Abstract
Invasive alien tree pathogens can cause significant economic losses as well as large-scale damage to natural ecosystems. Early detection to prevent their establishment and spread is an important approach used by several national plant protection organizations (NPPOs). Molecular detection tools targeting 10 of the most unwanted alien forest pathogens in Canada were developed as part of the TAIGA project (http://taigaforesthealth.com/). Forest pathogens were selected following an independent prioritization. Specific TaqMan real-time PCR detection assays were designed to function under homogeneous conditions so that they may be used in 96- or 384-well plate format arrays for high-throughput testing of large numbers of samples against multiple targets. Assays were validated for 1) specificity, 2) sensitivity, 3) precision, and 4) robustness on environmental samples. All assays were highly specific when evaluated against a panel of pure cultures of target and phylogenetically closely-related species. Sensitivity, evaluated by assessing the limit of detection (with a threshold of 95% of positive samples), was found to be between one and ten target gene region copies. Precision or repeatability of each assay revealed a mean coefficient of variation of 3.4%. All assays successfully allowed detection of target pathogen on positive environmental samples, without any non-specific amplification. These molecular detection tools will allow for rapid and reliable detection of 10 of the most unwanted alien forest pathogens in Canada.
Introduction
Invasive alien tree pathogens can cause significant economic losses as well as large-scale damage to natural ecosystems. Over the last century, Canada has experienced the dramatic consequences of introductions of alien forest pathogens. The pathogens responsible for white pine blister rust (Cronartium ribicola J.C. Fisch), beech bark disease (Cryptococcus fagisuga Lindinger), and Dutch elm disease (Ophiostoma ulmi (Buisman) Melin & Nannf. and O. novo-ulmi (Brasier)) were accidentally introduced into Canada and resulted in the death of millions of Pinus strobus, P. monticola, P. albicaulis, Fagus grandifolia Ehrh. and Ulmus americana L. trees throughout their distribution range. Despite public and institutional awareness of alien forest species, it is expected that their number and impact will keep increasing in the future [1, 2]. In order to implement quarantine and enforce mitigation measures following the introduction of exotic pathogens, national plant protection organizations (NPPOs) such as the Canadian Food and Inspection Agency (CFIA) need rapid, reliable, sensitive and accurate detection methods. The challenge for NPPOs is to be able to detect pathogens at their different life stages, including those that have the capacity to remain latent on asymptomatic tissues. Molecular detection using real-time PCR approaches allows for rapid, reliable and sensitive detection while simultaneously processing of large numbers of samples.
Real-time PCR has become the gold standard in pathogen detection in many fields, e.g. medicine, animal health, agriculture as well as forestry. It is sensitive enough to detect minute amounts of DNA from the target organism mixed with environmental material or host DNA, and the use of hydrolysis probes (TaqMan probes) offers an additional level of specificity, thereby enabling discrimination between closely related species with few polymorphic sites [3]. Real-time PCR also provides accurate quantification of target DNA in processed samples, which is directly proportional to the biomass of the targeted organism. For all these reasons, real-time PCR has been increasingly used to prevent and mitigate the introduction and dispersal of exotic and invasive plant pathogens.
So far, real-time PCR assays of forest pathogens have been mostly developed for single specific pathogens. However, real-time PCR using TaqMan probes offers the opportunity for multiplexing (multiple reactions in one tube [4]) and arraying (multiple reactions in separate tubes but on a single support), allowing for the simultaneous detection of a range of different pathogens in a large number of samples by performing a single real-time PCR run (e.g. [5–7]). Multiplexing usually requires extensive fine-tuning to avoid cross-reactivity and/or loss of sensitivity [8, 9]. This critical step can be circumvented by using arrays of assays operating under the same real-time PCR conditions, but running in as many tubes as there are assays included in the arrays.
The objective of the present study therefore was to develop and validate a set of sensitive, specific and precise real-time PCR assays for the rapid detection of 10 of the most unwanted alien forest fungal pathogens in Canada selected from a list of over 100 tree pathogens regulated by international, continental, and national phytosanitary organizations. Selection was based on the pathogen’s i) history of invasiveness, type and degree of damage/symptoms/pathogenicity, ii) host species and estimated economic impact, iii) dispersal pathways, establishment and adaptability, and iv) likelihood of establishment in Canada. Our goal was to develop assays that could be used either in simplex or in plate-based arrays.
Materials and Methods
Isolates selection
For each target pathogen, we built a panel of isolates encompassing multiple isolates of the target species and isolates from closely related species (sister species). Our selection of sister species was based on phylogenies found in recent scientific peer-reviewed studies [10–15] as well as on the advice of particular taxonomic group specialists. Cultures were obtained from collections (CBS, ATCC, as well as private ones) and stored in replicates at FPInnovations in Vancouver and the CFIA in Ottawa. When available, type cultures from referenced culture collections along with isolates provided by taxonomic authorities were preferred. To capture intraspecific genetic diversity, isolates from different hosts and different geographic origins were used when available. The list of isolates used to design the assays is presented in Table 1.
Table 1. Target and closely related species isolates used in this study.
| Target species | Species | Collection number | Host | Origin | Source a |
|---|---|---|---|---|---|
| Ceratocytis laricicola, C. polonica and C. fagacearum | Ambrosiella ferruginea | CBS 408.68 | - | WI, USA | CBS |
| A. ferruginea | CBS 460.82 | Fagus sylvatica | Germany | CBS | |
| Ceratocystis adiposa | UAMH 6973 | Picea sp. | QC, Canada | UAMH | |
| C. adiposa | UAMH 6974 | Picea sp. | QC, Canada | UAMH | |
| C. albifundus | CBS 128991 | Acacia mearnsii | - | CBS | |
| C. bhutanensis | CMW 8242; CBS 112907 | Picea sp. | Bhutan | M.J. Wingfield | |
| C. cacaofunesta | CBS 115169 | Theobroma cacao | Eucador | CBS | |
| C. cacaofunesta | CBS 152.62 | Theobroma cacao | Costa Rica | CBS | |
| C. caryae | CBS 114716 | Carya cordiformis | IA, USA | CBS | |
| C. caryae | CBS 115168 | Carya ovata | IA, USA | CBS | |
| C. coerulescens | C301 | Pinus banksiana | MN, USA | T.C. Harrington | |
| C. coerulescens | C313; CBS 140.37 | Picea abies | Germany | T. C. Harrington | |
| C. coerulescens | C693 | - | Finland | T.C. Harrington | |
| C. coerulescens | C1423 | Larix kaempferi | Japan | T. C. Harrington | |
| C. coerulescens | CPT9; CL1-2 | - | - | C. Breuil | |
| C. coerulescens | CPT11; CL2-15 | - | - | C. Breuil | |
| C. coerulescens | CPT12; CL2-25 | NA | NA | C. Breuil | |
| C. douglasii | C324; CBS 556.97 | Pseudotsuga menziesii | OR, USA | T. C. Harrington | |
| C. douglasii | C479 | NA | NA | T. C. Harrington | |
| C. eucalypti | CMW 3254 | Eucalyptus sieberi | Australia | M.J. Wingfield | |
| C. fagacearum | C460 | Quercus alba | IA, USA | T.C. Harrington | |
| C. fagacearum | C465 | Quercus macrocarpa | IA, USA | T.C. Harrington | |
| C. fagacearum | C505 | Quercus rubra | MN, USA | T.C. Harrington | |
| C. fagacearum | C520 | Quercus alba | MN, USA | T.C. Harrington | |
| C. fagacearum | C660 | Quercus macrocarpa | IA, USA | T.C. Harrington | |
| C. fagacearum | CMW 2039 | Quercus sp. | MN, USA | M.J. Wingfield | |
| C. fujiensis | CMW 1952 | Larix sp. | Japan | M.J. Wingfield | |
| C. fujiensis | CMW 1965 | Larix sp. | Japan | M.J. Wingfield | |
| C. fujiensis | CMW 1969 | Larix sp. | Japan | M.J. Wingfield | |
| C. laricicola | C181; CBS 100207 | Larix sp. | Scotland | T.C. Harrington | |
| C. laricicola | CMW 3212 | Larix sp. | Scotland | M.J. Wingfield | |
| C. moniliformis | CBS 118243 | Pinus mercusii | Indonesia | CBS | |
| C. norvegica | UAMH 11187 | Picea abies | Norway | UAMH | |
| C. norvegica | UAMH 11190 | Picea abies | Norway | UAMH | |
| C. paradoxa | UAMH 3314 | - | - | UAMH | |
| C. paradoxa | UAMH 8784 | Cocos nucifera | Jamaica | UAMH | |
| C. pinicola | C488; CMW 1311; CBS 100199 | Pinus sylvestris | United Kingdom | T.C. Harrington | |
| C. pinicola | C490; CMW 1323; CBS 100200 | Pinus nigra | United Kingdom | T.C. Harrington | |
| C. pinicola | C795; CBS 100201 | Pinus nigra | United Kingdom | T.C. Harrington | |
| C. platani | CBS 127662 | Platanus orientalis | Greece | CBS | |
| C. platani | CBS 129000 | Platanus sp. | USA | CBS | |
| C. polonica | C320; CBS 228.83 | Picea abies | Norway | T.C. Harrington | |
| C. polonica | CBS 133.38 | - | Poland | CBS | |
| C. polonica | CPT2; NISK 93-208/10 | Picea abies | Norway | C. Breuil | |
| C. polonica | CPT3; NISK 93-208/115; ATCC 201884 | Picea abies | Norway | C. Breuil | |
| C. polonica | CPT4; CBS 100205; CMW 2224 | Picea abies | Norway | C. Breuil | |
| C. polonica | CPT5; CBS 100206 | Picea jezoensis | Japan | C. Breuil | |
| C. polonica | CPT6; CBS 119236 | Picea jezoensis | Japan | C. Breuil | |
| C. radicicola | CMW 3186; CBS 114.47 | Phoenix sp. | CA, USA | M.J. Wingfield | |
| C. resinifera | C50 | Picea engelmannii | NM, USA | T. C. Harrington | |
| C. resinifera | Kasper | - | - | L. Bernier | |
| C. resinifera | PB 632 | Pinus banksiana | NB, Canada | L. Bernier | |
| C. rufipenni | C608; CBS 100209 | Picea engelmannii | BC, Canada | T.C. Harrington | |
| C. rufipenni | C613; 404/2 | Picea glauca | BC, Canada | T.C. Harrington | |
| C. smalleyi | CBS 114724 | Carya cordiformis | WI, USA | CBS | |
| C. variospora | CBS 114714 | Quercus robur | IA, USA | CBS | |
| C. variospora | CBS 114715 | Quercus alba | IA, USA | CBS | |
| C. virescens | CMW 11164 | Fagus americanum | USA | M.J. Wingfield | |
| Thielaviopsis australis | CMW 2333 | Nothofagus cunninghamii | Australia | M.J. Wingfield | |
| T. australis | CMW 2339 | Eucalyptus sp. | Australia | M.J. Wingfield | |
| T. basicola | CMW 7624 | Cichorium sp. | South Africa | M.J. Wingfield | |
| T. basicola | CMW 7625 | Cichorium sp. | South Africa | M.J. Wingfield | |
| Fusarium circinatum | Fusarium anthophilum | CBS 737.97; NRRL 13602 | Hippeastrum sp. | Germany | CBS |
| F. bactridioides | CBS 100057; NRRL 22201 | - | AZ, USA | CBS | |
| F. bulbicola | CBS 220.76; NRRL 13618 | Nerine bowdenii | Netherlands | CBS | |
| F. circinatum | CBS 405.97; NRRL 25331 | Pinus radiata | CA, USA | CBS | |
| F. circinatum | FCC1045; DAOM 238088 | Pinus patula | South Africa | K. Seifert | |
| F. circinatum | FCC2251; DAOM 238089 | Pinus patula | Mexico | K. Seifert | |
| F. circinatum | FCC2253; DAOM 238090 | Pinus greggii | Mexico | K. Seifert | |
