Abstract
Choline acetyltransferase catalyzes the synthesis of acetylcholine at cholinergic nerves. Mutations in human CHAT cause a congenital myasthenic syndrome (CMS) due to impaired synthesis of ACh; this severe variant of the disease is frequently associated with unexpected episodes of potentially fatal apnea. The severity of this condition varies remarkably, and the molecular factors determining this variability are poorly understood. Furthermore, genotype–phenotype correlations have been difficult to establish in patients with biallelic mutations. We analyzed the protein expression of seven ChAT mutations, p.Val136Met, p.Arg207His, p.Arg186Trp, p.Val194Leu, p.Pro211Ala, p.Arg566Cys and p.Ser694Cys, in HEK-293 cells to phosphorylated ChAT, determined their enzyme kinetics and thermal instability, and examined their structural changes. Three mutations, p.Arg207His, p.Arg186Trp and p.Arg566Cys, are novel, and p.Val136Met and p.Arg207His are homozygous in three families and associated with severe disease. The characterization of mutants showed a decrease in the overall catalytic efficiency of ChAT; in particular, those located near the active-site tunnel produced the most seriously disruptive phenotypic effects. On the other hand, p.Val136Met is located far from both active and substrate-binding sites produced the most drastic reduction of ChAT expression. Overall, CHAT mutations producing low enzyme expression and severe kinetic effects are associated with the most severe phenotypes.
Keywords: ChAT, phosphorylation, enzyme kinetics, genotype-phenotype correlations
INTRODUCTION
Deficiency of acetylcholine (ACh) synthesis is a variant of congenital myasthenic syndrome (CMS) caused by mutations in choline acetyltransferase (CHAT; MIM# 118490). This ChAT deficiency is also referred to as CMS associated with episodic apnea (Ohno et al., 2001; Barisic et al., 2005), and is one of the described types of CMS caused by a defect of an enzyme located in the nerve terminal (Engel et al., 2012). Other enzymes causing CMS are GFPT-1 (Senderek et al., 2011), DPAGT1 (Marek et al. 2015), ALG2 and ALG14 (Cossins et al., 2015), and PREPL (Régal et al., 2014); these enzymes are involved in the glycosylation pathways.
ChAT catalyzes the transfer of an acetyl group from acetyl coenzyme A (AcCoA) to choline to form the neurotransmitter ACh in cholinergic neurons (Nachmansohn and Machado, 1943). Synaptic transmission between cholinergic neurons and their target cells is dependent on functional ChAT, and although reduction of ChAT expression and activity has been found in several neurological and psychiatric disorders, such as Alzheimer disease (Whitehouse, 1998) and schizophrenia (Holt et al., 2005), the majority of CHAT mutations reported to date have manifested as CMS (Engel, 2012); however, sequence variants of CHAT have been described in association with Alzheimer disease (Mubumbila et al., 2002), and recurrent large-scale deletions have been reported in developmental delay (Stankiewicz et al., 2012).
The pathogenic mechanism responsible for failure of neuromuscular transmission in deficiency of ACh synthesis differs substantially from that of other variants of CMS, because in this syndrome neither the endplate neither potential quantal content nor the postsynaptic response to a single quantum of ACh is significantly diminished (Ohno et al., 2001; Maselli et al., 2003). Instead, there is a use-dependent reduction of the quantal size due to a decrease in the number of ACh molecules per synaptic vesicle (Engel and Lambert, 1987; Mora et al., 1987) with normal ultrastructure of the neuromuscular junction (NMJ) (Maselli et al., 2003).
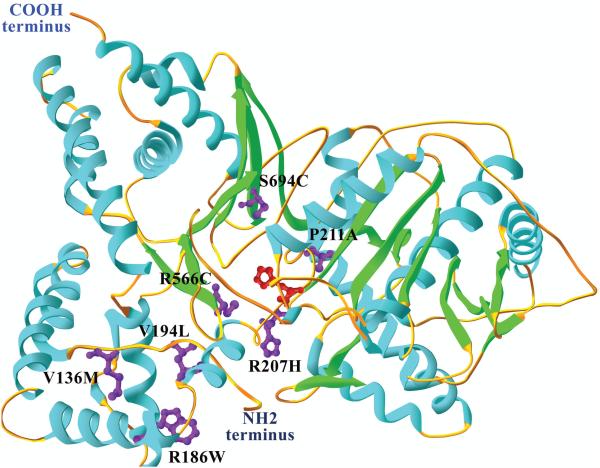
ChAT consists of a binding domain and a catalytic domain with an interfacial active-site tunnel and a key histidine for catalytic activity at the center of the tunnel (Cai et al., 2004; Kim et al., 2006), as shown in Fig. 1. The essential histidine residue, corresponding to His442 in human ChAT (Carbini and Hersh, 1993), is thought to act as a general base catalyst in the reaction. The activity of ChAT is decreased under oxidizing conditions and by modification with sulfhydryl-reactive agents (Hersh et al., 1979), indicating the presence of cysteines in the vicinity of the active site.
Fig 1.
Human ChAT structure. The tertiary structure of human ChAT is divided into two domains: a substrate binding domain comprising residues 118–207 and 510–733, and a catalytic domain with residues 208–509, based on the structure PDB 2FY2. This structure is missing the first 118 residues from the NH2 terminal domain, and the crystal coordinates start at residue 8. The analyzed tertiary structure corresponds to the 83-kDa ChAT isoform 2. The catalytic histidine, His442 (red), is located at β-sheet 8 at the active-site tunnel, which runs through the enzyme and is located at the domain interface. The structure is represented in ribbon diagram; the α-helices are in blue, the β-sheets in green and the coils in orange. The mutated residues in magenta are located as follows: p.Val136Met at coil 1, p.Arg186Trp at α-helix 3, p.Val194Leu at α-helix 4, p.Arg207His at coil 5, p.Pro211Ala at coil 5, p.Arg566Cys at β-sheet 12 and p.Ser694Cys at β-sheet 16. When the mutations are introduced, the Swiss-PdbViewer algorithm selects side chain rotamer from a backbone-dependent rotamer library, consequently introducing minimal steric violations and with the most favorable hydrogen-bond interactions.
Reaction of ChAT with butanedione, an arginine-modifying reagent, affects the activity of the enzyme (Carbini et al., 1990), and two arginine residues, Arg452 and Arg453, which correspond to human Arg560 and Arg561 respectively, that have been shown to be involved in CoA binding in rat ChAT (Wu and Hersh, 1995). In addition to these arginine residues, the binding of AcCoA and CoA is strongly affected by the enzyme location: the dissociation of CoA is the rate-limiting step at low ionic strength, but other steps become rate limiting at high ionic strength (Hersh et al., 1978; Hersh, 1979). The ChAT binding and catalytic domains have a six-stranded β-sheet surrounded by α-helices (Jogl and Tong, 2003; Cai et al., 2004).
