Abstract
IQGAP scaffolding proteins regulate many essential cellular processes including growth factor receptor signaling, cytoskeletal rearrangement, adhesion and proliferation, and are highly expressed in many cancers. Using genetically engineered human skin tissue in vivo, we demonstrate that diminished, sub-physiologic expression of IQGAP1 or IQGAP3 is sufficient to maintain normal epidermal homeostasis, while significantly higher levels are required to support tumorigenesis. To target this tumor-specific IQGAP requirement in vivo, we engineered epidermal keratinocytes to express individual IQGAP protein domains designed to compete with endogenous IQGAPs for effector protein binding. Expression of the IQGAP1-IQM decoy domain in epidermal tissue in vivo inhibits oncogenic Ras-driven MAPK signaling and antagonizes tumorigenesis, without disrupting normal epidermal proliferation or differentiation. These findings define essential non-redundant roles for IQGAP1 and IQGAP3 in epidermis, and demonstrate the potential of IQGAP antagonism for cancer therapy.
Introduction
The progression of invasive neoplasia frequently involves dysregulation of cellular signals that are integral to normal processes. Therapeutics against common oncogenic drivers often directly interfere with central components of core signaling pathways, and their clinical utility is limited by toxicity to normal cells. In this regard, blocking cell-signaling scaffolding molecules, in addition to oncogenic drivers, may be a promising strategy. As modulators of EGFR and MAPK signaling, IQGAPs may be useful therapeutic targets.
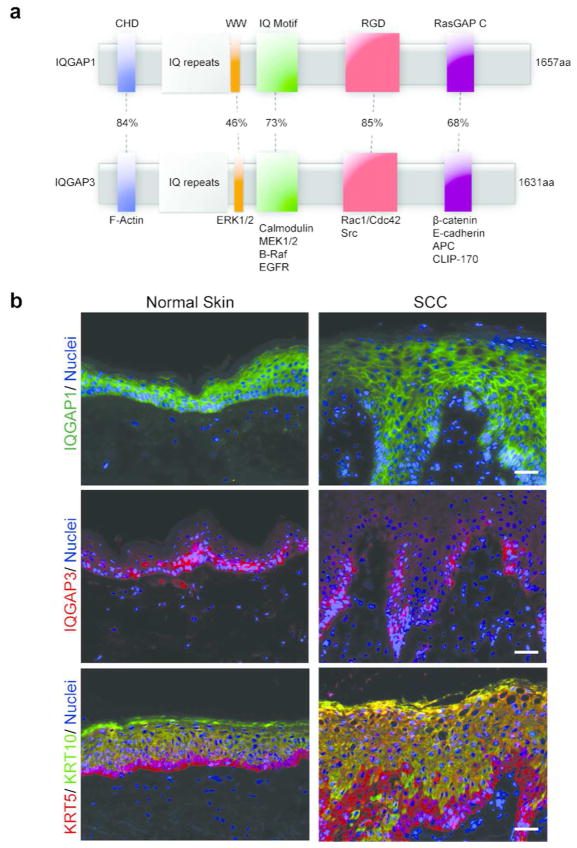
IQ motif-containing GTPase activating proteins (IQGAPs) are a family of ubiquitously-expressed scaffolding proteins that coordinate signaling pathways by facilitating physical interactions between effector proteins to enhance signal transduction (Brown and Sacks, 2009). The IQGAP family is comprised of three isoforms (IQGAP1, IQGAP2, and IQGAP3) which each contain multiple protein recognition domains: the F-actin-binding calponin homology domain (CHD); a polyproline-binding region (WW) that binds Erk 1/2; an IQ motif (IQM) that binds to MEK1/2, Raf, EGFR and Calmodulin; the Ras GAP related domain (RGD) which binds to Rac1 and Cdc42; and the C-terminal (RGCT) domain which interacts with E-cadherin, β-catenin, APC, and Clip-170 (Figure 1A) (Fukata et al., 2002; Johnson et al., 2009; White et al., 2009). Through these interactions, IQGAPs regulate growth factor receptor signaling, MAPK flux, cell-cell adhesion, cell cycle regulation and cytoskeletal rearrangements (Briggs and Sacks, 2003; White et al., 2012) – all critical cellular processes that are tightly coordinated both spatially and temporally for proper epidermal growth and differentiation.
Figure 1.
IQGAP1 and IQGAP3 are highly expressed in normal human epidermis and epidermal squamous cell carcinoma. (a) IQGAP1 and IQGAP3 are multidomain scaffolding proteins with highly homologous structural motifs and the ability to bind a range of cell signaling and cytostructrual molecules. (b) Cryosections of human normal and SCC tissue were prepared for immunofluorescence microscopy of IQGAP1 (green), IQGAP3 (red), and nuclei (blue), scale bar = 100 μm. IQGAP1 is expressed throughout the epidermal compartment of both normal and cancer tissue. IQGAP3 expression is concentrated within the proliferative basal layer. Keratin 5 (red) and Keratin 10 (green) differentiation markers are also shown in both normal and SCC tissue.
