Abstract
The use of small molecules to interfere with protein-protein interactions has tremendous therapeutic appeal and is an area of intense interest. Numerous techniques exist to assess these interactions and their disruption. Many, however, require large amounts of protein, do not allow interactions to be followed in real time, are technically demanding or require large capital expenditures and high levels of expertise. Surface plasmon resonance (SPR) represents a convenient alternative to these techniques with virtually none of their disadvantages. We have devised an SPR-based method that allows the heterodimeric association between the c-Myc (Myc) oncoprotein and its obligate partner Max to be quantified in a manner that agrees well with values obtained by other methods. We have adapted it to examine the ability of previously validated small molecules to interfere with Myc-Max heterodimerization and DNA binding. These inhibitors comprised two distinct classes of molecules that inhibit DNA binding by preventing Myc-Max interaction or distorting pre-formed heterodimers and rendering them incapable of DNA binding. Our studies also point out several potential artifacts and pitfalls to be considered when attempting to employ similar SPR-based methods. This technique should be readily adaptable to the study of other protein-protein interactions and their disruption by small molecules.
Keywords: c-Myc, Max, intrinsically disordered proteins, 10058-F4, 10074-G5, JKY-2-169
INTRODUCTION
Protein-protein interactions are critical for virtually all cellular functions ranging from the transmission of growth factor signals to the assembly of multi-protein complexes and organelles [1,2]. Because abnormal protein-protein interactions are also causally implicated in numerous disease states [2,3], considerable effort has been devoted to the development of small molecules capable of blocking or reversing these associations and their pathological consequences [4,5]. Although some notable successes have been reported, significant challenges remain in achieving these goals [6].
We have focused on the development of small, drug-like compounds to prevent or disrupt the association between the c-Myc (Myc) oncoprotein transcription factor and its obligate partner Max, which interact via their respective basic helix loop helix-leucine zipper (bHLH-ZIP) dimerization domains [7–9]. De-regulated expression of Myc is implicated in many different types of cancer [10] although even tumors without obvious Myc deregulation are dependent upon the oncoprotein for their proliferation and/or survival [11,12]. The Myc-Max heterodimer is required for the transcriptional regulation of numerous target genes whose encoded products determine most of the phenotypes that underlie the transformed state [13]. For these reasons, small molecules that directly interfere with Myc-Max dimerization (henceforth “Myc inhibitors”) present significant therapeutic potential [8,9].
Two mechanistically distinct classes of direct Myc inhibitors have been identified. The first, represented by the compounds 10058-F4 and 10074-G5 and their analogs [14–19] bind to distinct regions of monomeric Myc’s intrinsically disordered bHLH-ZIP domain and create a localized distortion that prevents heterodimerization with Max [20,21]. Due to the high free energy of Myc-Max association, these compounds are much less able to promote the dissociation of pre-formed heterodimers [8,9]. Members of the second, and more recently recognized, class of direct Myc inhibitors, represented here by the synthetic α-helix mimetic JKY-2-169, interact poorly with individual Myc and Max proteins [22]. Rather, by virtue of being designed to recognize key arginine and hydrophobic residues within the distinct conformational spaces they occupy in the α-helical Myc-Max heterodimer, JKY-2-169 distorts this structure, without causing its dissociation, and abrogates DNA binding [22].
Various biophysical methods have been used to measure the effects of these small molecules on Myc-Max association or their binding to DNA. They include fluorescence polarization, electrophoretic mobility shift assays (EMSA), circular dichroism, and NMR spectroscopy [15,18,20,21]. While these methods are specific and variably quantitative, each is subject to factors that can compromise or restrict its use. These include the need for large amounts of purified proteins, the requirement that small molecules of interest be fluorescent and an inability in some cases to follow protein-protein or protein-DNA interactions in real time [8,9,23]. Techniques such as NMR spectroscopy also require substantial expertise and capital investments in appropriate instrumentation.