The affinity for AcCoA is much higher than the affinity for choline, and the two-substrate reaction follows sequential kinetics with AcCoA as the leading substrate of the forward reaction (Hersh, 1979; Carbini and Hersh, 1993). His442 is proposed to function as a general base, enhancing the nucleophilicity of the choline hydroxyl for attack on the thioester bond in the forward reaction (Roskoski, 1974; Hersh, 1982).
ChAT is a member of the CoA-dependent acyltransferase superfamily, which also includes carnitine acetyltransferase, chloramphenicol acetyltransferase, dihydrolipoyl transacetylase, VibH, and carnitine palmitoyltransferase, enzymes involved in fatty acid metabolism and maintenance of acyl-CoA pools (Murzin et al., 1995; Ramsay et al., 2001). ChAT is a complex protein with alternative splicing that can produce six different mRNA isoforms, termed H, M, N1, N2, R, and S. Two of these transcripts, M and S, also encode 83- and 74-kDa isoforms of the protein through alternative translation start sites.
Previously, other CHAT mutations have been reported to cause CMS symptoms (Barisic et al., 2005; Maselli et al., 2006; Mallory et al., 2009). A first study by Ohno et al (2001) and a more recent study by Shen et al. (2011) describe CHAT mutations causing CMS and examine the biochemical effects of the mutant enzymes. However, genotype–phenotype correlations in these previous reports were difficult to establish due to the lack of homozygous patients. We describe here the result of expression studies analyzing the level of phosphorylated ChAT, kinetic properties, thermal instability and predicted H-bonding structural changes of seven CHAT mutants causing CMS. Three of these mutants are novel and two are unique because they are homozygous in three independent families.
MATERIALS AND METHODS
Cells and Reagents
HEK293 cells were grown in Dulbecco’s modified medium supplemented with 10% fetal bovine serum, L-glutamine, and penicillin/streptomycin, while being incubated at 37°C with 5% CO2.
Constructs
Mammalian expression vector
The full-length CHAT cDNA clone was obtained from Dr. David Rogers (Molecular and Cellular Biochemistry, University of Kentucky); we then subcloned the cDNA into a mammalian expression vector (pcDNA4/HisMax TOPO TA, Invitrogen). This wild-type CHAT cDNA was subjected to site-directed mutagenesis in order to create the CHAT p.Val136Met, p.Arg186Trp, p.Val194Leu, p.Pro211Ala, p.Arg207His, p.Arg566Cys and p.Ser694Cys mutations (QuikChange II, Agilent), and then transformed into XL1-Blue competent cells (Maselli et al., 2010). Presence of desired mutations and absence of unwanted mutations was confirmed using sequencing.
Mutational Analysis
The blood from 5 patients was used to extract genomic DNA using the QIAmp DNA Blood Kit (Qiagen). The genomic DNA was amplified by PCR and sequenced for all 20 expressed exons of the human CHAT gene. PCR products were sequenced on an ABI 3730 DNA Analyzer (Applied Biosystems). The sequences were analyzed with BioEdit software to find possible mutations, and the CHAT gene (NC_000010 and NM_020549.2) sequences were used as wild-type control. All DNA sequences showing differences from the control, whether pathogenic or not, were documented for later analysis. When possible, unaffected family members were genotyped as well. Our institutional review board approved the study. Patients were informed of their rights and the details of the research, and they all signed an informed consent form.
Expression Studies of Phosphorylated ChAT in HEK Cells
Expression in HEK293 cells
Since the ChAT S transcript transcribes all possible transcripts, it was used for the expression studies. It also contains all identified mutations, previously cloned from a spinal cord cDNA library (Ohno et al., 2001), and it is the predicted transcript used by motor neurons. Human CHAT wild-type and the p.Val136Met, p.Arg186Trp, p.Val194Leu, p.Pro21Ala, p.Arg207His, p.Arg566Cys and p.Ser694Cys CHAT mutant cDNAs, 19 μg per 75-cm2 flask were used to transfect HEK293 cells using DharmFECT1 transfection reagent as described previously (Arredondo et al., 2006, 2008), according to the manufacturer’s protocol. The cells were collected 48 h post-transfection and lysed with M-PER Mammalian Protein Extraction Reagent, including 10 μl Halt Protease Inhibitor per 1 ml of buffer; the extracts were used for determination of ChAT enzymatic activity (Ray et al., 2009) and affinity chromatography for purification of ChAT. Cells were also transfected with GFP to estimate transfection efficiency. HEK293 cells were used instead of bacteria cells to overexpress ChAT enzymes because we wanted the enzyme to undergo all post-translation modifications, such as phosphorylation, which is important for ChAT catalytic activity (Dobransky et al., 2000); (Dobransky and Rylett, 2005).
Purification of Phosphorylated ChAT by Affinity Chromatography
The HEK293 cells were transfected with human wild-type ChAT and mutant cDNAs as described above, and the cell lysates were used for enzyme purification. Briefly, NHS-activated Sepharose 4 Fast Flow (GE Healthcare) is a pre-activated agarose matrix that increases the capacity of coupling antigens, like ChAT antibody. Before coupling, the matrix was washed with 10-15 volumes of cold 1 mM HCl. A concentration of 10 mg/ml of ChAT antibodies was dissolved in the coupling buffer (0.2 M NaHCO3, 0.5 M NaCl pH 8.3), to achieve maximum efficiency; the coupling of matrix and ligand was at 40C overnight. After coupling, the matrix-ligand complex was washed with 0.1 M Tris-HCl buffer pH 8-9, and 0.1 M acetate buffer, 0.5 M NaCl pH 4-5. The procedure was 3 × 1 medium volumes of Tris buffer followed by 3 × 1 medium volumes of acetate buffer. This cycle was repeated 3-6 times.
After the affinity column equilibration, the cell lysates were loaded on to the NHS-activated Sepharose column for absorption; the column was washed with equilibration buffer (0.5 M ethanolamine, 0.1 M Tris-HCl, pH 8.5) followed by 0.2 M NaCl in equilibration buffer. The ChAT protein was eluted with 10 mM Tris-HCl buffer, pH 9.0, containing 2 mM β-mercaptoethanol, 2 nM EDTA, 10% glycerol, 0.3 M NaCl and 0.1 M choline chloride (Peng et al., 1983; Bruce et al., 1985). The purified ChAT enzyme was concentrated with Amicon filters.
SDS-PAGE and Western Blot Analysis
The purified wild-type and mutant ChAT proteins were quantified using a NanoDrop spectrophotometer (NanoDrop Technologies). The proteins were resuspended with a sample application buffer (1.0 mL of 0.5 M Tris-HCl pH 6.8, 1.9 g ultrapure urea, and 10% SDS), separated on a 4–20% SDS-PAGE gradient gel (Bio-Rad), and electroblotted as previously described (Arredondo et al., 2005). The polyclonal primary antibody, goat anti-ChAT (Millipore), was diluted 1:1000 in LI-COR Odyssey Blocking Buffer and incubated overnight at 4°C. The secondary antibody, donkey anti-goat IRDye800cw (LI-COR Biosciences), was diluted 1:5000. To normalize for protein content, the housekeeping protein β-actin was visualized in each sample with anti-β-actin monoclonal antibody (1:1000, Santa Cruz Biotechnology) and with a secondary antibody, goat anti-mouse IRDye800cw (1:5000, LI-COR Biosciences). The protein expression was then quantitated using the Odyssey Imaging System (LI-COR Biosciences).
