Abstract
Cancer treatments often lose their effectiveness due to the development of multiple drug resistance. Thus, identification of key proteins involved in the tumorigenic process and the survival mechanism(s), coupled with the design of novel therapeutic compounds (such as small molecule inhibitors), are essential steps towards the establishment of improved anticancer treatment strategies. DNA repair pathways and their proteins have been exposed as potential targets for combinatorial anticancer therapies that involve DNA-interactive cytotoxins, such as alkylating agents, because of their central role in providing resistance against DNA damage. In addition, an understanding of the tumor-specific genetics and associated DNA repair capacity has allowed research scientists and clinicians to begin to devise more targeted treatment strategies based on the concept of synthetic lethality. In this review, the repair mechanisms, as well as the links to cancer progression and treatment, of three key proteins that function in the base excision repair pathway, i.e. APE1, POLβ, and FEN1, are discussed.
Keywords: AP or abasic endonuclease, DNA polymerase, flap endonuclease, carcinogenesis, repair inhibitor
Clinical Anticancer Treatments: The Multi-Drug Resistance Problem
Cancer treatments have traditionally been divided into three categories: (1) chemotherapy, (2) radiation therapy, and (3) surgery. Chemotherapy treatments are based on the idea of targeting rapidly proliferating cells (the cancer) with one or more cytotoxic drugs. Unfortunately, the body contains rapidly dividing noncancerous cells that are also sensitive to this treatment strategy, causing side effects such as hair loss, leucopenia (immunosuppression), mouth/throat sores, nausea and vomiting. Chemotherapy is considered a systemic treatment, because it is not localized to a specific tissue or area of the body, whereas radiation and surgical techniques are targeted treatments used typically prior to metastasis of the cancer. Chemotherapy can also be used in concert with radiation or surgical methods to eradicate the primary tumor and attack any circulating cancer cells throughout the remainder of the body. Although the different treatment paradigms are quite rigorous, studies show that cancers continually evolve to develop drug resistance, making them difficult to completely destroy. As a result, anticancer treatments are not 100% effective.
The failure of a patient to respond to a specific anticancer therapy can result through two general routes: (1) the genetics and physiology of the patient (host factors) and (2) the genetics or epigenetics of the cancer itself. Some host factors include immediate drug excretion from the body that results in low serum levels, poor cellular absorption rates, and rapid metabolism once absorbed intracellularly [1]. The genetic and epigenetic changes that give rise the neoplastic disease often result in differential expression of oncogenes and consequent variability in the tumor phenotype, and such changes can differ greatly from patient to patient, by tissue type, and between cells within the tumor itself. Researchers for the past several decades have been trying to discern the various mechanisms of how cancer cells proliferate and become resistant to anticancer drugs. Some mechanisms include the loss of a cell surface receptor, a change in drug metabolism, or mutation of the drug target. It was therefore hypothesized that using multiple drugs with disparate entry and cytotoxic mechanisms would be effective and result in high cure rates. However, cancer cells appear to be able to develop simultaneous resistance to multiple anticancer remedies [2]. When a cancer reaches the point of multi-drug resistance (MDR), there are limited treatment options. It is thus critical to identify and target specific and selective processes that will immediately drive tumor cells into apoptosis, or that will allow clinicians to circumvent the various resistance mechanisms.
Studies that analyze and elucidate the molecular events of cancer cell progression and metastasis are often aimed at identifying novel targets for the development of new or synergistic treatment methods. Since many of the cytotoxins work by causing DNA lesions that induce cell death when unrepaired (Table 1), inhibiting DNA repair pathways, ultimately in the cancer itself, is of great interest in new drug development. In particular, many commonly used platinum-based drugs, like oxaliplatin and cisplatin [3], and the athracyclines, such as epirubicin, daunorubicin, doxorubicin and paclitaxel [4], aim to overwhelm a cancer cell’s DNA repair machinery by directly introducing DNA lesions and/or by elevating the production of reactive oxygen species (ROS). Other drugs, such as temozolomide (TMZ), melphalan, thioTEPA, methyl-lexitropsin, dacarbazine/procarbazine and streptozotocin, introduce alkylative lesions on DNA [5].
Table 1.
Common anticancer drugs and the modes of cytotoxicity.
| Drug Classification | Examples | Mechanism of Cytotoxicity |
|---|---|---|
| Alkylating agents | Nitrogen mustards, nitrosoureas, platinum | DNA damage: base monoadducts, intra- and inter-strand crosslinks |
| Anti-metabolites | Folic acid, purine and pyrimidine analogs | Inhibition of DNA synthesis: disruption of nucleotide pools, chain termination |
| Antibiotics | Anthracycline family (e.g. doxorubicin), bleomycin, hydroxyurea, mitomycin | DNA damage: trapped protein-DNA intermediates, free radical-induced damage, crosslinks |
| Topoisomerase inhibitors | Topotecan, irinotecan, etoposide | DNA damage: trapped protein-DNA intermediates |
| Radiotherapy | Ionizing radiation | DNA damage: single- and double-strand breaks, base damage, clustered lesions |
Though multiple DNA interactive drugs are often used in cancer treatment paradigms, cancer cells can develop MDR by increasing DNA repair efficiency. For example, the drug TMZ generates several DNA lesions, including O6-methylguanine [6], which can be repaired by the protein O6-methylguanine-DNA methyltransferase (MGMT). In the absence of MGMT, a persistent O6-methylguanine lesion can initiate a futile mismatch repair (MMR) response that triggers ataxia telangiectasia and Rad3-related (ATR) protein kinase activation leading to apoptosis. Thus, in many instances, a cancer cell’s resistance to TMZ therapy has been associated with high levels of MGMT (which effectively repairs the lesion) or loss of the MMR response (which prevents the initiation of apoptosis) [7,8,9]. Base Excision Repair (BER) pathways are responsible for the repair of oxidative and alkylative lesions, suggesting that BER-associated proteins would be effective prognostic markers and direct targets for relevant anticancer therapies. This review explores the connection between three key BER enzymes and cancer progression, and the potential for BER-targeted strategies to overcome MDR.
Predominant Components of BER
A range of DNA lesions arise spontaneously at a rate of at least 10,000 times per human genome per day under normal physiological conditions [10]. Specifically, the N-glycosylic bond can experience chemical instability that leads to base loss and apurinic/apyrimidinic (AP) site formation. AP sites can also occur through chemical modification of the base moiety, such as alkylation, oxidation, or ring saturation, which promotes base release. Normally, cells combat such genomic aberrations, i.e., base modifications, abasic sites, as well as single-strand breaks, via the BER pathway. In general, the core BER process requires only four or five proteins: a DNA glycosylase, an AP DNA lyase or endonuclease/phosphodiesterase, a DNA polymerase, and a DNA ligase.
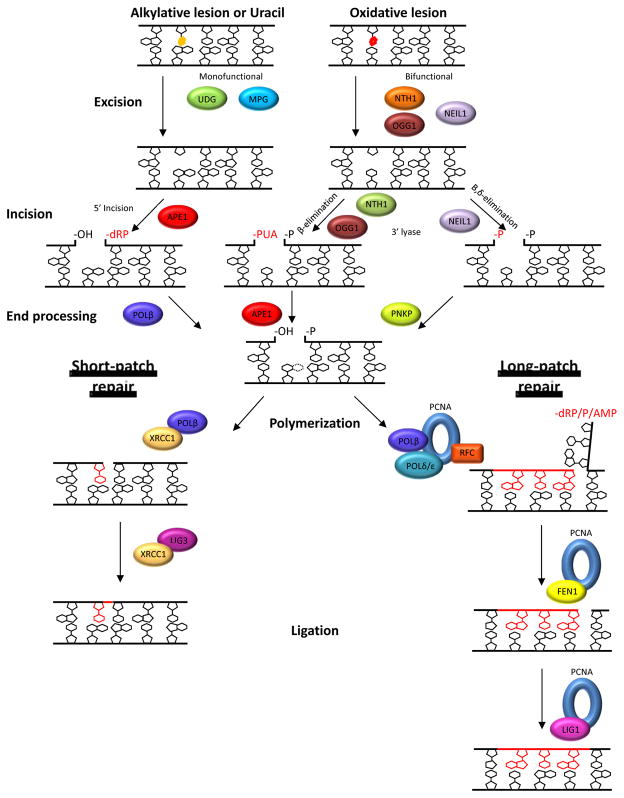
BER has been shown to diverge into two sub-pathways: short-patch (or single-nucleotide) and long-patch (Figure 1). Either sub-pathway can be initiated by a damage-specific DNA glycoslyase that recognizes a substrate base (e.g., 8-oxoguanine) and catalyzes hydrolysis of the N-glycosylic bond, producing an AP site. The glycosylase can be either monofunctional or bifunctional (reviewed in [11]). Monofunctional glycosylases, such as UDG and MPG, have only glycosylase activity, whereas bifunctional glycosylases, such as OGG1, NTH1 and NEIL1, possess an AP DNA lyase activity as well. In particular, a bifunctional glycosylase can catalyze AP site incision via either β-elimination (OGG1, NTH1) or β,δ-elimination (NEIL1), leading to the formation of a DNA strand break with a 3′-phospho-α,β-unsaturated aldehyde (3′-PUA) or 3′-phosphate (3′-P), respectively, and a 5′-phosphate (5′-P) end. The major mammalian AP endonuclease protein, APE1, can remove the 3′-PUA residue generated by β-elimination via its 3′-phosphodiesterase activity. The 3′-P moiety can be removed by the phosphatase activity of polynucleotide kinase phosphatase (PNKP) (reviewed in [12]). The subsequent steps of gap-filling and nick ligation are carried out as described below.
Figure 1.
The mammalian base excision repair and single-strand break repair pathways. Base excision repair (BER) begins by the removal of the damaged base by a monofunctional or bifunctional DNA glycosylase to leave an AP site (AP). After excision by a monofunctional glycosylase, APE1 cleaves the DNA backbone 5′ of the AP site. Base excision by a bifunctional DNA glycosylase is followed by incision 3′ to the AP site by either β- or β,δ-elimination via intrinsic 3′-AP lyase activity of NTH1, OGG1, or NEIL1. The 3′ or 5′ ends then undergo end processing by POLβ, APE1, or PNKP, depending on the obstructive termini. PARP1 recognizes such single-strand breaks, and end processing may utilize other factors such as TDP1 or APTX. After end processing, the BER pathway diverges into two sub-pathways: short-patch or long-patch. In short-patch BER, the single-nucleotide gap is repaired by POLβ in coordination with the scaffold protein/ligase complex of XRCC1/LIG3α. In long-patch BER, following strand-displacement synthesis of 2–13 nucleotides by POLβ and/or Polδ/ε (aided by PCNA and RFC), the 5′-DNA flap is removed by FEN1 and the backbone is subsequently ligated by LIG1. See text for additional details.
More commonly after excision of the damaged base by a DNA glycosylase (or following spontaneous base loss), the APE1 endonuclease enters and cleaves the DNA backbone 5′ to the abasic site product, creating a 5′-sugar phosphate (dRP) and a 3′-hydroxyl (3′-OH) end. DNA polymerase β (POLβ) then removes the 5′-dRP moiety through its lyase activity. In this scenario, BER primarily proceeds through short-patch repair synthesis (replacement of one nucleotide), with POLβ incorporating the complementary base. The DNA backbone is then resealed through the action of a DNA ligase 3α (LIG3) and XRCC1 protein complex (Figure 1).
In long-patch BER, repair synthesis proceeds via either a proliferating cell nuclear antigen (PCNA)-dependent pathway that utilizes DNA POLδ/ε or a PCNA-independent pathway that employs POLβ (reviewed in [13]). Both long-patch mechanisms involve the coordination of the DNA polymerase to displace the downstream strand and polymerize the complementary bases. The strand displaced by the polymerase creates a DNA-flap structure that is resolved by the 5′-flap structure-specific endonuclease I (FEN1). With the removal of the DNA flap, the repaired tract of DNA can be ligated by DNA ligase I (LIG1) to create an intact duplex [14,15] (Figure 1). What elements dictate which sub-pathway is selected by the cell are still being elucidated, but are thought to include the ability of the AP lyase activity of POLβ to remove the 5′-deoxyribose moiety and the intracellular levels of ATP [15,16,17].
If the initial damaged base or one of the repair intermediates is left unresolved, they pose a cytotoxic or mutagenic threat to the cell. Coordination of BER enzymes is therefore critical for the proper repair process. Indeed, dysregulation of these enzymes has been implicated in disease and cancer progression. For instance, up-regulation of the APE1 endonuclease has been linked to prostate cancer [18] and DNA POLβ variants can promote cellular transformation [19]. Thus, as expounded upon below, a comprehensive understanding of specific BER proteins and their coordination could assist in the development of novel anticancer therapies.
