Abstract
Genetic analysis of clinical phenotypes in consanguineous families is complicated by co-inheritance of large DNA regions carrying independent variants. Here we characterized a family with early onset cone-rod dystrophy (CRD) and muscular dystrophy. Homozygosity mapping followed by whole exome sequencing revealed a nonsense mutation, p.R270*, in ALMS1 and two novel potentially disease-causing missense variants, p.R1581C and p.Y2070C, in DYSF. ALMS1 and DYSF are genetically and physically linked on chromosome 2 in a genomic region suggested by homozygosity mapping and associated with Alström syndrome, which includes CRD, and with limb girdle muscular dystrophy, respectively. Affected family members lack additional systemic manifestations of Alström syndrome but exhibit mild muscular dystrophy. RNA-seq data did not reveal any significant variations in ALMS1 transcripts in the human retina. Our study thus implicates ALMS1 as a non-syndromic retinal disease gene and suggests a potential role of variants in interacting cilia genes in modifying clinical phenotypes.
Keywords: ALMS1, DYSF, retinal degeneration, pleiotropic phenotypes, photoreceptor, vision loss
The Israeli and Palestinian populations have a relatively high rate of consanguinity as well as marriages within semi-isolated sub-populations (Zlotogora, 1997). Offspring of first cousins have an increased risk of 2–4% of congenital genetic disorders and early mortality (Teeuw et al., 2010). Genetic mapping in consanguineous cohorts offers enhanced power to identify disease genes, especially with recessive inheritance, using homozygosity mapping (HM). One can track an identical-by-descent recessive mutation (arising from the same ancestor) by identifying blocks of homozygous markers in the genome of affected individuals (Alkuraya, 2010; Lander and Botstein, 1987). While HM is useful in localizing the disease gene to one or few chromosomal locations, genomic region(s) can still be relatively large and harbor dozens, if not hundreds, of genes. Next-generation sequencing (NGS) methods allow rapid genome-wide survey of variants and are transforming human genetics research (Schneeberger, 2014). Combining HM with NGS methods, such as whole exome sequencing (WES), can facilitate and expedite gene identification in families even with a single affected member.
Cone-rod dystrophies (CRD) are inherited retinal degenerative diseases that affect both cone and rod photoreceptor function in a cone > rod pattern, often exhibiting early cone and macular involvement (Hamel, 2007). The age of onset in CRDs is variable, but most patients demonstrate disease phenotype in the first two decades of life, with a prevalence of 1 in 40,000 (Hamel, 2007). Electroretinography (ERG) evaluation shows characteristic reduction of photopic cone b-wave amplitude with concurrent or subsequent scotopic rod dysfunction. Non-syndromic CRDs display autosomal dominant, recessive or X-linked inheritance and, like other retinal degenerations, present extensive genetic heterogeneity. To date, mutations in at least 30 different genes have been implicated in CRD phenotype (RetNet; https://sph.uth.edu/retnet/). Five major genes associated with CRDs are ABCA4 (MIM# 601691), KCNV2 (MIM# 607604), GUCY2D (MIM# 600179), CRX (MIM# 602225) and RPGR (MIM# 312610) (Roosing et al., 2014). We recently reported GUCY2D as a major gene associated with CRDs in Israel (Lazar et al., 2015).
As part of ongoing efforts to identify and catalog genetic defects in Israeli and Palestinian families with retinal and macular degeneration, we recruited a two-generation consanguineous Arab-Muslim family (MOL0339), which resided in the vicinity of Jerusalem and included two siblings affected with early-onset severe autosomal recessive CRD (arCRD) and muscular dystrophy phenotype (Fig. 1A). These two seemingly unrelated phenotypes in the affected individuals can be caused by either a single gene defect leading to a novel syndrome or two different mutations, one associated with the retinal disease and the other with the muscle phenotype. All human studies adhered to the tenets of the Declaration of Helsinki. After obtaining ethical approval from the Hadassah Medical Center institutional review board, we explained the study to all participants and obtained informed consent. Blood samples were collected from five members of the family, including two affected and two unaffected siblings and their mother. DNA was extracted using FlexiGene DNA kit (Qiagen, Venlo, The Netherlands). Family history was recorded, and full ophthalmologic evaluation was performed including visual function testing and retinal imaging, as described (Lazar et al., 2015). Whole genome single nucleotide polymorphism (SNP) analysis, using Affymetrix (Santa Clara, CA) SNP microarray platform 6.0, followed by whole exome capture, using Agilent SureSelectXT Human All Exon V5 Kit (Agilent Technologies, Santa Clara, CA) was performed on family members marked in Fig. 1A. Detailed description of experimental methods as well as data analysis and validation are provided in the Supp. Materials and Methods. Sanger sequencing was used for validation of candidate variants. Mutation nomenclature refers to GenBank reference sequence NM_015120.4 for ALMS1 and NM_003494.3 for DYSF (GRCh37). Identified genetic variants were submitted to the Leiden Open Variation Database (http://databases.lovd.nl/).
