defining its conformational geometry, and (iii) divalent cations can be localized along the stacked DNA duplexes and that these cations can affect the local and global geometry of the junction. It should be noted that although the cytosine C8 to phosphate hydrogen bond is seen as an intramolecular interaction in the junction, similar cytosine-phosphate hydrogen bonds were seen to provide sequence-dependent intermolecular interactions that `locked' two B-DNA double-helices together (Timsit et al., 1989). Finally, an interesting variation on this interaction is that this amino-phosphate hydrogen bond can be replaced, to some degree by, a halogen bond, an underappreciated interaction between a polarizable halogen (in this case a bromine of a 5-bromouracil) and a Lewis base (oxygen, nitrogen, sulfur, etc.) (Auffinger et al., 2004).
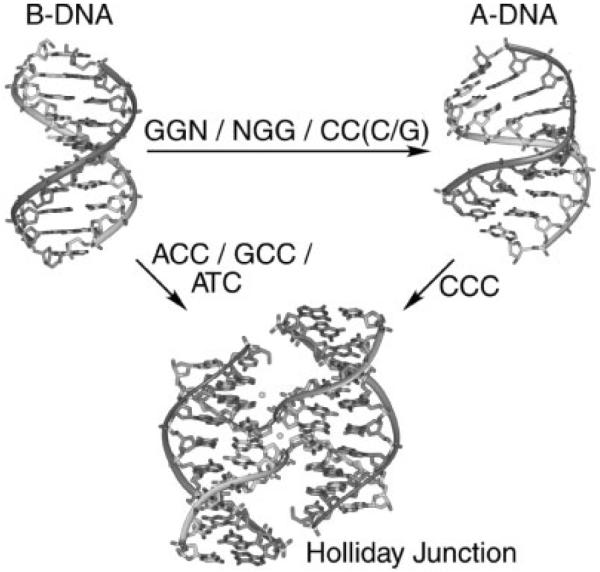
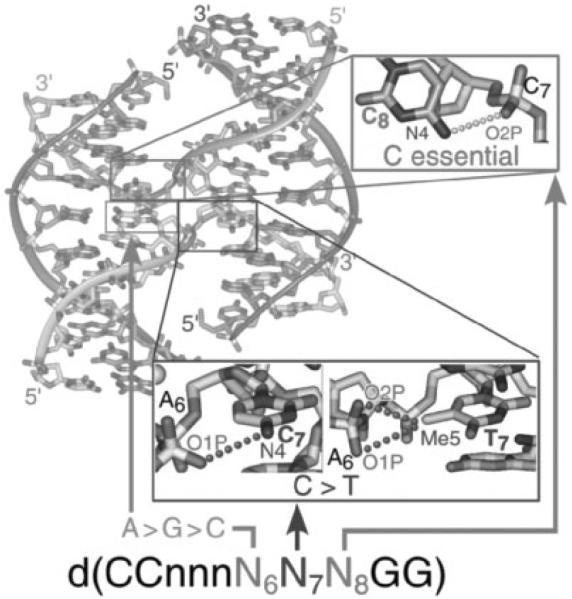
We have recently applied a crystallographic screen of the general IR sequence CCnnnN6N7N8GG (where N6N7N8 is a combination of any of the four standard nucleotides, and nnn are trinucleotides that maintain the overall inverted repeat pattern in the sequence) to search for junction forming sequences that do not contain the ACC-motif. At this point, 63 of the 64 possible sequence combinations have been crystallized and the structures of 29 of these have been determined (Hays et al., 2005). The screen to date has identified a set of sequences that relate the formation of junctions to B-DNA duplexes, junctions to A-DNA, and A-DNA to B-DNA (Fig. 3). Among the structures that resulted from the screen are three new junction-forming sequences, all of which, in contrast to the ACC-core sequence, are identified as amphimorphic (capable of adopting both junctions and double-helical structures). The sequence GCC (these sequences are referred to according to their unique N6N7N8 trinucleotides) was crystallized as a junction from Ca2+ solutions (Aymami et al., 2002; Hays et al., 2003a), but as B-DNA duplexes from Mg2+ solutions (Heinemann et al., 1992). ATC, however, forms both a junction and B-DNA in Ca2+, with high salt favoring the stacked-X junction. We propose that, in this case, the lower cation concentration allows for unstacking of the junction into the open-X form, which subsequently allows for migration and consequently the resolution of the junction into individual B-DNA duplexes. Finally, CCC was seen in Ca2+ solutions to form a junction at high salt and the altered A-DNA duplex at lower salt (Hays et al., 2005). With these amphimorphic sequences spanning the junction and duplex forms of the DNA, Holliday junctions could be related to B-DNA and A-DNA through a structural map constructed from 32 unique single-crystal structures from 29 different sequences (Hays et al., 2005). The structures from this map show (i) that the C8 to phosphate interaction is essential but not sufficient for formation of the junction, (ii) that the C7 position is favored by pyrimidines (C > T) because of electrostatic interactions from either the N4 amino of the cytosine or C5-methyl of the thymine base to a phosphate of an opposing arm, and (iii) that the N6 position shows the preference A > G > C, although the molecular rationale for the series has yet to be established (Fig. 4). In addition, the structural map supports the model that A-DNA is favored explicitly by GGN, NGG, and CC(C/G) trinucleotides within this sequence context, as expected. Thus, this structural map allows four-stranded junctions to be related to both B-DNA and A-DNA explicitly through sequence context. Finally, this sequence motif was seen to adopt structures and conformations relatively independent of crystal lattice and crystallization solutions effects, as evident from the broad range of crystal forms observed in the screen and the relatively limited crystallization solutions required to obtain these large variety of structural and crystal forms. We anticipate that additional junction forming sequences will be identified with the completion of the screen.