The Biacore™ biosensor, based on the surface plasmon resonance (SPR) detection principle, is an alternative method that permits the accurate and quantitative assessment of protein-protein and protein-DNA interactions. In addition to requiring relatively low amounts of material, real-time, label-free kinetics of interactions including measurements of weak interactions between small molecules and proteins can be determined [24–27]. The ability to operate the Biacore in an automated manner also permits medium to high-throughput sample analysis. The Biacore is a mass-based detection system and changes in mass on the sensor chip surface as a result of the binding interaction are correlated to changes in the refractive index. This is reflected in the SPR signal, which may impose some challenges, particularly when the binding partner in solution possesses a low mass [28]. Present SPR technology has overcome this limitation to a great extent by introducing systems like the Biacore™ T200 and the Biacore™ 4000 which are routinely used in the analysis of small molecule drugs and inhibitors (100 Daltons or below) as well as fragment libraries.
We have previously used SPR to measure the interaction between 10058-F4, 10074-G5 and a select group of their analogs with the bHLH-ZIP domain of N-Myc, a close relative of c-Myc, whose over-expression is associated with advanced-stage, poor prognosis neuroblastomas [24,25]. The interaction of these small molecules with N-Myc correlated quite well with their ability to prevent the in situ formation of N-Myc-Max heterodimers and to induce neuronal differentiation as previously described for 10058-F4 [24,26]. More recently, we have described an SPR-based method that allows for a more sensitive means of detecting the effects of these small molecules on Myc-Max heterodimer formation [24]. Rather than quantifying the direct interaction of the molecules with Myc, this alternative approach assesses their ability to prevent and/or disrupt the Myc-Max heterodimer’s binding to a biotinylated DNA target that is tethered to a streptavidin biosensor chip. The resultant loss of DNA binding is associated with a much more robust, reproducible and quantifiable signal. This technique also possesses greater versatility by virtue of its being amenable to the evaluation of compounds such as JKY-2-169 which induce non-DNA binding conformational distortions of the Myc-Max heterodimer without promoting its dissociation [22]. Although the methods described here are specifically aimed at assessing Myc inhibitors, they should be readily applicable to other protein-DNA interactions and their disruption with small molecules.
MATERIALS AND METHODS
Preparation of proteins
The bHLH-ZIP domain of human Myc (residues 353–437) along with full length Max(S) and Max(L) (151 and 160 residues, respectively, encoding the p20 and p21 isoforms, respectively) [7] were expressed in the pET151-D-TOPO vector as His6-N-terminally tagged fusion proteins in the E. coli strain BL21DE3(plysS) [15,20,21]. Bacterial cultures were grown at 37°C in L-Broth to an h ≈ 0.8 and then induced for 16 h with 1 mM isopropyl-L-thio-B-D-galactopyranoside (IPTG). Cultures were harvested, pelleted by centrifugation at 5,000 × g for 10 min and lysed in a buffer containing 8 M urea; 100 mM NaH2PO4 and 10 mM Tris-HCl, pH 8.0. Proteins were purified on an NTA nickel- agarose column (Qiagen, Inc., Valencia, CA) and eluted with a pH gradient according to the recommendations provided by the supplier. The purified proteins were then dialyzed against 150 mM NaCl, Tris-HCl pH 6.7 and cleaved with TEV protease at 25°C as previously described [20,27,28]. For larger amounts of protein, TEV protease:His6-tagged protein molar ratios were typically as low as 1:50 and were allowed to proceed for up to 72 h. The cleaved residues containing the His6 tag were then removed by an additional round of NTA-nickel-agarose chromatography. The final preparations of Myc, Max(S) and Max(L) were dialyzed against HBS-EP running buffer (10 mM HEPES, pH 7.4; 150 mM NaCl; 3 mM EDTA; 0.005% v/v Surfactant P20) and stored at −80°C in small aliquots.
Oligonucleotide synthesis
A biotin-tagged E-box-containing single-stranded oligonucleotide (5’-Biotin-TGAAGCAGAC CACGTGGTCGTCTTCA-3’, E-box sequence underlined) and its non-biotinylated complementary strand (both from IDT, Inc. Coralville, IA) were annealed at a 1:10 ratio in 100 mM NaCl; 10 mM EDTA, pH 7.5 and 1 mM EDTA. The resultant double-stranded DNA (hereafter referred to as the oligonucleotide) was then diluted to 500 nM in high salt HBS-EP buffer (10 mM HEPES, pH 7.4; 500 mM NaCl; 3 mM EDTA; 0.005% v/v Surfactant P20).