Despite the role that IQGAP scaffolds play in supporting these key cell functions, IQGAP1 null mice develop normally (Li et al., 2000). The lack of an overt phenotype suggests the possibility of functional rescue by another IQGAP isoform(s) in mice. In this regard, IQGAP3 seems the most likely candidate, as it shares 60% homology to IQGAP1 and has a relatively broad expression pattern (White et al., 2012). While IQGAP3 null murine tissue has not been generated, RNAi studies in the mouse epithelial cell line Eph4 demonstrate that, like IQGAP1, IQGAP3 also facilitates signaling through MAPK (Nojima et al., 2008). IQGAP2 expression is much more limited and expressed predominantly in the liver, gastro-intestinal tract, and platelets (Nojima et al., 2008). IQGAP2 is not expressed in skin. The effects of acute IQGAP loss in adult murine or human tissue have not been studied, but are potentially important to examine as phenotypes resulting from protein loss in developed tissues can vary significantly from those observed with loss during embryogenesis. Further, mouse and human tissues may differ with regard to their specific IQGAP requirements.
IQGAP1 is upregulated in many human cancers including lung, colon, liver, breast, glioblastoma, and melanoma (Clark et al., 2000), and regulates core cellular processes critical for malignant progression including proliferation, adhesion, and migration. Since progression to invasive neoplasia frequently involves dysregulation of normal cellular processes rather than activation of pathways completely lacking in the normal tissue, targeting central components of dysregulated pathways often results in high toxicity. As modulators rather than required core elements of key oncogenic signaling pathways, IQGAP1 or IQGAP3 may be uniquely poised therapeutic targets.
In this study, we define the roles of IQGAP1 and IQGAP3 in both human epidermal homeostasis and invasive epithelial cancer, by using genetically-defined human skin tissues maintained in their native, orthotopic location in vivo (Ridky et al., 2010). Tissues were generated using primary keratinocytes expressing shRNAs against IQGAP1 or IQGAP3. We show that the effects of IQGAP loss vary with the extent of the knockdown, as partial knockdown (50–85%) of either IQGAP1 or IQGAP3 was well tolerated in normal tissues, while more complete knockdown (85–95%) resulted in marked proliferation arrest and tissue failure in vivo. We also engineered human tissue grafts to express tumor-associated oncogenes sufficient to drive squamous cell carcinoma (SCC), and demonstrate a tumor-specific requirement for increased IQGAP1 and IQGAP3 in this cancer context. Finally, we developed an interfering decoy peptide based on the IQGAP-IQM domain that selectively inhibits oncogenic Ras activity in tissue. These findings establish the requirements of IQGAP1 and IQGAP3 in epidermal homeostasis and cutaneous tumor progression, and establish the potential therapeutic utility of targeting IQGAPs in human cancer.
RESULTS
IQGAP1 and IQGAP3 are expressed in normal human epidermis and in epidermal squamous cell carcinoma
Both normal human skin and spontaneous human SCC tumor tissues display robust IQGAP1 and IQGAP3 expression (Figure 1B). IQGAP1 is located throughout all layers of the epidermal compartment, whereas IQGAP3 expression is concentrated within the proliferative basal cells, and relatively absent in the post-mitotic suprabasal layers (Figure 1B, Figure S1). Within keratinocytes, IQGAP1 and IQGAP3 are localized throughout the cytoplasm, and are also concentrated at the plasma membrane. This expression and cellular distribution reflects the diversity of scaffolding activities and interacting effector proteins, and is consistent with IQGAP expression patterns in other tissues (Berglund et al., 2008; Nabeshima et al., 2002). We were unable to detect IQGAP2 protein in skin tissue or cultured primary keratinocytes (data not shown), consistent with its previously established limited tissue distribution (White et al., 2009; White et al., 2012).
IQGAP1 and IQGAP3 are both required for human epidermal keratinocyte proliferation
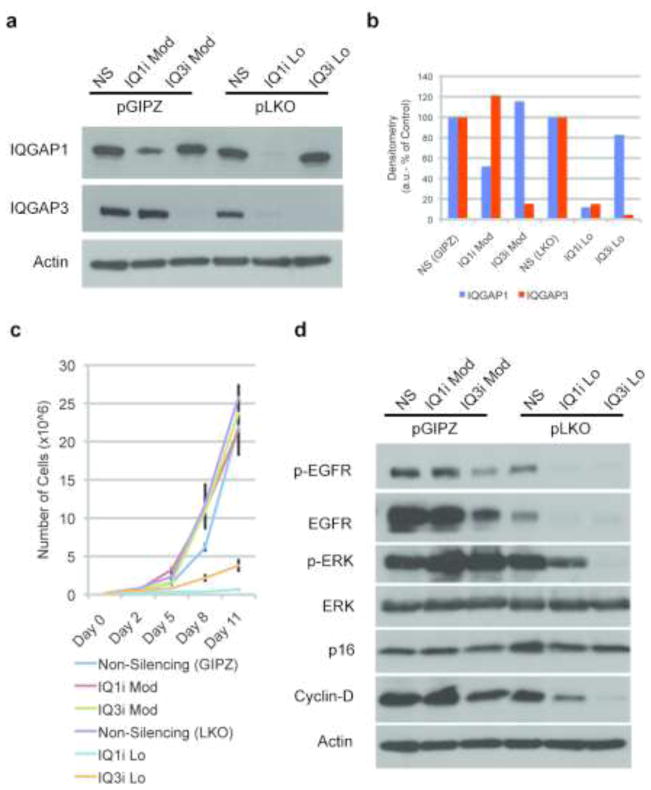
To determine the degree to which IQGAP1 and IQGAP3 are required for normal epidermal function, we generated stable populations of primary, early passage human IQGAP1 or IQGAP3 knockdown (IQGAP1i or IQGAP3i) keratinocytes utilizing lentiviral transduction to drive constitutive shRNA expression. Several lentivectors were used for each IQGAP to achieve varying levels of knockdown (Figure 2A, Figure S2). The pGIPZ, miR-30 based, IQGAP interference resulted in 50% IQGAP1 protein knockdown, and a 85% IQGAP3 protein knockdown, referred to here as IQ1iMod and IQ3iMod respectively. With these levels of knockdown, loss of either IQGAP1 or IQGAP3 does not impact the expression level of the other isoform. More complete knockdown was obtained with the pLKO U6-driven lentivectors, which resulted in IQGAP1 and IQGAP3 RNA knockdown of 90% and 96%, respectively, referred to as IQ1iLo and IQ3iLo (Figure 2A, B). In contrast to the moderate knockdowns, severe depletion of IQGAP1 also resulted in concurrent depletion of IQGAP3 protein. This is likely a consequence of the proliferative arrest in the IQ1iLo cells (discussed further below). IQGAP1 protein and transcript levels remained relatively normal in IQ3iLo cells (Figure 2A, B and Figure S3).