BER Proteins in Clinical Treatments
There are multiple potential protein targets in the different BER sub-pathways, including APE1, POLβ, XRCC1, and FEN1. Each of these proteins plays a critical role in the proper and efficient execution of BER. Notably, nullizygous knockout of APE1 [20], POLβ [21,22], FEN1 [23] or XRCC1 [24] in mice results in inviability, emphasizing the essential nature of their function(s) in organism development. It is presumed that this premature death stems from a defect in the processing of substrate DNA lesions, and thus, the accumulation of cytotoxic genomic damage. Additionally, decreased or low BER activity is associated with elevated cellular sensitivity to DNA-damaging agents, particularly oxidizing and alkylating compounds. It has therefore been hypothesized that strategic down-regulation of BER capacity could have significant clinical benefit in relevant combinatorial anticancer treatment paradigms involving DNA-interactive drugs (reviewed in [25] and [26]). Moreover, knowledge of an individual’s BER capacity, both global and tissue-specific, could provide insight into susceptibility to environmental exposures (reviewed in [27]), as well as response efficacy to certain therapeutic agents.
A method to target BER proteins for selective inactivation would be to use small molecule chemical inhibitors against a particular protein function. Combinatorial anticancer treatments would employ such inhibitors along with germane chemotherapeutic agents to eradicate tumors by driving the accumulation of substrate DNA damage. For instance, studies using poly(ADP-ribose) polymerase 1 (PARP1) inhibitors, which prevent effective processing of DNA strand breaks, in conjunction with DNA-damaging agents such as radiation or the alkylating agent TMZ, has been reported to improve treatment outcome [28]. Strategies employing anti-sense oligonucleotides or siRNAs to silence specific BER proteins could also be envisioned as part of the combinatorial arsenal.
Another method that has been proposed as an anticancer therapy strategy is called synthetic lethality. Synthetic lethality results when two or more non-allelic and non-essential mutations, which alone do not cause lethality, are cytotoxic in combination. This approach has received increased attention as it was recently shown that PARP1 inhibitors reduce the viability of homologous recombination-deficient BRCA mutant cancer cells [29,30]. Such targeted treatments are based on the principle that if cancer cells are dependent on one DNA repair pathway (in this case, PARP1-mediated strand break repair, a sub-pathway of BER), due to genetic loss of a compensatory or back-up pathway (in this case, BRCA-directed homologous recombination), then inhibition of the required pathway would cause DNA damage to exceed the cell’s tolerance capacity, resulting in cell death. Thus, by understanding the functional status of BER proteins in the context of cancer cell growth, clinicians would be able to devise better targeted anticancer therapies. Herein, three BER protein targets, APE1, POLβ, and FEN1, are discussed as current and potential clinical targets for anticancer research.
I. APE1
Human APE1 (a.k.a., APEX1, HAP1 and Ref-1) is a ubiquitous, multifunctional protein, which is part of the highly conserved exonuclease III family of AP endonucleases, named after the Escherichia coli ortholog [31,32,33]. The gene (spanning approximately 2.6 kb) is located on chromosome 14q11.2-12 and contains 4 introns and 5 exons. The coding region of the gene/transcript translates to a 318 amino acid protein that is approximately 35 kDa in size (Figure 2). APE1 is a globular α/β protein that contains redox and DNA repair activities. As will be discussed below, specific portions of the protein are still being assigned to particular function(s), but currently, the redox activity has been associated with the N-terminus of the protein, whereas the majority of the protein, the C-terminus, is essential for its DNA repair activities. Post-translational modifications of APE1, consisting of phosphorylation, acetylation, glutathionylation and ubiquitination, can regulate protein function, although the biological significance of these modifications is not yet fully understood [34,35,36].
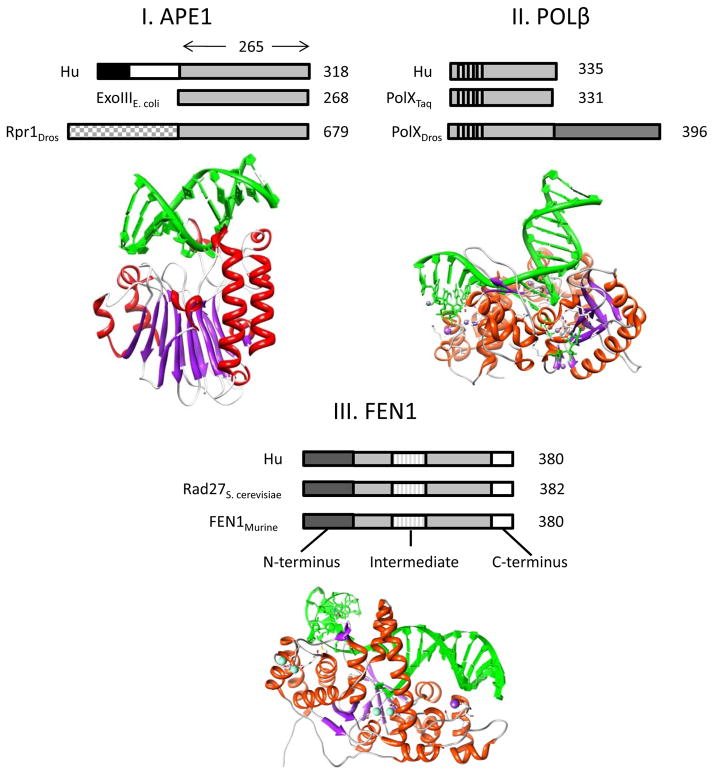
Figure 2.
Protein structure of base excision repair proteins: APE1, POLβ, and FEN1 I. The human (Hu) APE1 protein has a C-terminal sequence of about 265 amino acids that is conserved across species with E. coli exonuclease III and D. melanogaster (Dros) Rpr1. This region contains the DNA repair active site of the protein. The N-terminus of APE1 differs between species. The size of the protein is denoted to the right: Hu APE1 is 318 amino acids (aa), exonuclease III is 268 aa, and Rpr1 is 679 aa. The Hu APE1 3D protein structure (PDB ID # 1DE8) is shown below. II. Hu DNA POLβ is 335 aa (designated to right) and shares multiple helix-hairpin-helix motifs that are conserved between species, including in polymerase X (PolX) from T. aquaticus (Taq) and PolX from Dros. The 3D structure of Hu POLβ is shown below (PDB ID# 3RH4), and is commonly referred to as a hand, palm, and fingers structure. III. Hu FEN1 protein is 380 aa in length (designated to right), and the N-terminus, Intermediate, and C-terminal regions of the protein are conserved in S. cervisiae and in mice. Although eukaryotic FEN1 proteins harbor these 3 regions, prokaryotic versions only contain the N-terminus and Intermediate regions [152]. The 3D structure of the Hu FEN1 protein is shown below (PDB ID# 3Q8K). In each protein structure, helices are in red, β sheets in purple, and DNA in green.
APE1’s major function is as an AP endonuclease, cleaving the phosphodiester bond at AP sites in DNA in a magnesium-dependent manner to create a single-strand break with a 3′-OH group and a 5′-dRP residue (Figure 1). Over 95% of the total AP endonuclease function within mammalian cells is provided by the APE1 protein [37]. APE1 also has a 3′-phosphodiesterase activity that removes fragmented sugar groups found at the 3′ end of DNA breaks created by bifunctional DNA glycosylases (see above) or drugs such as bleomycin and ionizing radiation (3′-phosphoglycolates) [38]. A weaker function is its 3′-phosphatase activity, which removes 3′-phosphate blocking groups from DNA ends, though not with the efficiency of PNKP [39]. APE1 also possesses weak 3′-5′ exonuclease and RNase H activities [40,41,42,43]. Berquist et al. recently showed that APE1 is capable of cleaving abasic single-stranded RNA molecules, potentially as part of a defense system to remove damaged RNA templates [44]. Finally, APE1 is involved in a process termed nucleotide incision repair, where the enzyme initiates a long-patch-type repair response by cleaving 5′ to specific oxidized base modifications [45].
In addition to its multiple nucleic acid processing roles, APE1 has a seemingly unrelated function as a regulatory factor. Specifically, the N-terminal portion of the protein appears to be involved in redox modulation of various transcription factors, such as c-Jun, NF-κB, p53 and several others (reviewed in [46,25]). In the process, APE1 converts target transcription factors to an active state by reducing cysteine residues within their DNA binding/active site, allowing them to efficiently bind to cognate DNA sequences [47,48,49]. Thus, APE1 possesses the ability to indirectly regulate gene expression within the cell via a redox-dependent mechanism, which is still being defined at the molecular level.
An additional function of APE1 is its involvement in Ca2+-dependent repression of the parathyroid hormone (PTH) gene. The PTH gene is down-regulated when Ca2+ levels increase, due to elevated PTH protein levels. This negative feedback regulation is mediated by the trans-activating complex that contains the negative calcium response element proteins (nCaRe-A and -B), which were first detected at the PTH promoter. Studies have found APE1 to be part of this trans-activating complex [50,51], suggesting that APE1 may contribute directly to gene repression. Kuninger et al. more recently reported that the trans-activating complex, which contains the heterogeneous nuclear ribonucleoprotein L (hnRNP-L) protein, as well as APE1, associates with the APE1 promoter, indicating a potential for self-gene regulation [52]. Subsequent analysis has found that acetylation of APE1 at lysines 6 or 7 increases its affinity for the nCaRe proteins [53]. Thus, APE1 may have a role in PTH gene regulation that is modulated by post-translational modification, although the precise mechanism needs to be further elucidated.
APE1 has been associated with the immune response through binding to cytolytic T lymphocyte protease granzyme A (GzmA) and other proteins within the SET-complex. Granzymes are serine proteases within cytotoxic granules that are released from T lymphocytes and natural killer cells to eliminate viruses, intracellular bacteria, and tumors. Once the granules have entered the target cell, they release their content, allowing GzmA to inhibit the electron transport chain of mitochondria, thereby increasing the intracellular concentration of ROS. With increased ROS, an endoplasmic reticulum-associated oxidative stress response complex, called the SET-complex, localizes to the nucleus. The SET-complex is comprised of three nucleases, APE1, NM23-H1 and TREX1, as well as the chromatin modifying proteins SET and pp32, and the DNA binding protein HMGB2. Typically, when cells undergo oxidative stress, the relocalization of the SET-complex is thought to increase the repair of abasic DNA, due to the elevated level of nuclear APE1. However, GzmA can translocate to the nucleus and activate the SET-complex for abundant DNA cleavage that promotes apoptosis through an unknown mechanism (reviewed in [54]). The observation that GzmA binds and cleaves APE1 after Lys31, abolishing its repair function, indicates that the APE1 protein is associated with the GzmA-mediated apoptotic signaling cascade [55]. All told, APE1 has multiple roles in BER, as well as in gene regulation and apoptosis, that are still being defined.
APE1 in cancer
APE1 expression is ubiquitous and necessary for cell viability and embryonic development. Its essential role in cell proliferation and survival was demonstrated using both knock-down strategies in human cells and conditional knock-out mouse cell models [56,57]. Expression of APE1 has been shown to be cell cycle regulated in NIH3T3 mouse embryonic fibroblasts (MEFs). Similar to other DNA repair genes, the APE1 transcript was found to increase 2.5–3.5 fold in G1, remained high in S-phase, and returned to basal levels after mitosis [58]. APE1 expression is seen in all tissues, including of the immune, nervous, digestive, muscular, and secretory systems [59]. The APE1 protein normally localizes to the nucleus, consistent with its DNA repair function, although physiologically-relevant localization to the mitochondria (for mitochondrial DNA repair) and endoplasmic reticulum (as part of the SET-complex, see above) has also been observed (reviewed in [60]).
Increased levels of APE1 have been associated with multiple cancers. For example, studies using immunohistochemistry have revealed higher levels of APE1 in prostate cancer than in preneoplastic, prostatic lesions, or precancer cells [18]. Notably, APE1 overexpression in human embryonal carcinoma cell lines results in increased resistance to bleomycin and ionizing radiation, suggesting a role for the protein in dictating cellular DNA-damaging agent resistance [61]. When overexpressed as a chimeric protein with MGMT in HeLa cells, the APE1-MGMT fusion was found to confer a protective effect against alkylating agents [62]. It should be pointed out, however, that not all studies have observed an association of higher APE1 levels with resistance to external genotoxic insults ([63,64,65,66], reviewed in [67]). The variability in the results likely derives from the differential importance of the different DNA damage response pathways present in the diverse cell models studied.