SPR studies
All experiments were performed at 25°C using a Biacore™ 3000 instrument and streptavadin-coated biosensor chips (SA-Chip GE Healthcare, Inc. Piscataway, NJ). All buffers were freshly prepared and filtered using bottle top or syringe filters (0.22 µm, Corning, Inc. Corning, NY) and de-gassed. The instrument was first primed three times with HBS-EP running buffer and flow cell 1 (FC1) was used as the reference flow cell, which was unmodified and lacked the oligonucleotide ligand. Flow cell 2 (FC2) was used for immobilization of the oligonucleotide. It was conditioned with three consecutive 1 min injections of 50 mM NaOH in 1 M NaCl, in accordance with the manufacturer’s instructions. The oligonucleotide was then injected over a 30 min period at a flow rate 10 ml /min followed by the Extraclean feature. Oligonucleotide immobilization levels of 700–800 RU were routinely observed under these conditions.
Protein-DNA binding assays were performed in HBS-EP running buffer at the relatively high flow rate of 60 ml/min to avoid or minimize any mass-transport limitation effects. Protein solutions ranging from 3–100 nM were injected for 150 seconds followed by dissociation in running buffer for 100 seconds. At the end of the dissociation period the sensor chip was regenerated to remove any remaining bound material by injecting HBS-EP buffer containing 1 M NaCl and 0.002% SDS at 30 ml/min for 30 seconds.
Myc inhibitor evaluations
Stock solutions (1 mM) of 10074-G5 and JKY-2-169 were prepared in 100% DMSO. All dilutions were performed in buffers containing 5% DMSO. For protein binding studies, the compounds were pre-incubated for 30 min with monomeric Myc or Max(S) (20 nM each), with pre-formed Myc-Max(S) heterodimers (20 nM of each protein) or with Max(L) (40 nM). The mixture was then analyzed as described above for the protein-DNA binding assay except that the running buffer contained 5% DMSO. A solvent correction curve was generated by adding varying amounts of DMSO to running buffer to generate a range of concentrations ranging from 4.5–5.8%.
Data analyses
Data were analyzed with the BIA evaluation software 4.1 (GE Healthcare, Inc., Piscataway, NJ) using the 1:1 Langmuir model fit with mass transfer limitations for determination of the binding kinetics.
RESULTS AND DISCUSSION
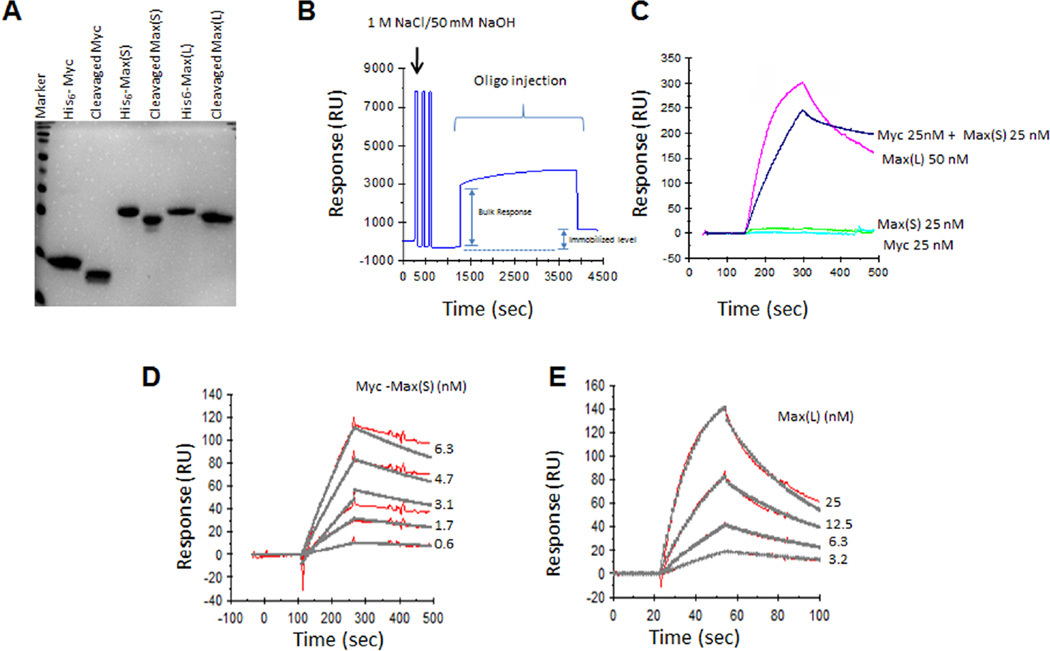
All experiments utilized His6-tagged recombinant Myc and Max proteins that were purified to near homogeneity using nickel-agarose affinity chromatography followed by cleavage of the His6 tag with TEV protease [15,20,21]. The final products were comprised only of endogenous sequences and were substantially free of other contaminating proteins (Fig. 1A). The Myc peptide consisted of the 84 residues comprising the bHLH-ZIP domain (amino acids 353–437) [15,20,21]. Two full-length isoforms of Max, termed Max(L) (160 residues) and Max(S) (151 residues), varying only by an N-terminal insertion of 9 residues in the former protein, were used for different purposes [7]. In the absence of Myc, Max(L) forms a DNA-binding homodimer that serves as an excellent control to test the specificity of small molecule Myc inhibitors [14,15,19]. Like Max(L), Max(S) also forms low-affinity homodimers [29] but binds DNA poorly, if at all. It can therefore be used to unequivocally assess the DNA binding activity of high affinity Myc-Max heterodimers without interference or competition from other DNA binding species [14,15,30,31].
