Abstract
Background
Prostatic ductal adenocarcinoma is an unusual and aggressive morphologic subtype of prostate cancer. PTEN gene deletion and ERG gene rearrangement are among the most common genomic changes in acinar prostate cancers. Though ductal adenocarcinoma most commonly occurs with synchronous usual-type acinar adenocarcinoma, little is known about the molecular phenotype of these mixed tumors.
Methods
We used genetically validated immunohistochemistry (IHC) assays to assess PTEN and ERG status in a group of 37 surgically treated ductal adenocarcinomas and 18 synchronous acinar adenocarcinomas where we have previously reported ERG gene rearrangement status by fluorescence in situ hybridization (FISH). A group of 34 stage and grade-matched pure acinar adenocarcinoma cases was studied as a control.
Results
ERG IHC was highly concordant with ERG FISH results, with 100% (36/36) concordance among ductal adenocarcinomas and 91% (31/34) concordance among 34 pure acinar carcinomas. Similar to previous FISH results, ERG expression by IHC was significantly less common among ductal adenocarcinomas (11% or 4/37) and their synchronous acinar tumors (6% or 1/18) compared to matched pure acinar adenocarcinoma cases (50% or 17/34; p= 0.0005 and 0.002, respectively). PTEN loss by IHC was also less common among ductal adenocarcinomas (18% or 6/34) and their synchronous acinar tumors (22% or 4/18) compared to matched pure acinar carcinomas (50% or 17/34; p = 0.01 and 0.08, respectively). As expected, PTEN loss was enriched among ERG positive compared to ERG-negative tumors in the pure acinar tumor control group (2.5-fold enrichment; p=0.04) however this was not observed among the ductal adenocarcinomas (1.5 fold enrichment; p=NS). Of ductal adenocarcinomas with an evaluable synchronous acinar component, ERG status was concordant in 94% (17/18) and PTEN status was concordant in 94% (16/17).
Conclusions
Based on PTEN and ERG, ductal adenocarcinomas and their concurrent acinar carcinomas may be clonally related in some cases and show important molecular differences from pure acinar carcinoma.
Keywords: Prostatic adenocarcinoma, ductal adenocarcinoma, endometrioid, PTEN, ERG, immunohistochemistry
Introduction
Ductal adenocarcinoma of the prostate is a rare morphologic subtype, accounting for less than 1% of all cases in its purest form and occurring intermixed with usual type acinar adenocarcinoma in as many as 5% of prostate cancer cases [1–4]. Classically arising in or involving the large periurethral ducts near the verumontanum, ductal adenocarcinoma frequently presents with urinary symptoms and is associated with significantly lower serum PSA levels compared to acinar prostate carcinoma cases [2, 4, 5]. Most contemporary studies have shown that ductal adenocarcinomas or mixed ductal and acinar tumors with >10% ductal component are associated with a higher stage at radical prostatectomy compared to acinar adenocarcinomas and may be less responsive to hormonal therapies [1, 3, 6–8]. Accordingly, ductal adenocarcinoma is more likely to present with distant metastasis than acinar tumors and shows a two-fold increase in risk of prostate cancer specific mortality in population-based studies [2, 4]. However, despite compelling data to suggest important clinical-pathologic differences between ductal and acinar prostatic adenocarcinoma, there have been relatively few molecular studies of the relationship between these two entities [9, 10].
With the advent of next generation sequencing, the genomic landscape of acinar prostate cancer has become increasingly clear. Although the mutation rate is relatively low compared to other epithelial tumor types, copy number alterations and rearrangements are common [11–14]. Among these, rearrangements involving the ERG gene or other ETS family members and deletion of the PTEN tumor suppressor gene are some of the most common alterations seen in primary acinar carcinomas. Though ERG rearrangements are not associated with altered prognosis in most surgically treated cohorts [15, 16], PTEN deletions are strongly associated with increased pathologic grade and stage and poor clinical outcomes [17–32]. Of note, PTEN loss likely occurs subsequent to ERG gene rearrangements in the majority of acinar adenocarcinomas [33–35] and is more common in ERG-rearranged tumors for reasons that remain unclear [13, 27–29, 33, 35–40].
To begin to molecularly subclassify prostate ductal adenocarcinoma, we used genetically validated immunohistochemistry assays to examine ERG and PTEN status in ductal tumors and their concurrent acinar components, as well as in a matched control group of pure acinar tumors. Previously, we used fluorescence in situ hybridization (FISH) in the same cohorts to demonstrate that ERG gene rearrangements occur significantly less frequently in prostatic ductal adenocarcinomas compared to pure acinar carcinomas [9]. Though ERG protein expression is highly correlated with ERG gene rearrangement in untreated acinar carcinomas, this concordance is less robust in subtypes of prostatic carcinoma with decreased androgen signaling and relatively low PSA levels, including small cell carcinoma [41, 42] and prostatic adenocarcinomas exposed to androgen deprivation therapy [43]. Because ductal carcinoma typically presents with lower levels of PSA, we hypothesized that androgen signaling might be decreased in this tumor subtype and thus there might be a greater discordance between ERG protein expression and gene rearrangement status than seen in acinar carcinoma.