Figure 3.
Sequence effects on B-DNA, A-DNA and Holliday junctions. The sequence dependent stabilization of DNA structures has been determined using a crystallographic screen of the decanucleotide sequence CCnnnN6N7N8GG, where each position in N6N7N8 is allowed to be any of the four standard nucleotides, and the trinucleotide nnn is specified to maintain the inverted repeat symmetry of the motif (Hays et al., 2005). The N6N7N8 trinucleotides that lead to formation of junctions from B-DNA, A-DNA from B-DNA, and junctions from A-DNA are indicated.
Figure 4.
Stabilizing interactions in the Holliday junction. The formation of four-stranded DNA junction in the inverted-repeat sequence type d(CCnnnN6N7N8GG) is dependent on the sequence and sequence dependent interactions at the N6N7N8 trinucleotide. A hydrogen bond from the N4 amino of a cytosine at N8 to the phosphate at the junction cross-over is seen to be essential, but not sufficient, to specify formation of the junction. Cytosine is favored over thymine at the N7 position (when N6=A and N8=C) because the electrostatic interaction from the phosphate oxygens of N6 to the N4 amino nitrogen of the cytosine base is stronger (distances range from 3.1 to 3.6Å) as compared to the C5 methyl group of the thymine base (distances range from 4.2 to 4.5Å). The preference is A > G > C (with N7 and N8=C) for the nucleotide at N6.
With the increasing number of crystal structures of DNA junctions, it became necessary to develop a set of definitions to accurately describe geometries of the four-stranded complexes relative to a set of defined planes (Vargason and Ho, 2002; Watson et al., 2004) in order to quantitatively analyze and compare the effects of various factors, including sequence, salt and drugs on their conformations. An analysis of the currently available DNA structures shows that stacked-X type junctions in the crystal exhibit a more shallow Jtwist (the angle relating the stacked duplex arms across the junction) as compared to the model derived from solution studies. In addition, the arms can be translated along their helix axes (characterized as Jslide) and rotated about their helical axes (measured by Jroll) to either bury or expose the major groove surfaces of the junction. These geometric perturbations are associated with the effect that sequence, salt, and substituents have on the intramolecular interactions at the junction core (Vargason and Ho, 2002; Hays et al., 2003a; Watson et al., 2004).
The obvious question is whether the properties of junctions seen in crystals have any relationship to the structure in solution. For example, all of the symmetric junctions in IR sequences have a more shallow twist angle relating the stacked duplex arms (40° −45°) compared to that determined for asymmetric junctions in non-IR sequence constructs (~60°). Studies using atomic force microscopy and hydroxyl radical foot-printing to probe the geometry of symmetric junctions that contain the ACC-core (Sha et al., 2002), however, show that this angle is ~40° and that the junction crosses between duplexes exactly between the A and C nucleotides of the core triplet, as seen in the crystal structures (Hays et al., 2003b). These results indicate that the sequence specific interactions identified within the ACC core are responsible for specifying the geometry of the DNA junction even outside the environment of the crystal lattice.