Figure 1. Characterization of purified proteins and quantification of their binding to DNA using SPR.
A. SDS-PAGE of purified Myc and Max proteins prior to or following cleavage of the His6 tags with TEV protease. 5 µg of each protein was resolved and stained with Coomassie Blue. B. Tethering of the oligonucleotide to the SA-Chip. C. After adjusting the relative response in (B) to 0, the indicated proteins were passed over the SA-Chip and their binding was assessed. Note that only Myc-Max(S) heterodimers and Max(L) homodimers were capable of binding the oligonucleotide as previously reported [15,30,31]. D. Kinetic analysis of Myc-Max(S) binding to immobilized oligonucleotide. Equimolar concentrations of Myc and Max(S) were allowed to dimerize for 30 min at room temperature and then passed over the biosensor chip containing bound oligonucleotide. The raw data curves are shown in red and the fitted curves in grey. Note that in each case, these curves overlap, indicating a near perfect correlation between theoretically calculated binding and observed binding. E. Kinetics of Max(L) binding to the oligonucleotide was performed as described in (D).
Initial experiments attempting to detect Myc-Max(S) association by tethering His6-Myc to an NTA Biosensor chip and using Max(S) as the analyte were hindered by probable His6-Myc protein surface aggregation as evidenced by the inability of the protein to bind Max(S) or to be subsequently removed from the chip with high concentrations of nickel salts or urea. These findings are in keeping with the poor solubility of His6-Myc that necessitates its being maintained in dilute form following purification (HW, unpublished results). Indeed, even when such dilute solutions of Myc were used for attachment, eventual aggregation occurred due to the progressive accumulation of high local concentrations of protein on the chip surface. Reciprocal experiments, in which His6-Max(S) was bound to the chip surface and Myc was used as the analyte, allowed us to overcome this problem and demonstrate both time- and dose-dependent Myc-Max(S) heterodimerization (see below). However, binding of a non-biotinylated double-stranded E-box-containing oligonucleotide to the resultant heterodimer was difficult to achieve, despite the ease with which it occurs in EMSA experiments [14–16]. We attribute this to probable steric hindrance arising from the proximity of the Myc and Max basic domains to the chip surface that precludes access to the oligonucleotide analyte. Additional interference with DNA binding under these conditions might arise as a result of conformation restraints imposed on the Myc bHLH-ZIP domain as a consequence of its surface attachment and/or to additional limitations to DNA binding arising from the proximity of the His6 tag to the basic domain.