Here, we show that ERG expression is, in fact, highly correlated with underlying ERG gene rearrangement status in ductal adenocarcinomas. Further, we demonstrate that PTEN loss occurs significantly less frequently in ductal adenocarcinoma and associated acinar carcinoma compared to pure acinar carcinoma and is not significantly enriched among ERG-positive ductal adenocarcinomas as it is in ERG-positive acinar carcinomas. Finally, using combined PTEN and ERG status, we provide additional evidence of a likely clonal relationship between concurrent ductal and acinar adenocarcinomas in some cases.
Materials and Methods
Tissue Selection
The tumor cohorts utilized in this study have been described in detail previously [9, 44] and the study was approved by the Johns Hopkins IRB. Briefly, two tissue microarrays (TMAs) were manually constructed from 39 radical prostatectomy specimens with ductal adenocarcinoma retrieved from the surgical pathology and consultation files of the Johns Hopkins Hospitals from 1984–2005. Overall, 95% (37/39) had evaluable tumor tissue present on the TMAs for ERG and/or PTEN immunohistochemistry. In 49% (18/37) of these cases, a concurrent acinar component was present and included and evaluable in the TMA. The ductal and acinar adenocarcinoma components were considered geographically separate primaries when the different morphologic components were separated by at least 5 mm in all dimensions. This was the case in 33% of tumors with a concurrent acinar component (6/18, Table 1). In 66% (12/18), the ductal and acinar tumor components were intermingled, or closer than 5 mm from one another (Table 1). In each case, quadruplicate 0.6 mm cores were punched from the ductal adenocarcinoma component, the acinar component (when present) and the surrounding benign prostatic tissue, with up to 16 total cores from each patient represented on the array.
Table 1.
| Case #: | Pathologic stage | Overall Gleason score | Spatial relationship | ERG FISH ductal component* | ERG FISH acinar component* | ERG IHC ductal component | ERG IHC acinar component | PTEN IHC ductal component | PTEN IHC acinar component |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TX NX MX | U | D | 1 | 1 | ||||
| 2 | TX NX MX | U | N | 0 | 1 | ||||
| 3 | TX NX MX | U | N | 0 | 1 | ||||
| 4 | T3A N0 MX | 7 | N | 0 | 1 | ||||
| 5 | T3A N0 MX | 7 | N | 0 | 0 | ||||
| 6 | T2 N0 MX | 7 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 7 | TX NX MX | U | Unknown | N | N | 0 | 0 | 1 | 1 |
| 8 | T3B N0 MX | 7 | Comingled | N | N | 0 | 0 | 0 | 0 |
| 9 | T2 N0 MX | 7 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 10 | T3A N0 MX | 7 | D | 1 | 1 | ||||
| 11 | T3B N0 MX | 8 | N | 0 | A | ||||
| 12 | T3A N0 MX | 8 | Unknown | N | N | 0 | 0 | 0 | 0 |
| 13 | T3A N0 MX | 8 | N | 0 | 1 | ||||
| 14 | T3B N0 MX | 7 | N | 0 | 1 | ||||
| 15 | T3A N0 MX | 7 | Separate | N | N | 0 | 0 | A | 1 |
| 16 | T3A N0 MX | 7 | Comingled | T | T | 1 | 1 | 0 | 0 |
| 17 | T2 N0 MX | 7 | Comingled | N | N | 0 | 0 | A | A |
| 18 | TX N0 MX | 7 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 19 | T3A N0 MX | 7 | N | 0 | 1 | ||||
| 20 | T2 N0 MX | 7 | N | 0 | 1 | ||||
| 21 | T2 N0 MX | 8 | Comingled | N | N | 0 | no tumor | 1 | 0 |
| 22 | T2 N0 MX | 8 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 23 | T2 N0 MX | 7 | N | no tumor | no tumor | ||||
| 24 | T2 N0 MX | 8 | Unknown | N | N | 0 | 0 | 1 | 1 |
| 25 | T3A N0 MX | 7 | N | 0 | 1 | ||||
| 26 | T3A N0 MX | 8 | N | 0 | 1 | ||||
| 27 | T3B N1 MX | 9 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 28 | TX N0 MX | 7 | N | no tumor | no tumor | ||||
| 29 | T3A N0 MX | 7 | Separate | N | N | 0 | 0 | 1 | 1 |
| 30 | T3A N0 MX | 8 | Separate | D | N | 1 | 0 | 1 | 1 |
| 31 | T2 N0 MX | 7 | Comingled | N | N | 0 | no tumor | 1 | no tumor |
| 32 | T2 N0 MX | 7 | Separate | N | N | 0 | no tumor | 1 | no tumor |
| 33 | T2 N0 MX | 7 | N | 0 | 1 | ||||
| 34 | T2 N0 MX | 8 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 35 | T2 N0 MX | 8 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 36 | T3B N1 MX | 8 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 37 | T3B N0 MX | 9 | Comingled | N | N | 0 | 0 | 1 | 1 |
| 38 | T3A N0 MX | 7 | N | 0 | 0 | ||||
| 39 | T3A N0 MX | 7 | NA | 0 | 0 |
FISH results presented for comparison purposes and previously published in Reference 9.
N= Normal ERG FISH; D=ERG gene rearrangement via deletion; T=ERG gene rearrangement via translocation; 0=absence of staining by IHC; 1=staining present by IHC; NA=not assessed; U=unknown.