| F. circinatum | FCC4869; DAOM 238091 | Pinus patula | USA | K. Seifert | |
| F. circinatum | FCC4873; DAOM 238092 | Pinus patula | USA | K. Seifert | |
| F. circinatum | FCC4874; DAOM 238093 | Pinus patula | USA | K. Seifert | |
| F. circinatum | FCC4878; DAOM 238094 | Pinus patula | USA | K. Seifert | |
| F. circinatum | FCC4880; DAOM 238095 | Pinus patula | South Africa | K. Seifert | |
| F. circinatum | FCC4881; DAOM 238096 | Pinus patula | Mexico | K. Seifert | |
| F. circinatum | FCC4885; DAOM 238097 | Pinus patula | Mexico | K. Seifert | |
| F. circinatum | FCC4913; DAOM 238098 | Pinus leiophylla | Mexico | K. Seifert | |
| F. guttiforme | CBS 409.97; NRRL 25295 | Ananas comosun | Brazil | CBS | |
| F. subglutinans | CBS 215.76; NRRL 20844 | Zea mays | Germany | CBS | |
| F. subglutinans | AAFC-Fcir-012 | - | - | K. Seifert | |
| F. sacchari | AAFC-Fcir-014 | - | - | K. Seifert | |
| F. succisae | AAFC-Fcir-001 | - | - | K. Seifert | |
| F. succisae | AAFC-Fcir-013 | - | - | K. Seifert | |
| Geosmithia morbida | Geosmithia argillacea | CBS 128034 | Xylosandrus mutilatus/Vitus rotundifolia | USA | CBS |
| G. argillacea | CBS 128787 | - | - | - | |
| G. fassatiae | CCF3334 | Quercus pubescens | Czech Republic | Miroslav Kolarik | |
| G. fassatiae | CCF4331 | Pityophthorus sp./ Pinus sabiniana | CA, USA | Miroslav Kolarik | |
| G. fassatiae | CCF4340 | Hylocurus hirtellus/Salix sp. | CA, USA | Miroslav Kolarik | |
| G. flava | CCF3333 | Xiphydria sp. /Castanea sativa | Czech Republic | Miroslav Kolarik | |
| G. flava | CCF4337 | Cerambycidae sp./Pseudotsuga douglasii | CA, USA | Miroslav Kolarik | |
| G. flava | CCF4341 | Cryphalus pubescens/Sequoia serpervirens | CA, USA | Miroslav Kolarik | |
| G. langdonii | CCF4326 | Phloeosinus cupressi/Cyperus groverianus | CA, USA | Miroslav Kolarik | |
| G. lavendula | CCF4336 | Bark beetle/Pinus longaeva | CA, USA | Miroslav Kolarik | |
| G. morbida | 1223 | Pityophthorus juglandis/Juglans nigra | UT, USA | Miroslav Kolarik | |
| G. morbida | 1256 | Pityophthorus juglandis/Juglans nigra | OR, USA | Miroslav Kolarik | |
| G. morbida | 1259 | Pityophthorus juglandis/Juglans nigra | OR, USA | Miroslav Kolarik | |
| G. morbida | 1268 | Pityophthorus juglandis/Juglans nigra | CA, USA | Miroslav Kolarik | |
| G. morbida | 1271 | Pityophthorus juglandis/Juglans nigra | CO, USA | Miroslav Kolarik | |
| G. morbida | 1272 | - | - | Miroslav Kolarik | |
| G. morbida | CCF3879; CBS 124664 | Pityophthorus juglandis/Juglans nigra | CO, USA | Miroslav Kolarik | |
| G. morbida | CCF3880 | Pityophthorus juglandis/Juglans nigra | AZ, USA | Miroslav Kolarik | |
| G. morbida | CCF3881; CBS 124663 | Pityophthorus juglandis/Juglans nigra | CO, USA | Miroslav Kolarik | |
| G. morbida | Gm6 | Juglans sp. | TN, USA | Ðenita Hadžiabdić Guerry | |
| G. morbida | Gm14 | Juglans sp. | TN, USA | Ðenita Hadžiabdić Guerry | |
| G. morbida | Gm19 | Juglans sp. | TN, USA | Ðenita Hadžiabdić Guerry | |
| G. morbida | Gm45 | Juglans sp. | TN, USA | Ðenita Hadžiabdić Guerry | |
| G. morbida | U19 | Pityophthorus juglandis/Juglans hindsii | CA, USA | Miroslav Kolarik | |
| G. obscura | CBS 121749 | - | USA | CBS | |
| G. pallida s.s. | CCF4279 | Platypus janosoni/Gymnacranthera paniculata | Papua New Guinea | Miroslav Kolarik | |
| G. pallida sp. 1 | MK1790 | Hypoborus ficus/Ficus carica | Azerbaijan | Miroslav Kolarik | |
| G. pallida sp. 2 | CCF4315 | Scolytus rugulosus, Pseudothysanoes hopkinsi/Prunus sp. | CA, USA | Miroslav Kolarik | |
| G. pallida sp. 5 | CCF4271 | Scolytus multistriatus/Ulmus laevis | Czech Republic | Miroslav Kolarik | |
| G. pallida sp. 23 | CCF3639 | Scolytus rugulosus/Prunus armeniaca | Turkey | Miroslav Kolarik | |
| G. pallida sp. MK1807 | MK1807 | Scolytid beetle/Acacia smithii | Australia | Miroslav Kolarik | |
| G. putterillii | CBS248.32 | Soil | Netherlands | CBS | |
| G. putterillii | CCF3342 | Scolytus rugulosus/Prunus sp. | Czech Republic | Miroslav Kolarik | |
| G. putterillii | CCF3442 | Liparthrum colchicum/Laurus nobilis | France | Miroslav Kolarik | |
| G. putterillii | CCF4204 | Bostrichid beetle/Umbellularia californica | CA, USA | Miroslav Kolarik | |
| G. rufescens | MK1821 | Cnesinus lecontei/Croton draco | Costa Rica | Miroslav Kolarik | |
| G. sp. 8 | CCF4277 | Scolytus intricatus/Quercus cerris | Bulgaria | Miroslav Kolarik | |
| G. sp. 9 | RJ0258 | Ips cembrae/Larix decidua | Poland | Miroslav Kolarik | |
| G. sp. 10 | CCF4282 | Hypoborus ficus/Ficus carica | Turkey | Miroslav Kolarik | |
| G. sp. 11 | CCF3555 | Scolytus intricatus/Quercus pubescens | Hungary | Miroslav Kolarik | |
| G. sp. 12 | CCF4320 | Hylesinus oregonus/Fraxinus sp. | CO, USA | Miroslav Kolarik | |
| G. sp. 13 | CCF3559 | Pteleobius vittatus/Ulmus minor | Czech Republic | Miroslav Kolarik | |
| G. sp. 16 | CCF4201 | Pityophthorus pityographus/Picea abies | Poland | Miroslav Kolarik | |
| G. sp. 16-like | CCF4322 | Pityophthorus sp., Scolytus oregoni, Cryphalus pubescens/Pseudotsuga douglasii | CO, USA | Miroslav Kolarik | |
| G. sp. 20 | CCF3641 | Hypoborus ficus/Ficus carica | France | Miroslav Kolarik | |
| G. sp. 20 | CCF4303 | Hypoborus ficus/Ficus carica | Syria | Miroslav Kolarik | |
| G. sp. 20 | CCF4316 | Ips plastographus/Calocedrus decurrens | CA, USA | Miroslav Kolarik | |
| G. sp. 20 | MK764 | Phloetribus scarabeoides /Olea europea | Syria | Miroslav Kolarik | |
| G. sp. 21 | CCF4321 | Pityophthorus sp./Pinus ponderosae | CO, USA | Miroslav Kolarik | |
| G. sp. 21 | CCF4334 | Phloesinus sp./Cyperus occidentalis var. australis | CA, USA | Miroslav Kolarik | |
| G. sp. 21 | MK1665 | Hypoborus ficus/Ficus carica | Spain | Miroslav Kolarik | |
| G. sp. 22 | CCF3645 | Phloetribus scarabeoides scarabeoides/Olea europea | Jordan | Miroslav Kolarik | |
| G. sp. 26 | CCF4330 | Nark beetle/Pinus monophylla | CA, USA | Miroslav Kolarik | |
| G. sp. 27 | CCF4206 | Pityogenes bidentatus/Pinus silvestris | Poland | Miroslav Kolarik | |
| G. sp. 29 | CCF4199 | Cryphalus piceae + Pityophthorus pityographus/Abies alba | Czech Republic | Miroslav Kolarik | |
| G. sp. 29 | CCF4221 | Cryphalus piceae + Pityophthorus pityographus/Abies alba | Czech Republic | Miroslav Kolarik | |
| G. sp. 30 | CCF4220 | Pityogenes chalcographus/Picea abies | Poland | Miroslav Kolarik | |
| G. sp. 31 | CCF4328 | Bark beetle/Pinus muricata | CA, USA | Miroslav Kolarik | |
| G. sp. 31 | RJ21k | Pityophthorus pityographus/Pinus sylivestris | Poland | Miroslav Kolarik | |
| G. sp. 35 | CCF4205 | Cryphalus piceae + Pityophthorus pityographus/Abies alba | Czech Republic | Miroslav Kolarik | |
| G. sp. MK1820 | CCF4292 | Cnesinus lecontei/Croton draco | Costa Rica | Miroslav Kolarik | |
| G. sp. U410 | CCF4324 | Pityophthorus sp./Pinus sabineana | CA, USA | Miroslav Kolarik | |
| G. sp. U410 | CCF4332 | Pityophthorus sp./Pinus sabineana | CA, USA | Miroslav Kolarik | |
| G. viridis | CBS 252.87 | - | Australia | CBS | |
| Gremmeniella abietina var. abietina (EU race) | Gremmeniella abietina var. abietina (EU race) | DAOM170389; ATCC34574; SN-2; 66.163/2 | Picea abies | Norway | DAOM |
| G. abietina var. abietina (EU race) | DAOM170402;SUS-9; 11-38D | Pinus resinosa | NY, USA | DAOM | |
| G. abietina var. abietina (EU race) | 83–043 | Pinus resinosa | QC, Canada | G. Laflamme | |
| G. abietina var. abietina (EU race) | DAOM170406; SW-2; ETH-7264 | Pinus cembrae | Switzerland | DAOM | |
| G. abietina var. abietina (EU race) | DAOM170407; SW-3; ETH-7269 | Pinus cembrae | Switzerland | DAOM | |
| G. abietina var. abietina (EU race) | DAOM170408; SW-4; ETH-7266 | Pinus cembrae | Switzerland | DAOM | |
| G. abietina var. abietina (EU race) | Oulanka | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Hedmark P.C.1.4 | - | Norway | M. Vuorinen | |
| G. abietina var. abietina (EU race) | Kai 1.5 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Hu 1.2X1.8 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Toro 2.8X1-A1.8 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Hedmark P.C.1.3 | - | Norway | M. Vuorinen | |
| G. abietina var. abietina (EU race) | YN 1.4 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Sup 1.2 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Sup 1.4 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | SIU 1.3 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Sup 1.6 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Sup 1.8 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Kai 1.7 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | KanKaan | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Kai 1.8X1.8 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Toro 2.6 X Sup 1.6 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Muistomä | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | SUO 2.1 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Sup1.1 X Sup 1.8 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Orivesi | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Kai 1.2 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Toro 2.7 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Pat 1.7 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Sup 1.7 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Sup 1.3 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Viheriäis | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Kai 1.8 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Hyytiälä | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Ahvenlampi | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Kai 1.3 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | MH 1.3 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (EU race) | Kai 1.6 | Pinus sylvestris | Finland | A. Uotila & J. Kaitera | |
| G. abietina var. abietina (NA race) | DAOM170372; SC-39; HF-1 | Pinus resinosa | - | DAOM | |
| G. abietina var. abietina (NA race) | DAOM170367; SC-25; WP-104 | Pinus strobus | Canada | DAOM | |
| G. abietina var. abietina (Asian race) | Asia5.1 | Abies sachalinensis | Japan | L. Bernier | |
| G. abietina var. balsamea | 84–301 | Abies balsamea | QC, Canada | G. Laflamme | |
| G. laricina | 81–857 | Larix laricina | QC, Canada | G. Laflamme | |
| Rosellinia necatrix | Rosellinia abscondita | CBS 450.89 | Driftwood | Switzerland | CBS |
| R. abscondita | CBS 447.89 | Alnus incana | Switzerland | CBS | |
| R. aquila | CBS 399.61 | - | South Africa | CBS | |
| R. britannica | CBS 446.89 | - | France | CBS | |
| R. limoniispora | CBS 382.86 | Triticum aestivum | Switzerland | CBS | |
| R. limoniispora | CBS 283.64 | - | - | CBS | |
| R. necatrix | CBS 349.36 | Malus sylvestris | Argentina | CBS | |
| R. necatrix | CBS 267.30 | Narcissus pseudonarcissus | Netherlands | CBS | |
| R. nectrioides | CBS 449.89 | - | Sweden | CBS | |
| R. thelena | CBS 400.61 | - | CA, USA | CBS | |
| Entoleuca mammata | CFL-2629 | Populus tremuloides | QC, Canada | - | |
| Sclerotinia pseudotuberosa (syn. Ciboria batschiana) | Botrytis cinerea | CBS 131.28 | Linum usitatissimum | Netherlands | CBS |
| B. cinerea | DAOM 166439 | - | - | DAOM | |
| B. cinerea | DAOM 192631 | - | - | DAOM | |
| B. cinerea | DAOM 193576 | - | - | DAOM | |
| B. cinerea | DAOM231368 | - | - | DAOM | |
| B. cinerea | DAOM231371 | - | - | DAOM | |
| B. cinerea | DAOM231372 | - | - | DAOM | |
| Ciboria americana | CBS 117.24 | Castanea sativa | - | CBS | |
| Pycnopeziza sympodialis | CBS 141.83 | Arctostaphylos uva-ursi | Switzerland | CBS | |