Along with expression, the localization pattern of APE1 can change during tumorigenesis as well. For example, while immunohistochemistry of normal colorectal mucosa found APE1 to be predominately nuclear, the distribution pattern was more cytoplasmic in adenomas and carcinomas [68]. A similar change in intracellular localization was seen in breast cancer [69], thyroid carcinomas [70], epithelial ovarian cancers [71], and non-small cell lung cancer [72]. Notably, in most cases, the altered distribution of APE1 correlated well with the aggressiveness and prognosis of the tumor: nuclear localization associates with a better prognosis, whereas combined cytoplasmic and nuclear staining correlates with a poor prognosis, defined as increased angiogenesis, positive lymph node status, and p53 positivity.
The dysregulation of APE1 intracellular localization is opposite in melanoma. Yang et al. found a global increase in APE1 levels, with predominately nuclear localization in melanoma tissue compared with normal skin [73]. Other studies examining cervical [74], non-small cell lung cancer [75], rhadomyoscarcomas [76], and squamous cell head-and-neck cancer [77] observed a strong up-regulation of nuclear APE1 as well. A study by Koukourakis et al. examined nuclear APE1 expression in advanced and inoperable squamous cell head-and-neck cancer treated with chemotherapy and radiotherapy. They found APE1 levels to be associated with resistance to chemoradiotherapy, poor survival, and an inverse correlation with nuclear p53 accumulation [77]. It seems to reason that the APE1 distribution pattern will differ due to cancer type and stage, as well as the overall intracellular environment and genetic composition of the tumor itself. Yet why the intracellular localization of APE1 changes and how this distribution disparity affects patient responsiveness still needs to be more thoroughly worked out.
APE1 has been associated with cancer through molecular epidemiology studies that have examined the common population variant Asp148Glu. Specifically, some investigations have suggested that the Asp148Glu variant is involved in cancer predisposition, such as for melanoma [78,79,80]. However, prior experimental work found that this amino acid substitution does not affect the repair endonuclease activity of APE1, raising doubt about the direct role of this variant in the etiology of the disease [81]. Pro112Leu and Arg237Cys variants have been identified as mutants specific to endometrial cancer [82]. Ongoing work is addressing whether these tumor-associated proteins are involved in promoting cellular transformation, as well as the possibility of targeting specific forms of APE1 in anticancer therapies.
There is mounting evidence that suppression of APE1 function increases cellular sensitivity to a number of relevant DNA-interactive drugs. For example, silencing of APE1 expression by siRNA in osteosarcoma cells results in enhanced sensitivity to the laboratory chemicals H2O2 and methyl-methane sulfonate (MMS), as well as the clinical agents ThioTEPA, etoposide, and ionizing radiation [72]. In addition, APE1 suppression or inhibition sensitizes human lymphoblastoid TK6 cells and the HCT116 colon cancer cell line to bleomycin and ionizing radiation, with the cell killing effect of bleomycin being more pronounced [83]. A lower level of APE1 in human glioma cell lines correlates with reduced survival following ionizing radiation exposure as well [84]. Moreover, a study using targeted antisense oligonucleotides found that reduced APE1 expression in the human glioma cell line, SNB19, resulted in decreased viability upon treatment with a range of alkylators, including TMZ, MMS or 1,3-(2-chloroethyl)-1-nitrosourea [85]. Given that APE1 protects against the cytotoxic effects of specific DNA-damaging agents, selective inhibition of APE1 activity in cancer cells could enhance the efficacy of relevant therapies.
Targeting APE 1
Several efforts have been initiated to screen various compound libraries for APE1 inhibitors [86]. In 2005, Madhusudan et al. identified the first biochemical and biological inhibitor of APE1 from a screen of about 5,000 structurally diverse molecules [87]. The lead compound, CRT0044876, was shown to inhibit the E. Coli exonuclease III family of AP endonucleases, and to enhance the cytotoxicity of relevant DNA-damaging agents, such as TMZ. Consistent with specificity for APE1, CRT0044876 treatment did not enhance the cytotoxic effects of non-BER related DNA-damaging agents, such as ultraviolet light. However, this compound did not have suitable tractability, and some in vitro studies were unable to reproduce its efficacy [88,89].
An independent effort identified multiple arylstibonic acid analogues as APE1 inhibitors [90]. However, these agents exhibit activity against other nucleic acid interacting enzymes [91,92], rendering their clinical development difficult. A pharmacophore screen was also employed to uncover potential small molecule inhibitors of APE1, using as a template 3D interactions between the protein active site and an AP-DNA substrate [93]. This effort identified several chemical species possessing an optimum sized central hydrophobic core with two negatively ionizable features in an arrangement similar to that of the abasic substrate. The best molecules exhibited inhibitory effects below 10 μM and were selective for APE1 among the different enzymes tested, but have yet to be tested in cell biology experiments. More recently, Mohammed et al. reported a virtual screen of 2.6 million compounds from which several APE1 inhibitors with promising pharmaceutical properties were found to display low μM IC50 potency [94]. Finally, Bapat et al. isolated additional small molecules that inhibit APE1 endonuclease activity [95]. In particular, compound AR03 (2,4,9-trimethylbenzo[b][1,8]-naphthyridin-5-amine), which was identified from a fluorescence-based high-throughput screen of 60,000 molecules, potentiated the cytotoxicity of MMS and TMZ in SF767 glioblastoma cells.
Our laboratory designed a 1536-well fluorescence-based, quantitative high-throughput screening (qHTS) assay to identify APE1 inhibitors [88]. This assay was used to screen the Library of Pharmacologically Active Compounds, where several small molecules, including hydroxyl-DL-DOPA, reactive blue and myricetin, were found to inhibit the AP site cleavage activity of purified APE1 (with IC50s in the nM range). In addition, these molecules inhibited the total AP endonuclease activity of HEK239T and HeLa whole cell extracts, and potentiated the cytotoxic effects of MMS in cell culture, although they likely have additional biological targets.
More recently, our laboratory has employed the qHTS assay to screen a collection of ~350,000 small molecules that comprise the NIH Molecular Libraries Small Molecule Repository (MLSMR) (manuscript in review). Following a series of selection and validation steps, structure-activity relationship (SAR) analysis of the parent compound, N-(3-benzo[d]thiazol-2-yl)-6-isoproyl-4,5,6,7,-tetrahydrotheno[2,3-c]pyridin-2-yl)acetamide, was carried out. This molecule, along with related analogs, were found to have low μM inhibition potency, as well as activity in HeLa whole cell extract assays and in potentiation experiments using MMS and TMZ. Studies with HeLa cells also found a hyper-accumulation of AP sites when one of the small molecules was co-incubated with MMS. Finally, intraperitoneal dosing of a few select compounds found good exposure levels in the plasma and brain of mice, suggesting that the molecules could be used in the treatment of brain cancers. The combination of the various efforts described above represents an important step towards the identification of potent and selective APE1 inhibitors with clinical utility, although further compound development is necessary.
There are two other compounds that have been reported to successfully inhibit APE1 endonuclease function: methoxyamine (MX) and lucanthone. MX forms a covalent alkoxyamine derivative that binds with the acyclic sugar of an AP site. This molecule thus creates a physical block in DNA for the activities of the downstream repair proteins APE1 and POLβ [96]. Unlike the other small molecules described above, MX does not directly bind to APE1, making it an indirect inhibitor of the protein. Studies by the Gerson laboratory have shown that MX enhances the pre-clinical efficacy of alkylating drugs such as carmustine and TMZ [87,97]. Consequently, MX is being tested in Phase I clinical trials by TRACON Pharmaceuticals in advanced solid tumors, such as malignant melanoma, glioma and lung cancer, and will continue into Phase II studies in patients with non-small cell lung cancer [98].
Lucanthone has a heterocyclic ring structure similar to actinomycin D, and like that compound, binds to DNA and inhibits RNA synthesis in bacteria [99]. In the 1970s, lucanthone was shown to inhibit topoisomerase II [100], and some years later, Luo and Kelly reported that this molecule is a direct inhibitor of APE1 in vitro [97]. Their studies also demonstrated that lucanthone enhanced the cell killing action of TMZ. More recently, lucanthone was shown to bind to the hydrophobic pocket of APE1, thereby preventing the enzyme from associating with and repairing substrate DNA [101]. Anticancer studies with lucanthone have entered Phase I clinical trials with Spectrum Pharmaceuticals for malignant glioma treatments. It is hypothesized that this compound will have therapeutic value when combined with TMZ, since lucanthone can cross the blood-brain barrier (see Spectrum Pharmaceuticals at http://www.sppirx.com/).
More recent investigations have begun to explore the potential of targeting the redox function of APE1. Studies have found that the small molecule APX3330 (E3330), which binds to APE1 and inhibits its redox function, suppresses growth of tumor endothelium and endothelial progenitor cells [102]. Moreover, this compound impairs the ability of human umbilical cord blood-derived endothelial colony forming cells to establish tubules, inhibits the proliferation and migration of mouse retinal vascular endothelial cells in culture, and reduces retinal neovascularization in a mouse model of age-related macular degeneration [103]. Such results suggest that the redox function of APE1 is required for angiogenesis, a finding that has significant implications regarding the transition of tumors from a dormant state to one of metastatic potential. Fishel et al. also found that APX3330 effectively induces apoptosis in myeloid leukemia cells treated with low doses of retinoic acid, a compound used clinically and in culture to promote differentiation [104], and blocks the growth of PaCa-2 and Panc-1 pancreatic cancer cell lines, as well as patient-derived and PaCa-2 xenografts [105]. The combination of above observations suggests that the redox function of APE1 regulates key transcription factors of the angiogenic, differentiation and carcinogenic processes (e.g., PAX, HIF-1α, NFκB, and AP-1 [106,89,60]), and thus, could be targeted in various therapeutic paradigms.
Notably, it was recently reported that APX3330 does not specifically bind to the presumed redox region of the APE1 protein, but rather to the DNA repair active site [107]. This conclusion was drawn from a combination of NMR spectroscopy and molecular docking studies. Moreover, APX3330 was found to inhibit the AP endonuclease activity of APE1, in addition to its redox function, suggesting that the molecule may not have the selectivity initially thought and may induce a conformational change in the protein to elicit its complex biochemical effects. As such, the finding raises questions about the previous conclusions drawn from experiments using APX3330, and demands further characterization of the molecular mechanism of this compound.
A collaborative effort between the Madhusudan laboratory and our group has detected a novel synthetic lethal relationship between APE1 and DNA double-strand break repair (manuscript in review). In light of the synthetic lethality of PARP inhibitors, which presumably affect some aspect of BER, in BRCA-deficient cells, it was reasoned that APE1 inhibition may induce a similar outcome in double-strand repair-deficient cell lines. Indeed, APE1 inhibitors reduced the cell viability of BRCA2- and ATM-deficient Chinese hamster fibroblasts as assessed by clonogenic survival assays. These results were recapitulated using ATM and DNA-PKcs inhibitors in Chinese hamster ovary (CHO) cells expressing a dominant-negative form of APE1 [108,109]. This is the first preclinical evidence that APE1 function is synthetically lethal with the double-strand break repair pathway, and may represent a novel approach for anticancer treatment paradigms.
II. DNA Polymerase β
DNA polymerases have been divided into 7 families based on sequence similarity. DNA POLβ belongs to the X-family, which includes the template-independent enzyme, terminal deoxynucleotidyl transferase (TdT), and the template-dependent DNA polymerases λ and μ. All of these proteins share similar conformations and subdomain organization (Figure 2) [110,111,112,113]. In eukaryotic cells, POLβ is the major gap-filling DNA polymerase, most frequently called upon during BER-type responses, and as discussed earlier, also possesses a dRP lyase activity. The POLβ gene is located on chromosome 8p11.2 and spans 33 kb that includes 14 exons and 13 introns. The gene encodes a 335 amino acid protein that is approximately 39 kDa in size and is restricted to the nucleus [114].
The full-length POLβ protein has two main functions separated into two independent domains. The lyase domain is approximately 8 kDa and is located at the N-terminus, whereas the larger C-terminal portion of the protein contains the polymerase domain. The lyase activity of POLβ functions in the BER pathway after APE1 incision, removing the 5′-sugar phosphate of the cleaved DNA intermediate to produce a ligatable 5′-phosphate on the downstream DNA strand (Figure 1). The C-terminal portion of the protein provides the nucleotidyl transferase activity, which typically incorporates a single-nucleotide to permit ligation by LIG3 or LIG1 in BER [115,116]. Nucleic acid binding studies have shown that the purified 8 kDa N-terminal fragment alone binds to single-stranded nucleic acids with the same affinity as the full-length POLβ enzyme, but has little affinity for double-stranded DNA [117]. In contrast, the 31 kDa C-terminus of the protein has affinity for double-stranded DNA, but not single-stranded DNA ([118], and reviewed in [116]). This physical and functional arrangement suggests that the distinct activities of POLβ work in concert during the BER cascade. Experimental studies indicate that the lyase step is most likely rate limiting during BER and follows the polymerization reaction [119,120].