As an internal control group, 14 “pure” acinar adenocarcinoma cases (Gleason pattern 4) were included and evaluable on the ductal adenocarcinoma tissue microarray. To provide additional pure acinar adenocarcinoma controls, an additional 20 acinar adenocarcinoma cases were selected for matched pathologic stage from a second tissue microarray constructed from a PSA-era prostatectomy cohort (cases dating from 1993–2000) [15]. As a group, these 34 pure acinar adenocarcinoma controls were matched with the ductal adenocarcinoma group for pathological stage and grade.
Immunohistochemistry
PTEN immunohistochemistry (IHC) was performed on the Ventana platform (Ventana Discovery Ultra, Ventana Medical Systems, Tucson, AZ) using a rabbit anti-human PTEN antibody (Clone D4.3 XP; Cell Signaling Technologies, Danvers, MA). We previously validated a manual version of this assay using the same primary antibody [30]. PTEN protein status was visually scored by a urologic pathologist (TLL). A tissue core was considered to have PTEN protein loss if the intensity of cytoplasmic and nuclear staining was markedly decreased or entirely negative across >10% of tumor cells compared to surrounding benign glands and/or stroma, which provide internal positive controls [30]. If PTEN was lost in >10% and <100% of the tumor cells sampled in a given core, the core was annotated as showing heterogeneous PTEN loss. Alternatively, if the core showed PTEN loss in 100% of sampled tumor glands, the core was annotated as showing homogeneous PTEN loss. Cores were scored as having ambiguous PTEN IHC results when the intensity of the tumor cell staining was light or absent in the absence of evaluable internal benign glands or stromal staining. For statistical analysis, each patient’s tumor sample was scored for the presence or absence of PTEN loss by summarizing the scores of the sampled cores. Based on a comparison with fluorescence in situ hybridization (FISH) data from 550 prostate cancer cases in an independent multi-institutional radical prostatectomy cohort, any loss of PTEN immunostaining (homogeneous or heterogeneous) using this assay was 98% sensitive for detecting homozygous PTEN gene deletion, 62% sensitive for detecting hemizygous PTEN gene deletion and 91% specific for intact PTEN gene [45].
ERG IHC was performed on the Ventana platform (Ventana Discovery Ultra, Ventana Medical Systems, Tucson, AZ) using a rabbit anti-human ERG antibody (Clone EPR3864; Ventana Medical Systems, Tucson, AZ). ERG protein status was visually scored by a urologic pathologist (TLL) blinded to the ERG FISH results. Cases were scored as positive if any tumor cells demonstrated nuclear ERG staining. Based on a comparison with FISH data from 408 prostate cancer cases at radical prostatectomy from Johns Hopkins, positive ERG immunostaining using this assay was 88% sensitive for detecting ERG gene rearrangement and negative ERG immunostaining was 87% specific for absence of ERG gene rearrangement ([15] and GJ Netto, AM De Marzo and TL Lotan, unpublished data). Further, the results of the automated ERG assay were highly concordant with results using a manual staining protocol and an independent ERG antibody clone (9FY, Biocare Medical), showing agreement in 96.2% of 289 cases (kappa = 0.923 or “very good agreement”) ([46] and GJ Netto, AM De Marzo and TL Lotan, unpublished data).
Statistical Analysis
Fisher’s exact test was used to compare frequency of PTEN loss and ERG expression fusion in ductal adenocarcinoma cases versus acinar adenocarcinoma controls.
Results
ERG expression
ERG protein was expressed in 11% (4/37) of evaluable ductal adenocarcinomas. Overall, ERG expression was highly concordant with presence of underlying ERG gene rearrangement by FISH, with 100% (36/36) of ductal adenocarcinomas with IHC and FISH results showing concordance. Three cases with ERG rearrangement via deletion showed ERG protein expression, one case with ERG gene rearrangement via translocation showed ERG protein expression and 32 cases lacking ERG gene rearrangements by FISH were all negative for ERG protein (Table 1, Figure 1).
Figure 1.
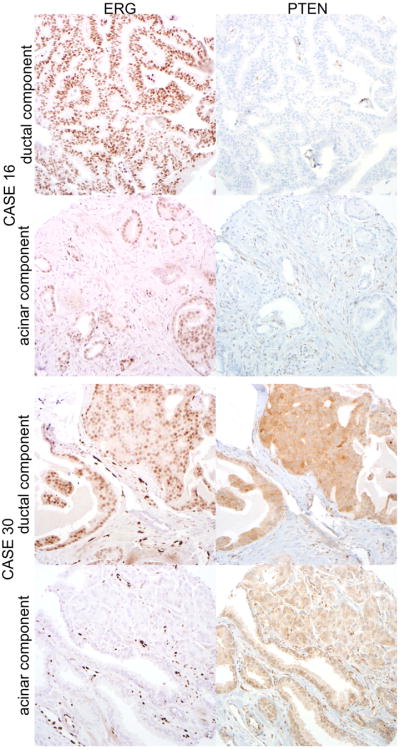
Top panels: ERG protein is expressed consistent with previously reported ERG gene rearrangement by translocation in the ductal and acinar components of Case 16. Similarly, PTEN protein is lost in both components, with positive internal control staining in the prostatic stroma and endothelial cells (all micrographs at 100× magnification). Bottom panels: ERG protein is expressed by the ductal component of Case 30 and absent from the spatially separate (>3mm away) acinar component, consistent with previously reported ERG gene rearrangement by deletion in the ductal component and absence of the rearrangement in the acinar component. PTEN expression is intact in both components (all micrographs at 100× magnification).