| P. sympodialis | CBS 332.39 | - | USA | CBS | |
| Sclerotinia bulborum | CBS 297.31 | - | USA | CBS | |
| S. minor | CBS 339.39 | Lactuca sativa | Italy | CBS | |
| S. minor | DAOM 191806 | - | - | DAOM | |
| S. pseudotuberosa | CBS 312.37 | Quercus sp. | Netherlands | CBS | |
| S. pseudotuberosa | CBS 327.75 | Quercus peduculata | France | CBS | |
| S. pseudotuberosa | CBS 331.35 | - | Italy | CBS | |
| S. pseudotuberosa | CBS 655.78 | Quercus robur | Netherlands | CBS | |
| S. sclerotiorum | CBS 499.50 | - | Netherlands | CBS | |
| S. sclerotiorum | DAOM 180751 | - | - | DAOM | |
| S. sclerotiorum | DAOM 241671 | - | - | DAOM | |
| S. trifoliorum | CBS 171.24 | Trifolium incarnatum | - | CBS | |
| Phytophthora kernoviae and P. ramorum | Phytophthora boehmeriae | CBS 100410 | - | Australia | CBS |
| P. boehmeriae | CBS 291.29; IMI180614 | Boehmeria nivea | Taiwan | CBS | |
| P. brassicae | CBS 179.87 | Brassica oleraceae | Netherlands | CBS | |
| P. brassicae | P10414; CBS113350 | Brassica oleraceae | Netherlands | CBS | |
| P. captiosa | CBS 119107 | Eucalyptus saligna | New Zealand | CBS | |
| P. cryptogea | CBS 113.19 | Lycopersicon esculentum | Ireland | CBS | |
| P. cryptogea | CBS 418.71 | Gerbera sp. | Netehrlands | CBS | |
| P. cryptogea | P1088; ATCC 46721; CBS 290.35; CBS 130866 | Aster sp. | USA | CBS | |
| P. drechsleri | CBS 292.35 | Beta vulgaris var. altissima | CA, USA | CBS | |
| P. erythroseptica | Br 664 | - | - | G. J. Bilodeau | |
| P. erythroseptica | DAOM 233917 | - | - | G. J. Bilodeau | |
| P. fallax | CBS 119109 | Eucalyptus delegatensis | New Zealand | CBS | |
| P. foliorum | CBS 121665; ATCC MYA-3638; CMW 31064 | Azalea | TN, USA | M. J. Wingfield | |
| P. gallica | CBS 117475 | - | Germany | CBS | |
| P. hibernalis | 1341320–3 | - | CA, USA | G. J. Bilodeau | |
| P. hibernalis | P3822; ATCC 56353; CBS 114104; IMI1 34760 | Citrus sinensis | Australia | CBS | |
| P. insolita | P6195; ATCC 56964; CBS 691.79; IMI 288805 | - | Taiwan | CBS | |
| P. kernoviae | CBS 122049; CMW 31066; PD 06/3121107 | Rododendron sp. | United Kingdom | CBS | |
| P. kernoviae | CBS 122208; CMW 31065; PD 0502010595 | Rhododendron ponticum | United Kingdom | CBS | |
| P. lateralis | CBS 102608 | - | CA, USA | G. J. Bilodeau | |
| P. lateralis | CBS 117106 | Chamaecyparis lawsoniana | Netherlands | G. J. Bilodeau | |
| P. lateralis | CBS 168.42 | Chamaecyparis lawsoniana | OR, USA | G. J. Bilodeau | |
| P. lateralis | Hansen 366 | Chamaecyparis lawsoniana | USA | G. J. Bilodeau | |
| P. lateralis | Hansen 368 | Chamaecyparis lawsoniana | USA | G. J. Bilodeau | |
| P. morindae | CBS 121982 | Morinda citrifolia | HI, USA | CBS | |
| P. porri | CBS 114101 | Parthenium argentatum | Australia | CBS | |
| P. primulae | CBS 114346 | Primula polyantha | New Zealand | CBS | |
| P. primulae | P10333; CBS 620.97 | Primula acaulis | Germany | CBS | |
| P. quininea | CBS 407.48 | Cinchona officinalis | Peru | CBS | |
| P. ramorum (EU1) | 03–0107 | Rhododendron sp. | Canada | G. J. Bilodeau | |
| P. ramorum (NA1) | 04–0002 | Camellia sp. | Canada | G. J. Bilodeau | |
| P. ramorum (NA2) | 04–0437 | Pyracantha koidzumii "Victory" | Canada | G. J. Bilodeau | |
| P. ramorum (NA2) | 10–3892 | Rhododendron sp. | Canada | G. J. Bilodeau | |
| P. ramorum (EU1) | BBA 14-98-a; CBS 101550 | Rhododendron catawbienses | Germany | G. J. Bilodeau | |
| P. ramorum (EU1) | BBA 9/95 | Rhododendron catawbienses | Germany | G. J. Bilodeau | |
| P. ramorum (EU1) | CBS 101553 | Rhododendron catawbienses | Germany | CBS | |
| P. ramorum (EU1) | P10301; CBS 101329 | Rhododendron sp. | Netherlands | CBS | |
| P. ramorum (NA1) | Pr 52; CBS 110537 | Rhododendron sp. | CA, USA | G. J. Bilodeau | |
| P. ramorum (NA2) | Pr1270626-1 | Peiris japonica | CA, USA | G. J. Bilodeau | |
| P. richardiae | CBS 240.30 | Zantedeschia aethiopica | USA | CBS | |
| P. sp. "sansomea" | CBS 117693 | Glycine max | Ireland | CBS | |
| P. sp. "sansomea" | P3163; CBS117692 | Silene latifolia subsp. alba | USA | CBS | |
| P. syringae | CBS 114107 | Prunus dulcis | CA, USA | CBS | |
| P. syringae | P10330; CBS110161 | Rhododendron sp. | Germany | CBS | |
| P. trifolii | CBS 117687 | Trofolium sp. | MS, USA | CBS |
a CBS: The Centraalbureau voor Schimmelcultures collection; DAOM: Agriculture and Agri-Food Canada Fungal collection; UAMH: University of Alberta Microfungus Collection and Herbarium
DNA extraction
For all isolates (except for Ceratocystis species), DNA was extracted using the Qiagen’s DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Ceratocystis DNA was extracted using a modified version of Zolan and Pukkila’s phenol/chloroform extraction protocol [16]. A small piece of mycelium was homogenized in 400 μl of extraction buffer (100mM Tris-HCl pH 9.5, 1.4 M NaCl, 20 mM EDTA, 2% CTAB, 1% PEG 8000, and 0.25% β-mercaptoethanol). Samples were then incubated at 65°C for 1h (vortexing every 15 minutes). Next, 400 μl of phenol:chloroform:isoamyl alcohol (25:24:1) were added to the homogenate, vortexed for 10 s and centrifuged at 13,000 x g for 10 min. Supernatant was mixed by inversion with 70 μl of 7.5M ammonium acetate and 600 μl of ice-cold isopropanol, incubated at -20°C for a minimum of 1h, and then centrifuged at 13,000 x g for 20 min (4°C). DNA was rinsed with 800 μl ice-cold 70% ethanol and centrifuged at 13,000 x g for 5 min (4°C). DNA was then incubated at 55°C to evaporate any remaining ethanol and re-suspended in 50 μl 10 mM Tris-HCl, pH 8.0. DNA was visualized on agarose gel stained with ethidium bromide, and DNA concentration was measured using the Qubit 2.0 Fluorometer with the dsDNA BR Assay Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions.
DNA sequencing and phylogenetic analyses
The internal transcribed spacer (ITS) gene, recognized as the universal DNA barcode for fungi [17], was systematically amplified and sequenced for all isolates. NCBI nucleotide blast of the ITS sequences was performed to detect misidentification and potential contamination of isolates. A list of the different gene regions sequenced along with the primers used is presented in Table 2. PCR reactions were performed in a final volume of 25 μl and contained 1X PCR buffer, 1.5 mM MgCl2, 200 μM of each dNTP (Invitrogen), 0.4 μM of each primer (Integrated DNA Technologies Inc., Coralville, IA, USA), 1 U of Platinum Taq DNA polymerase (Invitrogen), and 1 μl of template DNA. Sequencing of both DNA strands was performed by the Centre de recherche du Centre Hospitalier Universitaire de Québec (CHUQ) sequencing platform on an ABI 3730xl (Applied Biosystems, Foster City, CA, USA) using the specific forward and reverse primers.
Table 2. Primers used for DNA sequencing and genus general assays.
| NCBI Accession Number | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target gene | Primer name | Amplicon length (bp) | Sequence (5’→ 3’) | Reference | Ceratocystis | Fusarium | Geosmithia | Gremminiella | Rosellinia | Sclerotinia |
| DNA sequencing | ||||||||||
| ITS | ITS1F | ~ 600 | CTTGGTCATTTAGAGGAAGTAA | [70] | KC305097- KC305166 | KC464615-KC464634 | KF808295-KF808322 | KC352952-KC352997 | KF719196-KF719202 | KF859918-KF859936 |
| ITS4 | TCCTCCGCTTATTGATATGC | [71] | ||||||||
| β-tubulin | T10 | ~1300 | ACGATAGGTTCACCTCCAGAC | [72] | KC335975-KC336019; | |||||
| BT12 | GTTGTCAATGCAGAAGGTCTC | [72] | KC589388-KC589393 | |||||||
| EF1 | EF1F | ~ 900 | TGCGGTGGTATCGACAAGCGT | [73] | KC405262-KC405285; | |||||
| EF2R | AGCATGTTGTCGCCGTTGAAG | [73] | KC583303-KC583321 | |||||||
| Tsr1 | Tsr1_1453for | ~ 900 | CCIGAYGARATYGARCTICAYCC | [74] | KC405286-KC405319; | |||||
| Tsr1_2308rev | CTTRAARTAICCRTGIGTICC | [74] | KC590615-KC590632 | |||||||
| IGS | RU46.67 | ~ 900 | GTGTCGGCGTGCTTGTATT | [75] | KC147546-KC147564 | |||||
| CNS12 | GCACGCCAGGACTGCCTCGT | [75] | ||||||||
| TEF | Ef1 | ~ 675 | ATGGGTAAGGARGACAAGAC | [76] | KC514053-KC514067 | |||||
| Ef2 | GGARGTACCAGTSATCATGTT | [76] | ||||||||
| β-tubulin | T1 | ~ 850 | AACATGCGTGAGATTGTAAGT | [77] | KF853893-KF853956 | |||||
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC | [78] | ||||||||
| RPB2 | RPB2F5 | ~ 550 | CTATACTATCCCCAGAAGCCTCTTGCTACC | This study | KC533095-KC533140 | |||||
| RPB2R2 | CAATNGTWCCCTTYTGHCCGTGACG | This study | ||||||||
| LSU | LR0R | ~ 875 | ACCCGCTGAACTTAAGC | [79] | KF719203-KF719215 | |||||
| LR5 | TCCTGAGGGAAACTTCG | [79] | ||||||||
| Calmodulin | CAL_228F | ~ 500 | GAGTTCAAGGAGGCCTTCTCCC | [80] | KF871364-KF871386 | |||||
| CAL_737R | CATCTTTCTGGCCATCATGG | [80] | ||||||||
| G3PDH | G3PDH-Fbis | ~ 850 | GCTGTCAACGACCCTTTCAT | [81] | KF878354-KF878375 | |||||
| G3PDH-Rbis | ACCAGGAAACCAACTTGACG | [81] | ||||||||
| HSP60 | HSP60for-deg | ~ 975 | CAACAATTGAGATTYGCCCAYAAG | [81] | KF871387-KF871408 | |||||
| HSP60rev-deg | GATRGATCCAGTGGTACCGAGCAT | [81] | ||||||||
| Genus general assay | ||||||||||
| EF1 | Cerato_GEN_F510 | 166 | CGTGCTCGCCGGAAATAG | This study | ||||||
| Cerato_GEN_R612 | TGCCGCCTTTTGGTGC | This study | ||||||||
| IGS | Fus_GEN_F68 | 119 | GCCACCAAACCACAAAACC | This study | ||||||
| Fus_GEN_R186 | CCCACAGACCTCGCAC | This study | ||||||||
| β-tubulin | Geos_GEN_F479 | 168 | GTAGACGCTCATGCGCTC | This study | ||||||
| Geos_GEN_R646 | GTAACCAGATCGGTGCTGC | This study | ||||||||
| RPB2 | Gremm_GEN_F304 | 128 | CCAATCTGTGGAATCTTCGTGG | This study | ||||||
| Gremm_GEN_R431 | CGGGATGCTTCAACTCCTC | This study | ||||||||
| LSU | Rosel_GEN_F771 | 190 | CTACTCGACTCGTCGAAGGAG | This study | ||||||
| Rosel_GEN_R960 | GCGAGTGAAGCGGCAACAG | This study | ||||||||
| Hsp60 | Sclero_GEN_F193 | 178 | CTCCCCAAAGATCACCAAAGGTT | This study | ||||||
| Sclero_GEN_R371 | GGCAACATCTTGAATAAGTCTAGCACC | This study | ||||||||
| β-tubulin | Phyto_GEN_F736 | 80 | GGCTCGCAGCAGTACC | This study | ||||||
| Phyto_GEN_R815 | GCGGCGCACATCATGTTCT | This study | ||||||||
Alignments were used to guide the development of assays and were performed with the ClustalW algorithm implemented in BioEdit v7.1.3.0 [18]. Evolutionary relationships between targets and their sister species were inferred from DNA sequences of ITS. Phylogenetic trees were reconstructed by using the maximum likelihood method with the Tamura-Nei model implemented in MEGA5 [19]. Statistical support of nodes was assessed by performing 500 bootstrap replicates.
For the Phytophthora ramorum and Phytophthora kernoviae targets, we did not perform the gene region sequencing and phylogenetic analyses described above. Instead, the detection assays for these two species were designed in genes unique to these target species that were identified by using a comparative genomics approach developed in the TAIGA project (http://taigaforesthealth.com/Home.aspx) (S1 File).
SYBRGreen-based real-time PCR quantification for standardization of isolates’ DNA concentration
DNA concentration of all isolates was standardized following a qPCR quantification using genus general primers. To do so, we quantified the number of target gene copies that were initially present (before any PCR amplification) in the sample, which directly relates to the abundance of the pathogen prior to DNA extraction. This quantification allowed us to work with samples having a standardized DNA concentration for specificity validation. It assured us we had DNA in high enough concentration in all samples to confirm assay discrimination against closely related species.