Does this ACC core, however, help to stabilize the junction in solution? To address this question, we recently studied the parent (5′-CCGGTACCGG-3′)4 junction by analytical ultracentrifugation and determined dissociation constant for the junction to duplex equilibrium of 100–200 μM (Hays et al., 2006). Analytical ultracentrifugation studies showed that the similar sequence 5′-CCGCTAGCGG-3′ (which does not crystallize as a junction, but as B-DNA duplexes) exists only as double-helices in solution. Thus, the ACC core is seen to contribute ~5 kcal/mol of stabilization to the tetrameric junction. Moreover, the junction is dissociated at low Ca2+ concentrations even at high DNA concentrations, consistent with the general understanding that the stacked-X junction is stabilized by high concentrations of divalent cations (Duckett et al., 1990). It is clear, therefore, that the stacked-X DNA junction in solution is well described by the single-crystal structures, including their sequence dependent formation and the intramolecular interactions associated with their formation. One must ask, however, whether the compact stacked-X structure seen in isolated DNA constructs is at all relevant to the biological mechanisms of recombination where the DNA does not exist in isolation, but in the context of protein complexes.
JUNCTION BINDING PROTEINS
There are currently numerous junction binding proteins known, consistent with the variety of cellular mechanisms associated with homologous recombination (Aravind et al., 2000; Lilley and White, 2001); however, only a handful have been characterized in complex with their DNA substrates (Sharples, 2001). Although the DNA junctions seen in crystals structures of all current protein-DNA complexes are in the open-X form, a large number of junction binding proteins show high affinity for the stacked-X junction, including the BLM protein associated with Bloom's syndrome (Karow et al., 2000). The general forms of the DNA in complexes with several dimeric resolvases (enzymes that make symmetric cuts at the point of strand exchange in four-stranded junctions, (White and Lilley, 2001), including T7 endonuclease I (Declais et al., 2003) and Hjc resolvase (Fig. 2) (Fogg et al., 2001; Middleton et al., 2004), have been characterized biochemically by gel electrophoresis and fluorescence resonance energy transfer (reviewed in Lilley, 2000). The dimeric junction resolving enzyme RusA has been shown to bind stacked-X junctions and specifically cut homologous sequences at the phosphodiester bond 5′ of CC dinucleotides (Chan et al., 1997; Giraud-Panis and Lilley, 1998). Although, as with many resolvases, RusA distorts the junction upon binding, this sequence specificity, along with the observations that NCC trinucleotides favor junction formation in the absence of proteins both in crystals and in solution, suggests that the sequence context for junction formation may play a role in recognition.
Classifying the junction binding proteins according to whether they recognize the open-X or stacked-X form of the junction (those for which the structure of the DNA can be experimentally assigned, Table 1) provides some interesting insights into their structural specificity for the DNA substrate. Enzymes that recognize the two-fold symmetric stacked-X junction are all dimeric, while nearly all tetrameric proteins bind to the approximate four-fold symmetric open-X structure. Moreover, proteins that recognize open-X junctions also have some degree of sequence specificity (either a specific DNA sequence or damaged base pairs), while those that recognize stacked-X junctions are relatively non-specific at the sequence level (the lone exception in Table 1 is topoisomerase I from vaccinia, Liao et al., 2004). It seems reasonable, therefore, to think of the tetrameric proteins as a group that binds to the open-X form to allow for migration of the junction and provide a means for the protein to seek-out its target sequence.
Table 1.
Holliday Junction Binding Enzymes
| Enzyme | Organism | Oligomeric state | Sequence Specificity2 | References |
|---|---|---|---|---|
| Open-X Type DNA Junction Substrate | ||||
| Cre1 | Bacteriophage P1 | Tetramer | LoxP sequence | (Guo et al., 1997) |
| RuvAB1 | E. coli | Tetramer | Damaged DNA | (Hargreaves et al., 1998; Roe et al., 1998) |
| Flp1 | S. Cerevisiae | Tetramer | Flp Recombination target (FRT)3 | (Chen et al., 2000) |
| λ Integrase1 | Bacteriophage λ | Tetramer | TNNNTTNNTNNNANNAANNNG | (Biswas et al., 2005) |
| RecU | B. subtilis | Dimer | (G/t)G ↓ C(A/C) | (McGregor et al., 2005) |
| Induced Open-X Junction Substrate4 | ||||
| RuvC1 | E. coli | Dimer | (A/T)TT(G/C) | (Bennett and West, 1995b; Fogg et al., 2001) |
| CCE1 | S. Cerevisiae | Dimer | ACTA | (White and Lilley, 1997) |
| Ydc2 | S. Pombe | Dimer | CT and/or TT | (White and Lilley, 1998): |
| Stacked-X Type DNA Junction Substrate | ||||
| T4 nuclease VII | Bacteriophage T4 | Dimer | None | (White and Lilley, 1997; Raaijmakers et al., 1999) |
| T7 Endonuclease I | Bacteriophage T7 | Dimer | None, (C/T) ↓ (C/T) | (Declais et al., 2003) |
| Hjc | P. furiosu | Dimer | None | (Middleton et al., 2004) |
| Hjc | S. solfataricus | Dimer | None | (Fogg et al., 2001) |
| Hje | S. solfataricus | Dimer | None | (Middleton et al., 2004) |
| Vtopo I | Vaccinia | Dimer | CCCTT ↓ N | (Liao et al., 2004) |
| Tetrahedral DNA Junction Substrate | ||||
| RusA | E. coli | Dimer | ↓ CC | (Chan et al., 1997; Giraud-Panis, 1998) |
Crystallized DNA-complex.