Enzymatic Activity in Phosphorylated ChAT
ChAT activity was measured under steady-state conditions by the radioactive method of Fonnum (1969). The initial reaction velocities were determined at 37°C in a 40-μl reaction containing a range of 0.1–3.5 mM choline and 5–1000 μM [14C]acetyl-CoA concentrations, 0.2–10 ng ChAT protein, 50 mM sodium phosphate (pH 7.4), 250 mM NaCl, 1 mM EDTA and 0.5 mg/ml bovine serum albumin (BSA) (Ohno et al., 2001), in a 36-point assay.
The kinetic mechanism of the ChAT reaction followed a random Theorell-Chance mechanism in which a low but finite amount of ternary complex exists (Hersh and Peet, 1977). We first analyzed the two-substrate ChAT reaction by assuming an ordered random bi–bi mechanism described by the steady-state equation (Segel, 1975). The initial velocity of the reaction is defined in the equation below:
Where v is the initial reaction velocity, kcat represents the turnover number (Vmax/[ChAT]), KiA is the dissociation constant for the [ChAT] [Choline] complex, and are the Michaelis–Menten constants. The initial steps of the reaction are schematically represented by the scheme below, and according to the law of equilibrium, where is the dissociation constant for the [ChAT] [Choline] complex.
The catalytic efficiencies () were calculated by nonlinear regression using SIGMAPLOT ver. 12.
ChAT Structural Analysis
The Swiss-PdbViewer (Geneva, Switzerland) V4.1.0 graphics program (Guex and Peitsch, 1997) was used for analyzing the position of the identified human mutations in ChAT. Swiss-PdbViewer is an application that provides an interface allowing the analysis of several proteins at the same time (Schwede et al., 2003). The ChAT protein amino acid changes, p.Val136Met, p.Arg186Trp, p.Val194Leu, p.Pro211Ala, p.Arg207His, p.Arg566Cys and p.Ser694Cys, were superimposed in order to verify structural alignments and to identify H-bond changes resulting from the mutant residues in comparison with wild type. The amino acid mutations were introduced, and hydrogen-bonds and distance changes between atoms were measured for each mutation. Since the crystal structure of human ChAT is available (PDB 2FY2) (Kim et al., 2006); therefore, it is possible to predict the structural changes that occur when the mutations are introduced (protein sequence: NP_065574.1), which corresponds to the 82-83 kDa ChAT isoform 2. The best computed rotamer was selected for each mutation.
ChAT Thermal Inactivation Analysis
A different way to study the kinetic stability of ChAT mutant enzyme is to compare it with wild type by using the Fonnum method. Briefly, the enzymatic reactions were determined at 5°C, 15°C and 25°C in a 40-μl reaction containing 3.5 mM choline and 1000 μM [14C]acetyl-CoA concentrations, 10 ng ChAT protein, 50 mM sodium phosphate (pH 7.4), 250 mM NaCl, 1 mM EDTA and 0.5 mg/ml BSA. During the assays, enzyme mutants were incubated at different temperatures and different times, and the samples were diluted into the assay medium, or into ice-cold buffer to halt further inactivation; the activity of each sample was then measured at saturating substrate concentrations.
Quantification of ChAT Proteins Phosphorylation Residues by Tandem LC-MS/MS
The LC-MS method to identify ChAT phosphorylated residues is found in the Supp. Methods. Briefly, the proteins were digested into peptides to identify proteins with post-translation modifications. The peptides were analyzed by using tandem mass spectrometry (LC-MS/MS), and then searching for modifications using X!tandem. Scaffold Proteome Software was used to validate LC-MS/MS based peptide and protein identifications.
Statistical Analysis
All experiments were performed in triplicate and the results were expressed as mean ± standard deviation. Statistical significance was determined using Student’s t-test. Differences were deemed significant if the calculated p value was <0.05.
RESULTS
Clinical Material and Mutational Analysis
The expression studies were based on seven CHAT mutations identified in nine patients from seven independent families. The clinical presentation and severity of symptoms in these patients was markedly heterogeneous. Families 1–3 had been previously reported (Maselli et al., 2003).
Family 1
The two affected siblings of a non-consanguineous Caucasian family, a boy and a girl who were 14 and 16 respectively at the time of assessment, had a history of mild to moderate symptoms consisting of fatigable muscle weakness and repeated falls precipitated by exercise. In the girl, the symptoms were often triggered by contact with cold water. Both patients had transient respiratory difficulties that were attributed to asthma. The neurologic examination was normal in both patients. Repetitive stimulation showed decrement of compound muscle action potential (CMAP) amplitudes immediately after 5-minute stimulation at 10 Hz (Maselli et al., 2003). Anticholinesterase medication was not beneficial. Both patients had CHAT mutations p.Leu210Pro and p.Pro211Leu, each one inherited from each parent (Supp. Figure S1).
Family 2
Two sibling girls in a non-consanguineous Caucasian family experienced only mild to moderate symptoms. In the most affected patient, the first indication of fatiguing weakness was at age 18 months, when she was observed to lose leg control after physical activity or after exposure to cold water. She had two episodes of severe respiratory apnea. Examination of this patient at age 14 was normal, except for ptosis precipitated by sustained upward gaze. Repetitive nerve stimulation at 2 Hz showed no decrement, but stimulated single-fiber electromyography showed increased jitter (Maselli et al., 2003). The patients did not respond to either anticholinesterase medication or 3,4-diaminopyridine (DAP). Mutational analysis revealed that both patients carried CHAT mutations p.Val194Leu and p.Val506Leu, each one inherited from each parent (Supp. Figure S2).
Family 3
The only one affected of three siblings in this non-consanguineous Caucasian family was a girl age 4 at the time of clinical assessment. She had had recurrent episodes of respiratory distress and ptosis since birth. Her two older brothers were asymptomatic. Her neurological examination showed bilateral ptosis and mild proximal weakness. Anticholinesterase medication improved her weakness and fatigability. DNA analysis revealed CHAT mutations p.Arg548Ter and p.Ser694Cys, each one transmitted from each of her unaffected parents (Supp. Figure S3).
Family 4
The only one affected of three siblings in a non-consanguineous Caucasian family was a female aged 18 at the time of the consultation. The first indication of weakness noted by the parents was at the age of 18 months when she became weak after exposure to cold water. She had limited physical endurance and mild developmental delay. Later on she developed ptosis, diplopia, chewing difficulties and limb weakness. Neurologic examination revealed fatigable ptosis to upward gaze and mild proximal weakness. Repetitive stimulation at 2 Hz preceded by stimulation at 10 Hz elicited decrement of CMAP amplitudes. DNA testing revealed that she carried CHAT mutations p.Arg186Trp and p.Arg566Cys (Supp. Figure S4).