Figure 2.
Keratinocyte proliferation and mitogenic signaling require low-level expression of both IQGAP1 and IQGAP3. (a) Western analysis of primary human keratinocytes expressing constitutive pGIPZ-mediated shRNA interference (IQ1iMod and IQ3iMod) or pLKO-mediated knockdowns (IQ1iLo and IQ3iLo). (b) Quantification of the residual IQGAP1 and IQGAP3 protein expressions of IQ1iMod, IQ3iMod, IQ1iLo and IQ3iLo keratinocytes. (c) A proliferation assay reveals that a nearly-complete IQGAP knockdown in IQ1iLo and IQ3iLo keratinocytes completely arrests cell growth, while IQ1iMod and IQ3iMod cells proliferate at rates similar to that of the control (means ± SD). (d) Western analysis demonstrates IQ1iLo and IQ3iLo cells express markedly lower levels of both EGFR and p-EGFR compared to control or moderate knockdown cells. There is a corresponding reduction in phospho-ERK in IQ1iLo and IQ3iLo keratinocytes. (Stoll et al.)
While the IQ1iMod and IQ3iMod keratinocytes are morphologically indistinguishable from controls, and proliferate normally in culture, the growth of IQ1iLo and IQ3iLo keratinocytes was markedly inhibited (Figure 2C, Figure S2). This demonstrates that while normal cellular proliferation can be maintained with significantly diminished IQGAP1 or IQGAP3, they are not completely dispensable, nor functionally redundant in this context. The proliferative defect in primary human keratinocytes highlights a phenotypic difference between the human and mouse genetic systems. The observation that IQGAP3 expression is lost in the IQ1iLo cells is likely a consequence of the associated growth arrest, rather than a direct consequence of IQGAP1 loss itself, as IQGAP3 expression has been previously shown to be limited to proliferating cells (Nojima et al., 2008). To confirm that the decrease in IQGAP3 is not specifically dependent on IQGAP1 loss, primary keratinocytes were mitotically arrested with mitomycin-c. While IQGAP1 levels remained robust in mitomycin-c treated, non-proliferating cells, IQGAP3 levels dropped precipitously (Figure S1).
IQGAP1 and IQGAP3 are both required for EGFR activation and MAPK signaling
IQGAP scaffolds orchestrate a signaling axis from EGFR through the MAPK cascade, by physically binding to EGFR, BRaf, MEK1/2, and ERK1/2, and modulating their activities (Brown and Sacks, 2009; McNulty et al., 2011). In the absence of this scaffold, IQ1iLo and IQ3iLo cells displayed no detectable active phospho-EFGR, and also expressed less total EGFR protein (Figure 2D). This diminished EGFR activation is not the result of decreased EGFR transcription (Figure S4). It is also unlikey to result from decreased EGFR ligand expression, as the major keratinocyte EGFR ligands including amphiregulin (Stoll et al.) are only modestly affected by IQGAP antagonism (Figure S4), and exogenous EGF is routinely included in the cell culture media. In agreement with decreased EGFR activity, IQGAPi was associated with marked reduction in phospho-ERK. IQ1iLo and IQ3iLo keratinocytes also failed to maintain cyclin-D levels, consistent with loss of MAPK signaling (Diehl et al., 1997). As MAPK signaling flux downstream of EGFR is the primary mitogenic stimulus for epidermal keratinocytes, loss of this signaling is likely directly responsible for the proliferative arrest in IQ1iLo and IQ3iLo cells. Consistent with this, p16 protein was relatively unchanged (Figure 2D), indicating that the proliferative arrest likely results from loss of mitogenic signaling rather than cellular senescence. Further, the lack of proliferation was not a consequence of a prematurely engaged differentiation or apoptotic program (Figure S5). In contrast to IQ1iLo and IQ3iLo cells, IQ1iMod and IQ3iMod cells proliferated normally in culture without significant changes in MAPK signaling (Figure 2D).