Previous studies used synthetic primer-templates of different structures to study the efficiency of POLβ at single-nucleotide gaps, 6-nucleotide gaps, and mispairs [121]. These experiments showed that POLβ is most efficient at correct incorporation of a dNTP at a single-nucleotide gap, possibly due to its higher affinity for this DNA structure [122]. In particular, POLβ forms a doughnut-shaped conformation at the site of a single-nucleotide gap, as compared to an intact non-gapped DNA, allowing for greater binding affinity and repair [123]. Despite its greater affinity for and efficiency at single-nucleotide gaps, POLβ still has an error rate of 1×10−4, which is much higher than the replicative DNA polymerases [124]. This low fidelity is mainly due to the lack of an intrinsic proofreading activity. Thus, it would appear that POLβ depends on other extrinsic enzymes to remove a misincorporated nucleotide, which, if left uncorrected during the gap-filling step, would have the potential to introduce mutations and cause human disease [125].
DNA POLβ in cancer
Altered expression and/or functionality of DNA POLβ are often associated with different types of cancers. In normal tissue, POLβ is ubiquitously expressed at low levels, with the highest level being reported in the brain and testis [126]. Knock-out mice disrupted for both alleles of POLβ die shortly after birth due to impaired neurogenesis and consequent respiratory failure, indicating an essential role for the protein in proper development [21]. Wilson and colleagues have found elevated levels of POLβ in breast, colon, and prostate adenocarcinomas, relative to normal tissue [127]. It has been proposed that, since the fidelity of DNA POLβ is much lower than the replicative polymerases, increased levels of POLβ could interfere with replication, repair, and recombination, leading to a mutator phenotype [125]. Furthermore, it is possible that increased levels of POLβ would sequester binding partners, such as p53, XRCC1, and TRF2, preventing them from carrying out their important cellular functions.
Early experiments found that CHO cells engineered to overexpress POLβ acquire a spontaneous mutator phenotype, as assessed by three independent mutation assays [128]. This cell line also showed resistance to bifunctional DNA-damaging agents, such as cisplatin, melphalan and mechlorethamine, as well as increased accompanying mutagenesis, likely due to elevated translesion bypass synthesis. In a separate study, overexpression of POLβ in CHO cells was found to induce aneuploidy, abnormal spindle formation, and a defect in the mitotic checkpoint relative to control cells [129]. Furthermore, 72% of immunodeficient nude mice injected with the CHO POLβ-overexpressing cell line developed carinomas, as compared to 〈25% of mice injected with CHO control cells, consistent with the idea that POLβ overexpression drives tumorigenesis. However, a separate study found that overexpression of POLβ in mouse LN12 fibroblasts does not increase the mutation frequency [130]. This apparent discrepancy may stem from differences in the experimental set-up, such as the mutation assay or cell line (due to variability in unidentified genetic factors) employed. That said, the preponderance of evidence suggests that dysregulation of DNA POLβ expression adversely affects genome integrity.
Polymorphisms in POLβ, typically identified using RT-PCR, are a frequent phenomenon in cancers. In fact, POLβ mutations have been observed in approximately 30% of human tumors. Though studies have not found a common mutation within diseased tissue, of the tumor-associated sequence variants, 48% possess a single-amino acid change in the coding region [131]. In total, 44 amino acid substitution variants have been identified in at least 7 different cancer types, including gastric, colorectal, prostate, and lung. Cys239Arg is a POLβ variant associated with gastric cancer. The location of this residue is within the flexible loop region of the protein, where other substitutions (Glu249Lys and Asp246Val) were found to reduce the accuracy of DNA synthesis [132,133]. Moreover, Sweasy et al. demonstrated that expression of cancer-associated POLβ variants, such as Ile250Met and Lys289Met, can induce transformed phenotypes, including foci formation and anchorage-independent growth [19]. Such results suggest that over time, expression of disease-associated POLβ variant proteins has the capacity to induce mutations that drive tumorigenesis. Additionally, an Arg137Gln population variant has been shown to exhibit reduced polymerase fidelity, causing lower efficiencies of both short-patch and long-patch BER [134]. This substitution also results in a defective interaction with PCNA in cellular extracts, and the Arg137Gln variant (unlike the wild-type protein) is unable to protect POLβ −/− MEFs from MMS challenges. Overall, the data indicate that alterations in the expression or composition of DNA POLβ have the ability to interfere with BER in a way that promotes genomic instability and carcinogenesis.
Targeting POLβ could have beneficial impact on anticancer therapies. For instance, POLβ-null MEFs exhibit a severe hypersensitivity to monofunctional DNA alkylating agents, such as MMS, methylnitrosourea, and ethyl methane sulfonate (EMS) [22], as well as a more mild hypersensitivity to the radiomimetic antibiotic bleomycin [135]. After treatment with MMS, an accumulation of single-stranded DNA breaks is observed in POLβ-null MEFs [136], reflective of incomplete BER. Furthermore, it has been reported that POLβ-null MEFs are more sensitive to TMZ than isogenic wild type or methylpurine-DNA glycosylase (MPG, a.k.a. AAG)-deficient cells [137,138]. Notably, this alkylating agent hypersensitivity of POLβ-deficient cells is exacerbated by overexpression of MPG/AAG, indicating that BER DNA intermediates are necessary to drive cell death [139]. Knockdown of POLβ by RNAi techniques has been shown to increase cellular sensitivity to TMZ [138], MMS, and ionizing radiation [140], supporting the notion that POLβ deficiency may be a suitable strategy for combinatorial chemotherapies involving relevant DNA-interactive drugs.
Targeting DNA POLβ
Several small molecules, including long-chain fatty acids [141,142], have been identified that disrupt the biochemical function(s) of DNA POLβ. A natural product, koetjapic acid, interacts specifically with the 8 kDa dRP lyase domain of POLβ as shown by NMR chemical shift mapping, and inhibits both the lyase and polymerase activity of the protein [143]. Nine other structurally related synthetic compounds, including pamoic acid (PA), were characterized via NMR for their binding interactions with the 8 kDa domain as well. Of the nine compounds studied, PA, which like koetjapic acid inhibits both the lyase and polymerase activities of full length POLβ, was shown to sensitize wild type cells to MMS to a level similar to that seen for POLβ null cells. These findings indicate that binding of these small molecules to the 8 kDa domain can inhibit both functions of POLβ, resulting in impaired BER.
Recently, an in silico structure-based screen was performed to analyze the docking of approximately 140,000 small molecules to the adenomatous polyposis coli (APC)-binding site on DNA POLβ [144]. A compound termed NSC-124854 [4-amino-1-(2-deoxy-5-O-phosphonopentofuranosyl)-6-iodo-5,6-dihydropyrimidin-2(1H)-one] was identified as one potential binder. Subsequent in vitro biochemical studies showed that NSC-124854 inhibits the strand-displacement synthesis activity of POLβ at roughly 5 μM, as well as its single-nucleotide polymerization activity at higher concentrations. Moreover, this compound was found to enhance the growth inhibitory effect of TMZ on several colon cancer cell lines regardless of MMR status. This observation was substantiated in vivo by using xenographs of the various colon cancer cells lines, where it was found that NSC-124854 reduced tumor volume when combined with TMZ. Overall, the studies suggest a reasonably specific structure-based inhibitor of POLβ with potential utility in the treatment of colonic tumorigenesis.
To our knowledge, no published efforts have appeared that describe the use of high-throughput function-based screens to identify small molecule inhibitors of POLβ. This stems largely from the fact that most assays developed to date measure activity of highly efficient and processive polymerases and thus require the accumulation of double-stranded DNA products via multiple catalytic cycles, such as methods involving PicoGreen staining of duplex DNA, donor-acceptor FRET-type reporters, or molecular beacon-based strand displacement [145,146,147,148]. These methods are of course poorly suited for measuring the low processivity of DNA POLβ. Recently, Dorjsuren et al. described a real-time fluorescence-based method more appropriate for monitoring the enzymatic activity of DNA polymerases such as POLβ [149]. This assay uses a novel fluorogenic substrate designed to report strand displacement triggered by polymerase-dependent incorporation of 1 or 2 nucleotides. The method was optimized for a 1526-well format and successfully used in accessing the inhibitory effects of PA on POLβ. With the design of such techniques, future high-throughput screens can begin to identify novel POLβ inhibitor chemotypes.
Examining potential synthetic lethal relationships of the MMR proteins, MLH1 and MSH2, with various DNA polymerases, Martin et al. found that knockdown of DNA POLβ reduced the viability of human HEC59 endometrial cells that harbor compound heterozygous nonsense mutations in MSH2 [150]. Subsequent analysis revealed that silencing of POLβ in MSH2-deficient cells results in an accumulation of the oxidative base lesion 8-oxoguanine, not seen in MMR-proficient cells. Significantly, it was demonstrated that knockdown of OGG1, which is the predominant glycosylase for initiating the repair of 8-oxoguanine, also promoted lethality of HEC59 cells and that POLβ knockdown results in a concomitant decrease in the OGG1 protein level. These data suggest that the accumulation of 8-oxoguanine lesions in MSH2-deficient cells, as a result of reduced OGG1 function and the lack of an alternative MMR-dependent processing mechanism, drives cell death, indicating an alternative approach for the targeted treatment of cancers that arise from MSH2 defects.
III. Flap structure-specific endonuclease I (FEN1)
Two primary functions of FEN1 are to process the 5′-ends of Okazaki fragments during DNA replication and to remove 5′-overhanging DNA flaps during long-patch BER (reviewed in [151] and [152]). The FEN1 gene spans 4.6 kb and is located on chromosome 11q12.2. The translated protein consists of 380 amino acids and is approximately 38 kDa in mass. Eukaryotic FEN1 is composed of three major domains: the N-terminal (N), the intermediate (I), and the C-terminal (Figure 2). This protein architecture differs from that of the prokaryotic flap nuclease, Pol I, which contains only the N and I domains. The N and I domains of FEN1 are, in fact, conserved between eubacterial and viral systems, as seen in E. Coli Pol I and Taq DNA polymerase [153]. These three proteins, and other members of the RAD2 nuclease family, share sequence similarity and several conserved amino acids that play a critical role in substrate recognition and catalysis. The C-terminal portion of the yeast and mammalian FEN1 proteins contains the nuclear localization signal and a PCNA-interacting sequence [153]. Homozygous deletion of FEN1 results in mouse embryonic lethality, underscoring the significance of its DNA metabolic functions [154]. Notably, studies have found that FEN1−/− mouse blastocysts cannot enter S-phase to continue and complete DNA synthesis, and thus arrest in the endocycle.
FEN1 acts as both a 5′-flap endonuclease and a weak 5′-3′ exonuclease. As an endonuclease, FEN1 recognizes double-stranded DNA with a 5′-unannealed flap and cleaves near the base of the flap. As a 5′-3′ exonuclease, the enzyme degrades nucleotides from a nick or gap in duplex DNA [155]. FEN1 has been found to cleave RNA and DNA flap structures, but not single-stranded DNA, fully duplex DNA, heterologous loops, D loops (mimicking the structure found in telomeres or as a recombination intermediate), Holliday junctions (a recombination intermediate), or 3′- or 5′-overhangs [152]. The efficiency of cleavage is maintained in the presence of 1 mM to 10 mM Mg2+. Mn2+ accelerates the cleavage rate, whereas Ca2+ and Zn2+ cannot substitute for Mg2+ [156,157].
FEN1 is thought to commence its activity in vivo through recognition of a PCNA-bound short 5′-single-stranded DNA flap, although the protein can execute its cleavage activity in the absence of this replication factor. After initial recognition of the DNA substrate, models proposed that FEN1 tracks down the single-stranded DNA to the single-stranded/double-stranded DNA junction where incision occurs [158,159,152]. Whether FEN1 threads the free end of the 5′-single-stranded flap structure through its helical arch, a prominent structural feature of the protein, or whether the arch clamps onto the 5′-single-stranded DNA without threading has been argued for years. However, recent evidence from structural and functional analysis of human FEN1-DNA complexes suggests that the protein active site uses two helices to enforce single-stranded DNA threading and thus specificity for free 5′-ends for its endonuclease activity [160].
