Abstract
Importance
Evidence indicates a complex link between gut microbiome, immunity, and intestinal tumorigenesis. To target the microbiota and immunity for colorectal cancer prevention and therapy, a better understanding of the relationship between microorganisms and immune cells in the tumor microenvironment is needed. Experimental evidence suggests that Fusobacterium nucleatum may promote colonic neoplasia development by down-regulating antitumor T-cell-mediated adaptive immunity.
Objective
To test the hypothesis that higher amount of Fusobacterium nucleatum in colorectal carcinoma tissue is associated with lower density of T-cells in tumor tissue.
Design
A cross-sectional analysis was conducted on colorectal carcinoma cases in two U.S. nationwide prospective cohort studies. The amount of Fusobacterium nucleatum in colorectal carcinoma tissue was measured by quantitative polymerase chain reaction assay; we equally dichotomized positive cases (high versus low). Multivariable ordinal logistic regression analysis was conducted to assess associations of the amount of Fusobacterium nucleatum with densities (quartiles) of T-cells in tumor tissue, controlling for clinical and tumor molecular features, including microsatellite instability, CpG island methylator phenotype, LINE-1 methylation, and KRAS, BRAF, and PIK3CA mutation status. We adjusted two-sided α level to 0.013 for multiple hypothesis testing.
Setting
The Nurses’ Health Study and the Health Professionals Follow-up Study.
Participants
598 colon and rectal carcinoma cases.
Main outcomes and measures
Densities of CD3+, CD8+, CD45RO (PTPRC)+, and FOXP3+ T-cells in tumor tissue, determined by tissue microarray immunohistochemistry and computer-assisted image analysis.
Results
Fusobacterium nucleatum was detected in colorectal carcinoma tissue in 76 (13%) of 598 cases. Compared with Fusobacterium nucleatum-negative cases, Fusobacterium nucleatum-high cases were inversely associated with the density of CD3+ T-cells (for a unit increase in quartile categories of CD3+ T-cells as an outcome: multivariable odds ratio, 0.47; 95% confidence interval, 0.26 to 0.87; Ptrend = 0.006). The amount of Fusobacterium nucleatum was not significantly associated with the density of CD8+, CD45RO+, or FOXP3+ T-cells (Ptrend > 0.013).
Conclusions and relevance
The amount of tissue Fusobacterium nucleatum is inversely associated with CD3+ T-cell density in colorectal carcinoma tissue. Upon validation, our human population data may provide an impetus for further investigations on potential interactive roles of Fusobacterium and host immunity in carcinogenesis.
Introduction
Accumulating evidence attests to an important role of T-cell-mediated adaptive immunity in regulating tumor evolution, and immunotherapy has emerged as a promising strategy to treat various malignancies.1,2 In colorectal carcinoma, high-level infiltrates of CD3+, CD8+, CD45RO (PTPRC)+, and FOXP3+ T-cells have been associated with better clinical outcome.3–6 Evidence also indicates that molecular features of colorectal carcinoma, especially microsatellite instability (MSI), can influence antitumor T-cell-mediated adaptive immunity.7–11
The human intestinal microbiome encompasses at least 100 trillion (1014) microorganisms, which can influence the immune system and health conditions.12 A growing body of evidence indicates a complex link between gut microbiome, immunity, and intestinal tumorigenesis.13–17 Colorectal carcinogenesis represents heterogeneous processes with differing sets of genetic and epigenetic alterations, influenced by diet, environmental and microbial exposures, and host immunity.18–22 Metagenomic analyses have shown an enrichment of Fusobacterium nucleatum in colorectal carcinoma tissue, which has been confirmed by quantitative polymerase chain reaction (PCR) for the 16S ribosomal RNA gene DNA sequence of Fusobacterium nucleatum.23,24 Studies have shown that higher amount of Fusobacterium nucleatum in colorectal carcinoma tissue is associated with high degrees of MSI and CpG island methylator phenotype (CIMP).25 Experimental evidence suggests that virulence factors derived from Fusobacterium nucleatum inhibit T-cell activity,26,27 and that in the ApcMin/+ mouse model, Fusobacterium nucleatum promotes colonic neoplasia development by down-regulating antitumor T-cell-mediated adaptive immunity.28 Therefore, we hypothesized that higher amount of Fusobacterium nucleatum in colorectal carcinoma tissue might be associated with lower density of T-cells in tumor tissue. A better understanding of the relationship between Fusobacterium nucleatum and immune cells in the tumor microenvironment may open new opportunities to target the microbiota and immunity for colorectal cancer prevention and therapy.
To test our hypothesis, we utilized resources of two U.S. nationwide prospective cohort studies (the Nurses’ Health Study and the Health Professionals Follow-up Study), and examined the amount of Fusobacterium nucleatum in relation to densities of CD3+, CD8+, CD45RO (PTPRC)+, and FOXP3+ T-cells in tumor tissue of nearly 600 human colorectal carcinoma cases. To our knowledge, this is the first population-based study to examine the amount of Fusobacterium nucleatum in relation to the density of T-cells in human colorectal carcinoma tissue.