Invasive squamous cell carcinoma requires higher levels of IQGAP1 and IQGAP3 than does normal epidermis
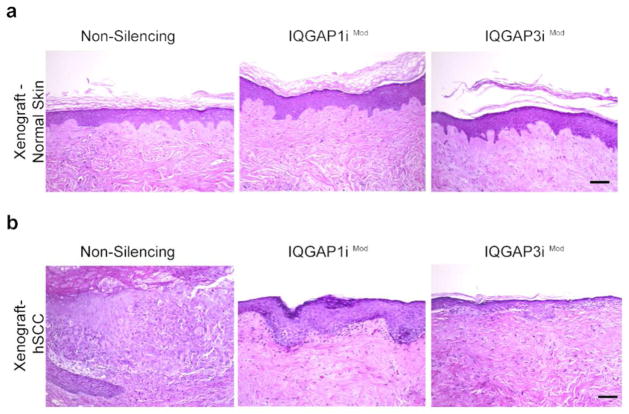
The profound proliferative defect in the IQGAPi cells was somewhat unexpected given the lack of overt phenotype in the IQGAP1 null mouse (Li et al., 2000). To explore the possibility that this difference was the result of in vitro 2-dimensional culture versus 3—dimensional tissue in vivo, we next used our human IQGAPi keratinocytes to regenerate 3–D organotypic skin tissues employing native human dermis with intact basement membrane (Ridky et al., 2010). These organotypic platforms support keratinocyte stratification and differentiation in vitro that recapitulate the structure and function of human skin. Engineered tissues can subsequently be grafted into the orthotopic location on the backs of immunodeficient mice for long-term in vivo study. Given that one of the hallmark features of epidermal tissues is a relatively high rate of basal cell proliferation and continual epidermal renewal, it was not surprising that attempts to establish 3–D in vitro organotypic, or in vivo xenograft skin using the nonproliferative IQ1iLo and IQ3iLo keratinocytes were unsuccessful. However, tissues generated using IQ1iMod and IQ3iMod keratinocytes, form functional epidermis both in vitro and in vivo. These tissues display normal basal proliferation and execute a normal stratification and differentiation program (Figure 3A). Thus, the phenotype of the 3–D organotypic experiments parallels that seen in 2–D cultures, and highlights the human requirement for basal maintenance of both IQGAP1 and IQGAP3 for normal epidermal homeostasis.
Figure 3.
Invasive epidermal SCC requires higher IQGAP1 and IQGAP3 expression than normal epidermis. (a) Moderate IQGAP1 and IQGAP3 knockdowns in normal human epidermal xenografts are well tolerated, with no discernable effects on tissue architecture or maintenance in vivo. (b) Moderate knockdowns of IQGAP1 and 3 markedly attenuate invasive SCC development in keratinocytes expressing H-RasG12V and CDK4R24C. Control cancer tissues form thick, hyperplastic, deeply invasive tumors, whereas IQ1iMod and IQ3iMod tissues are minimally hyperplastic, and lack significant invasion, Scale bar = 100 μm.
After establishing the IQGAP requirements in normal epidermis, we then explored the role of IQGAP in epidermal tumorigenesis using a genetically defined, in vivo SCC model (Lazarov et al., 2002). For this approach, stable populations of non-silencing or IQGAPiMod keratinocytes were engineered to express constitutively active oncogenic Ras and CDK4. These two oncogenic drivers alter centrally acting pathways frequently deregulated in spontaneous human SCC (Lazarov et al., 2002; Ridky et al., 2010). When used to generate surface skin grafts in vivo, oncodriver-expressing keratinocytes harboring non-silencing shRNA control formed hyperproliferative tumors that degraded the basement membrane and invaded deeply into supporting stroma – recapitulating hallmark features of spontaneous human SCC. In contrast, IQ1iMod and IQ3iMod grafts displayed marked attenuation of the tumor phenotype (Figure 3B). IQ1iMod and IQ3iMod tissues were only mildly hyperplastic, with only a few focal areas of early invasion. The baseline MAPK activation that persists in IQ1iMod and IQ3iMod keratinocytes (and is sufficient to support normal epidermis) is insufficient to support the full oncogenic effects of active Ras. These results indicate that significantly higher levels of IQGAP1 and IQGAP3 are necessary for the initiation and progression of invasive epidermal neoplasia than for the maintenance of normal human skin, and that IQGAP may therefore be a useful therapeutic target for human epidermal cancer. Consistent with this, IQGAP1 null mice are somewhat resistant to skin cancer induced by topical carcinogen (DMBA/TPA) application (Jameson et al., 2013).
Oncogenic Ras effects are blocked with decoy peptides based on the IQGAP IQM domain
The markedly attenuated tumor invasion and proliferation observed with IQGAP knockdown suggests that IQGAP may be a useful therapeutic target for cancer. To begin exploring this possibility we sought to antagonize IQGAP activity by targeting its interactions with protein effectors in a domain-specific fashion. To accomplish this, we constructed five lentiviruses, each designed to drive expression of a single IQGAP structural domain. We hypothesized that expression of these isolated IQGAP peptide domain “decoys” in keratinocytes may compete with endogenous IQGAP for effector binding and interfere with signaling.