Genus general primers were designed using Oligo Explorer v1.2 and Oligo Analyzer v1.2 (Gene Link, NY, USA) in a conserved gene region for all closely related species. The following criteria were also used to guide primer design: 1) length between 18 and 25 bp; 2) melting temperature (Tm) close to 60°C (using the nearest neighbor algorithm); 3) absence of polymorphism within targeted species; and 4) minimal secondary structure (especially dimer formation at the 3’ end). Primer pairs were designed such that PCR products were shorter than 200 bp (Table 2). Real-time PCR was performed with an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). All reactions were performed in a final volume of 10 μl and contained 1X QuantiTect SYBR Green PCR Master Mix (Qiagen, Valencia, CA, USA), 0.5 μM of each of the genus general primers (Table 2), and 1 μl of template DNA. Real-time PCR thermocycling conditions were set at 95°C for 15 min, followed by 50 cycles at 95°C for 15 s, 58°C (primer Tm-2°C) for 30 s, and 65°C for 90 s. Fluorescence was read at the end of the extension step.
Gene copy number quantification was then performed using a Java program based on linear regression of efficiency [20] and sample DNA concentration was adjusted to 5,000 gene copies per μl, whenever possible.
Target-specific TaqMan-based real-time PCR assays
All the molecular detection assays targeting prioritized tree pathogens are based on the TaqMan technology. The following strategies were used to design all of our detection assays. Based on the sequences recovered, we targeted the gene that allowed for the best discrimination at the species level, i.e. the gene that maximized the number of single nucleotide polymorphisms (SNPs) between species while keeping a low level of intraspecific variability. Primer and probe designs were performed using Oligo Explorer v1.2 and Oligo Analyzer v1.2. Each set of primer pair and probe was designed so that there was minimal secondary structure (especially dimer formation at the 3’ end) and amplicon length did not exceed 350 base pairs (Table 3). Primers and probes were also designed to ascertain that interspecific SNPs were preferentially localized at the 3’ end of the primers for maximum discrimination effect of the primer-template annealing [21]. The real-time PCR master mix used, QuantiTect Multiplex PCR NoROX Master Mix (Qiagen), possesses features that allow for the use of short oligonucleotides when necessary. By allowing the design of shorter primers and probes, these elements increase the SNP specificity of the primer and probe. All probes were labelled with fluorescein (6-FAM) at the 5’ end and with the quencher Iowa Black FQ (ZEN-IBFQ). All primers and TaqMan probes were manufactured by Integrated DNA Technologies Inc. All assays were designed to work under the same thermocycling conditions, offering the opportunity to array them into 96- or 384-well plates machine formats, based on the user’s needs.
Table 3. Primers used for the 10 tree pathogen species-specific TaqMan assays.
| Name | Target gene | Primer/Probe | Sequence (5’→ 3’) | Amplicon length (bp) |
|---|---|---|---|---|
| Ceratocystis laricicola | ||||
| Claricicola_F451 | β-tubulin | Forward | GCCCGCATCATGTTT | 88 |
| Claricicola_R538 | Reverse | GACGCTTGAGCGG | ||
| Claricicola_T505RC | Probe | 6-Fam/TGTGCCTGC/ZEN/TCTGATTCAT/3IABkFQ | ||
| Ceratocystis polonica | ||||
| Cpolonica_F527 | β-tubulin | Forward | CGTCCACGCCACAAT | 235 |
| Cpolonica_R761 | Reverse | CCTGAACACCAATTATGTTATATC | ||
| Cpolonica_T575 | Probe | 6-Fam/TGTATGATG/ZEN/AGACTAGACGATGC/3IABkFQ | ||
| Ceratocystis fagacearum | ||||
| Cfagacearum_F315 | EF1 | Forward | GTCTGTAGAAGGGGG | 92 |
| Cfagacearum_R406 | Reverse | CTCCATTCTTTACTACAACC | ||
| Cfagacearum_T357 | Probe | 6-Fam/AGAAGTAAC/ZEN/TGGACAACCGTCT/3IABkFQ | ||
| Fusarium circinatum | ||||
| Fcircinatum_F656 | IGS | Forward | CTATACAGCTTACATAATCATAC | 119 |
| Fcircinatum_R775 | Reverse | AGGGTAGGCTTGGAT | ||
| Fcircinatum_T717 | Probe | 6-Fam/TGTCCCTTC/ZEN/TCGAGCCA/3IABkFQ | ||
| Geosmithia morbida | ||||
| Gmorbida_F677 | β-tubulin | Forward | AGTCAGTGTTCTGACC | 202 |
| Gmorbida_R878 | Reverse | GAAGAAGAATAGGACGG | ||
| Gmorbida_T738 | Probe | 6-Fam/AATAGGCTG/ZEN/GACAGGAAGA/3IABkFQ | ||
| Gremmeniella abietina (EU race) | ||||
| Gabietina_F2b | RPB2 | Forward | GGCGCGGTCTTC | 216 |
| Gabietina_R4 | Reverse | GTATCGATCGTGGTCTA | ||
| Gabietina_T3 | Probe | 6-Fam/AATGATGTC/ZEN/CTCTCCAGATAC/3IABkFQ | ||
| Rosellinia necatrix | ||||
| Rnecatrix_F517 | ITS | Forward | GGTAGGGCACTTC | 102 |
| Rnecatrix_R618 | Reverse | GGGATCATTAAAGAGTTCTA | ||
| Rnecatrix_T551 | Probe | 6-Fam/AGGCAACGCGTGGTAT/3IABkFQ | ||
| Sclerotinia pseudotuberosa | ||||
| Spseudotuberosa_F218 | Hsp60 | Forward | TTGTAGAACTCCTAGTCGTA | 129 |
| Spseudotuberosa_R347 | Reverse | ACCGAGATTCTCGAATTTGTCTTTA | ||
| Spseudotuberosa_T269 | Probe | 6-Fam/ATCTCTAAT/ZEN/TGTTGTCGAACAGATGGT/3IABkFQ | ||
| Phytophthora ramorum | ||||
| Pram-C62-F | Cluster62 a | Forward | AACATGCTCGTGCTCAAGTG | 116 |
| Pram-C62-R | Reverse | CGGTGTTCTGGCGTTCTAGT | ||
| Pram-C62-P | Probe | 6-Fam/CAAGGGGAC/ZEN/CGGAACCGTAT/3IABKFQ | ||
| Phytophthora kernoviae | a | |||
| Pkernoviae_F97 | Cluster97 | Forward | GGACTGTGCAGCGCCTAT | 112 |
| Pkernoviae_R97 | Reverse | TCATCACCCCATTTCTTGC | ||
| Pkernoviae_T97 | Probe | 6-Fam/TGCCTCACC/ZEN/ACCAGATGG/3IABKFQ | ||
| Plant DNA extraction control | ||||
| PLCOIF57-74 | Cytochrome oxidase | Forward | TAAACATATGATGAGCCC | 184 |
| PLCOIR223-240 | Reverse | AGCATCTCTTTTGGTTCT | ||
| PLCOIT98-120 | Probe | 6-Fam/ATACTGATCATGGCATAAACCAT/3IABKFQ | ||
| Insect DNA extraction control | ||||
| InsectF1418 | 28S rRNA gene | Forward | CCAAGGAGTCTAGCAT | 264 |
| InsectR1681 | Reverse | GGTCCCAGCGTGT | ||
| InsectT1595 | Probe | 6-Fam/TTCCCGGGGCGTCTC/3IABKFQ |
a P. ramorum Cluster62 and P. kernoviae Cluster97 are both hypothetical proteins without any known function so far.
The validation principles and parameters followed the terminology and concepts described in Charlton (2000) [22] and Ederveen (2010) [23].
Validating the specificity of the tree pathogen TaqMan assays
Specificity validation of all the assays was performed using the panels of isolates presented in Table 1 and Fig 1. For target species belonging to same genera (Ceratocystis and Phytophthora), we used the whole genera panel to evaluate each of the assay’s specificity. Real-time PCR amplification was conducted using 1X QuantiTect Multiplex PCR NoROX Master Mix, with 0.6 μM of each primer, 0.1 μM of TaqMan probe, and 5,000 gene copies of template DNA, whenever possible, in a final reaction volume of 10 μl. Two technical replicates were performed for all reactions using an Applied Biosystems 7500 Fast Real-Time PCR System. Real-time PCR thermocycling conditions were set at 95°C for 15 min, followed by 50 cycles at 95°C for 15 s and 60°C for 90 s. Fluorescence was read at each cycle, at the end of the extension step.
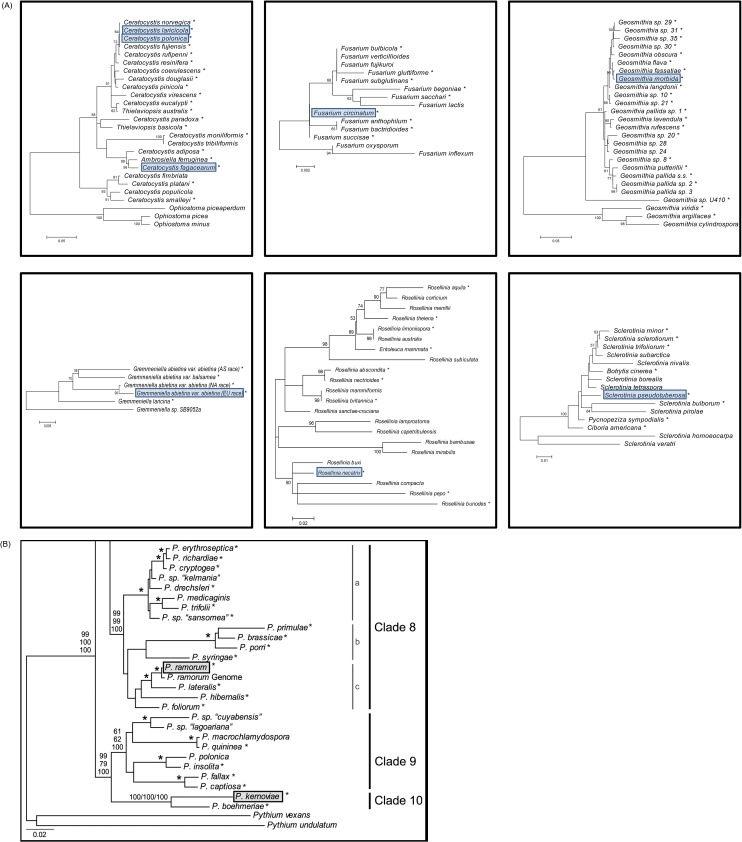
Fig 1. Phylogenetic trees of each genus, including target and closely related species.
For each tree, the target species is (are) shaded. Species followed by an asterisk (*) were used to perform specificity validation. (A) Maximum likelihood phylogenetic trees using internal transcribed spacer (ITS) sequences. (B) Maximum likelihood phylogenetic tree of Phytophthora clades 8–10 using seven nuclear loci (from Blair et al. [15]).
Validating the sensitivity of the tree pathogen TaqMan assays
Sensitivity of the TaqMan assays was evaluated in terms of both efficiency and limit of detection (LOD). For each target assay, experiments were conducted to 1) determine if Ct values were proportional to the amount of target template DNA (efficiency) and 2) evaluate the LOD, which is the smallest amount of target DNA that can be detected for each of the assays. One isolate for each of the target species was selected, and TaqMan assay sensitivity was assessed on parallel sets of serial dilutions from the DNA stock.
To assess efficiency of the amplification reaction, TaqMan assays were run with serial dilutions of template DNA from the target species, ranging from 1 to 15,000 copies of the target gene region, as quantified using the species-specific primers. Standard curves were obtained by plotting the values of Ct against the log value of the target gene region copy number. Amplification reaction efficiency was calculated using the following formula:
where E represents the amplification reaction efficiency and slope is the slope value of the line derived from the standard curve plot. Estimation of the LOD was done by performing 20 replicates of the TaqMan real-time PCR reactions for each of the following DNA concentrations: 1, 3, 5, and 10 copies per μL. The lowest DNA concentration with a level of 95% successful amplification was identified as the LOD.
Validating the precision of the tree pathogen TaqMan assays
Precision (or repeatability) of the assays refers to the robustness of the assay with the same samples repeatedly analyzed in the same manner [24]. Ct values from different real-time PCR runs on different isolates of target species, assessed with a standardized concentration of 5,000 gene copies, were compiled and used to determine the precision of the assays. For each assay, mean Ct value, standard deviation and coefficient of variation were calculated.
Validating the tree pathogen TaqMan assays on environmental samples
The complete list of all environmental samples, including the source, is presented in Table 4. Because of the phytosanitary risks of infected material, environmental samples were supplied by collaborators as purified DNA samples. Since the objective was to test the assays’ performance in a variety of different conditions, collaborators were free to use the routine DNA extraction protocols implemented in their respective laboratories instead of a unique standardized DNA extraction protocol. The efficiency of the DNA extraction was assessed for each sample by performing a control real-time TaqMan PCR reaction that targeted either the plant cytochrome oxidase gene (for primers and probes sequences, see Table 3). All reactions were performed in a final volume of 10 μL and contained 1X QuantiTect Multiplex PCR NoROX Master Mix, with 0.6 μM of each primer, 0.1 μM of TaqMan probe, and 1 μL of template DNA. Real-time PCR thermocycling conditions were set at 95°C for 15 min, followed by 50 cycles at 95°C for 15 s and 60°C for 90 s. Fluorescence was read at the end of each cycle.