Methods
Study population
We utilized the databases of two U.S. nationwide prospective cohort studies, the Nurses’ Health Study (NHS, with 121,700 women who enrolled in 1976) and the Health Professionals Follow-up Study (HPFS, with 51,529 men who enrolled in 1986).29,30 Every 2 years, participants were sent follow-up questionnaires to gather information on health and lifestyle factors, and to identify newly diagnosed cancers and other diseases. The National Death Index was used to ascertain deaths of study participants and identify unreported lethal colorectal carcinoma cases. Medical records were reviewed, and the cause of death was assigned by study physicians. Formalin-fixed paraffin-embedded (FFPE) tissue blocks were collected from hospitals where participants with colorectal carcinoma had undergone tumor resection. We included both colon and rectal carcinoma cases, considering the colorectal continuum model.31 A single pathologist (S.O.), who was unaware of other data, reviewed hematoxylin and eosin-stained tissue sections from all colorectal carcinoma cases, and recorded pathological features. Tumor differentiation was categorized as well to moderate or poor (>50% vs. ≤50% glandular area). Based on the availability of data on Fusobacterium nucleatum and T-cell densities, a total of 598 colorectal carcinoma cases were included. Written informed consent was obtained from all study participants. Tissue collection and analyses were approved by the human subjects committee at the Harvard T.H. Chan School of Public Health and the Brigham and Women’s Hospital (Boston, MA, USA).
Quantitative PCR for Fusobacterium nucleatum
Genomic DNA was extracted from colorectal carcinoma tissue and adjacent non-tumor tissue in whole-tissue sections of FFPE tissue blocks using QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA). Custom TaqMan primer/probe sets (Applied Biosystems, San Diego, CA) for the 16S ribosomal RNA gene DNA sequence of Fusobacterium nucleatum and for the reference gene, SLCO2A1 were used as previously described.24 The primer/probe set for Fusobacterium nucleatum was designed to target the nusG gene of Fusobacterium nucleatum, and it has been demonstrated that the amount of Fusobacterium nucleatum measured by the quantitative PCR assay highly correlates with that measured by using transcriptome sequencing data (Pearson’s r = 0.97).24 Each reaction contained 80 ng of genomic DNA and was assayed in 20 µL reactions containing 1× final concentration TaqMan Environmental Master Mix 2.0 (Applied Biosystems, San Diego, CA) and each TaqMan Gene Expression Assay (Applied Biosystems, San Diego, CA), in a 96-well optical PCR plate. Amplification and detection of DNA was performed with the StepOnePlus Real-Time PCR Systems (Applied Biosystems, San Diego, CA) using the following reaction conditions: 10 min at 95°C and 45 cycles of 15 sec at 95°C and 1 min at 60°C. The primer and probe sequences for each TaqMan Gene Expression Assay were as follows: Fusobacterium nucleatum forward primer, 5’-CAACCATTACTTTAACTCTACCATGTTCA-3’; Fusobacterium nucleatum reverse primer, 5’-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3’; Fusobacterium nucleatum FAM probe, 5’-GTTGACTTTACAGAAGGAGATTA-3’; SLCO2A1 forward primer, 5’-ATCCCCAAAGCACCTGGTTT-3’; SLCO2A1 reverse primer, 5’-AGAGGCCAAGATAGTCCTGGTAA-3’; SLCO2A1 VIC probe, 5’-CCATCCATGTCCTCATCTC-3’. In colorectal carcinoma cases with detectable Fusobacterium nucleatum, the cycle threshold (Ct) values in the quantitative PCR for Fusobacterium nucleatum and SLCO2A1 decreased linearly with the amount of input DNA (in a log scale) from the same specimen (r2 > 0.99; Figure 1A). The inter-assay coefficient of variation of Ct values from the same specimen in five different batches was 1% or less for all targets in our validation study using seven colorectal carcinomas (eTable 1 in the Supplement).
Figure 1. The amount of Fusobacterium nucleatum in colorectal cancer.
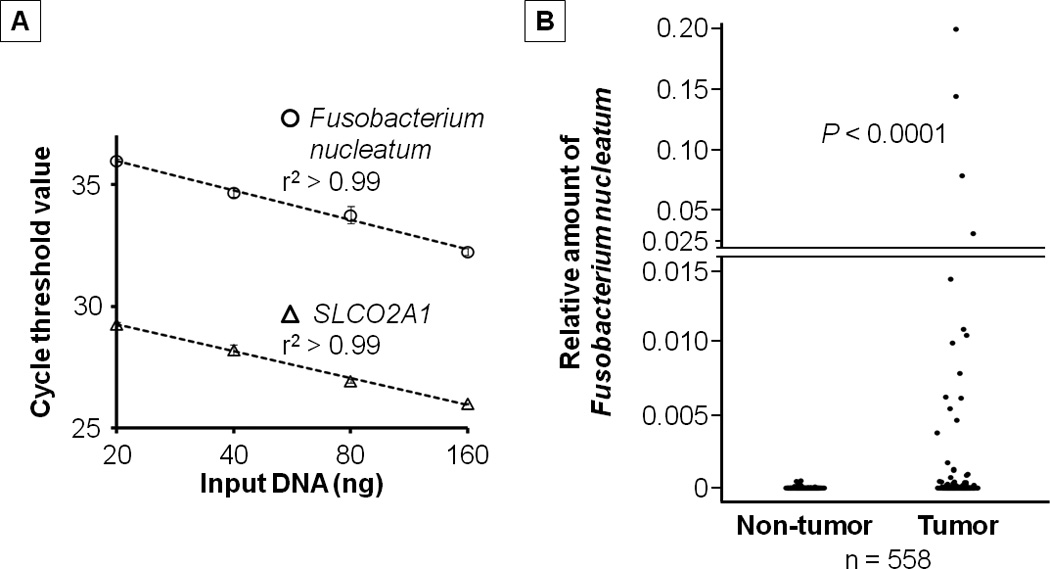
A, Quantitative polymerase chain reaction assays for Fusobacterium nucleatum and SLCO2A1 using 2-fold dilution series (20 ng, 40 ng, 80 ng, and 160 ng) from the same DNA specimen. Mean cycle threshold values (± standard deviation) of duplicate runs and the coefficient of determination (r2) in the assays for Fusobacterium nucleatum and SLCO2A1 are shown.