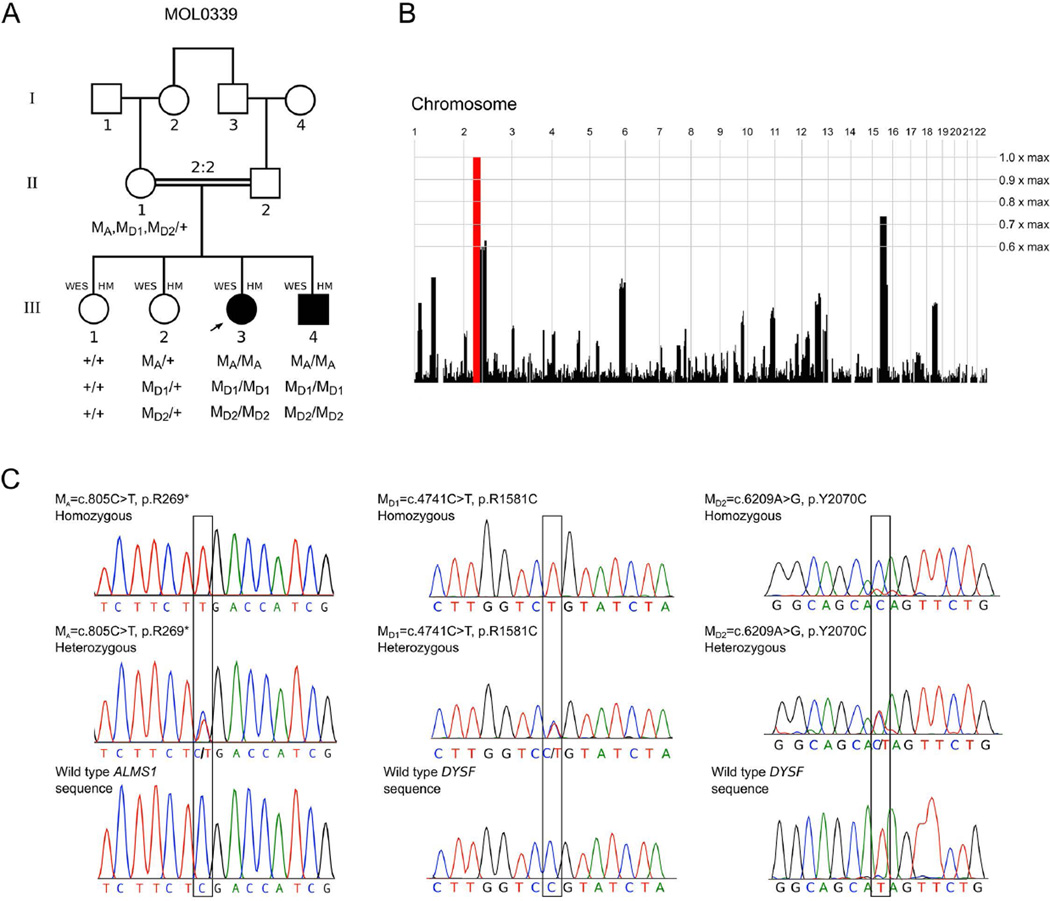
Figure 1. Genetic analysis of the family MOL0339.
(A) Pedigree structure. The segregation of three novel genetic variants is indicated as: M/M, homozygous for nucleotide change; M/+, heterozygous; +/+ homozygous for wild type allele. MA=c.808C>T (p.R270*) ALMS1 variant; MD1=c.4741C>T (p.R1581C) and MD2=c.6209A>G (p.Y2070C) DYSF variants. Individuals subjected to whole exome sequencing (WES) and homozygosity mapping (HM) are marked.