Family 5
The only one affected of three siblings in a non-consanguineous Native American family was a 4-year-old boy with severe hypotonia, limb and bulbar weakness, and respiratory insufficiency from birth. The patient had developmental delay and required a gastric tube, tracheotomy and mechanical ventilation. The examination showed ptosis, absent external ocular movements, severe facial and limb weakness and intact reflexes. Repetitive nerve stimulation at 2 Hz showed about 50% decrement of CMAP amplitudes, and a muscle biopsy was normal (Supp. Figure S5). Patient partially responded to combined treatment with an anticholinesterase drug and DAP. Mutational analysis demonstrated that the patient was homozygous and his parents heterozygous for CHAT mutation p.Arg207His.
Family 6
The only one affected of two siblings in a non-consanguineous Native American family was a 5-year-old girl with severe bulbar and limb weakness from birth. The patient required a gastric tube, tracheotomy and mechanical ventilation. She was nonverbal and had profound developmental delay. The examination showed ptosis, impaired external ocular movements and facial weakness, and she was unable to overcome gravity with the proximal muscles of the upper extremities. Repetitive stimulation showed moderate decrement of CMAP amplitudes, and the patient partially responded to anticholinesterase medication. Muscle biopsy, including electron microscopy of the NMJ, was normal (Supp. Figure S5). The patient was in the hospital for most of her life, and passed away from respiratory complications a few months after she was discharged home. Mutational analysis showed that she was homozygous and her parents heterozygous for CHAT mutation p.Arg207His (Supp. Figure S6).
Family 7
The only one affected of two siblings in a non-consanguineous Vietnamese American family was an 8-year-old girl who had had severe weakness as well as bulbar and respiratory deficit since birth. The examination showed ptosis, impaired external ocular movements and severe proximal limb weakness. She required a feeding gastrostomy tube, tracheotomy and continuous mechanical ventilation. Repetitive nerve stimulation showed severe decrement of CMAP amplitudes. Muscle biopsy, including electron microscopy of the NMJ, was normal. The patient, who responded partially to combined treatment with an anticholinesterase drug and DAP, died at age 8 of respiratory complications. Mutational analysis showed that she was homozygous and her parents heterozygous for CHAT mutation p.Val136Met (Supp. Figure S7). The electromyography findings are summarized in Supp. Table S1.
Expression Studies
The phosphorylated ChAT expression levels of p.Val136Met, p.Arg186Trp, p.Val194Leu, p.Pro211Ala, p.Arg207His, p.Arg566Cys and p.Ser694Cys mutants were examined in HEK-293 cells (Table 1). Mutation p.Leu210Pro had been previously reported and studied by another group (Ohno et al., 2001); p.Val506Leu was not biochemically characterized, as it is associated with a mild phenotype in the patient, and p.Arg548Ter expresses a truncated protein.
Table 1.
Identified ChAT mutations
| Family | Patient | Nucleotide changea |
Amino Acid changeb |
Protein Expression Levelsc |
|---|---|---|---|---|
| 7 | 7 | c.406G>A | p.Val136Met | 0.61 ± 0.04 |
| 4 | 4 | c.556C>T | p.Arg186Trp | 0.94 ± 0.05 |
| 2 | 2 | c.580G>T | p.Val194Leu | 0.99 ± 0.02 |
| 5 | 5 | c.620G>A | p.Arg207His | 0.81 ± 0.02 |
| 6 | 6 | c.620G>A | p.Arg207His | 0.81 ± 0.02 |
| 1 | 1 | c.631C>G | p.Pro211Ala | 1.12 ± 0.04 |
| 4 | 4 | c.1696C>T | p.Arg566Cys | 0.99 ± 0.03 |
| 3 | 3 | c.2081C>G | p.Ser694Cys | 1.16 ± 0.05 |
Nucleotide numbers according to reference sequence NM_ 020549.2 for isoform 2.
Codon numbers start from the translation initiation codon according to the reference sequence NP_065574.1, for the ChAT M variant encoding 83-kDa protein.
ChAT protein expression levels relative to wild type, which is equal to 1.
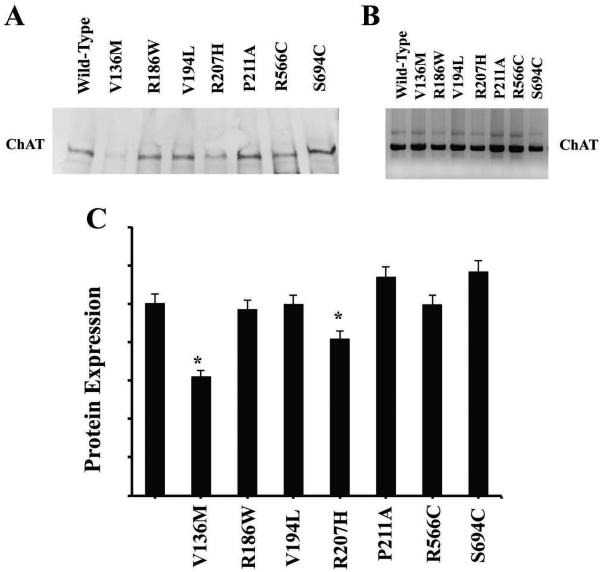
The wild-type and mutant CHAT cDNAs were transfected into HEK cells, and after 48 h post-transfection, the cell lysates were analyzed by Western blot (Fig. 2a). The CHAT S transcript has two alternative start sites that yield 70-kDa and 74-kDa proteins at a constant ratio (Ohno et al., 2001). Two mutants, p.Val136Met and p.Arg207His, were expressed at significantly lower levels than wild type, 61% and 81% respectively, and three mutants, p.Arg186Trp, p.Val194Leu and p.Arg566Cys, had a protein expression similar to that of wild type, as shown in Figure 2c and Table 1. Finally, p.Pro211Ala and p.Ser694Cys had an expression slightly higher than that of wild type, with 1.12% and 1.16% respectively. Although based on the immunoblots some mutants appear to express lower levels of protein at steady-state levels, an alternative explanation is that these mutant proteins degrade more readily or are less stable in HEK cells than the wild-type.
Fig 2.
Protein expression of phosphorylated ChAT. ChAT expression of wild type and mutants in HEK293 cells. a Overexpression of ChAT proteins on HEK cells transfected with wild-type and mutant cDNA. Crude cell lysates were immunoblotted with ChAT antibody. Western blot variations of transfection efficiency were corrected by monitoring the expression of β-actin in the immunoblots (data not shown). The housekeeping β-actin protein was also used as a control to verify that all the samples had the same amount of loaded protein. b Western blot shows the affinity purified phosphorylated ChAT protein from HEK293 cells. c Bar graph showing the quantification of ChAT protein shown on Western blots (*p < 0.05).