ERG IHC results were evaluable in 18 acinar carcinomas occurring concurrently with ductal adenocarcinoma. Among these, 6% (1/18) showed ERG protein expression. This acinar tumor showed ERG rearrangement via translocation on FISH and occurred with a ductal adenocarcinoma with an identical FISH and IHC result (Figure 1). The remaining 17 cases lacking ERG protein expression also lacked ERG gene rearrangement, such that there was 100% concordance (18/18) overall for ERG IHC and FISH among concurrent acinar adenocarcinomas.
Finally, ERG IHC was evaluable in 34 pure acinar adenocarcinomas. These tumors were roughly matched by stage and grade with the ductal adenocarcinomas as described previously [9]. Among these cases, 50% (17/34) showed ERG protein expression. Overall, there was 91% (31/34) concordance for ERG FISH and ERG IHC results in the pure acinar adenocarcinomas, including 9 cases rearranged by deletion, 6 cases rearranged by translocation and 16 cases without ERG gene rearrangement. Of the 3 discordant cases, one showed ERG gene rearrangement by deletion without ERG protein expression and two cases showing ERG protein expression without detection of underlying ERG gene rearrangement by FISH. Confirming the results of our prior FISH study, the lower rate of ERG protein expression among ductal adenocarcinomas (11%) and their associated acinar counterparts (6%) compared to pure acinar adenocarcinomas (50%) was statistically significant (p=0.0005 and 0.002, respectively by Fisher’s exact test).
PTEN loss
PTEN loss by IHC was evaluable in 34 ductal adenocarcinomas, of which 18% (6/34) showed loss (Figures 1 and 2). Though PTEN loss is frequently focal or heterogeneous among acinar adenocarcinomas [30, 33–35], the loss was homogeneous in all sampled tumor cells in all 6 ductal adenocarcinomas (Figure 1). Among acinar carcinomas occurring concurrently with ductal adenocarcinoma, 22% (4/18) showed PTEN loss, and the loss was homogeneous in all cases (Figures 1 and 2). In contrast, 50% (17/34) of pure acinar adenocarcinomas unassociated with a ductal component showed PTEN loss, with heterogeneous loss in 12% (2/17). The lower rate of PTEN loss among ductal adenocarcinomas and their associated acinar components compared to pure acinar adenocarcinomas was statistically significant or fell just short of conventional statistical significance (p = 0.01 and 0.08, respectively by Fisher’s exact test). As seen in prior FISH studies [13, 27–29, 33, 35–40], PTEN loss was about 2.5-fold enriched among ERG-positive compared to ERG-negative tumors in the pure acinar tumor control group (71% or 12/17 vs 29% or 5/17, respectively; p=0.04). However significant enrichment was not observed among the ductal adenocarcinomas (25% or 1/4 vs 17% or 5/30; p=0.55), a result that could potentially be due to the low numbers of cases with ERG expression among the ductal tumors.
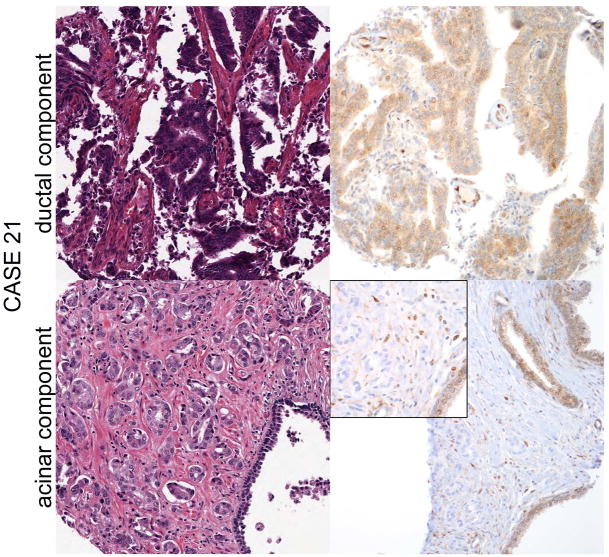
Figure 2.
Case 21 shows intact PTEN immunostaining in the ductal component, with loss of PTEN expression in the acinar component, while ERG is not expressed in either component. Adjacent benign glands provide an internal positive control for PTEN immunostaining.
Comparison of PTEN/ERG status in ductal adenocarcinoma and concurrent acinar tumors
Finally, of ductal adenocarcinomas with an evaluable concurrent acinar component, ERG status was concordant between the two components in 94% (17/18), including 16 ERG-negative cases and one ERG positive case with underlying ERG translocation in both components (Figure 1). The single discordant case showed ERG expression and ERG gene rearrangement by deletion in the ductal component and absence of ERG expression and gene rearrangement in the acinar component (Figure 1, case 30). PTEN expression was intact in both components. As previously reported, these components were spatially separated and may have represented synchronous but clonally distinct tumors. Similarly, PTEN status was concordant in 94% (16/17) of ductal adenocarcinomas with an evaluable concurrent acinar component, including 3 cases with PTEN loss and 13 cases with intact PTEN immunostaining. The single discordant case showed intact PTEN expression in the ductal component and loss of PTEN expression in the comingled acinar component, and lack of ERG protein expression or ERG gene rearrangement in both components (Figure 2).