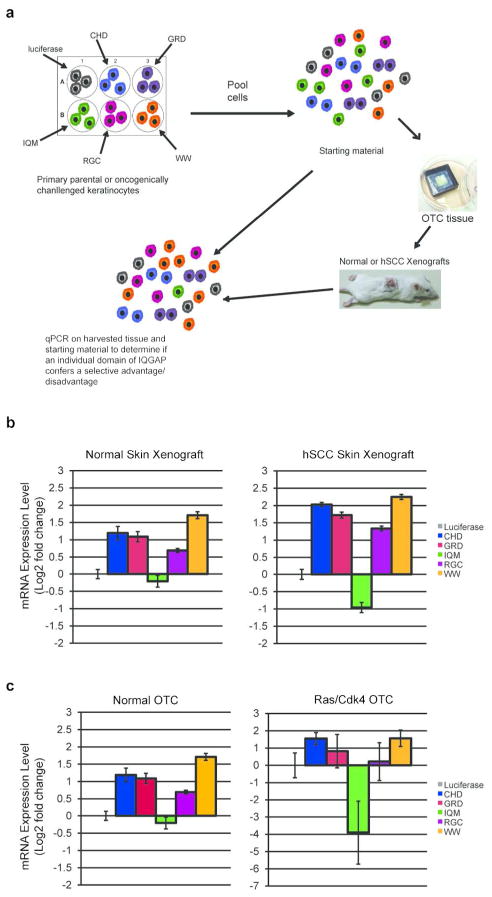
To quantitatively determine the degree of fitness advantage or disadvantage conferred by each of the domains in the context of intact epidermal tissue, we utilized a multiplexed competition assay (Figure 4A). In this approach, six different populations of keratinocytes, each engineered to express a single IQGAP domain (or luciferase control), along with a uniquely bar-coded fluorescent protein, were pooled in equal ratios and used to generate epidermis in vivo. After 4 weeks of in vivo selection, tissues were harvested and analyzed. Quantitative polymerase chain reaction (qPCR) was used to determine the relative representation of each cell population in the final resulting tissue, which was compared to that in the starting cell mixture. In the context of otherwise normal skin, none of the decoy peptides resulted in a fitness disadvantage, while four conferred a slight fitness advantage. However, in Ras-driven SCC tissue, cells expressing the isolated IQM domain displayed a significant fitness disadvantage (16-fold) (Figure 4B). Similar effects were observed in organotypic in vitro tissues with and without Ras oncogene challenge (Figure 4C). The expression of the IQM domain did not strongly alter endogenous expression of either IQGAP1 or IQGAP3, nor did it affect EGFR expression in primary keratinocytes (Figure S6). Similar inhibitory effects were not observed with any of the other 4 IQGAP domain peptides.
Figure 4.
Exogenous expression of IQGAP1-based decoy peptides alters the relative fitness of keratinocytes in organotypic tissues. (a) A multiplexed, internally-controlled, competition assay was utilized to assess relative cellular fitness of distinct, genetically-engineered keratinocyte populations. Keratinocytes expressing a single IQGAP domain (or luciferase control), were pooled in equal ratios and used to regenerate 3–D organotypic skin tissues. Tissues were either maintained in culture or xenografted onto immunodeficient mice. (b) Expression of individual IQGAP peptides does not adversely affect keratinocyte fitness in normal skin in vivo. However, keratinocytes expressing the IQM domain displayed a fitness disadvantage in tumor tissue. (c) In vitro organotypic cultures using the same cell populations from (b) showed similar selection against IQM expressing cells in Ras-expressing tissue (mean ± SD).
To more fully characterize the effects of IQGAP-IQM expression in human tissue, and to validate the effects seen in the multiplexed screen, we next expressed the IQM domain individually in engineered human skin xenografts. IQGAP-IQM expression in otherwise normal epidermis in vivo had no discernable effect on skin architecture or morphology (Figure 5A).
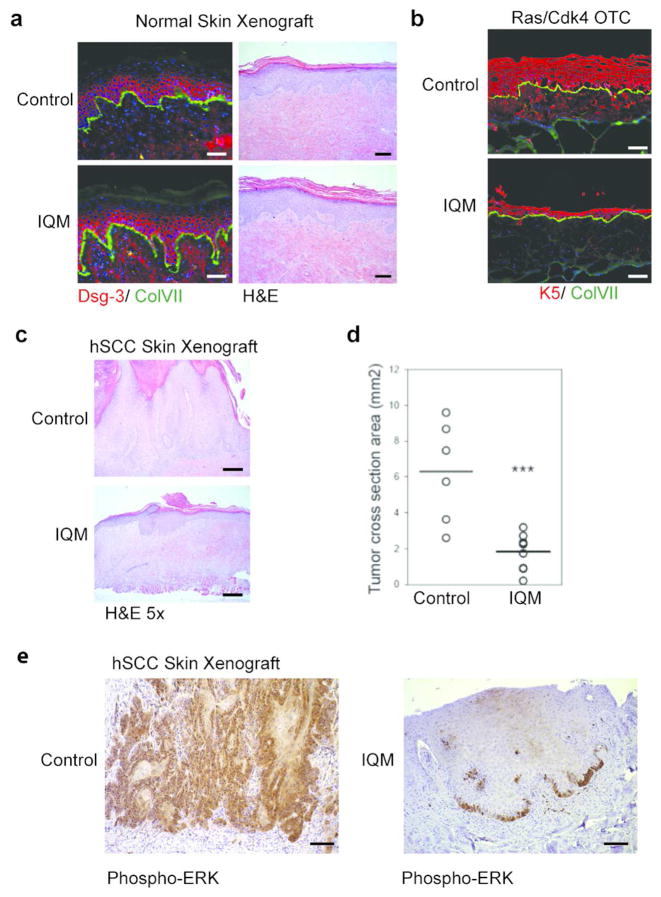
Figure 5.