(B) Homozygosity mapping analysis (using HomozygosityMapper) showing a single 35.3 Mb locus of homozygosity on chromosome 2 (between 49–84.3 Mb), containing both the ALMS1 and DYSF genes.
(C) Chromatograms of ALMS1 and DYSF DNA sequences in affected, carrier and unaffected individuals showing homozygous, heterozygous nucleotide changes and wild type sequence respectively. Affected nucleotide positions are marked by a black box. The nucleotide sequence surrounding p.Y2070C change is represented on the reverse complementary DNA strand.
The parents of the two affected siblings are first cousins, and therefore one can assume that the mutation(s) causing the two phenotypes are part of a large homozygous region in the affected individuals. To identify genomic regions shared by the two affected siblings but not by unaffected siblings (Fig. 1A), we performed HM using Affymetrix SNP arrays on these four samples. Mutations in the ABCA4 gene were reported previously (Maugeri et al., 2000) to be a common cause of arCRD. We therefore examined the ABCA4 genomic region in the HM data and found an identical homozygous region in the four siblings, thereby excluding ABCA4 as a possible causative gene in this family. Subsequent analysis revealed a single homozygous region on chromosome 2, encompassing 35.3 Mb (between 49–84.3 Mb; Fig. 1B), which contains 263 genes, none of which has been associated earlier with non-syndromic arCRD. We, therefore, performed WES analysis on the four individuals. Visual examination of sequencing reads from WES did not identify any rare coding variant or larger genetic aberration in several genes (e.g., FLVCR1, DTHD1, OPA3, ELOVL4, ABHD12, PRPS1, MT-ATP6 etc. (RetNet)) that have been associated with photoreceptor degeneration along with some form of muscular involvement.
WES generated approximately 3 Gb of sequence data per individual with 70X average depth, covering almost 90% of the targeted exons. We identified 20,000 – 23,000 exonic variants for each individual. Given that we are analyzing a consanguineous pedigree, we first presumed recessive inheritance of potential causative variants and selected homozygous changes shared by affected individuals and those missing or heterozygous in unaffected. We obtained a list of 23 rare (Minor Allele Frequency < 0.01) variants that were also absent in our extensive in-house Israeli and Caucasian exome dataset. Sequence coverage of the identified variants was visually inspected, and genes expressed in our retinal RNA-seq database as well as those previously associated with similar clinical phenotypes were evaluated for possible association with the disease in our family. By combining HM and WES, we discovered mutations in two independent yet linked genes, ALMS1 (MIM# 606844) and DYSF (MIM# 603009), mutations in which cause Alström syndrome (ALMS; MIM# 203800) (Marshall et al., 2015) and Miyoshi muscular dystrophy (MMD1; MIM# 254130) or limb-girdle muscular dystrophy (LGMD2B; MIM# 253601), respectively (Liu et al., 1998). We identified a novel nonsense variant c.808C>T (p.R270*) in exon 5 of the ALMS1 gene and two novel potentially disease causing missense variants c.4741C>T (p.R1581C) and c.6209A>G (p.Y2070C) in the DYSF gene (Supp. Table S1). The ALMS1 nonsense mutation was not detected in any of the following databases: 1000 Genome Project, dbSNP137, Exome Variant Server, Exome Aggregation Consortium (ExAC), and a local database of 408 Israeli exomes. The DYSF missense variants were observed in a heterozygous state only in some of these databases: c.4741C>T was seen in 5 out of 121410 in ExAC and 3 out of 408 at the local database and c.6209A>G was observed in 1 out of 121400 individuals in ExAC. All three variants were validated by Sanger sequencing (Fig. 1C).
To examine whether the c.808C>T change in ALMS1 is a founder mutation in the Arab-Muslim population, we screened a set of 87 index cases with inherited retinal degeneration for the exon 5 variant but no individual carried the mutation or any other potential disease-causing variant in this exon. In addition, we examined the expression of ALMS1 transcripts observed in our RNA-seq data of retinal tissues (Supp. Figure S1). No significant difference was observed in the expression of three transcripts, ALMS1-001, ALMS1-002 and ALMS1-003 (Ensemble version ENSG00000116127.13). Furthermore, the ALMS1-201 splice variant, containing only the first 9 exons of the gene was not expressed in any tissue (Supp. Figure S1).