The role of FEN1 in replication is to process Okazaki fragments by degrading the RNA primers found in lagging strand fragments. In vitro studies using Okazaki fragment primer-templates have shown that RNase H can remove most of the initiator RNA, but leaves residual ribonucleotides close to the RNA-DNA junction [161]. The 5′-nuclease activity of FEN1 removes the remainder of the ribonucleotides [162]. This cooperation between RNase H and FEN1 generates a nick, which is a substrate for DNA LIG1. Another endonuclease, DNA2, has been found to interact with FEN1 in Okazaki fragment processing through a coordinated mechanism involving single-stranded DNA binding protein RPA [163]. Studies in Saccharyomyces cerevisiae have found that RPA stimulates Dna2p (yeast DNA2 equivalent) cleavage on long (10–30 bp) DNA flaps, whereas it inhibits FEN1 endonucleolytic activity. Once Dna2p cleaves the flap substrate and removes RPA, FEN1 carries out cleavage of the shorter DNA flap, a critical step for proper DNA replication.
FEN1 plays an important role in the long-patch sub-pathway of BER, apparently both within the nuclear and mitochondrial compartments [164,165]. As alluded to earlier, when an oxidized or reduced 5′-abasic site fragment cannot be removed by the short-patch BER components, then long-patch BER must be employed [13,17]. This process involves strand displacement synthesis and exclusion of the downstream strand harboring the damaged 5′-sugar, creating a DNA flap intermediate of approximately 2 to 12 nt in length. This 5′-flap is removed by the endonucleolytic activity of FEN1 (Figure 1). DeMott et al. found that FEN1 cannot directly remove a 5′-abasic site, but can carry out excision if the 5′-AP site-terminated flap is at least one nucleotide long [166]. Thus, FEN1 relies on strand displacement synthesis to create a substrate for cleavage. Long-patch BER was initially found to be PCNA-dependent, as depletion of PCNA abolished the long-patch repair cascade [167,168,169]. The activity of FEN1 is also reduced with decreased levels of PCNA, thereby compromising the efficiency of the long-patch BER process [170]. However, recent studies have found that long-patch BER has a PCNA-independent mechanism facilitated by FEN1 and POLβ. FEN1 can simulate POLβ strand displacement synthesis to generate a 5′-dRP single-stranded DNA flap for cleavage [14,171]. Furthermore, PARP1, an enzyme involved in DNA strand break responses, stimulates both FEN1 and POLβ activity, suggesting that these three proteins may form a complex to scan DNA for repairable sites [172].
FEN1 in cancer and disease
Altered expression and functionality of FEN1 have been linked to tumor progression. As mentioned earlier, FEN1 is necessary for proper development, as deletion of both alleles results in mouse embryonic lethality [154]. Haploinsufficiency in FEN1 function, however, may play a role in tumorigenesis, apparently due to increased spontaneous genomic instability [154]. FEN1 is ubiquitously expressed in mammals, with the highest level found in testes, thymus, bone marrow, and other highly proliferative tissues, most likely due to its role in DNA replication [173]. Elevated FEN1 expression has been observed in metastatic prostate cancer [174], gastric cancer [175], neuroblastoma [176], pancreatic cancer [177], and lung cancer cell lines [178]. Recently, two single-nucleotide polymorphisms, one (c.-69G>A) within the gene promoter region and two (c.4150G>T) in the 3′ untranslated region of the transcript, have been found to associate with an approximately 2-fold decrease in FEN1 expression levels and may contribute to the risk of liver, esophageal, gastric, colorectal, and lung cancer [179,180]. These data suggest that FEN1 is a possible marker for certain cancer types, a probable contributor to cancer susceptibility, and a potential target for anticancer therapies.
FEN1 polymorphisms have been associated with cancer. Zheng et al. screened 253 human specimens from 12 common cancers for FEN1 mutations by direct sequencing of the gene coding region [181]. Interestingly, they found that several of the mutant proteins observed among the non-synonymous mutations detected displayed reduced exonuclease and gap-dependent endonuclease activities, but retained flap-endonuclease activity. Moreover, several of the cancer cells that contained FEN1 mutations exhibited an increased mutation rate. It was therefore hypothesized that mutations which result in a nuclease-deficient FEN1 protein product could drive cancer initiation and progression.
To demonstrate the significance of the above somatic mutations, a mouse model was created carrying a Glu160Asp substitution in FEN1 that abolishes more than 90% of the intrinsic exonuclease and gap-dependent endonuclease activities, yet retains the flap-endonuclease function. This mutant mouse spontaneously developed lung cancer in late life stages, and MEFs derived from these animals were sensitive to MMS, mitomycin C, methylnitrourea, and hydrogen peroxide [181,182]. In addition, upon treatment with benzo[α]pyrene, a common constituent of tobacco smoke, the mutant mouse exhibited a significant increase in DNA double-strand breaks and chromosomal aberrations [183]. These data suggest that FEN1 mutations play a role in cancer development, although more studies are necessary to determine the precise molecular mechanisms and cellular steps.
FEN1 has specifically been implicated in maintaining the stability of di- and tri-nucleotide repeat (TNR) sequences, which often show length alterations in human neurodegenerative disease and cancer (reviewed in [184]). In particular, mutations in yeast RAD27 (the human FEN1 equivalent) increase the frequency of TNR expansion and contraction during replication ([185] and reviewed in [186]). It is thought that this destabilization at TNRs stems from the propensity of repeat sequences to form DNA secondary structures, such as hairpins, which can inhibit DNA replication, repair, and/or the recombination events (reviewed in [187]). Consistently, it has been shown that hairpin conformations within a 5′-DNA flap impede FEN1 loading and proper processing [188,189]. However, a role for mammalian FEN1 in regulating repeat instability, such as the TNR in Huntington’s disease, remains unclear [190,191].
Longer CAG tandem repeats within the androgen receptor (AR) gene are associated with an increased risk of developing ovarian [192] and endometrial tumors [193], whereas shorter CAG repeat stretches correlate with a higher grade and more advanced stage of prostate cancer at diagnosis [194]. Haberman et al. recently described a genome-wide analysis that found that TNRs are five times more prevalent in cancer-related genes, such as those that encode kinases and cell cycle checkpoint proteins. The specific mechanisms of FEN1-mediated TNR expansions and contractions are still being explored, but in cancer and neurological disease, are currently being explored. However, this work highlights the role of TNR in cancer initiation and progression which can be influenced by the function(s) of FEN1.
Given its key role in several DNA metabolic processes, FEN1 could be a target for sensitizing cancer cells. FEN1−/− blastocysts exhibit an increased sensitivity to gamma radiation [23]. Matsuzaki et al. found that FEN1-null DT40 chicken cells are hypersensitive to the alkylators MMS and N-methyl-N′-nitro-N-nitrosoguanidine, as well as to the oxidizing agent H2O2, but not to ultraviolet light, X-rays or the DNA topoisomerase II inhibitor etoposide [195]. In studies where FEN1 levels were knocked-down by siRNA, inhibition of growth was observed for the prostate cancer cell line LNCaP, presumably due to impaired DNA replication [196]. Transient down-regulation of FEN1 by siRNA also sensitized human LN308 glioblastoma cells to MMS and the clinical drugs TMZ and cisplatin. Finally, Shibata and Nakamura found that stable expression of an Asp181Ala mutant FEN1 protein, which retains substrate binding, yet lacks enzymatic activity, in human bladder carcinoma cells inhibits cell growth when combined with MMS, indicating that interference of normal FEN1 function decreases the in vivo repair capacity for alkylation damage [197]. The compilation of these studies indicates that FEN1 is a rationale target for combinatorial anticancer therapies, such as those involving alkylating compounds [198].
Targeting FEN1
Recent inhibitors of FEN1 were designed to target critical amino acids within the protein active site. Panda et al. reasoned that if the Asp181 residue was vital to FEN1 function, as demonstrated by mutational analysis [199], then small molecule inhibitors should target the associated metal binding pocket. Using this criterion, they performed an in silico molecular docking screen and identified compound NSC-281680 [1,2,5,6-tetrazocine, octahydro-, dihydrochloride], which was seen to interact specifically with Asp181 [200]. Subsequent characterization found that NSC-281680 inhibited the endonuclease activity of wild type FEN1 protein in vitro, and enhanced the cytotoxic effect of TMZ in both MMR-deficient and MMR-proficient colon cancer cell lines. An earlier screening study had identified hydroxyurea-based compounds as potent and selective inhibitors of FEN1. During the structure-activity-relationship analysis, Tumey et al. synthesized 3-hydroxy-5-methyl-1-phenylthieno[2,3-d]pyrimidin-e-2,4(1H,3H)-dione (PTPD) and found that this molecule inhibited FEN1 cleavage activity at sub μM concentrations in vitro and induced apoptosis of bladder carcinoma cells in culture when combined with TMZ or MMS [201]. We have used PTPD in the development of a complementary pair of high-throughput fluorogenic screening assays to monitor FEN1 activity. In particular, in a 1536-well plate format, PTPD, as well as a broad-specificity enzyme inhibitor aurintricarboxylic acid (ATA), were found to effectively inhibit FEN1 flap endonuclease activity [92]. Future studies will need to develop the current small molecule inhibitors further, including examination of clinical usefulness, and employ the existing screening approaches to identify new lead compounds.
McManus et al. found that FEN1 could be a target for synthetic lethality in certain cancer cell types. In particular, this group demonstrated that RAD54B-deficient human colorectal cancer cells are sensitive to killing by reduced FEN1 expression or activity, while isogenic RAD54B-proficient cells are not [202]. Although the loss of FEN1 alone reduced cell viability, the enhanced cytotoxicity seen with the RAD54B-deficient cells likely stems from an accumulation of aberrant chromosome structures normally resolved by either FEN1 or a RAD54B-dependent recombination mechanism. Thus, as outlined earlier, inhibition of a specific DNA repair process in cancer cells, which are deficient in the compensatory back-up pathway, would seemingly drive damage accumulation and lead to selective tumor cell death through a mechanism involving synthetic lethality.
Summary of the potential of BER inhibition to overcome chemoresistance
BER is a major protective system against various forms of oxidative, alkylative and spontaneous DNA damage. In this review, we have described the biochemical and biological roles of APE1, POLβ, and FEN1, with a focus on their central contributions to the different sub-pathways of BER. Disruption in the expression level or composition of these proteins has been associated with increased genome instability, the tumorigenic process and possibly the MDR problem. Given the prominent function(s) of APE1, POLβ, and FEN1 in BER, their enzymatic activities are emerging as novel targets for sensitization of cancer cells to DNA-interactive drugs. Indeed, studies have shown that inactivating these proteins via small molecule inhibitors or gene-silencing disrupts the BER response to oxidative and alkylative DNA damage, suggesting a mechanism for improving the efficacy of relevant combinatorial therapeutic paradigms. While there is great potential in eventually eradicating cancer cell growth, careful targeting strategies, such as selective delivery or application of synthetic lethality, will need to be further developed to ultimately minimize systemic cellular consequences. In addition, determining the repair capacity of both normal and cancer cells and the genetics of the cancer cell population will allow for a more tumor-specific treatment tactic.
Table 2.
Summary of base excision repair proteins and relevance to anticancer therapies.
| Gene/Protein | Key Features | Biochemical Functions | Cancer Etiology | Anticancer Treatment |
|---|---|---|---|---|
| APE1 |
|
|
|
|
| POLβ |
|
|
|
|
| FEN1 |
|
|
|
|
Acknowledgments
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging. We thank Drs. Peter Sykora and Leslie Hoh for their constructive comments on the review.