More reproducible and robust results were obtained using an SA chip to which was attached a biotinylated, double-stranded E-box-containing oligonucleotide (hereafter referred to as the oligonucleotide) (Fig. 1B). As expected from previously published results [14,15,31], neither Myc nor Max(S) bound the oligonucleotide (Fig. 1C) whereas both Max(L) homodimers and Myc-Max heterodimers exhibited strong time- and dose-dependent binding (Fig. 1D and 1E). Additionally, in the absence of any proteins, the direct Myc inhibitors 10058-F4, 10074-G5 and JKY-2-169 [14,22] did not affect the response of the previously tethered oligonucleotide (data not shown). A 1:1 “Langmuir with Mass Transport” model, provided in the Biacore Evaluation software package and that takes into account the limitations of mass transfer, was used to fit the data to determine the binding kinetics. Because the actual percentage of Myc-Max(S) heterodimers in solution could not be directly determined, we relied on the association between Myc and a His6-tagged Max(S) protein that had been tethered to a nickel-impregnated biosensor chip (NTA Chip, Biacore, Inc). Testing serial two-fold dilutions of Myc on this chip allowed us to calculate a KD for Myc-Max(S) of ~6.08 nM (data not shown). Based on this value and using the equation: KD = [Myc]×[Max(S)]/[Myc-Max(S)], we calculated the true concentration of Myc-Max(S) heterodimers for each input concentration of their respective proteins and used these values to derive Figure 1D. This information allowed us to calculate DNA binding affinity for the Myc-Max(S) heterodimer using global fitting and we obtained a KD= 8.9 nM, which agreed well with a previous publication using independent means [32].
A similar calculation for the association of Max(L) homodimers with the oligonucleotide (Fig. 1E) could not be performed since the homodimers form spontaneously at the time of purification. However, even assuming the efficiency of this process to be 100%, the efficiency of DNA binding that we calculated was only about half that achieved by Myc-Max(S). The greater dependency of the KD for Myc-Max(S) heterodimers on their absolute concentration likely reflects the ongoing competition of Max(S) to participate in both heterodimeric as well as homodimeric interactions. Taken together, these and the foregoing results indicate that the order of addition of the assay components, their conformation at the sensor chip surface and additional rotational limitations imposed by their attachment can exert major influences over the experimental outcome. Of lesser importance are the amounts of input protein whose dimerization variability can be minimized by employing the maximal achievable concentrations.
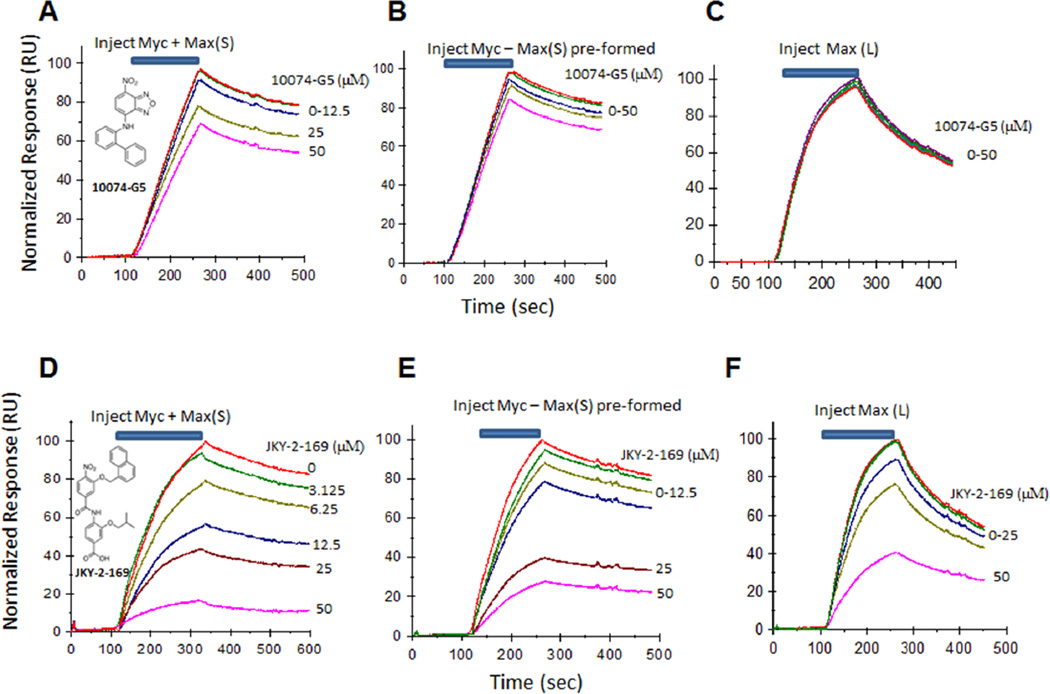