Phosphorylated ChAT Protein
To determine how ChAT mutants affect the enzymatic activity of ChAT, the wild type and mutant were expressed in HEK cells by using the S transcript that yields the 70-kDa ChAT. The advantage of expressing the enzymes in mammalian cells, rather than bacteria, was to produce the enzymes in their native forms. The benefit of purifying the enzyme from mammalian HEK cells is that all post-translational modifications occur, including ChAT phosphorylation (Dobransky and Rylett, 2005), and the phosphorylation of the enzyme is important to regulate its catalytic activity (Dobransky et al., 2000). The SDS-PAGE of purified phosphorylated ChAT enzymes is shown in Figure 2B. Even though HEK cells undergo protein phosphorylation, it is not clear whether the phosphorylation patterns in HEK cells are the same as the ones that occur in neurons.
ChAT Structural and Kinetic Analysis
To understand how each of the mutations affected ChAT enzymatic activity, we analyzed how each mutation residue altered the electrostatic dipole–dipole interaction of the hydrogen bonds (H-bonds) in the catalytic and binding domains. In Figures 3–6, we describe in more detail how the mutations affect normal Hydrogen-bonding interactions at the catalytic active-site and substrate binding sites. Because the residue side chain mutations usually affect the conformation of neighbor side chain residues, Table 2 lists the H-bonds before and after the residue mutation based on ChAT structure (PDB 2FY2). Figure 1 shows the location of each mutation, and a summary of the hydrogen-bonding interaction changes is described in Table 2, where the number of hydrogen-bonds changes for each mutation is shown. When a residue is mutated, the Swiss-Pdb software scans a backbone-dependent rotamer library and selects the conformation that minimizes the number of clashes with surrounding residues, as well as maximizes the H-bonding potential. Figures 3-6 show the best possible rotamer conformation for each of the mutations, which were computed to have the lowest energy. The summary of CMS ChAT mutations is shown in Table 3. Supp. Figures S8-S13 show all other possible conformations for each one of the mutations.
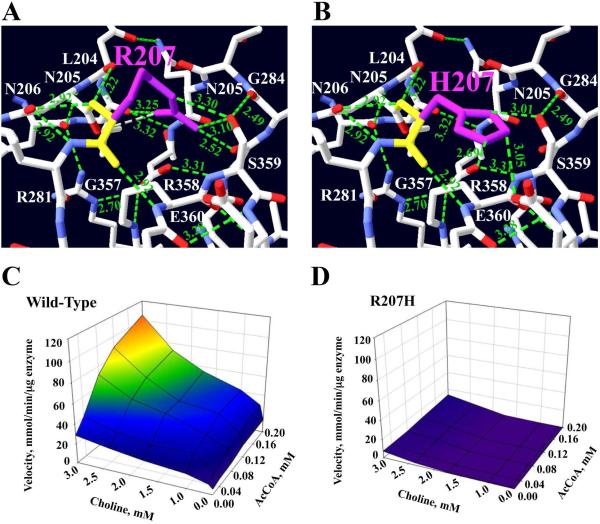
Fig 3.
Kinetic maps and position of p.Arg207His mutation at the active-site tunnel. The structure displays only hydrogen-bond interactions with neighbor residues that are close to Arg207 residue, and the hydrogen-bonding interactions are shown with green dotted lines. Hydrogen atoms are in blue, oxygen atoms in red, and Arg207 backbone in yellow. a Arg207 side chain (magenta) interacts with Gly284 backbone, Ser359, Asp206 and Asp288 to donate five H-bonds. Also, Arg207 backbone forms three hydrogen-bonds between Asn205, Arg358, and Asn206. b The mutated residue was best accommodated by rotamer #2. The mutation changes from the most hydrophilic polar residue (174 Da) to another, less hydrophilic polar, but smaller, His207 residue (155 Da) and changes all five H-bonds, but it forms three new H-bonds between its imidazole ring and Glu360 and Ser359. c 3D kinetic graph for the wild-type enzyme. d 3D kinetic graph for the mutant enzyme. This mutation produces the highest reduction in enzyme activity: the enzyme is basically inactive. Rotamer #2 has a score of −5 and p= 25%.
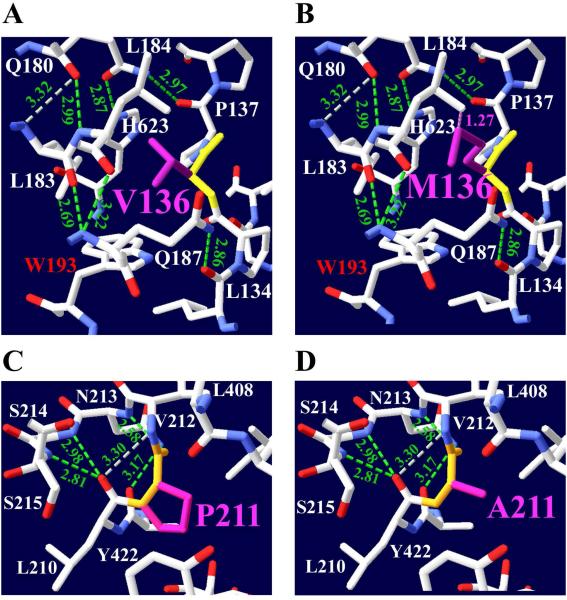
Fig 6.
Mutations p.Val136Met and p.Pro211Ala produce small structural changes. The structure shows hydrogen-bonding interactions with residues that are close to Val136 and Pro211. a p.Val136Met mutation changes from a smaller branched-chain residue (117 Da) with an aliphatic side chain to a larger hydrophobic residue (149 Da) with long alkyl side chain. b The mutated residue was best accommodated by rotamer #4. The mutation Met136, which is a sulfur-containing residue, creates a clash (pink dotted lines) between its sulfur atoms with the aliphatic residue Leu184 side chain. Val136 and Met136 side chains do not produce any hydrogen-bonding with neighbor residues. c Pro211 cyclic amino residue creates one hydrogen-bond with Asn213. Pro211 side chain does not produce any H-bonds. d The Ala211 residue is very close to His442. Ala211 also does not produce any hydrogen-bonds. This mutation has 59% and 56% reduction of catalytic efficiency and overall catalytic efficiency respectively. Met136 has rotamer #4 with a score of −1 and p=10%.
Table 2.