References
- 1.Pluen A, Boucher Y, Ramanujan S, McKee TD, Gohongi T, di Tomaso E, Brown EB, Izumi Y, Campbell RB, Berk DA, Jain RK. Role of tumor-host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. PNAS. 2001;98:4628–4633. doi: 10.1073/pnas.081626898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman MM, Ambudkar SV, Ni B, Aran JM, Sugimoto Y, Cardarelli CO, Pastan I. Exploiting multidrug resistance to treat cancer. Cold Spring Harb Symp on Quant Biol. 1994;59:667–683. doi: 10.1101/sqb.1994.059.01.078. [DOI] [PubMed] [Google Scholar]
- 3.Hannon MJ. Metal-based anticancer drugs: From a poast anchored in platinum chemistry to a post-genomic future of diverse chemistry and biology. Pure Appl Chem. 2007;79:2261. [Google Scholar]
- 4.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 5.Kelley MR, Fishel ML. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer Agents Med Chem. 2008;8:417–425. doi: 10.2174/187152008784220294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobol RW. In: Temozolomide. Schwab M, editor. Springer; 2009. [Google Scholar]
- 7.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyl-transferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 8.Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, Wick W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollack IF, Hamilton RL, Sobol RW, Burnham J, Yates AJ, Holmes EJ, Zhou T, Finlay JL. MGMT expression strongly correlates with outcome in childhood malignant gliomas: results from the CCG-945 cohort. J Clin Oncol. 2006;24:3431–3437. doi: 10.1200/JCO.2006.05.7265. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 11.Nemec AA, Wallace SS, Sweasy JB. Variant base excision repair proteins: Contributors to genome instability. Semin Cancer Biol. 2010;5:320–328. doi: 10.1016/j.semcancer.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphotase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- 15.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1) Embo Journal. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petermann E, Ziegler M, Oei SL. ATP-dependent selection between single nucleotie and long patch base excision repair. DNA Repair (Amst) 2003;2:1101–1114. doi: 10.1016/s1568-7864(03)00117-4. [DOI] [PubMed] [Google Scholar]
- 17.Sung JS, Demple B. Roles of base excision repair subpathways in oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 18.Kelley MR, Cheng L, Foster R, Tritt R, Broshears J, Koch M. Elevated and altered expression of the multifunction DNA base excision repair and redox enzyme APE1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7:824–830. [PubMed] [Google Scholar]
- 19.Sweasy JB, Lang T, Starcevic D, Sun KW, Lai CC, DiMaio D, Dalal S. Expression of DNA polymerase beta cancer associated variants in mouse cells results in cellular transformation. PNAS. 2005;102:14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein Ref-1: is essential for early embryonic development in mice. PNAS. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 23.Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the XRCC1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 25.Luo M, He H, Kelley MR, Georgiadis MM. Redox regulation of DNA repair: Implications for human health and cancer therapeutic development. Antioxid Redox Signal. 2010;12:1247–1269. doi: 10.1089/ars.2009.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gossage L, Perry C, Abbotts R, Madhusudan S. Base excision repair factors are promsing prognostic and predictive markers in cancer. Curr Mol Pharmocol. 2011 doi: 10.2174/1874467211205010115. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Wilson DM, III, Kim D, Berquist BR, Sigurdson AJ. Variation in base exision repair capacity. Mutat Res. 2011;711:100–112. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaremba T, Curtin NJ. PARP Inhibitor Development for Systemic Cancer Targeting. Anti-Cancer Agents Med Chem. 2007;7:515–523. doi: 10.2174/187152007781668715. [DOI] [PubMed] [Google Scholar]
- 29.Farmer M, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:914–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 30.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 31.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 32.Robson CN, Hochhauser D, Craig R, Rack K, Buckle VJ, Hickson ID. Structure of the human DNA repair gene HAP1 and its localization to chromosome 14q11. 2–12. Nucleic Acids Res. 1992;20:4417–4421. doi: 10.1093/nar/20.17.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hickson ID, Gorman MA, Freemont PS. Structure and functions of the major human AP endonuclease HAP1/Ref-1. Humana Press Inc; Totowa, NJ: 2000. [Google Scholar]
- 34.Busso CS, Lake MW, Izumi T. Posttranslational modification of mammalian AP endonuclease (APE1) Cell Mol Life Sci. 2010;67:3609–3620. doi: 10.1007/s00018-010-0487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YJ, Kim D, Illuzzi JL, Delaplane S, Su D, Bernier M, Gross ML, Georgiadis MM, Wilson DM., III S-glutathionylation of cysteine 99 in the APE1 protein impairs abasic endonuclease activity. J Mol Biol. 2011;414:313–326. doi: 10.1016/j.jmb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meisenberg C, Tait PS, Dianova II, Wright K, Edelmann MJ, Ternette N, Tasaki T, Kessler BM, Parson JL, Tae Kwon J, Dianov GL. Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and isrequired for genome stability. Nucleic Acids Res. 2012;40:701–711. doi: 10.1093/nar/gkr744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. PNAS. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh D, Wilson DM, III, Povirk LF. 3′-phophodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–210. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Wilson DM, III, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 41.Barzilay G, Walker LJ, Robson CN, Hickson ID. Site-directed mutagenesis of the human DNa repair enzyme HAP1: identification of residues important for AP endonulcease and RNase H activity. Nucleic Acids Res. 1995;23:1544–1550. doi: 10.1093/nar/23.9.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinc endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seki S, Ikeda S, Watanabe S, Hatsushika M, Tsutsui K, Akiyama K, Zhang B. A mouse DNA repair enzyme (APEX nuclease) having exonuclease and apurinic/apyrimidinic endonuclease activities: purification and characterization. Biochim Biophys Acta. 1991;1079:57–64. doi: 10.1016/0167-4838(91)90024-t. [DOI] [PubMed] [Google Scholar]
- 44.Berquist BR, McNeill DR, Wilson DM., III Characterization of abasic endonuclease activity of human APE1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J of Mol Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gros L, Ishchenko AA, Ide H, Elder RH, Saparbaev MK. The major human AP endonuclease (Ape1) is involvoed in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32:73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhakat KK, Mantha AK, Mitra S. Transcriptional Regulatory Functions of Mammalian AP-Endonuclease (APE1/Ref-1), an Essential Multifunctional Protein. Antioxid Redox Signal. 2009;11:621–637. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xanthoudakis S, Miao GG, Curran T. The redox and DNA-repair activites of Ref-1 are encoded by nonoverlapping domains. PNAS. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xanthoudakis S, Miao GG, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. A redox factor protein, ref1: is involved in negative gene regulation by extra cellular calcium. J Biol Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 51.Yamamoto M, Igarashi T, Muramatsu M, Fukagawa M, Motokura T, Ogata E. Hypocalcemia increases and hypercalcemia decreases the steady-state level of parathyroid hormone messenger RNA in the rat. J Clin Invest. 1989;83:1053–1056. doi: 10.1172/JCI113946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuninger DT, Izumi T, Papaconstantinou J, Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with nCaRe-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res. 2002;30:823–829. doi: 10.1093/nar/30.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieberman J. Granzyme A activates another way to die. Immunol Rev. 2010;235:93–104. doi: 10.1111/j.0105-2896.2010.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, Lieberman J. Cleaving the oxidative repair protein APE1 enhances cell death mediated by granzyme A. Nat Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 56.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 57.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential buy distinct functions of the mammalian abasic endonuclease. PNAS. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fung H, Bennett RA, Demple B. Key role of a downstream specificity protein 1 site in cell cycle-regulation trascription of the AP endonuclease gene APE1/APEX in NIH3T3 cells. J Biol Chem. 2001;276:42011–42017. doi: 10.1074/jbc.M106423200. [DOI] [PubMed] [Google Scholar]
- 59.Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 60.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon. Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 61.Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, Foster R, Elnhorn LH, Kelley MR. Altered expression of APE1/ref-1 in germ cell tumors and overexpresion in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61:2220–2225. [PubMed] [Google Scholar]
- 62.Hansen WK, Deutsch WA, Yacoub A, Xu Y, Williams DA, Kelley MR. Creation of a fully functional human chimeric DNA repair protein. Combining O6-methlyguanine DNA methyltrasnferase (MGMT) and AP endonuclease (APE/redox effector factor (Ref1)) DNA repair proteins. J Biol Chem. 1998;273:756–762. doi: 10.1074/jbc.273.2.756. [DOI] [PubMed] [Google Scholar]
- 63.Herring CJ, Deans B, Elder RH, Rafferty JA, MacKinnon J, Barzilay G, Hickson ID, Hendry JH, Margison GP. Expression levels of the DNA repair enzyme HAP1 do not correlate with the radiosensitivities of human or HAP1-transfected rat cell lines. Br J Cancer. 1999;80:940–945. doi: 10.1038/sj.bjc.6690447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kakolyris S, Kaklamanis L, Giatromanolaki A, Koukourakis M, Hickson ID, Gatter KC, Harris AL. Expression and subcellular localization of human AP endonuclease 1 (HAP1/Ref-1) protein: a basis ofr its role in human disease. Histopath. 1998;33:561–569. doi: 10.1046/j.1365-2559.1998.00541.x. [DOI] [PubMed] [Google Scholar]
- 65.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Machanic LE, Zurer I, Rotter V, Samson LD, Harris CC. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J Clin Invest. 2003;112:1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schild LJ, Brookman KW, Thompson LH, Wilson DM., III Effects of Ape1 overexpression on cellular resistance to DNA-damaging and anticancer agents. Somatic Cell Mol Genetics. 1999;25:253–262. doi: 10.1023/a:1019979613989. [DOI] [PubMed] [Google Scholar]
- 67.Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol Life Sci. 2010;67:3589–3608. doi: 10.1007/s00018-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kakolyris S, Kaklamanis L, Engels K, Turley H, Hickson ID, Gatter KC, Harris AL. Human pauinic endonuclease 1 expression in colorectal adenoma-carcinoma sequence. Cancer Res. 1997;57:1794–1797. [PubMed] [Google Scholar]
- 69.Kakolyris S, Kaklamanis L, Engels K, Fox SB, Taylor M, Hickson ID, Gatter KC, Harris AL. Human AP endonuclease 1 (HAP1) protein expression in breast cancer correlates with lymph node status and angiogenesis. Br J Cancer. 1998;77:1169–1173. doi: 10.1038/bjc.1998.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tell G, Pellizzari L, Pucillo C, Puglisi F, Cesselli D, Kelley MR, Di Loreto C, Damante G. TSH controls Ref-1 nuclear translocation in thyroid cells. J Mol Endocrinol. 2000;24:383–390. doi: 10.1677/jme.0.0240383. [DOI] [PubMed] [Google Scholar]
- 71.Moore DH, Michael H, Tritt R, Parsons SH, Kelley MR. Alternations in the expression of the DNA repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clin Cancer Res. 2000;6:602–609. [PubMed] [Google Scholar]
- 72.Wang D, Luo M, Kelley MR. Human apurinic endonuclease (APE1) expression and prognostic significance in osteosarcoma: enhance sensitivity of osteosarcoma to DNA damageing agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 73.Yang S, Irani K, Heffron SE, Jurnak F, Meyskens FL., Jr Alternations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol aa an APE/Ref-1 inhibitor. Mol Cancer Ther. 2005;4:1923–1935. doi: 10.1158/1535-7163.MCT-05-0229. [DOI] [PubMed] [Google Scholar]
- 74.Xu Y, Moore DH, Broshears J, Liu L, Wilson TM, Kelley MR. The apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme is elevated in premalignant and malignant cervical cancer. Anticancer Res. 1997;17:3713–3719. [PubMed] [Google Scholar]
- 75.Kakolyris S, Giatromanolaki A, Koukourakis M, Kaklamanis L, Kanavaros P, Hickson ID, Barzllay G, Georgoullas V, Gatter KC, Harris AL. Nuclear localization of human AP endonuclease 1 (HAP1/Ref-1) associates with prognosis in early operable non-small cell lung cancer. J Pathol. 1999;189:351–357. doi: 10.1002/(SICI)1096-9896(199911)189:3<351::AID-PATH435>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 76.Puglisi F, Aprile G, Minisini AM, Barbone F, Cataldi P, Tell G, Kelley MR, Damante G, Beltrami CA, Di Loreto C. Prognositc signficance of Ape1/Ref-1 subcellular lcoalization in non-small cell lung carcinomas. Anticancer Res. 2001;21:4041–4049. [PubMed] [Google Scholar]