B, The amount of Fusobacterium nucleatum in 558 pairs of colorectal carcinoma and adjacent non-tumor tissues. Dot plots represent the amount of Fusobacterium nucleatum in colorectal carcinoma tissue and paired adjacent non-tumor tissue. Statistical analyses were performed using two-sided Wilcoxon signed rank test.
Each specimen was analyzed in duplicate for each target in a single batch, and we used the average of the two Ct values for each target. Spearman’s rank-correlation coefficients between the two Ct values (in duplicated runs) in each of cases with detectable target amplification in the quantitative PCR assays for Fusobacterium nucleatum (n = 76) and SLCO2A1 (n = 598) were 0.95 and 0.92, respectively. The amount of Fusobacterium nucleatum in each specimen was calculated as a relative unitless value normalized with SLCO2A1 using the 2−ΔCt method (where ΔCt = “the average Ct value of Fusobacterium nucleatum” - “the average Ct value of SLCO2A1”) as previously described.32
Analyses of MSI, DNA methylation, and KRAS, BRAF, and PIK3CA mutations
Genomic DNA was extracted from colorectal carcinoma tissue in whole-tissue sections from FFPE tissue blocks. MSI status was analyzed with use of 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487) as previously described.33 We defined MSI-high as the presence of instability in ≥30% of the markers, and MSI-low/microsatellite stable (MSS) as instability in <30% of the markers. Methylation analyses of long interspersed nucleotide element-1 (LINE-1) and eight promoter CpG islands specific for CIMP (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) were performed as previously described.34–37 PCR reaction and pyrosequencing were performed for KRAS (codons 12, 13, 61, and 146), BRAF (codon 600), and PIK3CA (exons 9 and 20).38–40
Immunohistochemistry and quantification of the density of T-cells in tumor tissue
We constructed a tissue microarray, and conducted immunohistochemistry for CD3, CD8, CD45RO (PTPRC), and FOXP3. We used automated scanning microscope and the Ariol image analysis system (Genetix, San Jose, CA, USA) to measure densities (cells/mm2) of CD3+, CD8+, CD45RO (PTPRC)+, and FOXP3+ T-cells in tumor tissue. We evaluated up to four tissue cores from each tumor in tissue microarray, and calculated the average density of each T-cell subset as previously described.5
Statistical analysis
All statistical analyses were conducted using SAS (version 9.3, SAS Institute, Cary, NC) and all P values were two-sided. Neither the amount of Fusobacterium nucleatum, T-cell density, nor the log-transformed value of the amount of Fusobacterium nucleatum or T-cell density fit a normal distribution with the use of the Kolmogorov-Smirnov test for normality (P ≤ 0.022). Thus, our primary hypothesis testing was the linear trend test in an ordinal logistic regression model to assess associations of the amount of Fusobacterium nucleatum in colorectal carcinoma tissue (an ordinal predictor variable) with the density of CD3+, CD8+, CD45RO+, or FOXP3+ T-cells in tumor tissue (an ordinal quartile outcome variable). Cases with detectable Fusobacterium nucleatum were categorized as low or high based on the median cutpoint amount of Fusobacterium nucleatum, while cases without detectable Fusobacterium nucleatum were categorized as “negative”. The linear trend test was performed using the ordinal predictor variable of Fusobacterium nucleatum (negative, low, and high) as a continuous variable in an ordinal logistic regression model. Because we tested four primary hypotheses (for CD3+, CD8+, CD45RO+, and FOXP3+ T-cells as outcome variables), we adjusted two-sided α level to 0.013 (= 0.05/4) by simple Bonferroni correction. All other analyses on Fusobacterium nucleatum, including evaluation of individual odds ratio (OR) estimates represented secondary analyses. In those secondary analyses, in view of multiple comparisons, we interpreted our data cautiously, in addition to the use of the adjusted α level of 0.013.
We performed multivariable ordinal logistic regression analysis to adjust for potential confounders. The multivariable model initially included age (continuous), sex, year of diagnosis (continuous), family history of colorectal carcinoma in a first-degree relative (present vs. absent), tumor location (proximal colon vs. distal colon vs. rectum), tumor differentiation (well-moderate vs. poor), MSI (high vs. MSI-low/MSS), CIMP (high vs. low/negative), KRAS (mutant vs. wild-type), BRAF (mutant vs. wild-type), PIK3CA (mutant vs. wild-type), and LINE-1 methylation level (continuous). For cases with missing information in any of the covariates, we assigned a separate (“missing”) indicator variable. A backward stepwise elimination with a threshold of P < 0.05 was used to select variables in the final models. We assessed the proportional odds assumption in an ordinal logistic regression model, which was generally satisfied (P > 0.05).