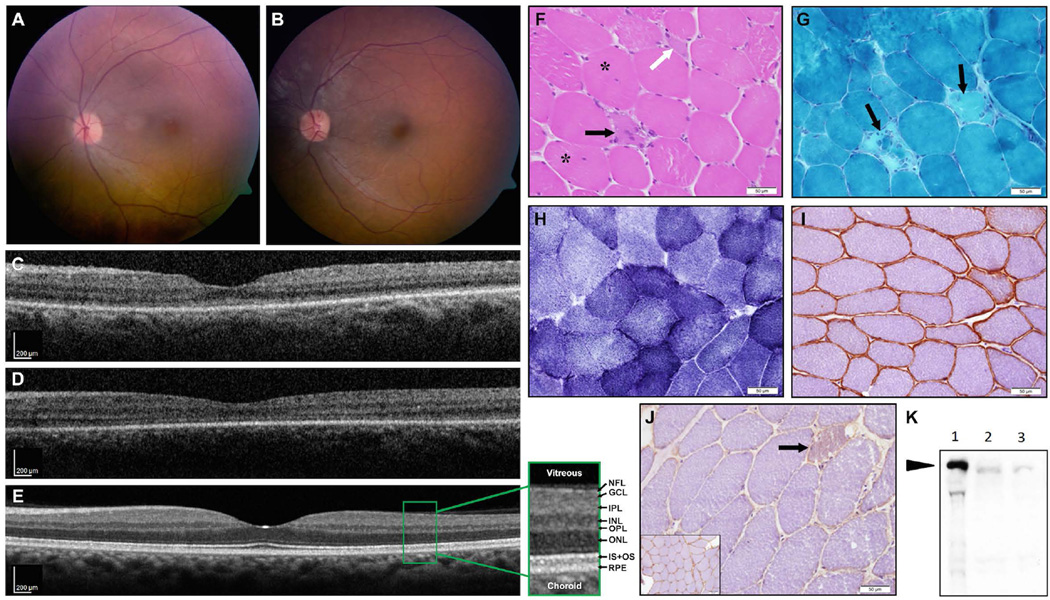
Clinical evaluations of the two affected siblings revealed vision loss and muscular dystrophy. The index case (III:3) suffered from photophobia and pendular nystagmus from early childhood, and at the age of 4 years visual acuity (using a picture cube) was 0.75/0.7.5 (i.e., 0.1) in each eye, with hypermetropic correction (spherical equivalent of +5.00 diopters in both eyes, Supp. Table S2). Fundus appearance was perceived as within normal limits on repeated exams between the age of 3 and 9 years. At the age of 23, visual acuity was limited to hand movements in each eye, and anterior segments were normal with no cataract. On fundus exam, hypermetropic discs with temporal pallor and mild narrowing of the retinal vessels were noted along with an abnormal macular reflex (Fig. 2A,B). No pigmentary retinal changes were observed. OCT imaging revealed that retinal structure in the macular area was largely preserved, with some thinning of the photoreceptor layer in the foveal area (Fig. 2C–D). Average central subfield thickness (CST) of the right and left eyes of this patient was 260µm as compared with 270±22.5µm in normal eyes (mean±SD, see Grover et. al., 2009). The average retinal thickness in the inner macular ring was 265µm in both eyes compared with a mean normal of 332µm. The patient underwent two ERG recordings, at the ages of 10 and 17, which indicated widespread cone > rod involvement (Supp. Table S2), that together with the clinical symptoms and findings are compatible with a generalized CRD phenotype. Her affected brother (III:4) presented very similar clinical findings, including photophobia and nystagmus, with a practically normal fundus on multiple examinations between ages 5 and 10. At age 19, he had visual acuity of hand movements in both eyes, and fundus, OCT and ERG findings largely resembled those of his sister (Fig. 2B,D and Supp. Table S2; average CST of the right and left eyes, 245µm; average inner macular ring thickness, 288 µm).
Figure 2. Fundus, OCT imaging, and muscle biopsy results of the affected patients.