- 77.Koukourakis M, Glatromanolakl A, Kakolyris S, Sivridis E, Georgoullas V, Funtzilas G, Hickson ID, Gatter KC, Harris AL. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int J Oncol Biol Phys. 2001;50:27–36. doi: 10.1016/s0360-3016(00)01561-3. [DOI] [PubMed] [Google Scholar]
- 78.Li C, Liu Z, Wang LE, Strom SS, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Cormier JN, Prieto VB, Duvic M, Grimm EA, Wei Q. Genetics variants of the ADPRT, XRCC1: and APE1 genes and risk of cutaneous melanoma. Carcinogenesis. 2006;27:1894–1901. doi: 10.1093/carcin/bgl042. [DOI] [PubMed] [Google Scholar]
- 79.Farkasova T, Gurska S, Witkovsky V, Gabelova A. Significance of amino acid substitution variants of DNA repair genes in radiosusceptibility. Neoplasm. 2008;55:330–337. [PubMed] [Google Scholar]
- 80.Gu D, Wang M, Wang M, Zhang Z, Chen J. The DNA repair gene APE1 T1349G polymorphism and cancer risk: a meta-analysis of 27 case-control studies. Mutagenesis. 2009;24:507–512. doi: 10.1093/mutage/gep036. [DOI] [PubMed] [Google Scholar]
- 81.Hadi MZ, Coleman MA, Fidelis K, Mohrensweiser HW, Wilson DM., III Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28:3871–3879. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pieretti M, Khattar NH, Smith SA. Common polymorphisms and somatic mutations in human base excision repair genes in ovarian and endometrial cancers. Mutat Res. 2001;432:53–59. doi: 10.1016/s1383-5726(00)00002-9. [DOI] [PubMed] [Google Scholar]
- 83.Fung H, Demple B. Distinct roles of Ape1 protein in the repair of DNA damage induced by ionizing radiation or bleomycin. J Biol Chem. 2011;286:4968–4977. doi: 10.1074/jbc.M110.146498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naidu MD, Mason JM, Pica RV, Fung H, Pena LA. Radiation resistance in glioma cells determine DNA damage repair activity of Ape1/Ref-1. J Radiat Res (Tokyo) 2010;51:393–404. doi: 10.1269/jrr.09077. [DOI] [PubMed] [Google Scholar]
- 85.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, Kostoe DD. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributers to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res. 2002;8:3008–3018. [PubMed] [Google Scholar]
- 86.Wilson DM, III, Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell Mol Life Sci. 2010;67:3621–3631. doi: 10.1007/s00018-010-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, Dianov GL, Hickson ID. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, Austin CP, Wilson DM., III Identification and characterization of inhibitors of human apurinic/apyrimidinic endonulcease APE1. PLoS ONE. 2009;4:e5740. doi: 10.1371/journal.pone.0005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventative target. Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 90.Seiple LA, Cardellina JH, 2nd, Akee R, Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol Pharmoacol. 2008;73:669–677. doi: 10.1124/mol.107.042622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim H, Cardellina JH, 2nd, Akee R, Champoux JJ, Stivers JT. Arylstibonic acids: novel inhibitors and activators of human topoisomerase IB. Bioorg Chem. 2008;36:190–197. doi: 10.1016/j.bioorg.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dorjsuren D, Kim D, Maloney DJ, Wilson DM, III, Simeonov A. Complementary non-radioactive assay for investigation of human flap endonuclease 1 activity. Nucleic Acids Res. 2010;39:e11. doi: 10.1093/nar/gkq1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zawahir Z, Dayam R, Deng J, Pereira C, Neamati N. Pharmacophore guided discovery of small-molecule human apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem. 2009;52:20–32. doi: 10.1021/jm800739m. [DOI] [PubMed] [Google Scholar]
- 94.Mohammed MZ, Vyjayanti VN, Laughton C, Dekker L, Fischer PM, Wilson DM, III, Abbotts R, Patel PM, Hickson ID, Madhusudan S. Development and evaluation of human AP endonuclease (APE1) inhibitors in Melanoma and Glioma cell lines. Br J Cancer. 2011;104:653–663. doi: 10.1038/sj.bjc.6606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bapat A, Glass L, Luo M, Fishel ML, Long EC, Georgiadis MM, Kelley MR. Novel small molecular inhibitor of APE1 endonuclease blocks proliferation and reduces viability of glioblastoma cells. J Pharmacol Exp Ther. 2010;334:988–998. doi: 10.1124/jpet.110.169128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu L, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 97.Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res. 2004;24:2127–2134. [PubMed] [Google Scholar]
- 98.Weiss GJ, Gordon MS, Rosen LS, Sawides P, Ramanathan RK, Mendelson DS, Goldman J, Liu L, Xu Y, Gerson SL, Adams BJ, Alvarez D, Theuer CP, Leigh BR. Final Results From a Phase 1 Study of Oral TRC102 (Methoxyamine HCl), an Inhibitor of Base-Excision Repair, to Potentiate the Activity of Pemetrexedin Patients with Refractory Cancer. ASCO; Alexandria, Virginia: 2010. [Google Scholar]
- 99.Hirschberg E, Weinstein IB, Gersten N, Marner E, Finkelstein T, Carchman R. Structure-activity studies on the mechanism of action of miracil D. Cancer Res. 1968;28:601–607. [PubMed] [Google Scholar]
- 100.Bases R. Enhancement of x-ray damage in HeLa cells by exposure of lucathone (Miracil D) following radiation. Cancer Res. 1970;30:2007–2011. [PubMed] [Google Scholar]
- 101.Naidu MD, Agarwal R, Pena LA, Cunha L, Mezei M, Shen M, Wilson DM, III, Liu Y, Sanchez Z, Chaudhary P, Wilson SH, Waring MJ. Lucanthone and its derivative hycanthone inhibit apurinic endonuclease-1 (APE1) by direct protein binding. PLoS ONE. 2011;6:e23679. doi: 10.1371/journal.pone.0023679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zou C, Karkari Y, Kabe H, Handa R, Anders A, Maitra A. The Ape-1/Ref-1 redox antagonis E3330 inhibits the growth of tumor endothelium and endothelia progenitor cells: Therapeutic implications in tumor angiogenesis. J Cell Physiol. 2008;219:209–218. doi: 10.1002/jcp.21666. [DOI] [PubMed] [Google Scholar]
- 103.Jiang A, Gao H, Kelley MR, Qiao X. Inhibition of APE1/Ref-1 redox activity with APX3330 block retinal angiogenesis in vitro and in vivo. Vision Res. 2010;51:93–100. doi: 10.1016/j.visres.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fishel ML, Colvin ES, Luo M, Kelley MR, Robertson KA. Inhibition of the redox function of APE1/Ref-1 in myeloid leukemia cell lines results in a hypersensitive response to retinoic acid-induced differentiation and apoptosis. Exp Hematol. 2010;38:1178–1188. doi: 10.1016/j.exphem.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fishel ML, Jiang Y, Rajeshkumar NV, Scandura G, Sinn AL, He Y, Shen C, Jones DR, Pollok KE, Ivan M, Mitra S, Kelley MR. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol Cancer Ther. 2011;10:1698–1708. doi: 10.1158/1535-7163.MCT-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:23–27. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 107.Manvilla BA, Wauchope O, Seley-Radtke KL, Drohat AC. NMR studies reveal an unexpected binding site for a redox inhibitor of AP endonuclease 1. Biochem. 2011;50:10540–10549. doi: 10.1021/bi201071g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McNeill DR, Wilson DM., III A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 109.McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM., III Impairment of APE1 function enhances cellular sensitivitry to clinically relevant alkylators and antimetabolites. Mol Cancer Res. 2009;7:897–906. doi: 10.1158/1541-7786.MCR-08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chang LM, Bollum FJ. Low molecular weight deoxyribonucleic acid polymerase in mammalian cells. J of Biol Chem. 1971;246:5835–5837. [PubMed] [Google Scholar]
- 111.Baril EF, Brown OE, Jenkins MD, Laszlo J. Deoxyribonucleic acid polymerase with rat liver riboxomes and smooth membranes. Purification and properties of the enzymes. Biochem. 1971;10:1981–1982. doi: 10.1021/bi00787a004. [DOI] [PubMed] [Google Scholar]
- 112.Garcia-Diaz M, Dominquez O, Lopez-Fernandez LA, de Lera LT, Sangier ML, Ruiz JF, Parraga M, Garcoa-Ortiz MJ, Kirchhoff T, del Mazo J, Bernad A, Blanco L. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 113.Duvauchelle JB, Blanco L, Fuchs RP, Cordonnier AM. Human DNA polymerase mu exhibits an unusual replication slippage ability at AAF lesion. Nucleic Acids Res. 2002;30:2061–2067. doi: 10.1093/nar/30.9.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chyan YJ, Ackerman S, Shepherd NS, McBride OW, Widen SG, Wilson SH, Wood TG. The human DNA polymerase beta gene structure. Evidence of alternative splicing in gene expression. Nucleic Acids Res. 1994;22:2719–2725. doi: 10.1093/nar/22.14.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beard WA, Wilson SH. Purificationand domain-mapping of mammalian DNA polymerase beta. Methods Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
- 116.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase B. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 117.Prasad R, Beard WA, Chyan YJ, Maciejewski MW, Mullen GP, Wilson SH. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase beta as revealted by site-directed mutagenesis. DNA binding and 5′-deoxyribose phosphate lyase activities. J Biol Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 118.Kumar A, Abbotts J, Karawya EM, Wilson SH. Identification and properties of the catalytic domain of mammlian DNA polymerase beta. Biochem. 1990;29:7156–7159. doi: 10.1021/bi00483a002. [DOI] [PubMed] [Google Scholar]
- 119.Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 120.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 121.Chagovetz AM, Sweasy JB, Preston BD. Increased activity and fidelity of DNA polymerase-B on single-nucleotide gapped DNA. J Biol Chem. 1997;272:27501–27504. doi: 10.1074/jbc.272.44.27501. [DOI] [PubMed] [Google Scholar]
- 122.Ahn J, Kraynov VS, Zhong X, Werneburg BG, Tsai MD. DNA polymerae B: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity, and conformational changes. J Biochem. 1998;311:79–87. doi: 10.1042/bj3310079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vande Berg BJ, Beard WA, Wilson SH. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase B. J Biol Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 124.Singhal RK, Prasad R, Wilson SH. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 125.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase B and Cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 126.Hirose F, Hotta Y, Yamaguchi M, Matsukage A. Difference in the expression level of DNA polymerase beta among mouse tissues: high expression in the pachytene spermatocyte. Exp Cell Res. 1989;181:169–180. doi: 10.1016/0014-4827(89)90191-2. [DOI] [PubMed] [Google Scholar]
- 127.Srivastava DK, Husain I, Arteaga C, Wilson SH. DNA polymerase B expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 128.Canitrot Y, Cazaux C, Frechet M, Bouayadi K, Lesca C, Salles B, Hoffman JS. Overexpression of DNA polymerase beta in cell results in a mutator phenotype and decreased sensitivity to anticancer drugs. PNAS. 1998;95:12586–12590. doi: 10.1073/pnas.95.21.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bergoglio V, Pillaire MJ, Lacroix-Triki M, Raynaud-Messina B, Canitrot Y, Beith A, Gates M, Wright M, Delsol G, Loeb LA, Cesselli D, Hoffman JS. Deregulated DNA polymerase beta induces chromosome instability and tumorigenesis. Cancer Res. 2002;30:3511–3514. [PubMed] [Google Scholar]
- 130.Clairmont CA, Narayanan L, Sun KW, Glazer PM, Sweasy JB. The Tyr-265-to-Cys mutator mutant of DNA polymerase beta induces a mutator phenotype in mouse LN12 cells. PNAS. 1999;96:9580–9585. doi: 10.1073/pnas.96.17.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sweasy JB. Fidelity mechansims of DNA polymerase beta. Prog Nucleic Acid Res Mol Biol. 2003;73:137–169. doi: 10.1016/s0079-6603(03)01005-5. [DOI] [PubMed] [Google Scholar]
- 132.Kosa JL, Sweasy JB. The E249K mutator mutant of DNA polymerase B extends mispaired termini. J Biol Chem. 1999;274:35866–35872. doi: 10.1074/jbc.274.50.35866. [DOI] [PubMed] [Google Scholar]
- 133.Dalal S, Kosa JL, Sweasy JB. The D246V mutant of DNA polymerase B is a mutator due to altered positioning of the DNA within its active site. J Biol Chem. 1997;279:577–584. doi: 10.1074/jbc.M309607200. [DOI] [PubMed] [Google Scholar]