Hydrogen-bond interactions of ChAT mutationsa predicted by Swiss-PdbViewer
| Mutation | H-bonds (Ǻ) before mutationb |
H-bonds (Ǻ) lost after mutationc |
H-bonds (Ǻ) gained after mutationd |
|---|---|---|---|
| V136M | M136-L184 (1.27)e | ||
| R186W | R186-T190 (2.95) R186-T190 (2.81) R186-K189 (3.29) R186-L182 (2.74) R186-L183 (3.07) R186-W193 (2.92) |
R186-W193 (2.92) | W186-W193e
1.3-2.09 (Ǻ)e W186-W193e 1.0-1.69 (Ǻ)e W186-L183e 1.0-1.69 (Ǻ)e |
| V194L | V194-W198 (2.72) V194-Y197 (3.15) V194-N192 (2.87) |
L194-W193e
1.30 and 1.18e L194-N192e (2.16) |
|
| R207H | R207-S359 (3.30) R207-S359 (3.10) R207-G284 (2.52) R207-L204 (3.25) R207-N206 (3.32) R207-N205 (3.22) R207-N206 (2.92) R207-R358 (2.92) |
R207-S359 (3.30) R207-S359 (3.10) R207-G284 (2.52) R207-L204 (3.25) R207-N206 (3.32) |
H207-E360 (3.05) H207-S359 (2,69) H207-S359 (3.01) |
| P211A | P211-N213 (2.58) | ||
| R566C | R566-S443 (2.65) R566-H442 (3.13) R566-D446 (2.90) R566-D446 (2.90) R566-Ile559 (3.24) R-566-E564 (3.32) |
R566-S443 (2.65) R566-H442 (3.13) R566-D446 (2.90) R566-D446 (2.90) |
C566-D446 (2.71) |
| S694C | S694-Y670 (2.71) S694-G679 (2.74) S694-G679 (2.65) |
S694-Y670 (2.71) | C694-P672 (3.24) C694-Y670 (2.41) |
Because the mutated residues affect the conformation of the residues in the neighborhood.
The interacting amino acids and the Hydrogen-bond distances found before the mutation.
The number of Hydrogen-bonds lost after introducing the mutation.
New Hydrogen-bonds formed by the mutation.
Mutated residues creating clashes. The clashes are not Hydrogen-bonds, because clashes involve distances that are much smaller than those in Hydrogen-bond interactions.
Table 3.
Summary of congenital ChAT mutationsa
| Mutation | Damaginga
SIFT Score |
ChAT activity affected |
Location on Sec. structure |
Mutation probable structural effect |
|---|---|---|---|---|
| p.Val136Met | yes 0 |
↓ Lowest enzyme expression |
Coil 1 before α-helix 1 binding domain |
Possible energetic interaction with AcCoA |
| p.Arg186Trp | yes 0 |
↓↓ kcat, ↓ | α-helix 3 binding domain |
Destabilize enzyme folding |
| p.Val194Leu | tolerated 1 |
↑↑kcat, ↑↑
↑ |
Coil 3 before α- helix 4 binding domain |
Alter AcCoA binding site |
| p.Arg207His | yes 0 |
↓↓kcat, ↓↓
Low ChAT expression |
Coil 5 between α-helix 4 and β-sheet 1 binding domain |
Modify active site, close to His424 |
| p.Pro211Ala | tolerated 0.39 |
↑ kcat, ↑↑ | Coil 5 before β-sheet 1 catalytic domain |
Change active site and choline-binding site |
| p.Arg566Cys | tolerated 1 |
↓ kcat, ↑
|
β-sheet 12 binding domain |
Disrupt H-bond with His424 at active site |
| p.Ser694Cys | Yes 0 |
↓ kcat, ↑↑ | β-sheet 16 binding domain |
Disrupt H-bond with Tyr670 involved in choline binding |
↑, increase in the Km value; ↑↑, large increase in the Km value; ↓, decrease in the Km value; ↓↓, large decrease in the Km value.
SIFT score predicts whether an amino acid substitution affects protein function based on sequence homology and the physical properties of amino acids.
Additional computational chemistry analysis was performed to confirm the results from the Swiss-Pdb algorithm. UCSF chimera software was performed in each of the mutations; the results are shown in Supp. Figures S14-S19.
Mutations Predicting Large Structural Changes
Arg207His
The mutation that was predicted to cause the largest structural changes, and consequently the highest decrease, in the enzyme activity is p.Arg207His. The Arg207 residue is located at the surface of coil 5 between α-helix 4 and β-sheet 1, and it is 15Ǻ from the His442 catalytic residue, as shown in Figure 1 and the summary in Table 3. This mutation has a very low Kcat and Km for AcCoA, having a catalytic efficiency and overall catalytic efficiency less than 3% (Table 4); kinetic maps of wild-type and mutant enzyme are shown in Figure 3c and d. The Arg207 side chain donates five hydrogen-bonds: The Arg207 amine group interacts with Gly284 carboxyl group to form a 2.52 Ǻ H-bond, and two interactions between its NH2 and NH groups and Ser359 carboxyl group create a 3.30Ǻ and a 3.10Ǻ bond. Two additional hydrogen-bonds are donated between the NH2 group of Arg207 and the carboxyl groups of Leu204 (3.25 Ǻ) and Asn206 (3.32 Ǻ). Also, the Arg207 backbone amino group forms two H-bonds with the carboxyl groups of Asn205 and Asn206 with distances of 3.22 Ǻ and 2.92 Ǻ respectively. Finally, another 2.92 Ǻ hydrogen-bond interaction takes place between the carboxyl group of Arg207 and the amino group of Asn206, Fig. 3a. The mutated residue changes from a basic hydrophilic large residue (174 Da) to a smaller bulky basic hydrophilic His207 residue (155 Da) with an imidazole ring. Rotamer #2 was the best structural conformation computed for this mutation (Fig. 3b), and it shows the following changes: His207 side chain cannot donated all five hydrogen-bonds like Arg207, but forms a new hydrogen-bond interaction between its imidazole side chain NE2 group and the Glu360 carboxyl group (3.0Ǻ). Two new H -bonds are donated between the His207 ND1 group and the Ser359 amino group, 2.69 and 3.0Ǻ respectively. All of these predicted hydrogen-bonding interaction changes would affect the ChAT structure during catalysis. The Arg207 residue is also close to the Tyr203 residue, which is 16 Ǻ from His442.
Table 4.
ChAT Kinetic parameters of wild type and mutant ChAT enzymesa
| kcat, s−1 | μM | μM | kcat/ |
kcat/ · |
|
|---|---|---|---|---|---|
| Wild-type | 281.9 ± 3.5 | 22.7 ± 1.8 | 396 ± 24 | 1.00 | 1.00 |
| p.Val136Met | 223.0 ± 2.6 | 27.4 ± 1.3 | 432 ± 31 | 0.65 | 0.71 |
| p.Arg186Trp | 1 3.7 ± 1.1 | 7.3 ± 1.3 | 224 ± 27 | 0.15 | 0.10 |
| p.Val194Leu | 608.2 ± 16 | 88 ± 7.4 | 565.8 ± 32 | 0.55 | 0.79 |
| p.Arg207His | 0.80 ± 0.05 | 4.53 ± 8.7 | 442 ± 21 | 0.02 | 0.03 |
| p.Pro211Ala | 488.7 ± 12.4 | 96.1 ± 5.3 | 425 ± 11 | 0.41 | 0.44 |
| p.Arg566Cys | 103.7 ± 6.9 | 37.1 ± 5.7 | 721 ± 23 | 0.23 | 0.45 |
| p.Ser694Cys | 135.8 ± 9.5 | 31.0 ± 2.3 | 831 ± 18 | 0.35 | 0.73 |
Values indicate estimate ± standard error. Catalytic efficiency (kcat/) is normalized with respect to wild type. The overall catalytic efficiency [kcat/·] is normalized with respect to wild type.