Expression of the IQGAP1-IQM decoy peptide inhibits invasive SCC. (a) IQGAP1-IQM expression does not adversely affect normal epidermis in vivo, scale bar = 100 μm. (b) IQGAP1-IQM expression inhibits keratinocyte proliferation and invasion in Ras-expressing neoplastic skin in vitro, scale bar = 100 μm, and in vivo(c), scale bar = 250 μm. (d) Relative tumor cross-section area of xenograft control versus IQGAP1-IQM SCC tumors. (e) MAPK signaling as indicated by phospho-ERK expression is markedly diminished in the IQM-expressing tissue, scale bar = 100 μm.
We then explored the effects of this single peptide in the SCC setting. IQGAP1-IQM keratinocytes were transformed with oncogenic Ras and CDK4, and used to establish both in vitro and in vivo organotypic skin tissues. In vitro, Ras expressing control tissues are hyperplastic, architecturally disordered, and degrade basement membrane with deep epithelial cell invasion into supporting stroma. The IQM decoy peptide significantly limits these oncogenic Ras effects, as tissues are markedly less hyperplastic, and lose invasive capacity (Figure 5B). Consistent with these in vitro results, IQM-expression also inhibits Ras oncogenic effects in xenografts in vivo. Resulting neoplastic IQM-tissues were markedly less hyperproliferative than controls (Figure 5C), displayed less architectural disorder, and contained only small focal areas of modest invasion. In contrast, control tumors were markedly larger and deeply invasive throughout the tumor (Figure 5D). Consistent with this significantly attenuated cancer phenotype, MAPK activity downstream of oncogenic Ras was also markedly reduced in the IQM expressing grafts, as evidenced by markedly decreased phospho-ERK (Figure 5E). The diminished size of IQM-expressing tumors is likely the direct result of the loss of MAPK flux. Correspondingly, IQM-expressing tumors display a significantly lower ki67+ proliferative index than control (Figure S6). Diminished IQM-tumor progression is not the result of increased apoptosis or premature differentiation (as determined by TUNEL, keratin 10, and filaggrin) (Figure S6).
Discussion
Proteins that are essential for neoplastic transformation, but relatively dispensable for normal tissue homeostasis, are ideal cancer targets. Here we demonstrate in vivo that IQGAP requirements are significantly higher in neoplastic epidermis, than in normal skin. Further, we demonstrate that IQGAP antagonism via IQM domain decoys may have potential as a therapeutic strategy for human cancer. As most human malignancies require increased Ras-Raf-MEK-ERK signaling, targeting the MAPK scaffolding function of IQGAPs may be effective as an orthogonal therapeutic approach to inhibiting the core Ras-MAPK elements directly.
Although IQGAP downregulation is well-tolerated, it seems that there is a critical minimum expression level of IQGAP1 and IQGAP3 that is required for maintaining primary keratinocyte proliferation. This demonstrates that endogenous IQGAP1 and IQGAP3 are not functionally redundant. There is no compensatory upregulation of one isoform with knockdown of the other. While our studies suggest that IQGAP1 and IQGAP3 function similarly in the maintainence of MAPK activity, they may differ with regard to their other tissue-context specific scaffolding roles, including regulation of cytoskeletal remodeling proteins. While IQGAP1 is expressed throughout the epidermal compartment and tumor, IQGAP3 expression is most prominent within the basal layer of normal skin and the advancing font of SCC tumors, where there may be a requirement for higher total levels of collective IQGAP activity.
The isolated IQM domain likely influences multiple IQGAP activities within keratinocytes that together results in less MAPK activity and attenuation of Ras-driven tumorigenesis. IQM was designed to interfere with oncogenic Ras signaling by competing with endogenous full-length IQGAP for binding to Raf and MEK, and thus limiting assembly of the coordinated signaling complex required for ERK phosphorylation. The IQM peptide may also inhibit Ras-induced paracrine signaling at the level of EGFR, a potent mitogenic signal for keratinocytes in culture or in tumor formation (Dlugosz et al., 1997) that signals through Ras-MAPK.
Currently available drugs targeting the MAPK pathway are small molecule, cell-permeable inhibitors. IQGAPs do not possess kinase activity and are not cell-surface proteins. Targeting them inside cells will therefore require a non-traditional approach. Engineering IQM domain with polyarginine or HIV TAT protein sequences would potentially allow exogenously supplied recombinant proteins to permeate tumor tissue. The feasibility of this strategy was recently demonstrated for an epitope tagged, polyarginine modified WW domain peptide, which when continually infused, slowed the growth of mouse pancreatic cancers, and tumor cell lines grown subcutaneously in immunodeficient mice (Jameson et al., 2013). While we included the unmodified IQGAP WW domain in our initial screen, we did not detect any anti-tumor effects in our system. The reasons for this are unclear, but may relate to the addition of polyarginine, or epitope sequences in the other study, or the use of different in vivo tumor models.
In summary, our findings define non-redundant roles for IQGAP1 and IQGAP3 in normal human skin, and establish an elevated tumor-specific IQGAP requirement that can be targeted to inhibit Ras-driven human cancer growth in vivo.
Materials and Methods
Cell culture
All experiments were conducted using primary keratinocytes isolated from normal human skin by methods delineated previously (Lazarov et al., 2002; Ridky et al., 2010). Keratinocytes were cultured in a 1:1 mix of Gibco Keratinocytes-SFM medium + L-glutamine + EGF + BPE and Gibco Cascade Biologics 154 medium with 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). Transduced keratinocytes were fully selected before the commencement of each experiment. CDK4/Ras challenged keratinocytes were transformed prior to the initiation of three-dimensional cultures.