(A-E) Fundus imaging: (A) Color fundus picture of the left eye of patient III:3 at age 22 showing a hypermetropic disc with temporal pallor and mild narrowing of the retinal vessels, along with an abnormal macular reflex. (B) Patient III:4 at age 19 shows similar findings with minimal funduscopic changes and preserved retinal structure in the macular area. No pigmentary retinal changes were present in either patient. (C,D) OCT imaging performed at the same time (C- patient III:3, D- patient III:4) revealed retinal structure in the macular area to be largely preserved, with some thinning of the photoreceptor layer (ONL, IS-OS) in the foveal area as compared to an OCT scan of an age-matched normal control (E, retinal layers as seen on OCT are detailed in an enlarged view: NFL- nerve fiber layer; GCL- ganglion cell layer; IPL- inner plexiform layer; INL- inner nuclear layer; OPL- outer plexiform layer; ONL- outer nuclear layer, containing nuclei of photoreceptors; IS+OS- inner +outer segments of the photoreceptors; RPE- retinal pigment epithelium).
(F-K) Muscle biopsy results: (F-J) Staining of muscle biopsy [H&E (F), Modified Gomori-TC (G), NADH (H), and immunohistochemical stains for caveolin-3 (I) and dysferlin (J)]. The results show mild dystrophic changes, including mild variation in myofiber diameter, mild increase of internally displaced nuclei (F, asterisks), and few necrotic (black arrows) as well as regenerating myofibers (F, white arrow). Neither endomysial fibrosis (F) nor significant changes in the cytoarchitecture (H) were observed. There was marked reduction in sarcolemmal staining on immunostain for dysferlin (J, inset-normal control), while immunostain for caveolin-3 (I) was normal. Immunoblot for dysferlin (K, NCL-Hamlet antibody, Novocastra) confirmed marked reduction in dysferlin (arrow indicates dysferlin band (lane 1 - normal control; lanes 2 and 3 - two different loadings of patient’s muscle extract), as opposed to normal immunoblot for calpain-3 (not shown), consistent with a diagnosis of dysferlinopathy.
Interestingly, both affected individuals were also diagnosed with limb-girdle muscular dystrophy. The index case had high levels of CPK (3114 at age 20, normal range 1–170 U/L), while her brother had even higher levels ranging from 14790 to 42,000 U/L, with lower limb weakness. The unaffected sisters and the mother were found to have normal CPK levels (ranging from 59 to 82 U/L) with no complaint of weakness. This prompted further evaluation, including MRI of crus leg muscles that showed areas of increased T2 relaxation times and muscle biopsy (Fig. 2F-J) that revealed mild dystrophic changes, associated with marked reduction in sarcolemmal staining on immunohistochemical stain for Dysferlin. The diagnosis of dysferlinopathy was confirmed by immunoblot (Fig. 2K). In addition, the index patient had mildly abnormal liver function tests at age 16 and a liver biopsy was then performed, which did not show specific findings except mild fibrotic dilation of the portal spaces and mild proliferation of the bile ducts. This mild abnormality of liver enzymes is likely secondary to the muscle disease; however, the possibility of combined effect of DYSF and ALMS1 abnormal protein function can’t be excluded. To date, neither of the two affected siblings have manifested any of the other typical systemic features of Alström syndrome (Supp. Table S3).
ALMS1 protein localizes to centrosome and basal body and is expressed in all pathologically affected tissues (Collin et al., 2002; Hearn et al., 2002; Hearn et al., 2005). To date, 239 ALMS1 mutations have been reported to cause Alström syndrome, a rare recessive disorder, characterized by CRD, hearing loss, childhood obesity, type 2 diabetes mellitus, cardiomyopathy, fibrosis and multiple organ failure (Marshall et al., 2011; Marshall et al., 2015). In some cases, affected children present with only a subset of features early in the course of the disease (Marshall et al., 2005), but typical features of disease eventually develop allowing a proper clinical diagnosis. In only one family so far, of Saudi Arabic origin, an ALMS1 mutation was reported to cause a non-syndromic retinopathy (Wang et al., 2011). The nonsense mutation, c.10945G>T (p.E3649*) in exon 16 of ALMS1 identified in this family (Wang et al., 2011), has been reported earlier in patients with Alström syndrome (Marshall et al., 2007). The clinical diagnosis in this family was described as Leber congenital amaurosis (LCA) rather than CRD, and the ocular findings included spoke-like cataracts, 3+/4 vascular attenuation, pigmentary maculopathy with central ellipse of gray silvery atrophy and coarse granular diffuse RPE peripheral atrophy, which differ from the findings in the two patients of the present study. ERG and imaging data of the Saudi family were not included in the published report; therefore, further detailed comparison of phenotypes is not possible, but given the diagnosis of LCA one would assume ERG responses were non-detectable as opposed to the cone > rod involvement observed in our patients.