Arg566Cys
Since p.Arg566Cys is exposed on the β-sheet 12 surface at the binding domain, next to His442, at the active-site tunnel, any structural changes in this region are expected to affect the enzymatic activity. The positively charged Arg566 NE and NH2 groups donate two H-bonds, both 2.90 Ǻ, with the Asp446 OD2 and OD1 groups. A third H-bond interaction is donated between the Arg566 NH2 group and the carboxyl group of His442, 3.13 Ǻ, and the fourth hydrogen -bond interaction forms a 2.65 Ǻ bond between the Arg566 amine group and the Ser443 carboxyl group. Two more H-bonds are formed between the Arg566 carboxyl group and the Ile559 amino group, 3.24 Ǻ, and the Arg566 amino group and the Glu564 carboxyl group, 3.32 Ǻ, Fig. 4a. The mutated residue changes from a hydrophilic positively charged residue to Cys566, which is a smaller (121 Da) nucleophilic non-polar residue with a thiol group. Rotamer #1 was the best conformation computed for the mutation structural changes, Fig. 4b. All four hydrogen-bonds interacting with the Arg566 side chain are changed, and a new 2.71 Ǻ H-bond is formed between the Cys566 thiol group and the side chain of Asp446. This mutation produces an increase of Km for AcCoA and choline, as well as reducing ChAT enzymatic efficiency by 77% and overall catalytic efficiency by 55% (Table 4); the kinetic map of the mutant enzyme is shown in Figure 4e. p.Arg566Cys is one of the three mutations that are predicted to cause severe structural changes; furthermore, it produces a 2.7-fold decrease of the enzyme Kcat.
Fig 4.
Mutation p.Arg566Cys close to His442 and p.Arg186Trp disrupts enzyme structure. The structure displays only hydrogen-bond interactions with neighbor residues that are close to Arg566 or Arg186. a Arg566 interacts with Asp446 backbone, His442 (red), Ser443, Arg566 backbone, Ile559 and Glu564, forming H-bonds. b The mutated residue was best accommodated by rotamer #1. The mutated residue changes from a large hydrophilic positive charge to Cys566, a smaller uncharged hydrophobic residue (121 Da). All four hydrogen-bonds interacting with Arg566 side chain are modified, and a hydrogen-bond is formed between Cys566 and Asp446. c Arg186, a polar very hydrophilic residue, changes to a Trp186 hydrophobic larger residue (205 Da). The Arg186 side chain donates hydrogen-bonds with Trp193 backbone, Thr190, Leu182, Leu183 and Lys189. d The mutated residue was best accommodated by rotamer #2. The Trp186 mutation changes the H-bond with Trp193, but forms many clash interactions (pink dotted lines) between the phenolic ring of Trp186 and Lys630, Lys183 and Tyr193. e 3D kinetic graph for p.Arg186Trp mutant enzyme. The mutation reduces the Kcat by 95% and the Km for both substrates. f 3D kinetic graph for p.Arg566Cys mutant enzyme shows a decrease of Kcat by 48% and an increase in Km for both substrates. Cys566 has rotamer #1 with a score of −3 and p = 45%. For Trp186 is rotamer #2 with a score of −3 and p = 32%
Arg186Trp
Even though the p.Arg186Trp mutation is partially surface exposed on α-helix 3 of the binding domain at the NH2 terminal domain, far from the active-site tunnel and binding substrate site (Fig. 1), it still produces large structural changes, as shown in Table 2. As a result of the mutation, Arg186, a polar hydrophilic residue with a positive charge side chain, changes to Trp186, a larger hydrophobic residue (204 Da) with an aromatic side chain. The Arg186 NE1 group has a 2.92 Ǻ H-bond with the Trp193 carboxyl group, and Arg186 carboxyl groups form two more H-bonds: one with the Thr190 amino group, 2.95 Ǻ, and one with the Lys189 amino group, 3.29 Ǻ. In addition, two H-bonds are donated between the Arg186 amino group and the carboxyl groups of Leu182 and Leu183, 2.74 and 3.07 Ǻ respectively, Fig. 4c. Lastly, one H-bond is formed between the Arg186 carboxyl group and the Thr190 OG1 group, 2.81 Ǻ (Table 2). Rotamer #2 was the best computed conformation for the mutation structural changes. The Trp186 mutation predicts modification of an important H-bond with Trp193, and since the Trp183 side chain is incapable of participating in hydrogen-bonding, only clash interactions (pink dotted lines) are created between the phenolic ring of Trp186 and Tyr197 (1.3–2.09 Ǻ), Lys183 (1–1.69 Ǻ) and Trp193 (1–1.69 Ǻ). The clashes are a result of the fact that Trp186 is quite large and may not fit in the pocket assigned for Arg186 without forcing the rearrangement of surrounding neighbor residues, Fig. 4d. This mutation reduces the catalytic efficiency and overall efficiency by 85% and 90%, respectively (Table 4); the kinetic map is shown in Figure 4f. Thus, p.Arg186Trp is an example of an NH2 terminal domain mutation that can cause severe structural changes affecting ChAT normal enzymatic activity.
Mutations affecting substrate binding conformation
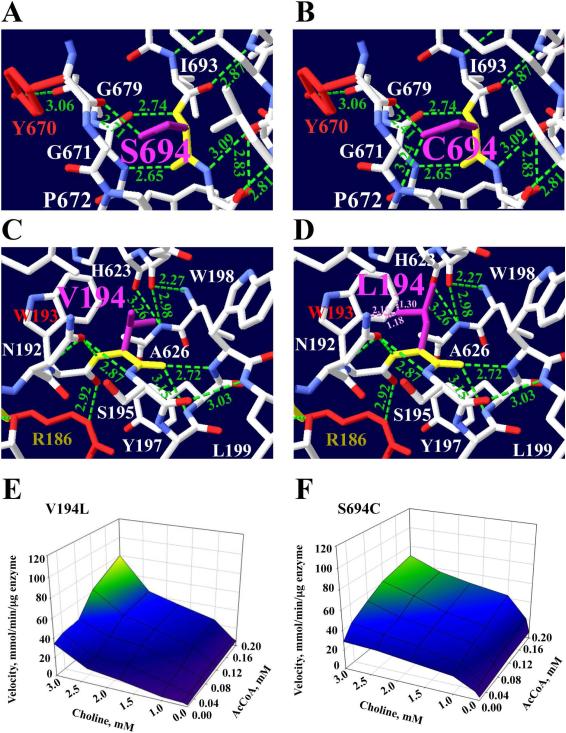
Ser694Cys
The p.Ser694Cys mutation is buried on β-strand 16, far from the catalytic active tunnel but close to the choline binding site (Fig. 1). Ser694 residue produces three H-bonds: one is between the Ser694 amino group and the Gly679 carboxyl group, 2.74 Ǻ; a second is between the Ser694 carboxyl group and the Gly679 amino group, 2.65 Ǻ; and a third results from the interaction of the S694 hydroxyl group with the carboxyl group of Tyr670, which is important for choline binding, 2.71 Ǻ (Table 2). Ser694 is a polar nucleophilic residue (105 Da) with a hydroxyl side chain (Fig. 5a), and changes to Cys694, which also has a nucleophilic side chain, but with a thiol group. Rotamer #3 was the best structural conformation computed for this mutation. The mutation forms two H-bonds, between the Cys694 thiol group and both the Pro672 amino group and the Tyr670 carboxyl group, 3.24 and 2.41 Ǻ respectively (Fig. 5b). This mutation produces an increase in Km choline, and reduces ChAT enzymatic efficiency by 65% and overall catalytic efficiency by 27% (Table 4).