All cross-sectional univariable analyses for clinical, pathological, and molecular associations (with variables listed in Table 1) were secondary exploratory analyses, and we adjusted two-sided α level to 0.003 (= 0.05/14) by simple Bonferroni correction for multiple hypothesis testing. To assess associations between the ordinal (negative, low, and high) categories of the amount of Fusobacterium nucleatum and categorical data, Fisher’s exact test was performed. To compare mean age and mean LINE-1 methylation levels, an analysis of variance assuming equal variances was performed.
Table 1.
Characteristics according to the amount of Fusobacterium nucleatum in colorectal carcinoma tissue
| Characteristica | All patients (n = 598) |
The amount of Fusobacterium nucleatum in colorectal carcinoma tissue |
P valueb | ||
|---|---|---|---|---|---|
| Negative (n = 522) |
Low (n = 38) |
High (n = 38) |
|||
| Mean age ± SD (year) | 67.2 ± 8.4 | 67.2 ± 8.5 | 67.7 ± 7.4 | 66.8 ± 7.3 | 0.89 |
| Sex | 0.08 | ||||
| Men | 202 (34%) | 184 (35%) | 11 (29%) | 7 (18%) | |
| Women | 396 (66%) | 338 (65%) | 27 (71%) | 31 (82%) | |
| Year of diagnosis | 0.14 | ||||
| Prior to 1995 | 256 (43%) | 232 (45%) | 10 (26%) | 14 (37%) | |
| 1996 to 2000 | 214 (36%) | 184 (36%) | 16 (42%) | 14 (37%) | |
| 2001 to 2008 | 124 (21%) | 102 (19%) | 12 (32%) | 10 (26%) | |
| Family history of colorectal carcinoma in a first-degree relative | 0.76 | ||||
| Absent | 465 (79%) | 404 (78%) | 32 (84%) | 29 (78%) | |
| Present | 125 (21%) | 111 (22%) | 6 (16%) | 8 (22%) | |
| Tumor location | 0.006 | ||||
| Cecum | 103 (17%) | 81 (16%) | 7 (18%) | 15 (39%) | |
| Ascending to transverse colon | 197 (33%) | 172 (33%) | 12 (32%) | 13 (34%) | |
| Splenic flexure to sigmoid | 168 (28%) | 154 (30%) | 7 (18%) | 7 (18%) | |
| Rectosigmoid and rectum | 128 (22%) | 113 (21%) | 12 (32%) | 3 (7.9%) | |
| Disease stage | 0.003 | ||||
| I | 123 (22%) | 116 (24%) | 4 (11%) | 3 (8.3%) | |
| II | 191 (34%) | 163 (33%) | 10 (27%) | 18 (50%) | |
| III | 174 (31%) | 145 (29%) | 21 (57%) | 8 (22%) | |
| IV | 77 (13%) | 68 (14%) | 2 (5.4%) | 7 (19%) | |
| Tumor differentiation | 0.002 | ||||
| Well to moderate | 544 (91%) | 484 (93%) | 30 (81%) | 30 (79%) | |
| Poor | 53 (8.9%) | 38 (7.3%) | 7 (19%) | 8 (21%) | |
| MSI status | < 0.0001 | ||||
| MSI-low/MSS | 493 (84%) | 447 (87%) | 24 (67%) | 22 (58%) | |
| MSI-high | 94 (16%) | 66 (13%) | 12 (33%) | 16 (42%) | |
| MLH1 hypermethylation | < 0.0001 | ||||
| Absent | 517 (87%) | 463 (90%) | 30 (81%) | 24 (63%) | |
| Present | 74 (13%) | 53 (10%) | 7 (19%) | 14 (37%) | |
| CIMP status | 0.0009 | ||||
| Low/negative | 494 (84%) | 441 (85%) | 30 (81%) | 23 (61%) | |
| High | 97 (16%) | 75 (15%) | 7 (19%) | 15 (39%) | |
| BRAF mutation | 0.08 | ||||
| Wild-type | 497 (84%) | 439 (85%) | 31 (86%) | 27 (71%) | |
| Mutant | 92 (16%) | 76 (15%) | 5 (14%) | 11 (29%) | |
| KRAS mutation | 0.48 | ||||
| Wild-type | 346 (59%) | 307 (60%) | 18 (50%) | 21 (55%) | |
| Mutant | 244 (41%) | 209 (40%) | 18 (50%) | 17 (45%) | |
| PIK3CA mutation | 0.83 | ||||
| Wild-type | 459 (84%) | 404 (84%) | 25 (81%) | 30 (86%) | |
| Mutant | 87 (16%) | 75 (16%) | 6 (19%) | 5 (14%) | |
| Mean LINE-1 methylation level (%) ± SD | 61.0 ± 9.4 | 60.9 ± 9.4 | 61.1 ± 9.9 | 63.3 ± 9.0 | 0.30 |
Abbreviation: CIMP, CpG island methylator phenotype; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; MSS, microsatellite stable; SD, standard deviation.
(%) indicate the proportion of cases with a specific clinical, pathological, or molecular feature according to the amount of Fusobacterium nucleatum in colorectal carcinoma tissue. There were cases that had missing values for any of the characteristics except for age and sex.
To assess associations between the ordinal (negative, low, and high) categories of the amount of Fusobacterium nucleatum and categorical data, Fisher’s exact test was performed. To compare mean age and mean LINE-1 methylation levels, an analysis of variance was performed. We adjusted two-sided α level to 0.003 (= 0.05/14) by simple Bonferroni correction for multiple hypothesis testing.