Binding or cutting sites (cut site specificity indicated by vertical arrow).
Natural Flp Recombination target is an A/T-rich 48 bp sequence (Chen et al., 2000).
Binds stacked-X, but induces an open-X structure in complex.
Enzymes are categorized according to conformation of the DNA substrate (open-X, stacked-X, or tetrahedral forms) as determined from crystal structures with junctions1, or inferred by biochemical data and/or molecular modeling (references are to studies that define the form of the DNA substrate).
One group of dimeric proteins that initially appears to violate these general trends includes RuvC (Bennett and West, 1995b; Fogg et al., 2001), and the resolvases CCE1 from S. cervisiae (White and Lilley, 1997) and Ydc2 from S. pombe (White and Lilley, 1998): these are all homodimers, but the DNA substrates in the complexes are seen to adopt the open-X structure. A more detailed analysis of this group suggests that these proteins are not so much exceptions to the general trends described above, but serve to bridge the dimeric stacked-X binding proteins with the tertrameric open-X binding proteins. These RuvC-related proteins are thought to initially recognize and bind to stacked-X junctions, but then to induce the DNA to adopt the more extended open-X form (Fogg et al., 2001). The induced structural perturbations to the DNA are associated with the dimerization of the protein—the monomers do not induce the open-X structure. Thus, the dimeric proteins all induce some structural perturbation to the stacked-X structure (from minor opening of the junction center and rotations of the stacked arms, to more dramatic changes to the open-X or even a possible tetrahedral form) presumably to allow the enzymes to gain access to the scissile bond or to stabilize a transition state (Sharples, 2001). We suggest here that the protein-induced open-X structure may also allow the junction to migrate and the protein to seek-out its specific recognition site, even if it is the immobile stacked-X form that is initially recognized.
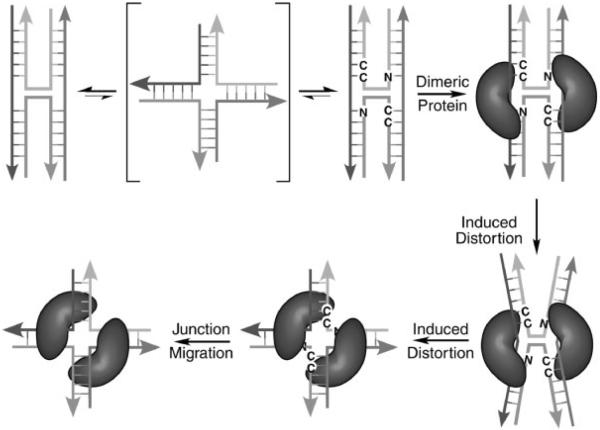
What then is the role of the stacked-X junction? We propose here a model in which the sequence dependent formation of this compact structure provides dimeric proteins, including nonspecific resolvases, with some degree of sequence specificity through an `indirect-readout' mechanism (Dickerson, 1983; Otwinowski et al., 1988; Olson et al., 1998; Lu et al., 2000; Arauzo-Bravo et al., 2005), as opposed to direct recognition of base pair identity (Fig. 5). In this model, inverted-repeat sequences that incorporate the ACC and, to lesser extents, the amphimorphic GCC, ATC and CCC trinucleotides help pause or fix junctions at specific sites along a genome. The evidence for sequence specific pausing during junction migration was first seen in immobilized symmetric junctions by Seeman's group (Sun, et al., 1998). It is then this stabilized junction that serves as the substrate for protein binding. In a classic example of an induced-fit model for enzymes, the protein subsequently induces distortions away from the intrinsic structure of the DNA junction in the course of its function. This perturbation can maintain the general topology and symmetry of the stacked-X junction, as with Hjc, or may induce the open-X structure if there is a need for the junction to migrate as the protein searches for a target sequence, as in the cases of CCE1 and Ydc2. Thus, in this model, sequence specificity is conferred at the initial point of recognition and binding, but through indirect sequence dependent stabilization of the stacked-X junction rather than by direct read-out of the base pairs that define the DNA sequence.