A vast majority of reported ALMS1 mutations are localized in exons 8, 10 and 16, and a recent report further extends the number of previously known mutations to a total of 239 that are distributed throughout most exons (Marshall et al., 2015). Only a few ALMS1 mutations have been identified in exons 1–7, and mutations in this region were hypothesized to be embryonic lethal (Marshall et al., 2011). However, the results presented here, as well as data on other ALMS1 mutations in this region, demonstrate that mutations in exons 1–7 can cause either nonsyndromic CRD (as in our patients) or Alström syndrome. To date, 4 mutations have been reported in exon 5 of ALMS1; two frameshift and two nonsense changes (Casey et al., 2014; Marshall et al., 2015). One of the exon 5 frameshift mutations (c.777delT, p.D260fs*26) was present in a compound heterozygous state with exon 20 mutation (c.12145_12146insC, p.S4049fs*36) and reported in a family with atypical Alström phenotype (Casey et al., 2014). We suggest that atypical phenotype is likely due to the localization of identified mutation to a specific ALMS1 domain. Alternatively, sequence changes in other genes, probably those encoding interacting ciliary proteins, might modulate disease expressivity. A larger study of patients with variable disease expressivity is needed to identify such modifier genes. Other ciliopathies (e.g., Bardet-Biedl syndrome) exhibit a similar phenomenon where different mutations in the same gene can cause either the typical syndromic form or a non-syndromic retinal disease. Another example is CEP290, a centrosomal-ciliary protein that has been associated with non-syndromic retinal phenotype in LCA patients (Cideciyan et al., 2011), while other mutations in the gene result in more severe phenotypes involving multiple tissues and are linked to various syndromes (Baala et al., 2007; Leitch et al., 2008; Sayer et al., 2006).
ALMS1 is ubiquitously expressed and alternative transcripts are observed as a result of splicing events (Collin et al., 2002; Hearn et al., 2002). ALMS1 gene is comprised of 23 coding exons, however the most abundant transcript does not contain exon 2 (Hearn et al., 2002). Evaluation of the most commonly observed transcripts in our in-house human retina expression data did not reveal significantly different expression of any specific transcript (Supp. Figure S1). Although several ALMS1 transcripts can be produced, a long transcript with the majority of coding exons is expressed in the retina as well as other human tissues. Functional or large-scale disease expressivity studies might be required to obtain additional insights and genotype-phenotype correlation.
We also identified two novel homozygous potentially disease-associated variants, c.4741C>T (p.R1581C) and c.6209A>G (p.Y2070C), in the DYSF gene, which encodes Dysferlin, a calcium-dependent phospholipid binding protein that plays a role in muscle membrane maintenance and repair (Bansal et al., 2003; Bashir et al., 1998). Previously described phenotypes of Dysferlin dysfunction include early onset muscle weakness that manifests in late teens to early twenties (Miyoshi et al., 1986). The dysferlin p.R1581C variant is part of the fifth C2 Ferlin domain, which is involved in vesicle fusion events. A different non-synonymous change in the same amino acid (p.R1581H) was reported as a polymorphism (Takahashi et al., 2003). The DYSF p.Y2070C variant is located at C terminus of Dysferlin (11 amino acids upstream the stop codon). A large number of disease-causing mutations have been identified in DYSF gene; over half of these are truncation and/or insertion-deletion mutations (Blandin et al., 2012). Homozygous missense changes represent about one-third of total observed variants. At this stage, we cannot conclude whether one or a combination of the two variants causes the muscular dystrophy phenotype. Nonetheless, the lack of Dysferlin in muscle pathology demonstrates that DYSF mutation(s) is indeed responsible for clinical findings. Mutations leading to new cysteine residues in Dysferlin are predicted to drastically alter its structure and stability; therefore, our results represent a novel paradigm in pathology of dysferlinopathies.