Lentiviral and Retroviral Production and Transduction
293T and 293-derivitive Phoenix cells (Swift et al., 2001) were cultured in Dulbecco’s Modified Eagles Medium (DMEM) supplemented with 5% FBS containing Antibiotic/Antimycotic. Lentiviral and retroviral particle were generated according to Thermoscientific specifications and as described previously (Chudnovsky et al., 2005; Lazarov et al., 2002; Ridky et al., 2010). Lentiviral shRNA constructs against IQGAP (OpenBiosystems, Lafayette, CO) were as follows: pGIPZ/sh-IQGAP1 (antisense: TGATATCCATGAGATTGAG), pGIPZ/sh-IQGAP3 (antisense: TGTCTTCTAGCACATCCTG), pLKO1/sh-IQGAP1 (antisense: AATGGAAATGTAGATTACTGG), pLKO1/sh-IQGAP3 (antisense: AACTTATCAGTCATGGCGAGG). Additional RNAi vectors targeting IQGAP1 at nucleotide position 864–884 (pLKO) and 4243–4261 (pGIPZ), and IQGAP3 at nucleotide 2848–2866 (pGIPZ) and 4120–4140 (pLKO) were used to validate findings, and displayed similar effects on keratinocyte growth (Figure S2). Individual IQGAP1 protein binding domains and luciferase were cloned into the pRRL lentivector. For the production of viral particles, lentiviral constructs were co-transfected with viral packaging plasmids pCMVΔR8.91 and pUC-MDG into 293T cells using Fugene 6 Transfection Reagent (Promega, Fitchburg, WI).
For oncogenic transformation, 293-derivitive Phoenix cells stably transfected with H-RasG12V or CDK4R24C were used to generate retroviral particles. Keratinocytes that were previously transduced with IQGAP knockdowns, non-silencing controls, or individual IQGAP domain expressing constructs, were then transformed with these retroviral particles as described previously (Chudnovsky et al., 2005; Ridky et al., 2010).
Proliferation assay
Cells were originally plated at 10,000 cells per cm2. At the indicated time points, cells were trypsinized and manually counted. Results are the means of three biological replicates (± s.d.).
Western Analysis
Cells were lysed in RIPA Buffer containing 1X protease inhibitors and 1X phosphatase inhibitors (Roche, Branchburg, NJ), and extracts analyzed by SDS-PAGE and western blotting. Primary antibodies used in this study include IQGAP1 (Millipore, Billerica, MA), IQGAP3 (Abcam, Cambridge, MA), Actin, pEGFR, EGFR, pERK, ERK, and cyclinD (Cell Signaling, Danvers, MA), and p16 (BD Pharmingen, San Diego, CA). After incubation with the appropriate secondary antibody, proteins were detected using either Luminata Crescendo Western HRP Substrate (Millipore, Billerica, MA) or ECL Western Blotting Analysis System (GE Healthcare, Lafayette, CO).
Organotypic culture and skin xenografts
Organotypic skin cultures were established using genetically engineered keratinocytes expressing IQGAP knockdowns, non-silencing controls, active Ras and CDK4 oncogenes, and individual IQGAP1 peptide domains. For each culture, between 8.0 × 105 and 1.0 × 106 keratinocytes were suspended in 80 μL KGM, seeded onto devitalized human dermis, and incubated at 37 °C at an air-liquid interface for three days prior to xenografting or for time indicated for in vitro OTC cultures according to previously established methods (Chudnovsky et al., 2005; Duperret et al., 2014; Ridky et al., 2010).
For xenografting, OTCs were surgically grafted onto the back skin of 5–7 week old female ICR SCID mice (Taconic, Centre Hudson, NY). For each in vivo experiment, 2–3 animals were used per condition, and results were verified over three experimental replicates. Engineered tissues were generated by methods delineated previously (Lazarov et al., 2002; Ridky et al., 2010).
For competition assays (Figure 4A), distinct populations of keratinocytes were engineered to express CHD, WW, IQM, GRD and RasGAP, or a luciferace control along with drug selectable markers and identifying barcodes, with or without oncogenic Ras/CDK4. These keratinocytes were then pooled in equal ratios and used to establish organotypic cultures, which were either maintained in vitro or xenografted on mice. To determine the relative fitness between the keratinocytes, transcript levels of the non-coding barcode for each of the domain decoys was analyzed against its relative abundance in the starting cell population by quantitative PCR. Three biological replicates were performed for each condition. All experiments involving animals were performed in accordance with a University of Pennsylvania IACUC-approved protocol.
Quantitative RT/PCR
RNA was extracted from cells and tissues according to the RNeasy Mini Kit protocol (Qiagen, Valencia, CA), and reverse transcribed to cDNA using the High Capacity RNA-to cDNA kit (Applied Biosystems, Grand Island, NY). Quantitative PCR of resulting cDNA was conducted using Power SYBR Green Master Mix (Applied Biosystems, Grand Island, NY) and gene-specific primers, in triplicate, on a ViiA 7 Real-Time PCR System (Life Technologies, Grand Island, NY). Relative expression was determined using the 2-[delta][delta] Ct method.