Results
Fusobacterium nucleatum in colorectal carcinoma tissue
We analyzed tumor tissues of 598 colorectal carcinoma cases within the Nurses’ Health Study and the Health Professionals Follow-up Study, using the quantitative PCR assay to detect the 16S ribosomal RNA gene DNA sequence of Fusobacterium nucleatum, as previously described.24 Fusobacterium nucleatum was detected in colorectal carcinoma in 76 (13%) of 598 cases, and in adjacent non-tumor tissue in 19 (3.4%) of 558 cases analyzed. In the 558 pairs of colorectal carcinoma and adjacent non-tumor tissues, the amount of Fusobacterium nucleatum was higher in colorectal carcinoma tissue than in paired adjacent non-tumor tissue (Wilcoxon signed rank test, P < 0.0001; Figure 1B).
We categorized colorectal carcinoma cases with detectable Fusobacterium nucleatum as low or high based on the median cutpoint amount of Fusobacterium nucleatum. Clinical, pathological, and molecular features are summarized according to the amount of Fusobacterium nucleatum in colorectal carcinoma tissue in Table 1. A higher amount of Fusobacterium nucleatum in colorectal carcinoma tissue was associated with stage II-IV disease, poor differentiation, MSI-high, MLH1 hypermethylation, and CIMP-high (P ≤ 0.003 for all comparisons by Fisher’s exact test; with adjusted α level of 0.003 for multiple hypothesis testing).
Associations of the amount of Fusobacterium nucleatum with T-cell densities in tumor tissue
Utilizing tissue microarray, we quantified densities of CD3+, CD8+, CD45RO+, and FOXP3+ T-cells in tumor tissue by immunohistochemistry and the Ariol image analysis system. Correlations between densities of CD3+, CD8+, CD45RO+, and FOXP3+ T-cells in tumor tissue (with Spearman’s rank-correlation coefficients ranging 0.14 to 0.42; P ≤ 0.002) are shown in eTable 2 in the Supplement.
Table 2 shows the distribution of colorectal carcinoma cases according to the amount of Fusobacterium nucleatum and densities of CD3+, CD8+, CD45RO+, and FOXP3+ T-cells. In our primary hypothesis testing, we conducted univariable and multivariable ordinal logistic regression analyses to assess associations of the amount of Fusobacterium nucleatum in colorectal carcinoma tissue (as an ordinal predictor variable) with the density of CD3+, CD8+, CD45RO+, or FOXP3+ T-cells in tumor tissue (as an ordinal quartile outcome variable) (Table 3, and eTable 3 in the Supplement). The amount of Fusobacterium nucleatum in colorectal carcinoma tissue was associated with lower density of CD3+ T-cells in univariable (Ptrend = 0.012) and multivariable ordinal logistic regression analysis (Ptrend = 0.006). Compared with Fusobacterium nucleatum-negative cases, Fusobacterium nucleatum-high cases were inversely associated with the density of CD3+ T-cells (for a unit increase in quartile categories of CD3+ T-cells as an outcome: multivariable OR, 0.47; 95% confidence interval, 0.26 to 0.87; Table 3). The association of the amount of Fusobacterium nucleatum with the density of CD8+, CD45RO+, or FOXP3+ T-cells in tumor tissue was not statistically significant (Ptrend > 0.013; with adjusted α level of 0.013; Table 3). We also used four ordinal categories of the amount of tissue Fusobacterium nucleatum (Fusobacterium nucleatum-negative, low, middle, and high), and observed similar findings in terms of the associations with the density of T-cells (eTable 4 in the Supplement).
Table 2.
Distribution of cases according to the amount of Fusobacterium nucleatum and the density of T-cells
| All patients (n = 598) |
The amount of Fusobacterium nucleatum in colorectal carcinoma tissue |
Ptrenda | |||
|---|---|---|---|---|---|
| Negative (n = 522) |
Low (n = 38) |
High (n = 38) |
|||
| CD3+ cell density (quartile) | 0.012 | ||||
| Q1 (0–85 cells/mm2) | 142 (25%) | 117 (24%) | 9 (26%) | 16 (44%) | |
| Q2 (86–210 cells/mm2) | 141 (25%) | 122 (25%) | 13 (37%) | 6 (16%) | |
| Q3 (211–425 cells/mm2) | 142 (25%) | 126 (25%) | 7 (20%) | 9 (24%) | |
| Q4 (≥426 cells/mm2) | 142 (25%) | 130 (26%) | 6 (17%) | 6 (16%) | |
| CD8+ cell density (quartile) | 0.11 | ||||
| Q1 (0–46 cells/mm2) | 140 (25%) | 117 (24%) | 12 (34%) | 11 (30%) | |
| Q2 (47–131 cells/mm2) | 140 (25%) | 118 (24%) | 11 (32%) | 11 (30%) | |
| Q3 (132–320 cells/mm2) | 140 (25%) | 129 (26%) | 4 (11%) | 7 (19%) | |
| Q4 (≥321 cells/mm2) | 140 (25%) | 124 (26%) | 8 (23%) | 8 (21%) | |
| CD45RO+ cell density (quartile) | 0.56 | ||||
| Q1 (0–184 cells/mm2) | 144 (25%) | 124 (25%) | 11 (31%) | 9 (24%) | |
| Q2 (185–427 cells/mm2) | 143 (25%) | 128 (26%) | 5 (14%) | 10 (27%) | |
| Q3 (428–895 cells/mm2) | 143 (25%) | 130 (26%) | 6 (17%) | 7 (19%) | |
| Q4 (≥896 cells/mm2) | 144 (25%) | 119 (23%) | 14 (38%) | 11 (30%) | |
| FOXP3+ cell density (quartile) | 0.038 | ||||
| Q1 (0–10 cells/mm2) | 137 (25%) | 117 (24%) | 7 (21%) | 13 (35%) | |
| Q2 (11–23 cells/mm2) | 137 (25%) | 118 (25%) | 6 (18%) | 13 (35%) | |
| Q3 (24–48 cells/mm2) | 137 (25%) | 117 (24%) | 11 (34%) | 9 (24%) | |
| Q4 (≥49 cells/mm2) | 137 (25%) | 126 (27%) | 9 (27%) | 2 (5.4%) | |
Abbreviation: Q1 to Q4, quartile 1 to quartile 4.