Discussion
ERG gene rearrangements have emerged as one of the earliest and most common genomic changes in acinar prostate cancer, occurring in roughly 50% of Caucasian cases in most surgical cohorts [16]. Of interest, ERG gene rearrangements are less common in acinar carcinomas occurring in African-American patients [47–49], slightly less common among older patients in low grade tumors [50] and infrequent among some unusual morphologic subtypes of prostate cancer such as p63-expressing acinar carcinoma [51]. In addition, the frequency of ERG-rearranged acinar tumors appears to depend on tumor location. Both anterior and transition zone carcinomas have been reported to have lower rates of ERG gene rearrangement compared to posterior peripheral zone tumors [52, 53], and lower rates have also been seen in studies where tumor tissue was sampled on transurethral resection of the prostate [54, 55]. Given that the occurrence of ERG gene rearrangements has been shown to be mediated in part by androgen receptor activity [56,57], these findings raise the important question of whether lower rates of ERG rearrangement may be due to lower basal levels of androgen signaling in some of these settings.
Perhaps more similar to centrally located acinar tumors occurring in the transition zone, prostatic ductal adenocarcinomas classically occur in or at least involve the periurethral region in a large fraction of cases [58] and show low rates of ERG rearrangement by both FISH and IHC compared to peripheral zone acinar carcinomas in the current study. Interestingly, ductal adenocarcinomas classically present with lower PSA levels than acinar tumors [2], suggesting that they may also have lower levels of basal androgen signaling, another potential explanation for their low rate of ERG gene rearrangement. Despite this possibility that they may have lower basal androgen signaling, all ductal tumors with ERG gene rearrangement by FISH did show ERG protein expression, confirming that some AR signaling is occurring in the rearranged tumors, where ERG oncogene expression is driven by an androgen regulated gene, TMPRSS2. In contrast, in prostate tumors with absent androgen receptor signaling due to androgen deprivation therapy or absence of the androgen receptor itself (such as small cell carcinoma), ERG IHC is relatively insensitive for detecting underlying ERG gene rearrangement [41–43]. Of interest, the only other study to investigate ERG protein expression in ductal adenocarcinomas did not find a lower rate of expression in these tumors versus acinar carcinomas [59]. Though this study did not do confirmatory FISH, this discrepancy with our current results raises the interesting question of whether variable criteria for diagnosing ductal adenocarcinoma (vs acinar carcinoma) may in part explain these differences [60]. Indeed, ductal adenocarcinoma and intraductal spread of acinar carcinoma of the prostate have overlapping morphologic features [3] and previous work by our group has suggested a relatively high rate of ERG expression among the latter entity [61].
Another interesting finding in current study is that PTEN loss occurs relatively uncommonly in ductal adenocarcinoma and acinar carcinomas associated with ductal carcinoma compared to acinar carcinomas occurring without a ductal component. In acinar carcinoma, we and others have shown that PTEN loss is more frequent among tumors with higher Gleason grade, occurring in more than 40% of Gleason score 8–10 tumors [30]. There is a similar association of PTEN loss with higher pathologic stage [30]. Given that ductal differentiation is considered roughly equivalent to Gleason score 8 acinar disease and typically presents at higher stage than acinar tumors [1–4], it is surprising that PTEN loss occurs in ductal adenocarcinomas at roughly half the rate of grade and stage-matched acinar carcinomas in the current series. This low rate of PTEN loss is also seen in acinar carcinomas occurring concurrently with ductal carcinomas, suggesting they may belong to a common, and molecularly distinct subclass from usual-type acinar carcinomas. Additional evidence that ductal adenocarcinomas are molecularly distinct from acinar tumors comes from the fact that PTEN loss does not appear to be enriched among ERG positive ductal tumors as is seen in acinar carcinoma. Although it remains unclear why this enrichment of PTEN loss occurs among ERG-positive acinar tumors in humans, genetically engineered mouse models have suggested these two genetic alterations may have synergistic effects on tumor cell growth, migration and androgen signaling, leading to selection for ERG-positive PTEN-negative tumor clones [37, 38, 62]. However, human studies examining the interaction between PTEN loss and ERG expression for disease outcomes have been mixed [29, 36, 50, 63, 64]. This raises the possibility that ERG rearrangement, which precedes PTEN deletion in most cases, may directly or indirectly predispose tumor cells to PTEN loss. In either case, the lack of enrichment of PTEN loss among ERG-positive ductal tumors may imply important biological and molecular differences between ductal and acinar tumors. However, one limitation of the current study is that since the rates of ERG expression and PTEN loss are both quite low among ductal tumors, we had relatively low power to detect an interaction in the ductal data. Thus larger studies of ductal carcinoma are required to substantiate this finding.
Finally, comparison of both PTEN and ERG status between ductal carcinomas and their concurrent acinar tumors adds additional evidence that when they co-exist, these two tumor types may be clonally related. Consistent with our prior study where we examined only ERG status on the two tumor components [9], the frequently concordant PTEN status between the two components also suggests a likely clonal relationship in some cases. Importantly, our PTEN IHC assay is highly genetically validated, enabling us to use it as a proxy for underlying PTEN gene status to infer a likely clonal relationship between two tumors. In a recent comparison with 4-color FISH for PTEN gene deletion, intact PTEN immunostaining was 91% specific for the absence of PTEN gene deletion by FISH and 98% and 62% sensitive for the detection of homozygous and hemizygous gene deletion, respectively, by FISH [45] (TL Lotan and Jeremy A. Squire et al, in preparation). Thus, in cases where there is concordant PTEN protein loss between the ductal and acinar tumor components, it is highly likely that this signifies a similar PTEN gene deletion in both components and, accordingly, a clonal relationship between the two. Clearly, however, the relatively rare occurrence of PTEN loss and ERG expression in the ductal cases means that it is impossible to assess the likely clonality of the majority of ductal tumors co-occuring with acinar carcinoma and thus additional whole genome or whole exome sequencing studies will be required to confirm these data.