Table 4. Description of environmental samples.
| Isolate | Type of material a | Host | Location | Year of collection | Collector/Provider |
|---|---|---|---|---|---|
| Ceratocystis laricicola b | |||||
| CEM5 | Juvenile or adult Ips cembrae collected from galleries | European larch (Larix decidua) | Austria | 2010 | T. Kirisits |
| CEM8 | Juvenile or adult Ips cembrae collected from galleries | European larch (Larix decidua) | Austria | 2010 | T. Kirisits |
| CEM10 | Juvenile or adult Ips cembrae collected from galleries | European larch (Larix decidua) | Austria | 2010 | T. Kirisits |
| CEM11 | Juvenile or adult Ips cembrae collected from galleries | European larch (Larix decidua) | Austria | 2010 | T. Kirisits |
| CEM13 | Juvenile or adult Ips cembrae collected from galleries | European larch (Larix decidua) | Austria | 2010 | T. Kirisits |
| CEM19 | Juvenile or adult Ips cembrae collected from galleries | European larch (Larix decidua) | Austria | 2010 | T. Kirisits |
| CEM25 | Juvenile or adult Ips cembrae collected from galleries | European larch (Larix decidua) | Austria | 2010 | T. Kirisits |
| Ceratocystis polonica b | |||||
| TYP1 | Adult Ips typographus collected from galleries | Norway spruce (Picea abies) | Austria | 2010 | T. Kirisits |
| TYP2 | Adult Ips typographus collected from galleries | Norway spruce (Picea abies) | Austria | 2010 | T. Kirisits |
| TYP3 | Adult Ips typographus collected from galleries | Norway spruce (Picea abies) | Austria | 2010 | T. Kirisits |
| TYP11 | Adult Ips typographus collected from galleries | Norway spruce (Picea abies) | Austria | 2010 | T. Kirisits |
| TYP16 | Adult Ips typographus collected from galleries | Norway spruce (Picea abies) | Austria | 2010 | T. Kirisits |
| TYP17 | Adult Ips typographus collected from galleries | Norway spruce (Picea abies) | Austria | 2010 | T. Kirisits |
| TYP19 | Adult Ips typographus collected from galleries | Norway spruce (Picea abies) | Austria | 2010 | T. Kirisits |
| Ceratocystis fagacearum | |||||
| SAP-1 | Sapwood of infected host | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| SAP-2 | Sapwood of infected host | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| SAP-3 | Sapwood of infected host | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| SAP-4 | Sapwood of infected host | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| SAP-5 | Sapwood of infected host | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| SAP-6 | Sapwood of infected host | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS1 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS2 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS3 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS4 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS5 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS6 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS7 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS8 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| CS9 | Carpophilus sayi collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| EC1 | Epuraea corticina collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| EC2 | Epuraea corticina collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| EC3 | Epuraea corticina collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| GS1 | Glischrochilus sanguinolentus collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| GS2 | Glischrochilus sanguinolentus collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| GS3 | Glischrochilus sanguinolentus collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| GS4 | Glischrochilus sanguinolentus collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| GS5 | Glischrochilus sanguinolentus collected from oak wilt mats | Red oak (Quercus rubra) | MN, USA | 2014 | J. Juzwik |
| Fusarium circinatum | |||||
| SB1a | Woody tissue of asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| SB3a | Woody tissue of asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| SB4a | Woody tissue of asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| 71-1A | Woody tissue | Loblolly pine (Pinus taeda) | USA | N/A | R. Ioos |
| 77-1A | Woody tissue | Ponderosa pine (Pinus ponderosa) | USA | N/A | R. Ioos |
| 124a | Woody tissue | Loblolly pine (Pinus taeda) | USA | N/A | R. Ioos |
| 819A | Woody tissue | Loblolly pine (Pinus taeda) | USA | N/A | R. Ioos |
| 860B | Woody tissue | Maritime pine (Pinus pinaster) | Spain | N/A | R. Ioos |
| MP1Ab | Woody tissue of symptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP1Ba | Woody tissue of asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP2A | Woody tissue of symptomatic and asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP3a | Woody tissue of symptomatic and asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP4Ba | Woody tissue of symptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP5Aa | Woody tissue of symptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP5Ba | Woody tissue of asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP6a | Woody tissue of symptomatic and asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| MP7a | Woody tissue of symptomatic and asymptomatic host | Monterey pine (Pinus radiata) | CA, USA | N/A | R. Ioos |
| S10-14 | Ips sexdentatus artificially inoculated with 10 spores of F. circinatum | - | - | - | R. Ioos |
| S50-13 | Ips sexdentatus artificially inoculated with 50 spores of F. circinatum | - | - | - | R. Ioos |
| S100-15 | Ips sexdentatus artificially inoculated with 100 spores of F. circinatum | - | - | - | R. Ioos |
| Geosmithia morbida | |||||
| JN2 Poz | Artificially-inoculated host (greenhouse) | Eastern black walnut (Juglans nigra) | - | - | M. Kolařík |
| JN3 Neg | Non-inoculated host (greenhouse) | Eastern black walnut (Juglans nigra) | - | - | M. Kolařík |
| WTB-G3-1 | Pityophthorus juglandis from TN, USA | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-2 | Pityophthorus juglandis from TN, USA | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-3 | Pityophthorus juglandis from TN, USA | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-4 | Pityophthorus juglandis from TN, USA | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-5 | Pityophthorus juglandis from TN, USA | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-6 | Pityophthorus juglandis from TN, USA | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-7 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-8 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-9 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G3-10 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-1 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-2 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-3 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-4 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-5 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-6 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-7 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-8 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-9 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| WTB-G10-10 | Pityophthorus juglandis collected on host | Eastern black walnut (Juglans nigra) | TN, USA | 2013 | J. Juzwik |
| Gremmeniella abietina (EU race) | |||||
| 64667 | Needles | Jack pine (Pinus banksianae) | QC, Canada | 2013 | MRNQ |
| 64668 | Needles | Jack pine (Pinus banksianae) | QC, Canada | 2013 | MRNQ |
| 64672 | Needles | Jack pine (Pinus banksianae) | QC, Canada | 2013 | MRNQ |
| 64673 | Needles | Jack pine (Pinus banksianae) | QC, Canada | 2013 | MRNQ |
| 65097 | Needles | Red pine (Pinus resinosa) | QC, Canada | 2013 | MRNQ |
| 65171 | Needles | Red pine (Pinus resinosa) | QC, Canada | 2013 | MRNQ |
| 65181 | Needles | Jack pine (Pinus banksianae) | QC, Canada | 2013 | MRNQ |
| 65539 | Needles | Jack pine (Pinus banksianae) | QC, Canada | 2013 | MRNQ |
| 67161 | Needles | Red pine (Pinus resinosa) | QC, Canada | 2013 | MRNQ |
| Rosellinia necatrix | M. Shishido | ||||
| A | Roots | Japanese pear (Pyrus pyrifolia var. culta) | Japan | N/A | M. Shishido |
| B | Roots | Japanese pear (Pyrus pyrifolia var. culta) | Japan | N/A | M. Shishido |
| D | Roots | Japanese pear (Pyrus pyrifolia var. culta) | Japan | N/A | M. Shishido |
| E | Roots | Japanese pear (Pyrus pyrifolia var. culta) | Japan | N/A | M. Shishido |
| H | Roots | Japanese pear (Pyrus pyrifolia var. culta) | Japan | N/A | M. Shishido |
| I | Roots | Japanese pear (Pyrus pyrifolia var. culta) | Japan | N/A | M. Shishido |
| Sclerotinia pseudotuberosa | |||||
| 1C | Nuts | Sweet chestnut (Castanea sativa) | Italy | N/A | G. Maresi |
| 2C | Nuts | Sweet chestnut (Castanea sativa) | Italy | N/A | G. Maresi |
| Phytophthora ramorum | |||||
| 16883 | N/A | N/A | UK | N/A | J. Tomlinson |
| 16885 | N/A | N/A | UK | 2007 | J. Tomlinson |
| 17085 | Leaves | Rhododendron (Rhododendron sp.) | UK | 2010 | J. Tomlinson |
| 17385 | Leaves | Chinese magnolia (Magnolia x soulangeana) | UK | 2008 | J. Tomlinson |
| 17358 | Leaves | Griselinia sp. | UK | 2008 | J. Tomlinson |
| 07-Qr3-2i | Leaf of artificially-inoculated host (greenhouse) | Red oak (Quercus rubra) | - | - | D. Rioux |
| 07-Ab3-1i | Leaf of artificially-inoculated host (greenhouse) | Balsam fir (Abies balsamea) | - | - | D. Rioux |
| 07-As2-4i | Leaf of artificially-inoculated host (greenhouse) | Sugar maple (Acer saccharum) | - | - | D. Rioux |
| 07-Ll1-3i | Leaf of artificially-inoculated host (greenhouse) | Tamarack (Larix laricina) | - | - | D. Rioux |
| 07-Fa3-1i | Leaf of artificially-inoculated host (greenhouse) | White ash (Fraxinus Americana) | - | - | D. Rioux |
| 07-Ba1-2i | Leaf of artificially-inoculated host (greenhouse) | Yellow birch (Betula alleghaniensis) | - | - | D. Rioux |
| 07-Rho1-4i | Leaf of artificially-inoculated host (greenhouse) | Rhododendron (Rhododendron catawbiense cv. Nova zembla) | - | - | D. Rioux |
| 07-Rho1-2c | Leaf of non-inoculated host (greenhouse) | Rhododendron (Rhododendron catawbiense cv. Nova zembla) | - | - | D. Rioux |
| 02045 | N/A | N/A | UK | 2011 | J. Tomlinson |
| 19347 | Leaf litter/soil | N/A | UK | 2011 | J. Tomlinson |
| 20181 | Water bait | N/A | UK | 2011 | J. Tomlinson |
| 20644 | Leaves | Rhododendron (Rhododendron sp.) | UK | 2011 | J. Tomlinson |
| 20816 | Water bait | N/A | UK | 2011 | J. Tomlinson |
| Phytophthora kernoviae | |||||
| 16833 | N/A | N/A | UK | N/A | J. Tomlinson |
| 16876 | Leaves | Rhododendron (Rhododendron sp.) | UK | 2007 | J. Tomlinson |
| 17072 | N/A | N/A | UK | N/A | J. Tomlinson |
| 02045 | N/A | N/A | UK | 2011 | J. Tomlinson |
| 19347 | Leaf litter/soil | N/A | UK | 2011 | J. Tomlinson |
| 20181 | Water bait | N/A | UK | 2011 | J. Tomlinson |
| 20644 | N/A | Rhododendron (Rhododendron sp.) | UK | 2011 | J. Tomlinson |
| 20816 | Water bait | N/A | UK | 2011 | J. Tomlinson |
a Type of material from which DNA was extracted.
b TYP samples were used as negative controls for C. laricicola specific assays, whereas CEM samples were used as negative controls for C. polonica specific assays.
TaqMan assays were performed with three technical replicates for each related environmental sample. Reactions were performed as described earlier, using 1 μL of environmental DNA. Positive (using target species’ DNA from pure culture) and negative (no template DNA) controls were included in all qPCR runs. Target gene region copy numbers were calculated by translating Ct values, using standard curve equations. Positive results were all confirmed by Sanger sequencing of the real-time TaqMan PCR product.
Results and Discussion
Assay design and development
Development of the detection assays was based on two strategies targeting 1) unique SNPs or 2) unique genes. The SNP-based approach uses alignment of genes present in all species, but exploits the presence of SNPs between the target species and the close relatives. It was used for all assays reported in this paper except for P. ramorum and P. kernoviae. For both of these species, comparative genomics was used to identify genes uniquely found in the target species to design the detection assay. Detail about this TAIGA strategy, the related genomic resources and bioinformatics pipeline are available on the TAIGA project website (http://taigaforesthealth.com/).
A crucial step in the development of the SNP-based detection assays is the identification of appropriate target DNA regions. Readily amplified genes across taxa of a group were sequenced, and genes showing interspecific variability were selected for assay development (Table 2). As a result, different genes were selected for each target species, some of them being single- or low-copy genes (e.g. β-tubulin, EF1, RPB2 and Hsp60), and others being multi-copy genes (e.g. IGS, ITS, Cluster62 and Cluster97).
In order to standardize the DNA concentration of all isolates, a genus general real-time PCR SYBRGreen I assay targeting the selected DNA region was designed and validated for each target group. Using the linear regression of efficiency (LRE) quantification approach [20], DNA concentration of isolates was determined and standardized to 5,000 target gene region copies, which usually translates into a Ct value ranging between 20 and 25.
To ensure the repeatability of the qPCR experiments described by other teams and to ease the interpretation of results, most of the Minimum Information for Quantitative Real-Time PCR Experiments (MIQE), as described by Bustin et al. (2009) [24], is presented in this paper.
Specificity of the tree pathogen TaqMan assays
Probes’ and primers’ specificity was first tested in silico using BLAST on the NCBI nucleotide collection (nr/nt) database. A wet lab was then performed to assess candidate sets’ panel specificity on DNA samples from target and sister species isolates. During the first round of specificity validation, whenever an unexpected amplification was observed, two hypotheses were explored. First, it could be due to trace contamination of the DNA sample with target species’ DNA, which sometimes happens during sample manipulation. This was suspected when cycle threshold (Ct) values were much higher (around 35–37) than those of the target species isolates. When contamination was suspected, a SYBRGreen real-time PCR reaction along with a melting curve gradient was performed. When melting curves of positive and suspected false positive samples were identical, the SYBRGreen real-time PCR reaction product of the false positive was sequenced and aligned with reference sequences to confirm the contamination. In such cases, the contaminated DNA sample was discarded and fresh DNA was re-extracted from a pure culture of the isolate. If the first hypothesis was confirmed, we concluded we had a real false positive reaction due to a lack of specificity. In such cases, further screening of primers and probes was conducted.