- 134.Guo Z, Zheng L, Dai H, Zhou M, Xu H, Shen B. Human DNA polyerase beta polymorphism, Arg137Gln, impairs its polymerase activity and itneraction with PCNA and the cellular base excision repair capacity. Nucleic Acids Res. 2009;37:3431–3441. doi: 10.1093/nar/gkp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu S, Lai Y, Zhao W, Wu M, Zhang Z. Links between DNA polymerase beta expression and sensitivity to bleomycin. Toxicology. 2011;281:63–69. doi: 10.1016/j.tox.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 136.Horton JK, Watson M, Stefanick DF, Snaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst) 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 138.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 139.Trivedi RN, Wang XH, Jelezcova E, Goellner EM, Tang JB, Sobol RW. Human methyl purine DNA plycosylase and DNA polymerase beta expression collectively predict sensitivity to temozolomide. Mol Pharmacol. 2008;74:505–516. doi: 10.1124/mol.108.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Polosina YY, Rosenquist TA, Grollman AP, Miller H. ‘Knock down’ of DNA polymerase beta by RNA inferference: recapitulation of null phenotype. DNA Repair (Amst) 2004;3:1469–1474. doi: 10.1016/j.dnarep.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 141.Mizushina Y, Tanaka N, Yagi H, Kurosawa T, Onoue M, Seto H, Horie T, Aoyagi M, Yamaoka A, Matsukage S, Yoshida S, Sakaguchi K. Fatty acids selectively inhibit eukaryotic DNA polymerase activities in vitro. Biochim Biophys Acta. 1996;1308:256–262. doi: 10.1016/0167-4781(96)00121-2. [DOI] [PubMed] [Google Scholar]
- 142.Mizushina Y, Ohkubo T, Date T, Yamaguchi M, Saneyoshi M, Sugawara F, Sakaguchi K. Mode analysis of a fatty acid molecule biniding to the N-terminal 8-kDa domain of the DNA polymerase beta. A 1:1 complex and binding surface. J Biol Chem. 1999;274:25599–25607. doi: 10.1074/jbc.274.36.25599. [DOI] [PubMed] [Google Scholar]
- 143.Hu HY, Horton JK, Gryk MR, Prasad R, Naron JM, Sun DA, Hecht SM, Wilson SH, Mullen GP. Identification of small molecule sunthetic inhibitors of DNA polymerase beta by NMR chemical shift mapping. J Biol Chem. 2004;279:39736–39744. doi: 10.1074/jbc.M402842200. [DOI] [PubMed] [Google Scholar]
- 144.Jaiswal AS, Banerjee S, Aneja R, Sarkar FH, Ostrov DA, Narayan S. DNA Polymerase beta as a novel target for chemotherapeutic intervention of colorectal cancer. PLoS ONE. 2011;6:e16691. doi: 10.1371/journal.pone.0016691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Summerer D, Marx A. A molecular beacon for quantitative monitoring of the DNA polymerase reaction in real-time. Angew Chem Intl Ed. 2002;41:3620–3622. doi: 10.1002/1521-3773(20021004)41:19<3620::AID-ANIE3620>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 146.Seville M, West AB, Cull MG, McHenry CS. Fluorometric assay for DNA polymerases and reverse transcripase. Biotechniques. 2006;21:664–672. doi: 10.2144/96214st04. [DOI] [PubMed] [Google Scholar]
- 147.Kozlov M, Bergendahl V, Burgess R, Goldfarb A, Mustaev A. Homogenous fluorescent assay for RNA polymerase. Anal Biochem. 2005;342:206–213. doi: 10.1016/j.ab.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 148.Ma C, Tang Z, Wang K, Tan W, Li J, Li W, Li Z, Yang W, Li H, Liu L. Real-time monitoring of DNA polymerase activity using molecular beacon. Anal Biochem. 2006;353:141–143. doi: 10.1016/j.ab.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 149.Dorjsuren D, Wilson DM, III, Beard WA, McDonald JP, Austin CP, Woodgate R, Wilson SH, Simeonov A. A real-time fluorescence method for enzymatic characterization of specialized human DNA polymerases. Nucleic Acids Res. 2009;37:e128. doi: 10.1093/nar/gkp641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zheng L, Jia J, Finger D, Guo Z, Zer C, Shen B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011;39:781–794. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Ann Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 153.Shen B, Qiu J, Hosfield D, Tainer JA. Flap endonuclease homologs in archaebacteria exist as independent proteins. Trends Biochem Sci. 1998;23:171–173. doi: 10.1016/s0968-0004(98)01199-2. [DOI] [PubMed] [Google Scholar]
- 154.Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J, Kane MF, Fan K, Russel R, Brown AM, Kneitz B, Edelmann W, Kolodner RD, Lopkin M, Kucherlapati R. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. PNAS. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Murante RS, Huang L, Turchi JJ, Bambara RA. The calf 5′- to 3′-exonuclease is also an endonuclease with both activities dependent on primers annealed upstream of the point of cleavage. J Biol Chem. 1994;269:1191–1196. [PubMed] [Google Scholar]
- 156.Harrington JJ, Lieber MR. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lindahl T. The action pattern of mammalian deoxyribonuclease IV. Eur J Biochem. 1971;18:415–421. doi: 10.1111/j.1432-1033.1971.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 158.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 159.Devos JM, Tomanicek SJ, Jones CE, Nossal NG, Mueser TC. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J Biol Chem. 2007;282:31713–31724. doi: 10.1074/jbc.M703209200. [DOI] [PubMed] [Google Scholar]
- 160.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Turchi JJ, Huang L, Murante RS, Kim Y, Bambara RA. Enzymatic completion of mammalian lagging-strand DNA replication. PNAS. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang L, Rumbaugh JA, Murante RS, Lin RJ, Rust L, Bambara RA. Role of calf RTH-1 nuclease in removal of 5′-ribonucleotides during Okazaki fragment processing. Biochem. 1996;28:9266–9277. doi: 10.1021/bi9603074. [DOI] [PubMed] [Google Scholar]
- 163.Kao HO, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J Biol Chem. 2004;279:15014–15024. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
- 164.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM., III Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochodria DNA intergrity. DNA Repair (Amst) 2009;8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.DeMott MS, Shen B, Park MS, Bambara RA, Zigman S. Human RAD2 homolog 1 5′-to 3′-exo/endonucleae can efficiently excise a displaced DNA fragment containing a 5′-terminal abasic lesion by endonuclease activity. J Biol Chem. 1996;271:30068–30076. doi: 10.1074/jbc.271.47.30068. [DOI] [PubMed] [Google Scholar]
- 167.Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in short- and long-patch base excision repair in mammalian cells. Biochem. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 168.Matsumoto Y, Kim K, Bogenhagen DF. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol Cell Biol. 1994;14:6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Biade S, Sobol RW, Wilson SH, Matsumoto Y. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J Biol Chem. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]
- 170.Gary R, Park MS, Nolan JP, Cornelius HL, Kozyreva OG, Tran HT, Lobachev KS, Resnick MA, Gordenin DA. A novel role in DNA metabolism for the binding of FEN1/Rad27 to PCNA and implications for genetic risk. Mol Cell Biol. 1999;19:5373–5382. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J Biol Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- 172.Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evience for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem. 2001;276:25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 173.Otto CJ, Amqvist E, Hayden MR, Andrews SE. The “flap” endonuclease gene FEN1 is exculded as a candidate gene implicated in the CAG repeat expansion underlying Huntington disease. Clin Genet. 2001;59:122–127. doi: 10.1034/j.1399-0004.2001.590210.x. [DOI] [PubMed] [Google Scholar]
- 174.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 175.Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, Song KS, Rho SM, Yoo HS, Kim YS, Kim JO, Kim JG, Kim NS. Identification of gastric cancer-related genes using a cDNA microarray containing novel exprssed sequence tags expressed in gastric cancer cells. Clin Cancer Res. 2005;11:473–482. [PubMed] [Google Scholar]
- 176.Krause A, Combaret V, Iacono I, Lacroix B, Compagnon C, Bergeron C, Valsesia-Wittmann S, Leissner P, Mougin B, Puisieux A. Genome -wide analysis of gene expression in neuroblastomas detected by mass screening. Cancer Lett. 2005;225:111–120. doi: 10.1016/j.canlet.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 177.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfag R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene exprsesion patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C, Minna JD. Increased expression and no mutation of the flap endonuclease (FEN1) gene in human lung cancer. Oncogene. 2003;22:7243–7246. doi: 10.1038/sj.onc.1206977. [DOI] [PubMed] [Google Scholar]
- 179.Yang M, Guo H, Wu C, He Y, Yu D, Zhou L, Wang F, Xu J, Tan W, Wang G, Shen B, Yuan J, Wu T, Lin D. Functional FEN1 polymorphisms are associated with DNA damge levels and lung cancer risk. Hum Mutat. 2009;30:1320–1328. doi: 10.1002/humu.21060. [DOI] [PubMed] [Google Scholar]
- 180.Liu L, Zhou C, Zhou L, Peng L, Li D, Zhang X, Zhou M, Kuang P, Yuan Q, Song X, Yang M. Functional FEN1 genetic variants contribute to rish of hepatocellular carcinoma, esophageal cancer, gastric cancer, and colorectal cancer. Carcinogenesis. 2012;33:119–123. doi: 10.1093/carcin/bgr250. [DOI] [PubMed] [Google Scholar]
- 181.Zheng L, Dai H, Zhou M, Li M, Singh P, Tsark W, Huang Q, Kernstine K, Zhang X, Lin D, Shen B. Fen1 mutations results in autoimmunity, chronic inflammation and cancers. Nature Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 182.Xu H, Zheng H, Dai M, Zhou Y, Hua Y, Shen B. Chemical-induced cancer incidence and underyling mechanisms in Fen1 mutant mice. Oncogene. 2011;30:1072–1081. doi: 10.1038/onc.2010.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wu Z, Lin Y, Xu H, Dai H, Zhou M, Tsao S, Zheng L, Shen B. High risk of benzo[alpha]pyrene-induced lung cancer in E160D FEN1 mutant mice. Mutat Res. 2012;731:85–91. doi: 10.1016/j.mrfmmm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 185.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 186.Mirkin SM. Molecular models for repeat expansions. Biochem Mol Biol. 2004;17:639–662. [Google Scholar]
- 187.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 188.Spiro C, Pelletier R, Rolfsmeier ML, Dixon MJ, Lahue RS, Gupta G, Park MS, Chen X, Mariappan SVS, McMurray CT. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 189.Henrickesen LA, Tom S, Liu Y, Bambara RA. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J Biol Chem. 2000;275:16420–16427. doi: 10.1074/jbc.M909635199. [DOI] [PubMed] [Google Scholar]
- 190.Goula AV, Berquist BR, Wilson DM, III, Wheeler VC, Trottier Y, Merienne K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu Y, Prasad R, Beard WA, Hou EW, Horton JK, McMurrary CT, Wilson SH. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J Biol Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Terry KL, De Vivo I, Titus-Ernstoff L, Shih MC, Cramer DW. Androgen receptor cytosine, adenine, guanine repeats, and haplotypes in relationship to ovarian cancer risk. Cancer Res. 2005;65:5981. doi: 10.1158/0008-5472.CAN-04-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yaron M, Levy T, Chetrit A, Levavi H, Sabah G, Schneider D, Halperin R, Ben-Rafael Z, Friedman E. The polymorphic CAG repeat in the androgen receptor gene in Jewish Israeli women with endometrial carcinoma. Cancer. 2001;92:1190–1194. doi: 10.1002/1097-0142(20010901)92:5<1190::aid-cncr1437>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 194.Giovannucci E, Stampfer MH, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, Hennekens CH, Kantoff PW. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. PNAS. 1997;94:3320–3323. doi: 10.1073/pnas.94.7.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Matsuzaki Y, Adachi N, Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30:3273–3277. doi: 10.1093/nar/gkf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Urbanucci A, Sahu B, Seppala J, Larjo A, Latonen LM, Waltering KK, Tammella TLJ, Vessella RL, Lahdesmaki H, Janne OA, Visakorpi T. Overexpression of androgen receptor enhances the binding of the receptor to the chromatin in prostate cancer. Oncogene. 2011 doi: 10.1038/onc.2011.401. Epud ahead of print. [DOI] [PubMed] [Google Scholar]
- 197.Shibata Y, Nakamura T. Defective flap endonuclease 1 activity in mammalian cells is associated with impared DNA repair and prolonged S phase delay. J Biol Chem. 2002;277:746–754. doi: 10.1074/jbc.M109461200. [DOI] [PubMed] [Google Scholar]
- 198.Nikolova T, Christmann M, Kaina B. FEN1 is overexpressed in testis, lung, and brain tumors. Anticancer Res. 2009;29:2453–2459. [PubMed] [Google Scholar]
- 199.Shen B, Nolan JP, Sklar LA, Park MS. Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J Biol Chem. 1996;271:9173–9176. doi: 10.1074/jbc.271.16.9173. [DOI] [PubMed] [Google Scholar]
- 200.Panda H, Jaiswal AS, Corsino PE, Armas ML, Law BK, Narayan S. Amino acid Asp181 of 5′-flap endonuclease 1 is useful target for chmotherapeutic development. Biochem. 2009;48:9952–9958. doi: 10.1021/bi9010754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tumey LN, Bom D, Huck B, Gleason E, Wang J, Silver D, Brunden K, Boozer S, Rundlett S, Sherf B, Murphy S, Dent T, Leventhal C, Bailey A, Harrington J, Bennani YL. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg Med Chem Lett. 2005;15:277–281. doi: 10.1016/j.bmcl.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 202.McManus KJ, Barrett IJ, Nouhi Y, Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. PNAS. 2009;106:3276–3281. doi: 10.1073/pnas.0813414106. [DOI] [PMC free article] [PubMed] [Google Scholar]