In summary, our studies implicate ALMS1 as a gene for non-syndromic CRD and reinforce the value of WES in improving molecular genetic diagnosis and genotype-phenotype correlation. We conclude that two independent mutations in ALMS1 and DYSF cause CRD and muscular dystrophy in the studied consanguineous Israeli Arab family, without manifestation of additional Alström syndrome features, at least up to the age of 22. Our study further demonstrates the contribution of consanguineous marriages to autosomal recessive disease that can, in some cases, lead to a chromosomal region harboring mutations in two (or more) linked genes resulting in distinct unrelated phenotypes. Patients who inherit such chromosomal regions may have a complex clinical phenotype that could be mistakenly considered as a single syndrome. We thus advocate a careful clinical evaluation and genetic diagnosis for disease management in families with complex phenotypes.
Supplementary Material
ACKNOWLEDGMENTS
Grant Sponsors: United States - Israel Binational Science Foundation (# 2011202, to DS and AS), Yedidut Research Grant (to EB), and Intramural Research Program (# EY000473) of the National Eye Institute, National Institutes of Health (to AS).
We are grateful to the family for participation. We thank Alexis Boleda and Matthew Brooks for support of NGS data analysis, and to Alexey Obolensky, Shelly Stika, Israel Barzel and Inbar Erdinest for assistance in clinical evaluation of the patients.
REFERENCES
- Alkuraya FS. Homozygosity mapping: one more tool in the clinical geneticist's toolbox. Genet Med. 2010;12:236–239. doi: 10.1097/GIM.0b013e3181ceb95d. [DOI] [PubMed] [Google Scholar]
- Baala L, Audollent S, Martinovic J, Ozilou C, Babron MC, Sivanandamoorthy S, Saunier S, Salomon R, Gonzales M, Rattenberry E, Esculpavit C, Toutain A, et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet. 2007;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal D, Miyake K, Vogel SS, Groh S, Chen CC, Williamson R, McNeil PL, Campbell KP. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, Sadeh M, Mahjneh I, et al. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- Blandin G, Beroud C, Labelle V, Nguyen K, Wein N, Hamroun D, Williams B, Monnier N, Rufibach LE, Urtizberea JA, Cau P, Bartoli M, et al. UMD-DYSF, a novel locus specific database for the compilation and interactive analysis of mutations in the dysferlin gene. Hum Mutat. 2012;33:E2317–E2331. doi: 10.1002/humu.22015. [DOI] [PubMed] [Google Scholar]
- Casey J, McGettigan P, Brosnahan D, Curtis E, Treacy E, Ennis S, Lynch SA. Atypical Alstrom syndrome with novel ALMS1 mutations precluded by current diagnostic criteria. Eur J Med Genet. 2014;57:55–59. doi: 10.1016/j.ejmg.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Collin GB, Marshall JD, Ikeda A, So WV, Russell-Eggitt I, Maffei P, Beck S, Boerkoel CF, Sicolo N, Martin M, Nishina PM, Naggert JK. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat Genet. 2002;31:74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Rachel RA, Aleman TS, Swider M, Schwartz SB, Sumaroka A, Roman AJ, Stone EM, Jacobson SG, Swaroop A. Cone photoreceptors are the main targets for gene therapy of NPHP5 (IQCB1) or NPHP6 (CEP290) blindness: generation of an all-cone Nphp6 hypomorph mouse that mimics the human retinal ciliopathy. Hum Mol Genet. 2011;20:1411–1423. doi: 10.1093/hmg/ddr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover S, Murthy RK, Brar VS, Chalam KV. Normative data for macular thickness by high-definition spectral-domain optical coherence tomography (spectralis) Am J Ophtalmol. 2009;148:266–271. doi: 10.1016/j.ajo.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn T, Renforth GL, Spalluto C, Hanley NA, Piper K, Brickwood S, White C, Connolly V, Taylor JF, Russell-Eggitt I, Bonneau D, Walker M, et al. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alstrom syndrome. Nat Genet. 2002;31:79–83. doi: 10.1038/ng874. [DOI] [PubMed] [Google Scholar]