Having identified conditions under which DNA binding by Myc-Max-(S) and Max(L) could be reproducibly quantified, we next asked whether this could be prevented or reversed by two mechanistically distinct types of direct Myc inhibitors previously described by our group. An example of the first type of Myc inhibitor, 10074-G5, binds to a segment of Myc’s helix 1 domain lying immediately adjacent to the basic region [20,21]. The binding of 10074-G5 was confirmed by its ability to recognize an 18 residue synthetic peptide encompassing this site with an affinity nearly equal to that obtained with the full-length bHLH-ZIP domain [20,21]. This interaction results in a local conformational distortion that interferes with the protein’s ability to interact with Max. The second type of Myc inhibitor, exemplified by the synthetic α-helix mimetic JKY-2-169, recognizes neither Myc monomers nor Max(L) homodimers but instead alters the structure of the Myc-Max(S) heterodimer by interacting with arginine and hydrophobic residues located within the α-helical helix 1 domain in a manner that leads to loss of DNA binding but not heterodimerization [22].
When 10074-G5 was pre-incubated with Myc, exposed to Max(S) and then passed over the Biosensor chip, a dose-dependent reduction in DNA binding of the heterodimer was observed (Fig. 2A). In contrast, 10074-G5 was much less effective at reversing DNA binding by pre-formed Myc-Max(S) heterodimers (Fig. 2B). These results are consistent with the idea that compounds such as 10074-G5, while readily preventing Myc-Max interaction, are less efficient at disrupting pre-formed heterodimers due to their high free energy of association and the inherent difficulty in overcoming this with a small molecule [8,9]. That 10074-G5 also did not interfere with DNA binding by Max(L), a relatively low-affinity association (Fig. 2C) [29], is consistent with our previous results [14,20,21].
Figure 2. Differential effect of 10074-G5 and JKY-2-169 on oligonucleotide binding by Myc-Max(S).
A. 10074-G5 prevents DNA binding by Myc-Max-(S) heterodimers if added prior to their formation. 10074-G5 was pre-incubated with Myc for 30 min before the addition of Max(S). The entire mixture was then tested for DNA binding by passing over the SA-Chip containing the tethered oligonucleotide. B. 10074-G5 is much less effective at disrupting DNA binding by pre-formed Myc-Max(S) heterodimers. Pre-formed Myc-Max(S) heterodimers were incubated with the indicated concentrations of 10074-G5 for 30 min and then passed over the SA-Chip as described in (A). C. 10074-G5 does not prevent DNA binding by Max(L) homodimers. D. The experiment described in A was repeated using JKY-2-169. E. The experiment described in B was repeated using JKY-2-169. F. The experiment described in C was repeated with JKY-2-169. For (B) and (E), change the label Myc-Max(S) to pre-formed Myc-Max(S) for clarification.