Figure 5.
Proposed model for indirect sequence recognition of stacked-X junctions. In this model, migration of the Holliday junction in free DNA requires a transition to the open-X form. This is consistent with models proposed from single-molecule studies on junction isomerization (McKinney et al., 2003) and translocation (Lushnikov et al., 2003). However, certain sequences such as the ACC-trinucleotide in an inverted repeat help to stabilize and stall the junction, thereby presenting a defined structure for recognition by a dimeric junction binding protein. In the complex, the protein induces a structural perturbation that either maintains the topology and symmetry of the stacked-X junction, or induces an open-X junction that can then migrate to a protein specific target site.
SUMMARY AND PERSPECTIVES
We now have a very detailed understanding of the DNA Holliday junction alone in the compact stacked-X form, at least within the context of inverted-repeat DNA sequences. Specific intramolecular interactions are seen to direct the formation and conformation of the DNA junction both in solution and in crystals. We suggest, therefore, that sequences that favor junction formation may provide a stable substrate for recognition and binding by proteins involved in recombination and DNA integration processes. This seems to be particularly important for dimeric enzymes that typically are not highly specific for a particular DNA sequence. Thus, specificity may be conferred by the ability of certain DNA sequences, particularly in inverted repeat sequences, to fix the junction and, thereby, indirectly confer sequence specificity through structure specificity. We recognize that the model proposed here is based on a small subset of known junction binding proteins, and the two general observations on which the model is based may need to be revised as the structures of the DNA substrates that are recognized and bound become characterized for additional proteins.
Where do we go from here in terms of the structure of the Holliday junction? For the DNA itself, we are challenged to determine the structures of the stacked-X junction in non-symmetric sequences to help bridge the conceptual gap between junctions in crystals and the wealth of information on their behavior in solution (as reviewed by Lilley, Lilley, 2000). In addition, it would be informative to determine the structure of the open-X junction in the absence of protein (in order to understand how, by comparison, the protein affects this form) and perhaps other possible junction forms, including a potential tetrahedral four-stranded junction and three-way junctions. For junction binding proteins, the challenge has been to determine the structure of a resolvase in complex with a stacked-X substrate, particularly one that does not bind to or induce an open-X junction. Together, structural studies on junction binding proteins and their DNA substrates will provide us with an understanding for how sequence directed conformations contribute to `indirect read-out' of genomic information, particularly for the ever growing class of biological functions that rely on the mechanism of genetic exchange first elucidated by R. Holliday over 40 years ago.
Acknowledgements
Work in the lab of PSH was funded by grants from the National Institutes of Health (R1GM62957) and the National Science Foundation (MCB0090615). PSH would like to thank the Franco-American Fulbright Commission and Prof. Eric Westhof at the CNRS at the Université Louis Pasteur in Strasbourg, France for support and help in the writing of this manuscript.