All our final assays were 100% specific successfully discriminating the target species of tree pathogen from the closely related species. In cases where we were unable to obtain culture or DNA of some closely related species, we still performed in silico specificity validation using sequences obtained from the public domain. Despite the current results, we cannot rule out potential cross reactivity of the present assays with evolutionarily related species that have not been described yet. Some of the target species belong to what can be considered as orphan and poorly resolved taxonomic groups (such as Mycosphaerella, with newly described species M. musivoides P.E. Busby & G. Newc and M. wasatchii P.E. Busby & G. Newc [25]). Instead of being a drawback, molecular cross reactivity with cryptic species can represent an opportunity to isolate and describe novel fungal species with similar or different pathogenic behaviors [26].
Sensitivity of the tree pathogen TaqMan assay
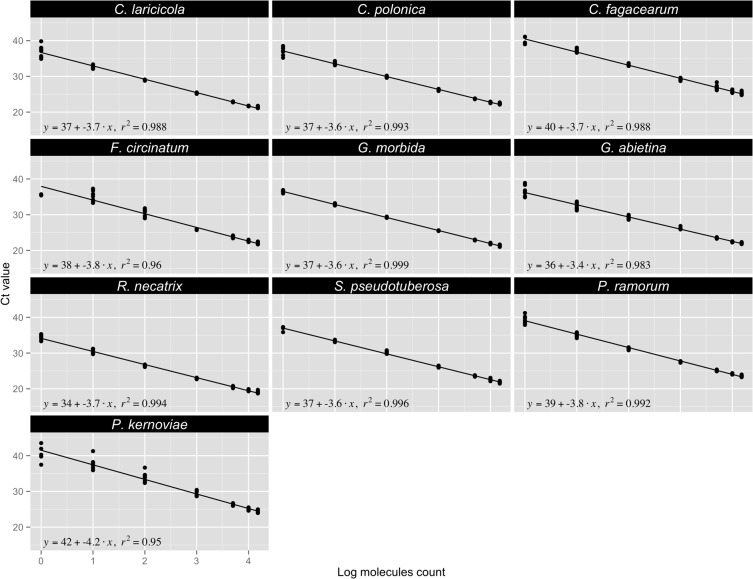
Overall, the assays we developed have high efficiency and sensitivity, with limits of detection varying between 1 and 10 target gene region copies (Table 5). In the present study, no difference in sensitivity values was observed between assays targeting single- or low-copy genes and those targeting multi-copy genes. For all tested species, Ct values were proportional to the amount of template DNA used for the real-time PCR reaction. The standard curves generated by plotting the log of DNA (copies) against the Ct value determined by qPCR display linearity across the whole range of dilutions assessed, with a correlation coefficient (r 2) ranging from 0.950 for the P. kernoviae assay to 0.999 for the G. morbida assay (Fig 2). Moreover, PCR amplification efficiencies ranged between 83 and 97%, which is considered to be an acceptable range [27], except for P. kernoviae. The low amplification efficiency (73%) of the P. kernoviae assay could be due to a number of factors, such as the presence of inhibitors in the DNA samples, suboptimal primer and probe design (e.g. presence of non-specific products and primer dimers), and pipetting errors. Experimental investigations ruled out the presence of non-specific products and dimers, sample contamination, inappropriate dilution series and pipetting errors. Another possible explanation for this reduced efficiency is the presence of a secondary structure in the region targeted by this assay. Amplification efficiency can vary across a genome [28, 29]. Genomic regions resistant to amplification by PCR correlate with high GC contents [30, 31] that do not denature efficiently under routine amplification conditions. However, the GC content of that region was around 55%, which is not considered a problem.
Table 5. Limit of detection for the 10 tree pathogen TaqMan assays.
| Species | Target gene | Limit of detection (LOD) a |
|---|---|---|
| Ceratocystis laricicola | β-tubulin | 3 |
| Ceratocystis polonica | β-tubulin | 3 |
| Ceratocystis fagacearum | EF1 | 10 |
| Fusarium circinatum | IGS | 10 |
| Geosmithia morbida | β-tubulin | 3 |
| Gremmeniella abietina (EU race) | RPB2 | 1 |
| Rosellinia necatrix | ITS | 1 |
| Sclerotinia pseudotuberosa | Hsp60 | 5 |
| Phytophthora ramorum | Cluster62 | 5 |
| Phytophthora kernoviae | Cluster97 | 3 |
a Represented as the copy number of the target gene region.
Fig 2. Standard curve for each of the 10 tree pathogen assays.
Ct values are plotted against the log value of the target gene region copy number. Curve equations and the squared correlation coefficient are presented.
The limit of detection (LOD) was defined as the lowest concentration of target DNA at which 95% of the positive samples were detected [24]. According to Bustin et al. (2009), and assuming an even Poisson distribution, the lowest theoretically possible detection limit is three DNA copies per PCR reaction to provide a positive signal in 95% of the PCR reactions performed. Our assays revealed LOD values varying between 1 and 10 target gene region copies (Table 5). Those values are comparable to what has been reported by others working on trace detection for regulatory or public health applications, looking either for the presence of genetically modified DNA [32–35], virus DNA in blood samples [36, 37], or human DNA [38, 39], where LOD values varying between 1 and 25 copies were obtained.
As an additional validation step, we were able to compare results from our F. circinatum assay with those published for a similar qPCR test developed by Ioos et al. (2009) [40]. The published assay was not compliant with our set of real-time PCR conditions and had to be redesigned to suit qPCR standardized parameters. Our assay targets a different segment of the intergenic spacer region than the one used elsewhere [40]. To conduct a fair comparison, a subset of the environmental samples used by Ioos et al. (2009) [40] was obtained and tested with our assay. The results we obtained were actually very similar. Although our assay had a slightly delayed detection threshold of approximately 3 Ct values compared with that of Ioos et al. (2009), these values were not significantly different.
Precision of the tree pathogen TaqMan assays
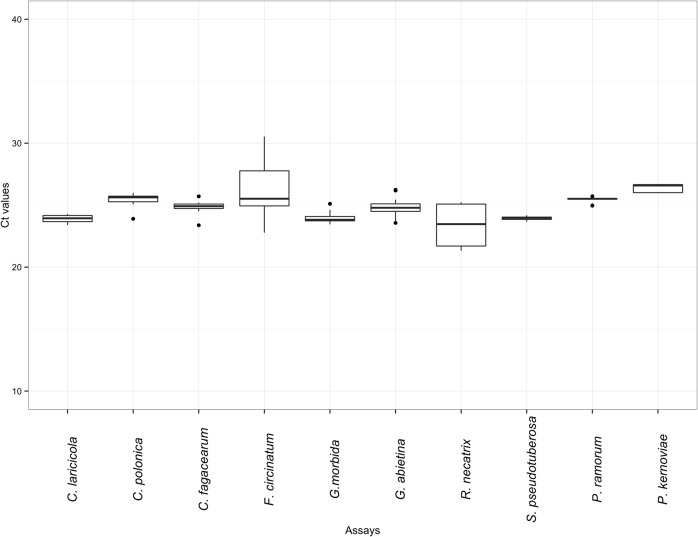
The precision of our ten tree pathogen TaqMan assays is shown in Fig 3. All assays have a mean Ct value ranging between 23 and 26 for 5,000 target gene region copies. This value depends on amplicon size and primers and probe properties. Using the mean Ct value and the standard deviation, we also calculated a coefficient of variation for each assay, which varied between 0.7% for the S. pseudotuberosa assay and 8.2% for the F. circinatum assay. Those values clearly demonstrate that our assays have a high degree of repeatability, an important advantage when dealing with possible regulatory issues.
Fig 3. Precision of each of the 10 tree pathogen assays.
Box plot representing variation of the Ct value between technical replicates of various isolates from target species (mean Ct value ± coefficient of variation). DNA samples at a concentration of 5,000 copies of the target gene region were used.
Validation of the tree pathogen TaqMan assays on environmental samples
All tree pathogen assays successfully detected target pathogens from the positive environmental samples provided by collaborators (Tables 4 and 6). Negative environmental DNA samples were available for 6 out of the 10 assays; no false positive results were obtained with any of these.
Table 6. Results from species-specific TaqMan real-time PCR assays using environmental samples.
| Isolate | Expected result a | Genus gene copy number b | Specific TaqMan assay Ct value (± SD) | Target gene region copy number c |
|---|---|---|---|---|
| Ceratocystis laricicola | + | |||
| CEM5 | + | 712 | 29.3 (0.1) | 121 |
| CEM8 | + | 1,701 | 29.7 (0.1) | 97 |
| CEM10 | + | 542 | 29.4 (0.2) | 117 |
| CEM11 | + | 1,621 | 27.9 (0.1) | 284 |
| CEM13 | + | 1,638 | 28.0 (0.2) | 271 |
| CEM19 | + | 2,227 | 29.4 (0.1) | 111 |
| CEM25 | + | 964 | 28.3 (0.1) | 220 |
| TYP1 | - | 910 | None | - |
| TYP2 | - | 818 | None | - |
| TYP3 | - | 751 | None | - |
| TYP11 | - | 555 | None | - |
| TYP16 | - | 373 | None | - |
| TYP17 | - | 613 | None | - |
| TYP19 | - | 176 | None | - |
| Ceratocystis polonica | ||||
| TYP1 | + | 910 | 34.7 (0.4) | 4 |
| TYP2 | + | 818 | 35.0 (0.5) | 4 |
| TYP3 | + | 751 | 35.3 (0.3) | 3 |
| TYP11 | + | 555 | 34.6 (0.1) | 5 |
| TYP16 | + | 373 | 31.8 (ND) d | 28 |
| TYP17 | + | 613 | 34.6 (0.1) | 5 |
| TYP19 | + | 176 | 36.1 (ND) d | 2 |
| CEM5 | - | 712 | None | - |
| CEM8 | - | 1,701 | None | - |
| CEM10 | - | 542 | None | - |
| CEM11 | - | 1,621 | None | - |
| CEM13 | - | 1,638 | None | - |
| CEM19 | - | 2,227 | None | - |
| CEM25 | - | 964 | None | - |
| Ceratocystis fagacearum | ||||
| SAP-1 | + | 148 | 36.8 (1.9) | 7 |
| SAP-2 | + | 5 | 37.4 (0.9) | 5 |
| SAP-3 | + | 91 | 36.5 (0.3) | 9 |
| SAP-4 | + | 54 | 39.1 (ND) d | 2 |
| SAP-5 | + | 2 | 37.1 (0.6) | 6 |
| SAP-6 | + | 1 | 38.1 (0.3) | 3 |
| CS1 | + | 4,626 | 26.5 (0.2) | 4,326 |
| CS2 | + | 3,467 | 25.8 (0.2) | 6,805 |
| CS3 | + | 2,593 | 27.5 (0.0) | 2,409 |
| CS4 | + | 3,342 | 27.4 (0.1) | 2,518 |
| CS5 | + | 4,872 | 27.3 (0.0) | 2,682 |
| CS6 | + | 2,261 | 28.1 (0.1) | 1,694 |
| CS7 | + | 3,293 | 25.6 (0.1) | 8,041 |
| CS8 | + | 2,677 | 24.7 (0.3) | 12,421 |
| CS9 | + | 1,334 | 24.5 (0.1) | 15,351 |
| EC1 | + | 7,081 | 27.0 (0.1) | 3,195 |
| EC2 | + | 8,918 | 26.9 (0.0) | 3,478 |
| EC3 | + | 13,140 | 28.3 (0.1) | 1,476 |
| GS1 | + | 6,724 | 25.8 (0.1) | 6,794 |
| GS2 | + | 2,838 | 27.5 (0.1) | 2,361 |
| GS3 | + | 3,313 | 27.2 (0.1) | 2,940 |
| GS4 | + | 6,087 | 27.2 (0.0) | 2,902 |
| GS5 | + | 41,882 | 22.7 (0.1) | 48,405 |
| Fusarium circinatum | ||||
| SB1a | + | 1 | None | - |
| SB3a | + | 6 | 34.0 (0.8) | 11 |
| SB4a | + | 300,332 | 18.1 (0.0) | 170,090 |
| 71-1A | + | 1 | None | - |
| 77-1A | + | 45 | 34.5 (0.1) | 8 |
| 124a | + | 1,936 | 28.0 (0.1) | 434 |
| 819A | + | 8 | 35.9 (ND) d | 4 |
| 860B | + | 262 | 28.4 (0.1) | 332 |
| MP1Ab | + | 6,096 | 24.0 (0.1) | 4,944 |
| MP1Ba | + | 737 | 27.0 (0.3) | 771 |
| MP2A | + | 200 | 30.1 (0.2) | 118 |
| MP3a | + | 700 | 27.0 (0.1) | 795 |
| MP4Ba | + | 346 | 27.9 (0.4) | 455 |
| MP5Aa | + | 49 | 31.2 (0.0) | 63 |
| MP5Ba | + | 35 | 31.6 (0.9) | 48 |
| MP6a | + | 1,399 | 26.5 (0.1) | 1,087 |
| MP7a | + | 551 | 27.5 (0.0) | 588 |
| S10-14 | + | 1 | 34.8 (ND) d | 7 |
| S50-13 | + | 1 | 36.0 (ND) d | 3 |
| S100-15 | + | 5 | 32.7 (0.4) | 24 |
| Geosmithia morbida | ||||
| JN2 Poz | + | 4 | 31.7 (0.3) | 29 |
| JN3 Neg | - | 23 | None | - |
| WTB-G3-1 | + | 1 | 34.5 (0.3) | 5 |
| WTB-G3-2 | + | 4 | 34.5 (0.2) | 5 |
| WTB-G3-3 | + | 1 | 35.7 (1.0) | 2 |
| WTB-G3-4 | + | 3 | 34.2 (0.2) | 6 |
| WTB-G3-5 | + | 1 | 34.9 (0.4) | 4 |
| WTB-G3-6 | + | 13 | 31.7 (0.1) | 30 |
| WTB-G3-7 | + | 3 | 34.7 (0.2) | 4 |
| WTB-G3-8 | + | 11 | 32.5 (0.1) | 17 |
| WTB-G3-9 | + | 2 | 35.2 (0.7) | 3 |
| WTB-G3-10 | + | 6 | 33.2 (0.5) | 11 |
| WTB-G8-1 | + | 3 | 34.3 (0.2) | 5 |
| WTB-G8-2 | + | 3 | 34.3 (0.0) | 5 |
| WTB-G8-3 | + | 2 | 34.8 (0.4) | 4 |
| WTB-G8-4 | + | 1 | 36.7 (0.7) | 1 |
| WTB-G8-5 | + | 1 | 36.1 (0.4) | 2 |
| WTB-G8-6 | + | 0 | 37.4 (0.8) | 1 |
| WTB-G8-7 | + | 1 | 35.7 (0.0) | 2 |
| WTB-G8-8 | + | 0 | 36.8 (0.8) | 1 |
| WTB-G8-9 | + | 0 | 37.8 (ND) d | 1 |
| WTB-G8-10 | + | 2 | 34.0 (0.1) | 7 |
| Gremmeniella abietina (EU race) | ||||
| 64667 | - | 7,370 | None | - |
| 64668 | - | 5,509 | None | - |
| 64672 | - | 1,820 | None | - |
| 64673 | - | 9,569 | None | - |
| 65097 | + | 5912 | 25.4 (0.3) | 1,608 |
| 65171 | + | 65,081 | 21.9 (0.2) | 19,176 |
| 65181 | - | 916 | None | - |
| 65539 | - | 26,396 | None | - |
| 67161 | + | 16,236 | 24.1 (0.0) | 4,139 |
| Rosellinia necatrix | ||||
| A | + | 15,891 | 24.6 (0.1) | 340 |
| B | + | 2,064 | 25.1 (0.1) | 250 |
| D | + | 5,842 | 28.8 (0.0) | 25 |
| E | + | 3,974 | 26.9 (0.1) | 84 |
| H | + | 24,804 | 25.9 (0.0) | 153 |
| I | + | 3,091 | 25.4 (0.2) | 211 |
| Sclerotinia pseudotuberosa | ||||
| 1C | + | 6,492,200 | 21.3 (0.1) | 23,180 |
| 2C | + | 5,530,494 | 21.6 (0.1) | 18,600 |
| Phytophthora ramorum | ||||
| 16883 | + | 12,377 | 33.6 (0.2) | < 1 |
| 16885 | + | 17,646 | 33.2 (0.2) | 3 |
| 17085 | + | 3,883 | 39.1 (0.2) | <1 |
| 17385 | + | 10,515 | 35.8 (0.1) | 1 |
| 17358 | + | 10,131 | 37.4 (0.0) | < 1 |
| 07-Qr3-2i | + | 10 | None | - |
| 07-Ab3-1i | + | 4 | 34.2 (0.3) | 2 |
| 07-As2-4i | + | 44 | 38.3 (0.7) | < 1 |
| 07-Ll1-3i | + | 15 | 34.6 (0.2) | 1 |
| 07-Fa3-1i | + | 686 | 28.2 (0.1) | 63 |
| 07-Ba1-2i | + | 2,254 | 26.5 (0.1) | 170 |
| 07-Rho1-4i | + | 2,743 | 26.0 (0.4) | 241 |
| 07-Rho1-2c | - | 0 | None | - |
| 02045 | - | 0 | None | - |
| 19347 | - | 9 | None | - |
| 20181 | - | 263 | None | - |
| 20644 | - | 10 | None | - |
| 20816 | - | 758 | None | - |
| Phytophthora kernoviae | ||||
| 16833 | + | 7,911 | 30.5 (0.4) | 570 |
| 16876 | + | 983 | 31.7 (0.2) | 292 |
| 17072 | + | 76 | 35.0 (0.4) | 47 |
| 02045 | - | 0 | None | - |
| 19347 | - | 9 | None | - |
| 20644 | - | 10 | None | - |
a According to the environmental samples’ provider.