Our findings with 10074-G5 contrasted with those obtained with JKY-2-169 which promoted DNA-protein dissociation irrespective of whether it was added prior to or after Myc-Max(S) heterodimer formation (Fig. 2D and 2E). This molecule would be expected to bind to the Myc-Max(S) heterodimer immediately after its formation thereupon altering its structure and preventing its interaction with the oligonucleotide. We have previously used NMR spectroscopy to demonstrate the formation of such an altered structure following the addition of JKY-2-169 [22]. JKY-2-169 had much less of an effect on DNA binding by Max(L) homodimers (Fig. 2F), thus attesting to the specificity of the compound as previously noted and documented by both NMR spectroscopy and other methods [22]. Finally, although numerous factors can influence the cellular effects of Myc inhibitors [15,18], we would note that the results obtained with SPR were in good agreement with JKY-2-169 being a more potent inhibitor of Myc-over-expressing cancer cells than 10074-G5 [18,22].
The foregoing studies provide a relatively simple, rapid and quantitative means of assessing DNA binding by Myc-Max(S) heterodimers and Max(L) homodimers in real-time. The method provides robust results that are in good agreement with those obtained by more traditional methods such as EMSAs or NMR [14,15,22] and that require much greater amounts of the protein reagents and/or highly specialized equipment and expertise. The method is also suitable for evaluating the capacity for Myc inhibitors to either prevent the formation of these dimeric proteins or disrupt pre-existing associations. It seems likely that this approach can be adapted to other types of protein-protein interactions and that the principles learned here will be applicable to these future studies.
Acknowledgements
This work was supported by NIH grant R01CA140624 awarded to EVP.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
References
- 1.Sudha G, Nussinov R, Srinivasan N. An overview of recent advances in structural bioinformatics of protein-protein interactions and a guide to their principles. Prog Biophys Mol Biol. 2014;116:141–150. doi: 10.1016/j.pbiomolbio.2014.07.004. PMID: 25077409. [DOI] [PubMed] [Google Scholar]
- 2.Chakraborty C, Doss C, George P, Chen L, Zhu H. Evaluating protein-protein interaction (PPI) networks for diseases pathway, target discovery, and drug-design using 'in silico pharmacology'. Curr Protein Pept Sci. 2014;15:561–571. doi: 10.2174/1389203715666140724090153. PMID: 25059326. [DOI] [PubMed] [Google Scholar]
- 3.Lage K. Protein-protein interactions and genetic diseases: The interactome. Biochim Biophys Acta. 2014;1842:1971–1980. doi: 10.1016/j.bbadis.2014.05.028. PMID: 24892209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fletcher S, Hamilton AD. Protein-protein interaction inhibitors: small molecules from screening techniques. Curr Top Med Chem. 2007;7:922–927. doi: 10.2174/156802607780906735. PMID: 17508923. [DOI] [PubMed] [Google Scholar]
- 5.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Curr Opin Chem Biol. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. PMID: 20395165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. PMID: 18075579. [DOI] [PubMed] [Google Scholar]
- 7.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. PMID: 2006410. [DOI] [PubMed] [Google Scholar]
- 8.Prochownik EV, Vogt PK. Therapeutic Targeting of Myc. Genes Cancer. 2010;1:650–659. doi: 10.1177/1947601910377494. PMID: 21132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher S, Prochownik EV. Small-molecule inhibitors of the Myc oncoprotein. Biochim Biophys Acta. 2015;1849:525–543. doi: 10.1016/j.bbagrm.2014.03.005. PMID: 24657798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. PMID: 10378696. [DOI] [PubMed] [Google Scholar]
- 11.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. PMID: 18716624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, et al. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene. 2007;27:1905–1915. doi: 10.1038/sj.onc.1210823. PMID: 17906696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu WB, Helander S, Pilstål R, Hickman KA, Lourenco C, et al. Myc and its interactors take shape. Biochim Biophys Acta. 2014;1849:469–483. doi: 10.1016/j.bbagrm.2014.06.002. PMID: 24933113. [DOI] [PubMed] [Google Scholar]