REFERENCES
- Arauzo-Bravo MJ, Fujii S, Kono H, Ahmad S, Sarai A. Sequence-dependent conformational energy of DNA derived from molecular dynamics simulations: toward understanding the indirect readout mechanism in protein-DNA recognition. J. Am. Chem. Soc. 2005;127(46):16074–16089. doi: 10.1021/ja053241l. [DOI] [PubMed] [Google Scholar]
- Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28(18):3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, Hays FA, Westhof E, Ho PS. Halogen bonds in biological molecules. Proc. Natl. Acad. Sci. USA. 2004;101(48):16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymami J, Pous J, Lisgarten JN, Coll M. Crystallization and preliminary X-ray analysis of the DNA decamers d(CCGGATCCGG) and d(CCGGCGCCGG) Acta Crystallogr. D. Biol. Crystallogr. 2002;58(Pt 2):310–311. doi: 10.1107/s0907444901018959. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, West SC. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J. Mol. Biol. 1995a;252:213–226. doi: 10.1006/jmbi.1995.0489. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, West SC. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J. Mol. Biol. 1995b;252(2):213–226. doi: 10.1006/jmbi.1995.0489. [DOI] [PubMed] [Google Scholar]
- Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435(7045):1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater RP, Wells RD. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:159–202. doi: 10.1016/s0079-6603(00)66029-4. [DOI] [PubMed] [Google Scholar]
- Chan SN, Harris L, Bolt EL, Whitby MC, Lloyd RG. Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J. Biol. Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6(4):885–897. [PubMed] [Google Scholar]
- Cooper JP, Hagerman PJ. Gel electrophoretic analysis of the geometry of a DNA four-way junction. J. Mol. Biol. 1987;198:711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Hagerman PJ. Geometry of a branched DNA structure in solution. Proc. Natl. Acad. Sci. USA. 1989;86:7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 2001;35:53–82. doi: 10.1146/annurev.genet.35.102401.090016. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Declais AC, Fogg JM, Freeman AD, Coste F, Hadden JM, Phillips SE, Lilley DM. The complex between a four-way DNA junction and T7 endonuclease I. EMBO J. 2003;22(6):1398–1409. doi: 10.1093/emboj/cdg132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson RE. The DNA helix and how it is read. Sci. Am. 1983;249(6):94–98. [Google Scholar]
- Duckett DR, Murchie AI, Lilley DM. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990;9(2):583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett DR, Murchie AIH, Diekmann S, von Kitzing E, Kemper B, Lilley DMJ. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Eichman BF, Mooers BHM, Alberti M, Hearst JE, Ho PS. The crystal structures of psoralen cross-linked DNAs: drug dependent formation of Holliday junctions. J. Mol. Biol. 2001;301:15–26. doi: 10.1006/jmbi.2001.4567. [DOI] [PubMed] [Google Scholar]
- Eichman BF, Vargason JM, Mooers BHM, Ho PS. The Holliday junction in an inverted repeat sequence: sequence effects on the structure of four-way junctions. Proc. Natl. Acad. Sci. USA. 2000;97:3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming K, Riser DK, Kumari D, Usdin K. Instability of the fragile X syndrome repeat in mice: the effect of age, diet and mutations in genes that affect DNA replication, recombination and repair proficiency. Cytogenet. Genome Res. 2003;100(1–4):140–146. doi: 10.1159/000072848. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H, Kolodner RD. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci. 2000;292:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg JM, Kvaratskhelia M, White MF, Lilley DM. Distortion of DNA junctions imposed by the binding of resolving enzymes: a fluorescence study. J. Mol. Biol. 2001;313(4):751–764. doi: 10.1006/jmbi.2001.5081. [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Lilley DM. Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J. Mol. Biol. 1998;278:117–133. doi: 10.1006/jmbi.1998.1681. [DOI] [PubMed] [Google Scholar]
- Grady WM. Genomic instability and colon cancer. Cancer Metastasis Rev. 2004;23(1–2):11–27. doi: 10.1023/a:1025861527711. [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389(6646):40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- Hargreaves D, Rice DW, Sedelnikova SE, Artymiuk PJ, Lloyd RG, Rafferty JB. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6Å resolution. Nat. Struct. Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- Hays FA, Jones ZJ, Ho PS. Influence of minor groove substituents on the structure of DNA Holliday junctions. Biochemistry. 2004;43(30):9813–9822. doi: 10.1021/bi049461d. [DOI] [PubMed] [Google Scholar]