Immunofluorescence microscopy
Whole mount cryosections were prepared for immunofluorescence microscopy as previously described (Ridky et al., 2010). Briefly, slides were fixed in 4% paraformaldehyde or −20°C methanol, permeabilized as required and blocked with 5% goat serum/PBS, followed by incubation with primary antibodies, and secondary antibodies conjugated to fluorophores. Slides were mounted with Prolong Gold Antifade Reagent with DAPI (Life Technologies, Grand Island, NY). The primary antibodies used in this study were IQGAP1 and Collagen VII (Millipore, Billerica, MA), IQGAP3 (Abcam, Cambridge, MA), Desmogelin-3 (Invitrogen, Carlsbad, CA), and Keratin-5 (Covance, Conshohocken, PA).
Immunohistochemistry
Formalin fixed paraffin embedded (FFPE) tissue sections from xenografts were stained for pERK protein expression using peroxidase/DAB (diaminobenzidine)-complex method (EXPOSE biotin free detection kit, Abcam, Cambridge, MA). Tissue sections were deparaffinized and rehydrated. To retrieve the antigens, tissue sections were microwaved in 10mM Citrate buffer pH 6.0. To quench endogenous peroxidase, tissues were incubated in 3% H2O2. Afterward tissues were incubated with primary antibody to phospho-ERK (Cell Signaling) for three hours at room temperature. The secondary antibody, HRP conjugated goat anti-rabbit, was supplied with the kit and incubated 18 min at room temperature. Following multiple washes in TBS the antigen-antibody complex was visualized with 3.3% DAB-solution. The tissues were counterstained with hematoxylin and dehydrated prior to mounting coverslips.
Supplementary Material
Acknowledgments
The authors thank the University of Pennsylvania Skin Disease Research Center (SDRC) for primary keratinocytes and immunohistochemistry, and John Seykora, Sarah Millar, and George Cotsarelis, for critical pre-submission manuscript review. This work was supported by The National Institutes of Health (NIH) (R01CA163566, T.W.R.), and the University of Pennsylvania Undergraduate Research Mentoring Program (E.S.). EKD is supported by an NIH/NIAMS training grant (5-T32-AR0007465-30)
Abbreviations
- SCC
squamous cell carcinoma
- IQGAPs
IQ motif-containing GTPase activating proteins
- IQM
IQ motif
- NS
Non-silencing
Footnotes
Conflict of Interest
The authors state no conflict of interest.
References
- Berglund L, Bjorling E, Oksvold P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Molecular & cellular proteomics: MCP. 2008;7:2019–27. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO reports. 2003;4:571–4. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cellular signalling. 2009;21:462–9. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovsky Y, Adams AE, Robbins PB, et al. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nature genetics. 2005;37:745–9. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, Golub TR, Lander ES, et al. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–72. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- Dlugosz AA, Hansen L, Cheng C, et al. Targeted disruption of the epidermal growth factor receptor impairs growth of squamous papillomas expressing the v-ras(Ha) oncogene but does not block in vitro keratinocyte responses to oncogenic ras. Cancer Res. 1997;57:3180–8. [PubMed] [Google Scholar]
- Duperret EK, Oh SJ, McNeal A, et al. Activating FGFR3 mutations cause mild hyperplasia in human skin, but are insufficient to drive benign or malignant skin tumors. Cell cycle (Georgetown, Tex) 2014;13 doi: 10.4161/cc.28492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Watanabe T, Noritake J, et al. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–85. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- Jameson KL, Mazur PK, Zehnder AM, et al. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med. 2013;19:626–30. doi: 10.1038/nm.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Sharma M, Henderson BR. IQGAP1 regulation and roles in cancer. Cellular signalling. 2009;21:1471–8. doi: 10.1016/j.cellsig.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Lazarov M, Kubo Y, Cai T, et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat Med. 2002;8:1105–14. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- Li S, Wang Q, Chakladar A, et al. Gastric hyperplasia in mice lacking the putative Cdc42 effector IQGAP1. Molecular and cellular biology. 2000;20:697–701. doi: 10.1128/mcb.20.2.697-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty DE, Li Z, White CD, et al. MAPK scaffold IQGAP1 binds the EGF receptor and modulates its activation. J Biol Chem. 2011;286:15010–21. doi: 10.1074/jbc.M111.227694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Shimao Y, Inoue T, et al. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: its overexpression in carcinomas and association with invasion fronts. Cancer letters. 2002;176:101–9. doi: 10.1016/s0304-3835(01)00742-x. [DOI] [PubMed] [Google Scholar]
- Nojima H, Adachi M, Matsui T, et al. IQGAP3 regulates cell proliferation through the Ras/ERK signalling cascade. Nature cell biology. 2008;10:971–8. doi: 10.1038/ncb1757. [DOI] [PubMed] [Google Scholar]
- Ridky TW, Chow JM, Wong DJ, et al. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat Med. 2010;16:1450–5. doi: 10.1038/nm.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll SW, Johnson Jl, Bhasin A, Johnston A, et al. Metalloproteinase-mediated, context-dependent function of amphiregulin and HB-EGF in human keratinocytes and skin. doi: 10.1038/jid.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, Lorens J, Achacoso P, et al. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 10.7C. Chapter 10. 2001. [DOI] [PubMed] [Google Scholar]
- White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS letters. 2009;583:1817–24. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cellular signalling. 2012;24:826–34. doi: 10.1016/j.cellsig.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.