In summary, we have found that ERG protein expression and ERG FISH are highly concordant in ductal adenocarcinomas of the prostate, just as seen in acinar carcinomas. Using ERG IHC, we have confirmed that ductal adenocarcinomas have relatively lower rates of ERG gene rearrangement compared to peripherally-located acinar tumors, and more consistent with rates seen in centrally-located acinar tumors. In addition, we find that PTEN loss is relatively less common in ductal adenocarcinoma than in stage- and grade-matched acinar tumors, and PTEN status is highly concordant between concurrently occurring ductal and acinar tumors in the same patient, additional evidence for a clonal relationship between the two in at least some cases.
Acknowledgments
Funding: Funding for this research was provided by a generous gift from David H. Koch and the NIH/NCI Prostate SPORE P50CA58236.
Footnotes
Disclosure/Conflicts of Interest: The authors have no disclosures/conflicts of interest to declare.
References
- 1.Brinker DA, Potter SR, Epstein JI. Ductal adenocarcinoma of the prostate diagnosed on needle biopsy: correlation with clinical and radical prostatectomy findings and progression. Am J Surg Pathol. 1999;23:1471–9. doi: 10.1097/00000478-199912000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Morgan TM, Welty CJ, Vakar-Lopez F, Lin DW, Wright JL. Ductal adenocarcinoma of the prostate: increased mortality risk and decreased serum prostate specific antigen. J Urol. 2010;184:2303–7. doi: 10.1016/j.juro.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amin A, Epstein JI. Pathologic stage of prostatic ductal adenocarcinoma at radical prostatectomy: effect of percentage of the ductal component and associated grade of acinar adenocarcinoma. Am J Surg Pathol. 2011;35:615–9. doi: 10.1097/PAS.0b013e31820eb25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meeks JJ, Zhao LC, Cashy J, Kundu S. Incidence and outcomes of ductal carcinoma of the prostate in the USA: analysis of data from the Surveillance, Epidemiology, and End Results program. BJU Int. 2012;109:831–4. doi: 10.1111/j.1464-410X.2011.10520.x. [DOI] [PubMed] [Google Scholar]
- 5.Melicow MM, Pachter MR. Endometrial carcinoma of proxtatic utricle (uterus masculinus) Cancer. 1967;20:1715–22. doi: 10.1002/1097-0142(196710)20:10<1715::aid-cncr2820201022>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 6.Greene LF, Farrow GM, Ravits JM, Tomera FM. Prostatic adenocarcinoma of ductal origin. J Urol. 1979;121:303–5. doi: 10.1016/s0022-5347(17)56763-4. [DOI] [PubMed] [Google Scholar]
- 7.Bostwick DG, Kindrachuk RW, Rouse RV. Prostatic adenocarcinoma with endometrioid features. Clinical, pathologic, and ultrastructural findings. Am J Surg Pathol. 1985;9:595–609. doi: 10.1097/00000478-198508000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Christensen WN, Steinberg G, Walsh PC, Epstein JI. Prostatic duct adenocarcinoma. Findings at radical prostatectomy. Cancer. 1991;67:2118–24. doi: 10.1002/1097-0142(19910415)67:8<2118::aid-cncr2820670818>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 9.Lotan TL, Toubaji A, Albadine R, Latour M, Herawi M, Meeker AK, DeMarzo AM, Platz EA, Epstein JI, Netto GJ. TMPRSS2-ERG gene fusions are infrequent in prostatic ductal adenocarcinomas. Mod Pathol. 2009;22:359–65. doi: 10.1038/modpathol.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanati S, Watson MA, Salavaggione AL, Humphrey PA. Gene expression profiles of ductal versus acinar adenocarcinoma of the prostate. Mod Pathol. 2009;22:1273–9. doi: 10.1038/modpathol.2009.103. [DOI] [PubMed] [Google Scholar]
- 11.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toubaji A, Albadine R, Meeker AK, Isaacs WB, Lotan T, Haffner MC, Chaux A, Epstein JI, Han M, Walsh PC, Partin AW, De Marzo AM, Platz EA, Netto GJ. Increased gene copy number of ERG on chromosome 21 but not TMPRSS2-ERG fusion predicts outcome in prostatic adenocarcinomas. Mod Pathol. 2011;24:1511–20. doi: 10.1038/modpathol.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon AH, Suppan C, Flavin R, Sesso HD, Rider JR, Sweeney C, Stampfer MJ, Fiorentino M, Kantoff PW, Sanda MG, Giovannucci EL, Ding EL, Loda M, Mucci LA. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 18.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 19.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman JG, Jen J, Isaacs WB, Bova GS, Sidransky D. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 20.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–6. [PubMed] [Google Scholar]
- 21.Halvorsen OJ, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res. 2003;9:1474–9. [PubMed] [Google Scholar]
- 22.Verhagen PC, van Duijn PW, Hermans KG, Looijenga LH, van Gurp RJ, Stoop H, van der Kwast TH, Trapman J. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol. 2006;208:699–707. doi: 10.1002/path.1929. [DOI] [PubMed] [Google Scholar]