- 14.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. PMID: 13679853. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, et al. Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther. 2007;6:2399–2408. doi: 10.1158/1535-7163.MCT-07-0005. PMID: 17876039. [DOI] [PubMed] [Google Scholar]
- 16.Mustata G, Follis AV, Hammoudeh DI, Metallo SJ, Wang H, et al. Discovery of novel Myc-Max heterodimer disruptors with a three-dimensional pharmacophore model. J Med Chem. 2009;52:1247–1250. doi: 10.1021/jm801278g. PMID: 19215087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap JL, Wang H, Hu A, Chauhan J, Jung K, et al. Pharmacophore identification of c-Myc inhibitor 10074-G5. Bioorg Med Chem Lett. 2012;23:370–374. doi: 10.1016/j.bmcl.2012.10.013. PMID: 23177256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Chauhan J, Hu A, Pendleton K, Yap JL, et al. Disruption of Myc-Max heterodimerization with improved cell-penetrating analogs of the small molecule 10074-G5. Oncotarget. 2013;4:936–947. doi: 10.18632/oncotarget.1108. PMID: 23801058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chauhan J, Wang H, Yap JL, Sabato PE, Hu A, et al. Discovery of methyl 4'-methyl-5-(7-nitrobenzo[c][1,2,5]oxadiazol-4-yl)-[1,1'-biphenyl]-3- carboxylate, an improved small-molecule inhibitor of c-Myc-max dimerization. ChemMedChem. 2014;9:2274–2285. doi: 10.1002/cmdc.201402189. PMID: 24976143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol. 2008;15:1149–1155. doi: 10.1016/j.chembiol.2008.09.011. PMID: 19022175. [DOI] [PubMed] [Google Scholar]
- 21.Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J Am Chem Soc. 2009;131:7390–7401. doi: 10.1021/ja900616b. PMID: 19432426. [DOI] [PubMed] [Google Scholar]
- 22.Jung KY, Wang H, Teriete P, Yap JL, Chen L. Perturbation of the c-Myc- Max Protein-Protein Interaction via Synthetic alpha-Helix Mimetics. J Med Chem. 2015;58:3002–3024. doi: 10.1021/jm501440q. PMID: 25734936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg T. Small-molecule modulators of c-Myc/Max and Max/ Max interactions. Curr Top Microbiol Immunol. 2011;348:139–149. doi: 10.1007/82_2010_90. PMID: 20680803. [DOI] [PubMed] [Google Scholar]
- 24.Müller I, Larsson K, Frenzel A, Oliynyk G, Zirath H, et al. Targeting of the MYCN protein with small molecule c-MYC inhibitors. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097285. PMID: 24859015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beltran H. The N-myc Oncogene: Maximizing its Targets, Regulation, and Therapeutic Potential. Mol Cancer Res. 2014;12:815–822. doi: 10.1158/1541-7786.MCR-13-0536. PMID: 24589438. [DOI] [PubMed] [Google Scholar]
- 26.Zirath H, Frenzel A, Oliynyk G, Segerström L, Westermark UK, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc Natl Acad Sci U S A. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. PMID: 23733953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferre-D&apos, Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. PMID: 8479534. [DOI] [PubMed] [Google Scholar]
- 28.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. PMID: 12553908. [DOI] [PubMed] [Google Scholar]
- 29.Fieber W, Schneider ML, Matt T, Kräutler B, Konrat R, et al. Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J Mol Biol. 2001;307:1395–1410. doi: 10.1006/jmbi.2001.4537. PMID: 11292350. [DOI] [PubMed] [Google Scholar]
- 30.Prochownik EV, VanAntwerp ME. Differential patterns of DNA binding by myc and max proteins. Proc Natl Acad Sci U S A. 1993;90:960–964. doi: 10.1073/pnas.90.3.960. PMID: 8430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Fan S, Prochownik EV. Distinct roles for MAX protein isoforms in proliferation and apoptosis. J Biol Chem. 1997;272:17416–17424. doi: 10.1074/jbc.272.28.17416. PMID: 9211884. [DOI] [PubMed] [Google Scholar]
- 32.Follis AV, Hammoudeh DI, Daab AT, Metallo SJ. Small-molecule perturbation of competing interactions between c-Myc and Max. Bioorg Med Chem Lett. 2008;19:807–810. doi: 10.1016/j.bmcl.2008.12.025. PMID: 19114306. [DOI] [PubMed] [Google Scholar]