b Calculated with the results obtained from a SYBRGreen real-time PCR reaction.
c Values obtained by plotting Ct values from the species-specific TaqMan assay into the standard curve-derived equation (Fig 2).
d ND: one of the two replicates did not amplify.
One of the most critical steps when dealing with environmental samples is the quality of the DNA sample, which may vary with the extraction protocol used. In spite of this, detection limits as low as one target gene region copy were obtained (Table 6). For some samples, we obtained a positive result, i.e. a detectable Ct value using the TaqMan specific assays, but the calculated target gene region copy number was less than one. These results might be explained by the fact that we used standard curve equations to extrapolate those copy number values. Therefore, there exists a certain level of imprecision that has a more important effect on samples with a low level of target pathogen DNA. This imprecision caused by the extrapolation of target gene region copy values can also explain why, in some other cases (e.g. C. fagacearum, G. morbida, F. circinatum and S. pseudotuberosa), we obtained a slightly higher value for the target gene region than the one obtained with the genus assay (Table 6). The opposite result was also seen; for some environmental samples (e.g. C. laricicola, C. polonica, G. abietina (EU race), R. necatrix, S. pseudotuberosa and P. ramorum), the genus gene copy number was much higher than the target gene region copy number. This is most probably due to the presence of more than one species from the targeted genus in the environmental samples. For example, G. abietina (EU race) positive environmental samples were obtained from infected Pinus resinosa samples from the province of Québec, Canada. We know that the North American race of G. abietina, which is detected and counted with the genus assay but not with the EU race specific assay, might also be present on red pines [41]. Other examples are Rhododendon sp., Griselinia sp., and Magnolia x soulangeana plant tissues infected with P. ramorum. Those plants are well known to be hosts for other Phytophthora species [42, 43], which might explain the high level of quantification at the genus level, concomitant with a low level of P. ramorum itself.
Inhibitors from plant material or insect specimens co-extracted with DNA are a source of contamination that can impact PCR amplification accuracy [44–46], which is the variation between observed and expected data [24]. This property was evaluated for some of those assays (C. polonica and C. laricola) in a previous study [47]. In those specific cases, the presence of environmental DNA or any other co-extracted compound had no effect on the performance of the TaqMan assays. However, we are aware that assay performance could vary slightly depending on the material it is tested against. In fact, inhibition may be caused by the presence of different compounds such as acidic plant polysaccharides [48, 49], plant phenolics [50], the contamination of DNA samples with co-extracted polyphenol-bound proteins from the insect cuticle [51], or with phenolics and tannins found in the digestive tracts of xylophagous insects [45]. Accuracy evaluation should therefore be one of the initial steps for any user of those assays dealing with new environmental material.
We developed sensitive and specific molecular assays for ten alien tree pathogens identified as high priority potential threats for Canadian forests: Ceratocystis fagacearum, Ceratocystis laricicola, Ceratocystis polonica, Fusarium circinatum, Gremmeniella abietina (EU race), Geosmithia morbida, Phytophthora kernoviae, Phytophthora ramorum, Rosellinia necatrix and Sclerotinia pseudotuberosa. All of these assays are specific, i.e. they have the ability to amplify a unique DNA fragment of interest without amplifying or detecting non-target sequences. Detection assays were already available for some of the target tree pathogens selected. In some cases, they were included in our tree pathogen TaqMan assay panel (e.g. C. polonica and C. laricicola [47]). However, in most cases, existing assays had to be redesigned either because 1) they were not compliant with our real-time PCR conditions (C. fagacearum [52], F. circinatum [40, 53], P. kernoviae [54], P. ramorum [9, 54–59], S. pseudotuberosa [60]), 2) they were not tested against all closely related species (F. circinatum [61], P. kernoviae [7, 62], P. ramorum [7, 55, 63–65], R. necatrix [66–68]), or 3) they did not target the specific race of interest (G. abietina EU race [69]).
All assays were designed to be used under the same real-time PCR conditions, using the same chemistry and the same thermocycling parameters. Therefore, they can be performed in micro-well plates arrayed in any machine format to suit individual users’ needs and to increase throughput. Reactions for multiple samples, targeting multiple pathogens, can be performed in a single real-time PCR run, which is an important advantage under operational conditions where testing a large number of samples against of large number of targets is required. Molecular detection of these pathogenic species directly from plant material or insect vectors represents a powerful tool to prevent their introduction and establishment as potential invasive species.
Supporting Information
(DOCX)
Acknowledgments
We would like to acknowledge all the members of the TAIGA team for their collaboration to the project. We also thank C. Breuil, Ð.H. Guerry, T.C. Harrington, C. Husson, R. Ioos, J. Juzwik, T. Kirisits, M. Kolařík, G. Laflamme, G. Maresi, H. Nakamura, D. Rioux, K. Seifert, M. Shishido, J. Tomlinson, M.J. Wingfield, and the Ministère des Forêts, de la Faune et des Parcs du Québec for sharing DNA, fungal isolates and/or environmental samples from their collections. We also thank Marie-Josée Bergeron and Isabelle Lamarre for the revision of the manuscript, as well as the three anonymous reviewers.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from Genome Canada and Genome British Columbia (Large-Scale Applied Research Program, Grant #164) as well as by the Genomics Research and Development Initiative (GRDI) of Natural Resources Canada, FPInnovations and its government and industry members, and the Canadian Food and Inspection Agency (CFIA).
References
- 1. Santini A, Ghelardini L, De Pace C, Desprez-Loustau ML, Capretti P, et al. (2013) Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytologist 197: 238–250. 10.1111/j.1469-8137.2012.04364.x [DOI] [PubMed] [Google Scholar]
- 2. Desprez-Loustau M-L (2009) The alien fungi of Europe In: Drake JA, editor. Handbook of alien species in Europe: Dordrecht: Springer; pp. 15–28. [Google Scholar]
- 3. Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Research 6: 986–994. [DOI] [PubMed] [Google Scholar]
- 4. Bilodeau G, Pelletier G, Pelletier F, Hamelin RC, Lévesque CA (2009) Multiplex real-time polymerase chain reaction (PCR) for detection of Phytophthora ramorum, the causal agent of sudden oak death. Canadian Journal of Plant Pathology 31: 195–210. [Google Scholar]
- 5. Qu XS, Wanner LA, Christ BJ (2011) Multiplex real-time PCR (TaqMan) assay for the simultaneous detection and discrimination of potato powdery and common scab diseases and pathogens. Journal of Applied Microbiology 110: 769–777. 10.1111/j.1365-2672.2010.04930.x [DOI] [PubMed] [Google Scholar]
- 6. López-Fabuel I, Wetzel T, Bertolini E, Bassler A, Vidal E, et al. (2013) Real-time multiplex RT-PCR for the simultaneous detection of the five main grapevine viruses. Journal of Virological Methods 188: 21–24. 10.1016/j.jviromet.2012.11.034 [DOI] [PubMed] [Google Scholar]
- 7. Schena L, Hughes KJD, Cooke DEL (2006) Detection and quantification of Phytophthora ramorum, P. kernoviae, P. citricola and P. quercina in symptomatic leaves by multiplex real-time PCR. Molecular Plant Pathology 7: 365–379. 10.1111/j.1364-3703.2006.00345.x [DOI] [PubMed] [Google Scholar]
- 8. Ippolito A, Schena L, Nigro F, Soleti ligorio V, Yaseen T (2004) Real-time detection of Phytophthora nicotianae and P. citrophthorain citrus roots and soil. European Journal of Plant Pathology 110: 833–843. [Google Scholar]
- 9. Tooley PW, Martin FN, Carras MM, Frederick RD (2006) Real-time fluorescent polymerase chain reaction detection of Phytophthora ramorum and Phytophthora pseudosyringae using mitochondrial gene regions. Phytopathology 96: 336–345. 10.1094/PHYTO-96-0336 [DOI] [PubMed] [Google Scholar]
- 10.Harrington TC. The genus Ceratocystis. Where does the oak wilt fungus fit? In: Appel DN, Billings RF, editors; 2009; Austin, TX.
- 11. Witthuhn RC, Harrington TC, Steimel JP, Wingfield BD, Wingfield MJ (2000) Comparison of isozymes, rDNA spacer regions and MAT-2 DNA sequences as phylogenetic characters in the analysis of the Ceratocystis coerulescens complex. Mycologia 92: 447–452. [Google Scholar]
- 12. Marin M, Preisig O, Wingfield BD, Kirisits T, Yamaoka Y, et al. (2005) Phenotypic and DNA sequence data comparisons reveal three discrete species in the Ceratocystis polonica species complex. Mycological Research 109: 1137–1148. [DOI] [PubMed] [Google Scholar]
- 13. O’Donnell K, Nirenberg H, Aoki T, Cigelnik E (2000) A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- 14. Kolařík M, Freeland E, Utley C, Tisserat N (2011) Geosmithia morbida sp. nov., a new phytopathogenic species living in symbiosis with the walnut twig beetle (Pityophthorus juglandis) on Juglans in USA. Mycologia 103: 325–332. 10.3852/10-124 [DOI] [PubMed] [Google Scholar]
- 15. Blair JE, Coffey MD, Park S-Y, Geiser DM, Kang S (2008) A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45: 266–277. [DOI] [PubMed] [Google Scholar]
- 16. Zolan ME, Pukkila PJ (1986) Inheritance of DNA methylation in Coprinus cinereus . Molecular and Cellular Biology 6: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- 19. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rutledge RG (2011) A Java program for LRE-based real-time qPCR that enables large-scale absolute quantification. PLoS ONE 6: e17636 10.1371/journal.pone.0017636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, et al. (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Research 17: 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Charlton S, Giroux R, Hondred D, Lipton C, Worden K (2000) PCR validation and performance characteristics. AEIC Biotech Consensus Paper The Analytical Environmental Immunochemical Consortium (AEIC) Secretariat. Dow AgroSciences, Indianapolis, IN. [Google Scholar]
- 23.Ederveen J (2010) A practical approach to biological assay validation. Progress Report No. 08090.: Project Management and Engineering.