- Hays FA, Teegarden A, Jones ZJ, Harms M, Raup D, Watson J, Cavaliere E, Ho PS. How sequence defines structure: a crystallographic map of DNA structure and conformation. Proc. Natl. Acad. Sci. USA. 2005;102(20):7157–7162. doi: 10.1073/pnas.0409455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays FA, Vargason JM, Ho PS. Effect of sequence on the conformation of DNA holliday junctions. Biochemistry. 2003a;42(32):9586–9597. doi: 10.1021/bi0346603. [DOI] [PubMed] [Google Scholar]
- Hays FA, Watson J, Ho PS. Caution! DNA crossing: crystal structures of Holliday junctions. J. Biol. Chem. 2003b;278(50):49663–49666. doi: 10.1074/jbc.R300033200. [DOI] [PubMed] [Google Scholar]
- Hays FA, Schirf V, Ho PS, Demeler B. Solution formation of Holliday juncions in inverted-repeat DNA sequences. Biochemistry. 2006;45(8):2461–2471. doi: 10.1021/bi052129x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U, Alings C, Bansal M. Double helix conformation, groove dimensions and ligand binding potential of a G/C stretch in B-DNA. EMBO J. 1992;11:1931–1939. doi: 10.1002/j.1460-2075.1992.tb05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PS, Eichman BF. The crystal structures of DNA Holliday junctions. Current Opin. Struct. Biol. 2001;11:302–308. doi: 10.1016/s0959-440x(00)00219-0. [DOI] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Holliday R. Molecular aspects of genetic exchange and gene conversion. Genetics. 1974;78:273–287. doi: 10.1093/genetics/78.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach NR, Ma RI, Wand AJ, Veeneman GH, van Boom JH, Seeman NC. Fourth rank immobile nucleic acid junctions. J. Biomol. Struct. Dyn. 1983;1(1):159–168. doi: 10.1080/07391102.1983.10507432. [DOI] [PubMed] [Google Scholar]
- Kamstra SA, Kuipers AG, De Jeu MJ, Ramanna MS, Jacobsen E. The extent and position of homoeologous recombination in a distant hybrid of Alstroemeria: a molecular cytogenetic assessment of first generation backcross progenies. Chromosoma. 1999;108(1):52–63. doi: 10.1007/s004120050351. [DOI] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li J-L, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci., USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN. Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 2004;59:43–67. doi: 10.1146/annurev.micro.59.030804.121255. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Moens PB, Wang JC. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III beta. Proc. Natl. Acad. Sci. USA. 2003;100(5):2526–2531. doi: 10.1073/pnas.0437998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Mao C, Birktoft JJ, Shuman S, Seeman NC. Resolution of undistorted symmetric immobile DNA junctions by vaccinia topoisomerase I. Biochemistry. 2004;43(6):1520–1531. doi: 10.1021/bi0358061. [DOI] [PubMed] [Google Scholar]
- Lilley DM, White MF. The junction-resolving enzymes. Nat. Rev. Mol. Cell. Biol. 2001;2(6):433–443. doi: 10.1038/35073057. [DOI] [PubMed] [Google Scholar]
- Lilley DMJ. Structures and interactions of helical junctions in nucleic acids. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structure. Oxford University Press; New York: 1999. pp. 471–498. [Google Scholar]
- Lilley DMJ. Structures of helical junctions in nucleic acids. Quart. Rev. Biochem. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120(4):497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Lu X-J, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- Lushnikov AY, Bogdanov A, Lyubchenko YL. DNA recombination: holliday junctions dynamics and branch migration. J. Biol. Chem. 2003;278(44):43130–43134. doi: 10.1074/jbc.M308228200. [DOI] [PubMed] [Google Scholar]
- MacDonald M, Hassold T, Harvey J, Wang LH, Morton NE, Jacobs P. The origin of 47,XXY and 47,XXX aneuploidy: heterogeneous mechanisms and role of aberrant recombination. Hum. Mol. Genet. 1994;3(8):1365–1371. doi: 10.1093/hmg/3.8.1365. [DOI] [PubMed] [Google Scholar]
- McGregor N, Ayora S, Sedelnikova S, Carrasco B, Alonso JC, Thaw P, Rafferty J. The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure (Camb) 2005;13(9):1341–1351. doi: 10.1016/j.str.2005.05.011. [DOI] [PubMed] [Google Scholar]
- McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta. 2004;1677(1–3):165–180. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- McKim KS, Jang JK, Manheim EA. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 2002;36:205–232. doi: 10.1146/annurev.genet.36.041102.113929. [DOI] [PubMed] [Google Scholar]
- McKinney SA, Declais AC, Lilley DM, Ha T. Structural dynamics of individual Holliday junctions. Nat. Struct. Biol. 2003;10(2):93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- Middleton CL, Parker JL, Richard DJ, White MF, Bond CS. Substrate recognition and catalysis by the Holliday junction resolving enzyme Hje. Nucleic Acids Res. 2004;32(18):5442–5451. doi: 10.1093/nar/gkh869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC. Sister chromatid cohesion and genome stability in vertebrate cells. Biochem. Soc. Trans. 2003;31(Pt 1):263–265. doi: 10.1042/bst0310263. [DOI] [PubMed] [Google Scholar]