Ptrend value was calculated by the linear trend test across the ordinal (negative, low, and high) categories of the amount of Fusobacterium nucleatum as a continuous variable in univariable ordinal logistic regression model for the density of CD3+, CD8+, CD45RO+, or FOXP3+ T-cells (an ordinal quartile outcome variable). Because we assessed four primary outcome variables, we adjusted two-sided α level to 0.013 (= 0.05/4) by simple Bonferroni correction.
Table 3.
The amount of Fusobacterium nucleatum in colorectal carcinoma tissue and the density of T-cells
| Univariable OR (95% CI) |
Multivariable OR (95% CI)a |
|||
|---|---|---|---|---|
| Model for CD3+ cell density (n = 567, as an outcome variable) | ||||
| The amount of Fusobacterium nucleatum | Negative | 1 (reference) | 1 (reference) | |
| Low | 0.68 (0.37–1.25) | 0.63 (0.34–1.17) | ||
| High | 0.50 (0.27–0.91) | 0.47 (0.26–0.87) | ||
| Ptrendb | 0.012 | 0.006 | ||
| Model for CD8+ cell density (n = 560, as an outcome variable) | ||||
| The amount of Fusobacterium nucleatum | Negative | 1 (reference) | 1 (reference) | |
| Low | 0.60 (0.32–1.12) | 0.66 (0.35–1.23) | ||
| High | 0.71 (0.39–1.30) | 0.79 (0.43–1.44) | ||
| Ptrendb | 0.11 | 0.24 | ||
| Model for CD45RO+ cell density (n = 574, as an outcome variable) | ||||
| The amount of Fusobacterium nucleatum | Negative | 1 (reference) | 1 (reference) | |
| Low | 1.33 (0.72–2.43) | 1.25 (0.67–2.31) | ||
| High | 1.09 (0.60–1.98) | 0.97 (0.53–1.79) | ||
| Ptrendb | 0.56 | 0.88 | ||
| Model for FOXP3+ cell density (n = 548, as an outcome variable) | ||||
| The amount of Fusobacterium nucleatum | Negative | 1 (reference) | 1 (reference) | |
| Low | 1.25 (0.66–2.35) | 1.14 (0.60–2.15) | ||
| High | 0.46 (0.25–0.84) | 0.41 (0.22–0.76) | ||
| Ptrendb | 0.038 | 0.014 | ||
Abbreviation: CI, confidence interval; OR, odds ratio.
The ordinal logistic regression analysis model initially included age, sex, year of diagnosis, family history of colorectal carcinoma in parent or sibling, tumor location, tumor differentiation, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and LINE-1 methylation level. A backward stepwise elimination with a threshold of P < 0.05 was used to select variables in the final models. Variables remaining in the final multivariable ordinal logistic regression models are shown in eTable 3 in the Supplement.
Ptrend value was calculated by the linear trend across the ordinal (negative, low, and high) categories of the amount of Fusobacterium nucleatum as a continuous variable in the ordinal logistic regression model for the density of CD3+, CD8+, CD45RO+, or FOXP3+ T-cells (an ordinal quartile outcome variable). Because we assessed four primary outcome variables, we adjusted two-sided α level to 0.013 (= 0.05/4) by simple Bonferroni correction.
In an exploratory analysis, we did not observe a significant association of tissue Fusobacterium nucleatum with Crohn’s-like reaction, peritumoral lymphocytic reaction, intratumoral periglandular reaction, or tumor infiltrating lymphocytes (P ≥ 0.11; eTable 5 in the Supplement).
Tissue Fusobacterium nucleatum and colorectal cancer mortality
In our exploratory analysis, we did not observe a significant association of tissue Fusobacterium nucleatum with colorectal cancer-specific mortality (Ptrend = 0.45) or overall mortality (Ptrend = 0.64; eTable 6 in the Supplement).
Discussion
We conducted this study to test the hypothesis that the amount of Fusobacterium nucleatum in colorectal carcinoma tissue might be inversely associated with the density of T-cells in tumor tissue. Utilizing the database of the 598 colorectal carcinoma cases in the two U.S. nationwide prospective cohort studies, we found that higher amount of Fusobacterium nucleatum was associated with lower density of CD3+ T-cells in colorectal carcinoma tissue.