- 24. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55: 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 25.Busby PE (2012) Causes and consequences of fungal disease in cottonwoods [Ph. D. Thesis]. Standford, CA: Standford University. 129 p.
- 26. Hawksworth DL, Rossman AY (1997) Where Are All the Undescribed Fungi? Phytopathology 87: 888–891. 10.1094/PHYTO.1997.87.9.888 [DOI] [PubMed] [Google Scholar]
- 27. D’haene B, Vandesompele J, Hellemans J (2010) Accurate and objective copy number profiling using real-time quantitative PCR. Methods 50: 262–270. 10.1016/j.ymeth.2009.12.007 [DOI] [PubMed] [Google Scholar]
- 28. Veal C, Freeman P, Jacobs K, Lancaster O, Jamain S, et al. (2012) A mechanistic basis for amplification differences between samples and between genome regions. BMC Genomics 13: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gallup JM (2011) qPCR inhibition and amplification of difficult templates In: Kennedy S, Oswald N, editors. PCR troubleshooting and optimization: the essential guide. Norfolk, UK: Caister Academic Press; pp. 23–66. [Google Scholar]
- 30. Wintzingerode FV, Göbel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiology Reviews 21: 213–229. [DOI] [PubMed] [Google Scholar]
- 31. McDowell DG, Burns NA, Parkes HC (1998) Localised sequence regions possessing high melting temperatures prevent the amplification of a DNA mimic in competitive PCR. Nucleic Acids Research 26: 3340–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nielsen C, Berdal K, Holst-Jensen A (2004) Characterisation of the 5′ integration site and development of an event-specific real-time PCR assay for NK603 maize from a low starting copy number. European Food Research and Technology 219: 421–427. [Google Scholar]
- 33. Berdal K, Holst-Jensen A (2001) Roundup Ready soybean event-specific real-time quantitative PCR assay and estimation of the practical detection and quantification limits in GMO analyses. European Food Research and Technology 213: 432–438. [Google Scholar]
- 34. Rønning S, Vaïtilingom M, Berdal K, Holst-Jensen A (2003) Event specific real-time quantitative PCR for genetically modified Bt11 maize (Zea mays). European Food Research and Technology 216: 347–354. [Google Scholar]
- 35. Hübner P, Waiblinger H-U, Pietsch K, Brodmann P (2001) Validation of PCR methods for quantitation of genetically modified plants in food. Journal of AOAC International 84: 1855–1864. [PubMed] [Google Scholar]
- 36. Sofi Ibrahim M, Kulesh DA, Saleh SS, Damon IK, Esposito JJ, et al. (2003) Real-time PCR assay to detect smallpox virus. Journal of Clinical Microbiology 41: 3835–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kulesh DA, Baker RO, Loveless BM, Norwood D, Zwiers SH, et al. (2004) Smallpox and pan-orthopox virus detection by real-time 3′-minor groove binder TaqMan assays on the Roche LightCycler and the Cepheid Smart Cycler platforms. Journal of Clinical Microbiology 42: 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zimmermann B, El-Sheikhah A, Nicolaides K, Holzgreve W, Hahn S (2005) Optimized real-time quantitative PCR measurement of male fetal DNA in maternal plasma. Clinical Chemistry 51: 1598–1604. [DOI] [PubMed] [Google Scholar]
- 39. Ellison S, English C, Burns M, Keer J (2006) Routes to improving the reliability of low level DNA analysis using real-time PCR. BMC Biotechnology 6: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ioos R, Fourrier C, Iancu G, Gordon TR (2009) Sensitive detection of Fusarium circinatum in pine seed by combining an enrichment procedure with a real-time polymerase chain reaction using dual-labeled probe chemistry. Phytopathology 99: 582–590. 10.1094/PHYTO-99-5-0582 [DOI] [PubMed] [Google Scholar]
- 41. Hamelin RC, Lecours N, Laflamme G (1998) Molecular evidence of distinct introductions of the European race of Gremmeniella abietina into North America. Phytopathology 88: 582–588. 10.1094/PHYTO.1998.88.6.582 [DOI] [PubMed] [Google Scholar]
- 42. Hantula J, Lilja A, Nuorteva H, Parikka P, Werres S (2000) Pathogenicity, morphology and genetic variation of Phytophthora cactorum from strawberry, apple, rhododendron, and silver birch. Mycological Research 104: 1062–1068. [Google Scholar]
- 43. Brasier CM, Beales PA, Kirk SA, Denman S, Rose J (2005) Phytophthora kernoviae sp. nov., an invasive pathogen causing bleeding stem lesions on forest trees and foliar necrosis of ornamentals in the UK. Mycological Research 109: 853–859. [DOI] [PubMed] [Google Scholar]
- 44. Juen A, Traugott M (2006) Amplification facilitators and multiplex PCR: Tools to overcome PCR-inhibition in DNA-gut-content analysis of soil-living invertebrates. Soil Biology and Biochemistry 38: 1872–1879. [Google Scholar]
- 45. Calderon-Cortés N, Quesada M, Cano-Camacho H, Zavala-Paramo G (2010) A simple and rapid method for DNA isolation from xylophagous insects. International Journal of Molecular Sciences 11: 5056–5064. 10.3390/ijms11125056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palacio-Bielsa A, Cubero J, Cambra MA, Collados R, Berruete IM, et al. (2011) Development of an efficient real-time quantitative PCR protocol for detection of Xanthomonas arboricola pv. pruni in Prunus species. Applied and Environmental Microbiology 77: 89–97. 10.1128/AEM.01593-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lamarche J, Stewart D, Pelletier G, Hamelin RC, Tanguay P (2014) Real-time PCR detection and discrimination of the Ceratocystis coerulescens complex and of the fungal species from the Ceratocystis polonica complex validated on pure cultures and bark beetle vectors. Canadian Journal of Forest Research 44: 1103–1111. [Google Scholar]
- 48. Holden MJ, Blasic JR, Bussjaeger L, Kao C, Shokere LA, et al. (2003) Evaluation of extraction methodologies for corn kernel (Zea mays) DNA for detection of trace amounts of biotechnology-derived DNA. Journal of Agricultural and Food Chemistry 51: 2468–2474. [DOI] [PubMed] [Google Scholar]
- 49. Demeke T, Adams RP (1992) The effects of plant polysaccharides and buffer additives on PCR BioTechniques 12: 332–334. [PubMed] [Google Scholar]
- 50. Rohn S, Rawel HM, Kroll J (2002) Inhibitory effects of plant phenols on the activity of selected enzymes. Journal of Agricultural and Food Chemistry 50: 3566–3571. [DOI] [PubMed] [Google Scholar]
- 51. Margam V, Gachomo E, Shukle J, Ariyo O, Seufferheld M, et al. (2010) A simplified arthropod genomic-DNA extraction protocol for polymerase chain reaction (PCR)-based specimen identification through barcoding. Molecular Biology Reports 37: 3631–3635. 10.1007/s11033-010-0014-5 [DOI] [PubMed] [Google Scholar]
- 52. Wu CP, Chen GY, Li B, Su H, An YL, et al. (2011) Rapid and accurate detection of Ceratocystis fagacearum from stained wood and soil by nested and real-time PCR. Forest Pathology 41: 15–21. [Google Scholar]
- 53. Schweigkofler W, O'Donnell K, Garbelotto M (2004) Detection and quantification of airborne conidia of Fusarium circinatum, the causal agent of pine pitch canker, from two California sites by using a real-time PCR approach combined with a simple spore trapping method. Applied and Environmental Microbiology 70: 3512–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tomlinson JA, Dickinson MJ, Boonham N (2010) Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathology 100: 143–149. 10.1094/PHYTO-100-2-0143 [DOI] [PubMed] [Google Scholar]
- 55. Hayden KJ, Rizzo D, Tse J, Garbelotto M (2004) Detection and quantification of Phytophthora ramorum from California forests using a real-time polymerase chain reaction assay. Phytopathology 94: 1075–1083. 10.1094/PHYTO.2004.94.10.1075 [DOI] [PubMed] [Google Scholar]
- 56. Tomlinson JA, Boonham N, Hughes KJD, Griffin RL, Barker I (2005) On-site DNA extraction and real-time PCR for detection of Phytophthora ramorum in the field. Applied and Environmental Microbiology 71: 6702–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chandelier A, Ivors K, Garbelotto M, Zini J, Laurent F, et al. (2006) Validation of a real-time PCR method for the detection of Phytophthora ramorum . EPPO Bulletin 36: 409–414. [Google Scholar]
- 58. Belbahri L, Calmin G, Wagner S, Moralejo E, Woodward S, et al. (2007) Specific hybridization real-time PCR probes for Phytophthora ramorum detection and diagnosis. Forest Pathology 37: 403–408. [Google Scholar]
- 59. Bilodeau GJ, Martin FN, Coffey MD, Blomquist CL (2014) Development of a multiplex assay for genus- and species-specific detection of Phytophthora based on differences in mitochondrial gene order. Phytopathology 104: 733–748. 10.1094/PHYTO-09-13-0263-R [DOI] [PubMed] [Google Scholar]
- 60. Vettraino AM, Paolacci A, Vannini A (2005) Endophytism of Sclerotinia pseudotuberosa: PCR assay for specific detection in chestnut tissues. Mycological Research 109: 96–102. [DOI] [PubMed] [Google Scholar]
- 61. Dreaden TJ, Smith JA, Barnard EL, Blakeslee G (2012) Development and evaluation of a real-time PCR seed lot screening method for Fusarium circinatum, causal agent of pitch canker disease. Forest Pathology 42: 405–411. [Google Scholar]
- 62. Hughes KD, Tomlinson J, Giltrap P, Barton V, Hobden E, et al. (2011) Development of a real-time PCR assay for detection of Phytophthora kernoviae and comparison of this method with a conventional culturing technique. European Journal of Plant Pathology 131: 695–703. [Google Scholar]
- 63. Hughes KJD, Tomlinson JA, Griffin RL, Boonham N, Inman AJ, et al. (2006) Development of a one-step real-time polymerase chain reaction assay for diagnosis of Phytophthora ramorum . Phytopathology 96: 975–981. 10.1094/PHYTO-96-0975 [DOI] [PubMed] [Google Scholar]
- 64. Bilodeau GJ, Lévesque CA, de Cock AWAM, Duchaine C, Brière S, et al. (2007) Molecular detection of Phytophthora ramorum by real-time polymerase chain reaction using TaqMan, SYBR Green, and molecular beacons. Phytopathology 97: 632–642. 10.1094/PHYTO-97-5-0632 [DOI] [PubMed] [Google Scholar]
- 65. Schena L, Duncan JM, Cooke DEL (2008) Development and application of a PCR-based ‘molecular tool box’ for the identification of Phytophthora species damaging forests and natural ecosystems. Plant Pathology 57: 64–75. [Google Scholar]
- 66. Schena L, Nigro F, Ippolito A (2002) Identification and detection of Rosellinia necatrix by conventional and real-time Scorpion-PCR. European Journal of Plant Pathology 108: 355–366. [Google Scholar]
- 67. Schena L, Ippolito A (2003) Rapid and sensitive detection of Rosellinia necatrix in roots and soils by real-time Scorpion-PCR. Journal of Plant Pathology 108: 355–366. [Google Scholar]
- 68. Shishido M, Kubota I, Nakamura H (2012) Development of real-time PCR assay using TaqMan probe for detection and quantification of Rosellinia necatrix in plant and soil. Journal of General Plant Pathology 78: 115–120. [Google Scholar]
- 69. Børja I, Solheim H, Hietala AM, Fossdal CG (2006) Etiology and real-time polymerase chain reaction-based detection of Gremmeniella- and Phomopsis-associated disease in Norway spruce seedlings. Phytopathology 96: 1305–1314. 10.1094/PHYTO-96-1305 [DOI] [PubMed] [Google Scholar]
- 70. Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- 71. White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc; pp. 315–322. [Google Scholar]
- 72. Loppnau PA, Breuil C (2003) Species level identification of conifer associated Ceratocystis sapstain fungi by PCR-RFLP on a β-tubulin gene fragment. FEMS Microbiology Letters 222: 143–147. [DOI] [PubMed] [Google Scholar]
- 73. Jacobs K, Bergdahl DR, Wingfield MJ, Halik S, Seifert KA, et al. (2004) Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycological Research 108: 411–418. [DOI] [PubMed] [Google Scholar]
- 74. Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, et al. (2009) New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23: 35–40. 10.3767/003158509X470602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mbofung GY, Hong SG, Pryor BM (2007) Phylogeny of Fusarium oxysporum f. sp. lactucae inferred from mitochondrial small subunit, elongation factor 1-α, and nuclear ribosomal intergenic spacer sequence data. Phytopathology 97: 87–98. 10.1094/PHYTO-97-0087 [DOI] [PubMed] [Google Scholar]
- 76. O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. O'Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7: 103–116. [DOI] [PubMed] [Google Scholar]
- 78. Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species . Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- 81. Andrew M, Barua R, Short SM, Kohn LM (2012) Evidence for a common toolbox based on necrotrophy in a fungal lineage spanning necrotrophs, biotrophs, endophytes, host generalists and specialists. PLoS ONE 7: e29943 10.1371/journal.pone.0029943 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.