- Hearn T, Spalluto C, Phillips VJ, Renforth GL, Copin N, Hanley NA, Wilson DI. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54:1581–1587. doi: 10.2337/diabetes.54.5.1581. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- Lazar CH, Mutsuddi M, Kimchi A, Zelinger L, Mizrahi-Meissonnier L, Marks-Ohana D, Boleda A, Ratnapriya R, Sharon D, Swaroop A, Banin E. Whole exome sequencing reveals GUCY2D as a major gene associated with cone and cone-rod dystrophy in Israel. Invest Ophthalmol Vis Sci. 2015;56:420–430. doi: 10.1167/iovs.14-15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, Alfadhel M, Lewis RA, Eyaid W, Banin E, Dolifus H, Beales PL, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Hamida MB, Bohlega S, Culper EJ, et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Bronson RT, Collin GB, Nordstrom AD, Maffei P, Paisey RB, Carey C, Macdermott S, Russell-Eggitt I, Shea SE, Davis J, Beck S, et al. New Alstrom syndrome phenotypes based on the evaluation of 182 cases. Arch Intern Med. 2005;165:675–683. doi: 10.1001/archinte.165.6.675. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Hinman EG, Collin GB, Beck S, Cerqueira R, Maffei P, Milan G, Zhang W, Wilson DI, Hearn T, Tavares P, Vettor R, et al. Spectrum of ALMS1 variants and evaluation of genotype-phenotype correlations in Alstrom syndrome. Hum Mutat. 2007;28:1114–1123. doi: 10.1002/humu.20577. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Maffei P, Collin GB, Naggert JK. Alstrom syndrome: genetics and clinical overview. Curr Genomics. 2011;12:225–235. doi: 10.2174/138920211795677912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JD, Muller J, Collin GB, Milan G, Kingsmore SF, Dinwiddie D, Farrow EG, Miller NA, Favaretto F, Maffei P, Dolifus H, Vettor R, et al. Alstrom Syndrome: Mutation Spectrum of ALMS1. Hum Mutat. 2015 doi: 10.1002/humu.22796. 2015 Apr 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet. 2000;67:960–966. doi: 10.1086/303079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Kawai H, Iwasa M, Kusaka K, Nishino H. Autosomal recessive distal muscular dystrophy as a new type of progressive muscular dystrophy. Seventeen cases in eight families including an autopsied case. Brain. 1986;109(Pt 1):31–54. doi: 10.1093/brain/109.1.31. [DOI] [PubMed] [Google Scholar]
- Roosing S, Thiadens AA, Hoyng CB, Klaver CC, den Hollander AI, Cremers FP. Causes and consequences of inherited cone disorders. Prog Retin Eye Res. 2014;42:1–26. doi: 10.1016/j.preteyeres.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna Y, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Schneeberger K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat Rev Genet. 2014;15:662–676. doi: 10.1038/nrg3745. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Aoki M, Tateyama M, Kondo E, Mizuno T, Onodera Y, Takano R, Kawai H, Kamakura K, Mochizuki H, Shizuka-Ikeda M, Nakagawa M, et al. Dysferlin mutations in Japanese Miyoshi myopathy: relationship to phenotype. Neurology. 2003;60:1799–1804. doi: 10.1212/01.wnl.0000068333.43005.12. [DOI] [PubMed] [Google Scholar]
- Teeuw ME, Henneman L, Bochdanovits Z, Heutink P, Kuik DJ, Cornel MC, Ten Kate LP. Do consanguineous parents of a child affected by an autosomal recessive disease have more DNA identical-by-descent than similarly-related parents with healthy offspring? Design of a case-control study. BMC Med Genet. 2010;11:113. doi: 10.1186/1471-2350-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Cao M, Li Z, Chen X, Patenia C, Gore A, Abboud EB, Al-Rajhi AA, Lewis RA, Lupski JR, Mardon G, et al. Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3, and MYO7A mutations in patients with Leber congenital amaurosis. Hum Mutat. 2011;32:1450–1459. doi: 10.1002/humu.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotogora J. Genetic disorders among Palestinian Arabs: 1. Effects of consanguinity. Am J Med Genet. 1997;68:472–475. doi: 10.1002/(sici)1096-8628(19970211)68:4<472::aid-ajmg20>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.