High densities of CD3+ pan-T-cells and T-cell subpopulations (CD8+, CD45RO+, and FOXP3+ T-cells) in colorectal carcinoma have been associated with better patient survival, indicating a major role of T-cell-mediated adaptive immunity in inhibiting colorectal tumor progression.41 Virulence factors derived from Fusobacterium nucleatum have been shown to inhibit T-cell activity.26,27 Previous studies have shown that a virulence factor derived from Helicobacter pylori, which has been shown to cause gastric adenocarcinoma, can inhibit T-cell activity.42 This immunosuppressive effect may be similar to the potential immunosuppressive effect of Fusobacterium nucleatum. In the ApcMin/+ mouse model, Fusobacterium nucleatum promotes colonic neoplasia development through the recruitment of myeloid-derived suppressor cells into the tumor microenvironment.28 Myeloid-derived suppressor cells inhibit T-cell proliferation and induce T-cell apoptosis.43 These lines of experimental evidence are consistent with our finding of the inverse association between the amount of Fusobacterium nucleatum and the density of CD3+ T-cells in tumor tissue. Further studies are needed to investigate myeloid-derived suppressor cells in relation to Fusobacterium nucleatum in colorectal carcinoma tissue.
Studies have shown significant differences in the composition of intestinal microbiota along bowel subsites, likely leading to differences in colonic mucosal immunity.44,45 Consistent with a continuum of changes in intestinal microbiota and luminal contents, proportions of specific molecular features in colorectal cancer (namely, MSI-high, CIMP-high, and BRAF and PIK3CA mutations) continuously decrease from ascending colon to rectum.46,47 In the current study, the amount of Fusobacterium nucleatum in colorectal carcinoma tissue appeared to be associated with proximal tumor location, consistent with the previous study.25 Because epidemiologic evidence suggests that colonoscopy screening may be less effective for preventing proximal colon cancer than distal colon cancer,30,48 more effective prevention strategies may need to be developed for proximal colon cancer. Diet, antibiotics, and pro- and prebiotics have been shown to influence the composition of intestinal microbiota.49 In light of our findings, it would be intriguing for future investigations to explore potential influence of diet, lifestyle factors, and environmental exposures on Fusobacterium nucleatum and its immunosuppressive effect, which may have important implications for the development of colorectal cancer prevention strategies through targeting microbiota and immune cells.
We acknowledge limitations of our study. With the use of FFPE tissue specimens, routine histopathology procedures including tissue fixation, paraffin embedding, and storage may influence the quantitative PCR assay to detect microorganisms. However, technical artifacts, if any, would have affected our results likely towards the null hypothesis. In our limited validation study, we did detect Fusobacterium nucleatum in both of paired FFPE and frozen tissue specimens in two colorectal carcinoma cases by the quantitative PCR assay. Our validation study also showed a high linearity (r2 > 0.99) and a high reproducibility (inter-assay coefficient of variation ≤ 1%) of the quantitative PCR assay for Fusobacterium nucleatum with the use of FFPE tissue specimens. In addition, our data on the relationship between Fusobacterium nucleatum and tumor MSI and CIMP status are in agreement with the recent study which used a quantitative PCR assay for Fusobacterium in frozen tissue specimens.25 These results suggest an acceptable performance of the quantitative PCR assay for Fusobacterium nucleatum in FFPE tissue specimens. Another limitation is the cross-sectional nature of our study. Hence, we cannot exclude a possibility of reverse causation. Although it is possible that immune cells may eradicate Fusobacterium nucleatum, experimental evidence indicating an immunosuppressive effect of Fusobacterium nucleatum on T-cell activity26–28 formed a basis for our specific hypothesis. Because any experimental system cannot perfectly recapitulate the complex nature of human tumor or immune system, analyses of human cancer tissue in a large population are useful in elucidating the relationship between microbiota and immunity in cancer.
Strengths of this study include the use of our molecular pathological epidemiology database (of nearly 600 colorectal carcinoma cases in the two U.S. nationwide, prospective cohort studies) which integrates key tumor molecular features, the amount of Fusobacterium nucleatum, and immune reaction status in colorectal carcinoma tissue. The sample size and comprehensiveness of this population-based colorectal cancer database enabled us to assess the association between the amount of Fusobacterium nucleatum and the density of T-cells, controlling for potential confounders.
Conclusions
In this cross-sectional analysis of the U.S. nationwide prospective cohort studies, higher amount of Fusobacterium nucleatum was associated with lower density of CD3+ T-cells in colorectal carcinoma tissue. These findings need to be validated by further studies. Upon validation, our findings may provide insights for strategies to target microbiota and immune cells for colorectal cancer prevention and treatment.
Supplementary Material
Acknowledgments
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Funding: This work was supported by U.S. National Institutes of Health (NIH) grants [P01 CA87969 to S.E. Hankinson; UM1 CA186107 to M.J. Stampfer; P01 CA55075 to W.C. Willett; UM1 CA167552 to W.C. Willett; P50 CA127003 to C.S.F.; R01 CA137178 to A.T.C.; R01 CA151993 to S.O.; and K07 CA190673 to R.N.]; and by grants from The Paula and Russell Agrusa Fund for Colorectal Cancer Research, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. K.M. is supported by a fellowship grant from Uehara Memorial Foundation. S.A.K. is supported by Early Exchange Postdoctoral Fellowship Grant from Asan Medical Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Andrew T. Chan previously served as a consultant for Bayer Healthcare, Millennium Pharmaceuticals, Pozen Inc, and Pfizer Inc. This study was not funded by Bayer Healthcare, Millennium Pharmaceuticals, Pozen Inc, or Pfizer Inc. No other conflict of interest exists.