- Nowakowski J, Shim PJ, Prasad GS, Stout CD, Joyce GF. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10–23 DNA enzyme. Nat. Struct. Biol. 1999;6:151–156. doi: 10.1038/5839. [DOI] [PubMed] [Google Scholar]
- Nowakowski J, Shim PJ, Stout CD, Joyce GF. Alternative conformations of a nucleic acid four-way junction. J. Mol. Biol. 2000;300:93–102. doi: 10.1006/jmbi.2000.3826. [DOI] [PubMed] [Google Scholar]
- Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. USA. 1998;95(19):11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Lombardía M, González A, Eritja R, Aymamí J, Azorín F, Coll M. Crystal structure of a DNA Holliday junction. Nat. Struct. Biol. 1999;6:913–917. doi: 10.1038/13277. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Raaijmakers H, Vix O, Tör I, Golz S, Kemper B, Suck D. X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 1999;18:1447–1458. doi: 10.1093/emboj/18.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Kim SH, Nijjar T, Yaswen P, Campisi J. Cancer and aging: the importance of telomeres in genome maintenance. Int. J. Biochem. Cell. Biol. 2005;37(5):977–990. doi: 10.1016/j.biocel.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Roe SM, Barlow T, Brown T, Oram M, Keeley A, Tsaneva IR, Pearl LH. Crystal structure of an octameric RuvA-Holliday junction complex. Cell. 1998;2:361–372. doi: 10.1016/s1097-2765(00)80280-4. [DOI] [PubMed] [Google Scholar]
- Seeman NC, Kallenbach NR. Design of immobile nucleic acid junctions. Biophys. J. 1983;44(2):201–209. doi: 10.1016/S0006-3495(83)84292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC, Maestre MF, Ma RI, Kallenbach NR. Physical characterization of a nucleic acid junction. Prog. Clin. Biol. Res. 1985;172A:99–108. [PubMed] [Google Scholar]
- Sha R, Liu F, Seeman NC. Atomic force microscopic measurement of the interdomain angle in symmetric Holliday junctions. Biochemistry. 2002;41(19):5950–5955. doi: 10.1021/bi020001z. [DOI] [PubMed] [Google Scholar]
- Sharples GJ. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 2001;39(4):823–834. doi: 10.1046/j.1365-2958.2001.02284.x. [DOI] [PubMed] [Google Scholar]
- Sherratt DJ, Soballe B, Barre FX, Filipe S, Lau I, Massey T, Yates J. Recombination and chromosome segregation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2004;359(1441):61–69. doi: 10.1098/rstb.2003.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KC. Recombinational DNA repair: the ignored repair systems. Bioessays. 2004;26(12):1322–1326. doi: 10.1002/bies.20109. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Tewari AK, Nunes-Duby SE, Foster MP. Dynamics and DNA substrate recognition by the catalytic domain of lambda integrase. J. Mol. Biol. 2003;329(3):423–439. doi: 10.1016/s0022-2836(03)00469-8. [DOI] [PubMed] [Google Scholar]
- Sun W, Mao C, Liu F, Seeman NC. Sequence dependence of branch migratory minima. J. Mol. Biol. 1998;282:59–70. doi: 10.1006/jmbi.1998.1991. [DOI] [PubMed] [Google Scholar]
- Thorpe JH, Gale BC, Teixeira SC, Cardin CJ. Conformational and Hydration Effects of Site-selective Sodium, Calcium and Strontium Ion Binding to the DNA Holliday Junction Structure d(TCGGTACCGA)(4) J. Mol. Biol. 2003;327(1):97–109. doi: 10.1016/s0022-2836(03)00088-3. [DOI] [PubMed] [Google Scholar]
- Timsit Y, Westhof E, Fuchs RPP, Moras D. Unusual helical packing in crystals of DNA bearing a mutation hot spot. Nature. 1989;341:459–462. doi: 10.1038/341459a0. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Vargason JM, Ho PS. The effect of cytosine methylation on the structure and geometry of the Holliday junction: the structure of d(CCGGTACm5CGG) at 1.5 A resolution. J. Biol. Chem. 2002;277(23):21041–21049. doi: 10.1074/jbc.M201357200. [DOI] [PubMed] [Google Scholar]
- Watson J, Hays FA, Ho PS. Definitions and analysis of DNA Holliday junction geometry. Nucleic Acids Res. 2004;32(10):3017–3027. doi: 10.1093/nar/gkh631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF, Giraud-Panis M-JE, Pöhler JRG, Lilley DMJ. Recognition and manipulation of branched DNA structure by junction-resolving enzymes. J. Mol. Biol. 1997;269:647–664. doi: 10.1006/jmbi.1997.1097. [DOI] [PubMed] [Google Scholar]
- White MF, Lilley DM. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J. Mol. Biol. 1997;266(1):122–134. doi: 10.1006/jmbi.1996.0795. [DOI] [PubMed] [Google Scholar]
- White MF, Lilley DM. Interaction of the resolving enzyme YDC2 with the four-way DNA junction. Nucleic Acids Res. 1998;26(24):5609–5616. doi: 10.1093/nar/26.24.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]