- 23.Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, Ghosh PM. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res. 2007;13:3860–7. doi: 10.1158/1078-0432.CCR-07-0091. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz M, Grignard G, Margue C, Dippel W, Capesius C, Mossong J, Nathan M, Giacchi S, Scheiden R, Kieffer N. Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int J Cancer. 2007;120:1284–92. doi: 10.1002/ijc.22359. [DOI] [PubMed] [Google Scholar]
- 25.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, Squire JA. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–85. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen G, Darnel AD, Aprikian AG, Saad F, Bismar TA, Squire JA. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol. 2009;218:505–13. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- 27.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–93. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, Clark J, Flohr P, Edwards S, Berney DM, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter VE, Scardino PT, Cuzick J, de Bono JS, Cooper CS Transatlantic Prostate Group. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–84. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krohn A, Diedler T, Burkhardt L, Mayer PS, De Silva C, Meyer-Kornblum M, Kotschau D, Tennstedt P, Huang J, Gerhauser C, Mader M, Kurtz S, Sirma H, Saad F, Steuber T, Graefen M, Plass C, Sauter G, Simon R, Minner S, Schlomm T. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181:401–12. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 30.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, Hicks JL, Park BH, Humphreys E, Partin AW, Han M, Netto GJ, Isaacs WB, De Marzo AM. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–73. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonarakis ES, Keizman D, Zhang Z, Gurel B, Lotan TL, Hicks JL, Fedor HL, Carducci MA, De Marzo AM, Eisenberger MA. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer. 2012;118:6063–71. doi: 10.1002/cncr.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaux A, Peskoe SB, Gonzalez-Roibon N, Schultz L, Albadine R, Hicks J, De Marzo AM, Platz EA, Netto GJ. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol. 2012;25:1543–9. doi: 10.1038/modpathol.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krohn A, Freudenthaler F, Harasimowicz S, Kluth M, Fuchs S, Burkhardt L, Stahl P, Tsourlakis CM, Bauer M, Tennstedt P, Graefen M, Steurer S, Sirma H, Sauter G, Schlomm T, Simon R, Minner S. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.70. [DOI] [PubMed] [Google Scholar]
- 34.Gumuskaya B, Gurel B, Fedor H, Tan HL, Weier CA, Hicks JL, Haffner MC, Lotan TL, De Marzo AM. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis. 2013;16:209–15. doi: 10.1038/pcan.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bismar TA, Yoshimoto M, Duan Q, Liu S, Sircar K, Squire JA. Interactions and relationships of PTEN, ERG, SPINK1 and AR in castration-resistant prostate cancer. Histopathology. 2012;60:645–52. doi: 10.1111/j.1365-2559.2011.04116.x. [DOI] [PubMed] [Google Scholar]
- 36.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, Zielenska M, Soares FA, Squire JA. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 37.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, Gerald WL, Sawyers CL. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bismar TA, Yoshimoto M, Vollmer RT, Duan Q, Firszt M, Corcos J, Squire JA. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–85. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
- 40.Reid AH, Attard G, Brewer D, Miranda S, Riisnaes R, Clark J, Hylands L, Merson S, Vergis R, Jameson C, Hoyer S, Sorenson KD, Borre M, Jones C, de Bono JS, Cooper CS. Novel, gross chromosomal alterations involving PTEN cooperate with allelic loss in prostate cancer. Mod Pathol. 2012;25:902–10. doi: 10.1038/modpathol.2011.207. [DOI] [PubMed] [Google Scholar]
- 41.Lotan TL, Gupta NS, Wang W, Toubaji A, Haffner MC, Chaux A, Hicks JL, Meeker AK, Bieberich CJ, De Marzo AM, Epstein JI, Netto GJ. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24:820–8. doi: 10.1038/modpathol.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, Dhir R, Nelson JB, de la Taille A, Allory Y, Gerstein MB, Perner S, Pienta KJ, Chinnaiyan AM, Wang Y, Collins CC, Gleave ME, Demichelis F, Nanus DM, Rubin MA. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–95. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Udager AM, Shi Y, Tomlins SA, Alva A, Siddiqui J, Cao X, Pienta KJ, Jiang H, Chinnaiyan AM, Mehra R. Frequent discordance between ERG gene rearrangement and ERG protein expression in a rapid autopsy cohort of patients with lethal, metastatic, castration-resistant prostate cancer. Prostate. 2014;74:1199–208. doi: 10.1002/pros.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herawi M, Epstein JI. Immunohistochemical antibody cocktail staining (p63/HMWCK/AMACR) of ductal adenocarcinoma and Gleason pattern 4 cribriform and noncribriform acinar adenocarcinomas of the prostate. Am J Surg Pathol. 2007;31:889–94. doi: 10.1097/01.pas.0000213447.16526.7f. [DOI] [PubMed] [Google Scholar]