Abbreviations
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- Ct
cycle threshold
- FFPE
formalin-fixed paraffin-embedded
- HPFS
Health Professionals Follow-up Study
- LINE-1
long interspersed nucleotide element-1
- MSI
microsatellite instability
- MSS
microsatellite stable
- NHS
Nurses’ Health Study
- OR
odds ratio
- PCR
polymerase chain reaction
- SD
standard deviation
Footnotes
Author contributions: All authors contributed to review and revision. Danny A. Milner, Curtis C. Harris, Edward Giovannucci, Levi A. Garraway, Glenn Dranoff, Wendy S. Garrett, Curtis Huttenhower, Charles S. Fuchs, and Shuji Ogino developed the main concept and designed the study. Charles S. Fuchs, Andrew T. Chan, and Shuji Ogino wrote grant applications. Kosuke Mima, Yasutaka Sukawa, Reiko Nishihara, Zhi Rong Qian, Mai Yamauchi, Kentaro Inamura, Sun A Kim, Atsuhiro Masuda, Katsuhiko Nosho, Andrew T. Chan, Charles S. Fuchs, and Shuji Ogino were responsible for collection of tumor tissue, and acquisition of epidemiologic, clinical and tumor tissue data, including histopathological and immunohistochemical characteristics. Kosuke Mima, Yasutaka Sukawa, Alecsandar D. Kostic, Marios Giannakis, Hideo Watanabe, Susan Bullman, Danny A. Milner, Curtis Huttenhower, Charles S. Fuchs, and Shuji Ogino performed data analysis and interpretation. Kosuke Mima, Reiko Nishihara, and Shuji Ogino drafted the manuscript. Jonathan A. Nowak, Susan Bullman, Marios Giannakis, Edward Giovannucci, Gordon J. Freeman, Glenn Dranoff, Andrew T. Chan, Wendy S. Garrett, Charles S. Fuchs, and Shuji Ogino contributed to editing and critical revision for important intellectual contents.
Conflict of interest: The other authors declare that they have no conflicts of interest.
Use of standardized official symbols: We use HUGO (Human Genome Organisation)-approved official symbols for genes and gene products, including APC, BRAF, CACNA1G, CD3, CD8, CDKN2A, CRABP1, FOXP3, IGF2, KRAS, MLH1, NEUROG1, PIK3CA, PTPRC, RUNX3, SLCO2A1, and SOCS1; all of which are described at www.genenames.org.
References
- 1.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 2.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12(4):237–251. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mlecnik B, Bindea G, Angell HK, et al. Functional network pipeline reveals genetic determinants associated with in situ lymphocyte proliferation and survival of cancer patients. Sci Transl Med. 2014;6(228):228ra237. doi: 10.1126/scitranslmed.3007240. [DOI] [PubMed] [Google Scholar]
- 4.Di Caro G, Bergomas F, Grizzi F, et al. Occurrence of tertiary lymphoid tissue is associated with T-cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin Cancer Res. 2014;20(8):2147–2158. doi: 10.1158/1078-0432.CCR-13-2590. [DOI] [PubMed] [Google Scholar]
- 5.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222(4):350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salama P, Phillips M, Grieu F, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27(2):186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 7.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Y, Freeman GJ. The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov. 2015;5(1):16–18. doi: 10.1158/2159-8290.CD-14-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander J, Watanabe T, Wu TT, Rashid A, Li S, Hamilton SR. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158(2):527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tougeron D, Fauquembergue E, Rouquette A, et al. Tumor-infiltrating lymphocytes in colorectal cancers with microsatellite instability are correlated with the number and spectrum of frameshift mutations. Mod Pathol. 2009;22(9):1186–1195. doi: 10.1038/modpathol.2009.80. [DOI] [PubMed] [Google Scholar]
- 11.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15(20):6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 13.Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grivennikov SI, Wang K, Mucida D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belcheva A, Irrazabal T, Robertson SJ, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158(2):288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 16.Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105(24):1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnet M, Buc E, Sauvanet P, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20(4):859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 18.Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111(51):18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell. 2014;157(4):776–783. doi: 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Irrazabal T, Belcheva A, Girardin SE, Martin A, Philpott DJ. The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell. 2014;54(2):309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 21.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 22.Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology--analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8(12):711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan CW, Ma X, Paranjpe A, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. 2010;78(11):4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369(12):1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61(6):794–797. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 33.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122(12):2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8(2):209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12(2):177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3(11):e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7(3):413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura Y, Lochhead P, Yamauchi M, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the 'Immunoscore' in the classification of malignant tumours. J Pathol. 2014;232(2):199–209. doi: 10.1002/path.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301(5636):1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- 43.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 45.Li X, LeBlanc J, Truong A, et al. A metaproteomic approach to study human-microbial ecosystems at the mucosal luminal interface. PLoS One. 2011;6(11):e26542. doi: 10.1371/journal.pone.0026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148(1):77–87. e72. doi: 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samadder NJ, Curtin K, Tuohy TM, et al. Characteristics of missed or interval colorectal cancer and patient survival: a population-based study. Gastroenterology. 2014;146(4):950–960. doi: 10.1053/j.gastro.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Zitvogel L, Galluzzi L, Viaud S, et al. Cancer and the gut microbiota: An unexpected link. Sci Transl Med. 2015;7(271):271ps271. doi: 10.1126/scitranslmed.3010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