- 45.Lotan T, Morais C, Wei W, Troyer D, Jamaspishvili T, Fend Z, McKenney J, Simko J, Brooks J, Squire J. PTEN Status Determination in Prostate Cancer: Comparison of IHC and FISH in a Large Multi-Center Cohort. Mod Pathol. 2015;28:241A. [Google Scholar]
- 46.Chaux A, Albadine R, Toubaji A, Hicks J, Meeker A, Platz EA, De Marzo AM, Netto GJ. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–20. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, Klein E, Rubin MA, Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 48.Rosen P, Pfister D, Young D, Petrovics G, Chen Y, Cullen J, Bohm D, Perner S, Dobi A, McLeod DG, Sesterhenn IA, Srivastava S. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology. 2012;80:749–53. doi: 10.1016/j.urology.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khani F, Mosquera JM, Park K, Blattner M, O’Reilly C, MacDonald TY, Chen Z, Srivastava A, Tewari AK, Barbieri CE, Rubin MA, Robinson BD. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res. 2014;20:4925–34. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steurer S, Mayer PS, Adam M, Krohn A, Koop C, Ospina-Klinck D, Tehrani AA, Simon R, Tennstedt P, Graefen M, Wittmer C, Brors B, Plass C, Korbel J, Weischenfeldt J, Sauter G, Huland H, Tsourlakis MC, Minner S, Schlomm T. TMPRSS2-ERG fusions are strongly linked to young patient age in low-grade prostate cancer. Eur Urol. 2014;66:978–81. doi: 10.1016/j.eururo.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 51.Tan HL, Haffner MC, Esopi DM, Vaghasia AM, Giannico GA, Ross HM, Ghosh S, Hicks JL, Zheng Q, Sangoi AR, Yegnasubramanian S, Osunkoya AO, De Marzo AM, Epstein JI, Lotan TL. Prostate adenocarcinomas aberrantly expressing p63 are molecularly distinct from usual-type prostatic adenocarcinomas. Mod Pathol. 2015;28:446–56. doi: 10.1038/modpathol.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo CC, Zuo G, Cao D, Troncoso P, Czerniak BA. Prostate cancer of transition zone origin lacks TMPRSS2-ERG gene fusion. Mod Pathol. 2009;22:866–71. doi: 10.1038/modpathol.2009.57. [DOI] [PubMed] [Google Scholar]
- 53.Gopalan A, Leversha MA, Dudas ME, Maschino AC, Chang J, Al-Ahmadie HA, Chen YB, Tickoo SK, Reuter VE, Fine SW. TMPRSS2-ERG rearrangement in dominant anterior prostatic tumours: incidence and correlation with ERG immunohistochemistry. Histopathology. 2013;63:279–86. doi: 10.1111/his.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bismar TA, Trpkov K. TMPRSS2-ERG gene fusion in transition zone prostate cancer. Mod Pathol. 2010;23:1040–1. doi: 10.1038/modpathol.2010.89. author reply 1041–2. [DOI] [PubMed] [Google Scholar]
- 55.Liu S, Yoshimoto M, Trpkov K, Duan Q, Firszt M, Corcos J, Squire JA, Bismar TA. Detection of ERG gene rearrangements and PTEN deletions in unsuspected prostate cancer of the transition zone. Cancer Biol Ther. 2011;11:562–6. doi: 10.4161/cbt.11.6.14376. [DOI] [PubMed] [Google Scholar]
- 56.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42(8):668–75. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326(5957):1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seipel AH, Wiklund F, Wiklund NP, Egevad L. Histopathological features of ductal adenocarcinoma of the prostate in 1,051 radical prostatectomy specimens. Virchows Arch. 2013;462:429–36. doi: 10.1007/s00428-013-1385-5. [DOI] [PubMed] [Google Scholar]
- 59.Seipel AH, Samaratunga H, Delahunt B, Wiklund F, Wiklund P, Lindberg J, Gronberg H, Egevad L. Immunohistochemical profile of ductal adenocarcinoma of the prostate. Virchows Arch. 2014;465:559–65. doi: 10.1007/s00428-014-1636-0. [DOI] [PubMed] [Google Scholar]
- 60.Seipel AH, Delahunt B, Samaratunga H, Amin M, Barton J, Berney DM, Billis A, Cheng L, Comperat E, Evans A, Fine SW, Grignon D, Humphrey PA, Magi-Galluzzi C, Montironi R, Sesterhenn I, Srigley JR, Trpkov K, van der Kwast T, Varma M, Zhou M, Ahmad A, Moss S, Egevad L. Diagnostic criteria for ductal adenocarcinoma of the prostate: interobserver variability among 20 expert uropathologists. Histopathology. 2014;65:216–27. doi: 10.1111/his.12382. [DOI] [PubMed] [Google Scholar]
- 61.Lotan TL, Gumuskaya B, Rahimi H, Hicks JL, Iwata T, Robinson BD, Epstein JI, De Marzo AM. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol. 2013;26:587–603. doi: 10.1038/modpathol.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, Scher HI, Zheng D, Sawyers CL. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leinonen KA, Saramaki OR, Furusato B, Kimura T, Takahashi H, Egawa S, Suzuki H, Keiger K, Ho Hahm S, Isaacs WB, Tolonen TT, Stenman UH, Tammela TL, Nykter M, Bova GS, Visakorpi T. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:2333–44. doi: 10.1158/1055-9965.EPI-13-0333-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fontugne J, Lee D, Cantaloni C, Barbieri CE, Caffo O, Hanspeter E, Mazzoleni G, Dalla Palma P, Rubin MA, Fellin G, Mosquera JM, Barbareschi M, Demichelis F. Recurrent prostate cancer genomic alterations predict response to brachytherapy treatment. Cancer Epidemiol Biomarkers Prev. 2014;23:594–600. doi: 10.1158/1055-9965.EPI-13-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]