Fig 5.
Mutations p.Ser694Cys and p.Val194Leu affect substrate binding site. The structure shows interactions with residues that are close to Ser694 or Val194. a Ser694 forms H-bonds with Ser694 backbone, Gly679 amino group, Gly679 and Tyr670 (red) backbone. b The mutated residue was best accommodated by rotamer #3. The Ser694 (105 Da) with a small polar hydroxyl side chain changes to Cys694 (121 Da), which has a larger hydrophobic side chain. The mutation forms a new H-bond between the Cys694 side chain NH2 group and the Pro672 amino group. c The p.Val194Leu mutation is close to the acetyl-CoA binding site at α-helix 4. Val194 forms three H-bonds, with Ile559, Tyr197 and Asn192. The Val194 aliphatic side chain changes to Leu194 with a larger aliphatic side chain. d The mutated residue was best accommodated by rotamer #3. The Leu194 mutation creates clash interactions (pink dotted lines) with the Thr193 phenolic ring and with Asn192. e This mutation produces an increase of Km for choline by 1.4-fold, for AcCoA by 3.8-fold, and for Kcat by 2-fold. f The 3D kinetic graph shows a decrease of 48% for Kcat and a 2-fold increase in Km for choline. Cys694 has rotamer #3 with score of −4 and p= 22%. For Leu194 is rotamer #3 with a score of −4 and p = 40%
Val194Leu
The p.Val194Leu mutation is close to the binding site on a surface of coil 3 next to α-helix 4, as shown in Figure 1. The Val194 backbone creates three H-bonds: the first is between the Val194 carboxyl group and the Trp198 amino group, 2.72 Ǻ; the second is with the Tyr197 amino group, 3.15 Ǻ; and the third is between the Val194 amino group and the Asn192 OD1 group, 2.87 Ǻ (Fig. 5c, Table 2). Because of this mutation, the Val194 is a medium residue (140 Da) with an aliphatic side chain changes to Leu194, which is a larger residue (166 Da), also with an aliphatic side chain and also incapable of donating H-bonds. The Leu194 mutation creates clash interactions (pink dotted lines): one with Asp192 (2.16 Ǻ), and two with the Trp193 phenolic ring (1.30 and 1.18 Ǻ). Rotamer #3 was the best structural conformation computed for this mutation, Fig. 5d. This mutation increases the Km of AcCoA and choline, and consequently, the catalytic efficiency and overall efficiencies are reduced by 45% and 21% respectively (Table 4); the kinetic maps of p.Val194Leu and p.Ser694Cys enzymes are shown in Figure 5e and f.
Mutations causing small structural changes
Val136Met
The p.Val136Met mutation is located at the NH2 terminal domain on the first coil before α-helix 1 (Fig. 1). The p.Val136Met mutation changes from a smaller hydrophobic residue with an aliphatic side chain to a larger hydrophobic residue (149 Da) with a long alkyl side chain with a sulfur atom (Fig. 6a and b). The mutation Met136 forms a 1.27 Ǻ clash between its sulfur atom and the Leu184 side chain (Table 2). This NH2 terminal domain mutation is capable of inducing structural changes that not only affect its overall efficiency, which is reduced by 29% (Table 4), but may also affect the enzyme stability. This mutant expressed the lowest protein levels of the seven mutants, and less than 39% of that of wild type. Although the p.Val136Met mutation is found far from substrate binding and catalytic sites, it still is able to produce a mild effect on enzyme efficiency. A possible energetic interaction between Val136 and Trp193-Asn622-Val567-Ser558 residues has been reported, which contributes to the acetyl-CoA binding site (Shen, et al., 2011). Trp193 is 3.4 Ǻ from Val136; furthermore, Ser558 residue is involved in the choline and acetyl-CoA binding, as well as contributing to the negative charge on the binding tunnel at the P-loop. Rotamer #4 was predicted the best structural conformation computed for this mutation.
Pro211Ala
The only mutation in the catalytic domain is p.Pro211Ala, which is on coil 5 between β-helix 4 close to β-sheet 1 (Table 3). The side chain of aliphatic Pro211 (115 Da) does not donate any H-bonds, but its backbone forms one H-bond with Asn213, 2.58 Ǻ. The p.Pro211Ala mutation changes from an aliphatic cyclic amino acid to a smaller aliphatic residue (89 Da), and neither side chain create any H-bonds with surrounding neighbor residues (Fig. 6c). The Ala211 residue is only 9.7 Ǻ from catalytic His442 (Fig. 1). This mutation produces 59% and 56% reductions of catalytic efficiency and overall catalytic efficiency respectively (Table 4). This mutation produces only one rotamer, as shown in Fig. 6d.
Thermal Inactivation
The calculated thermal inactivation showed that none of the mutations altered the stability of the mutant enzymes, (data in Supp. Table S2).
Identification of ChAT Phosphorylation
The following phosphorylated residues were identified in ChAT protein: Threonine 225 and 456, and Serine 346, 347, 440 and 476 residues (data in Supp. Table S3). The reported ChAT phosphorylated residues have been identified previously by another group (Dobransky and Rylett, 2005), and showed that CaM and MAP phosphorylate the threonine residues and PKC phosphorylates the serine and threonine resides.
DISCUSSION
In our study, we identify and functionally characterize seven missense mutations in CHAT as the cause of a highly fatal CMS in humans (Table 1). Three mutations, p.Arg186Trp, p.Arg207His and p.Arg566Cys, are novel. Two others, p.Val136Met and p.Pro211Ala, were previously reported and functionally characterized by Shen et al. (2011) and Ohno et al. (2001) respectively; however, neither enzyme was phosphorylated because they were expressed in bacteria. Mutations p.Val194Leu and p.Ser694Cys were first reported by our lab (Maselli et al., 2003), but had never been biochemically characterized. None of the novel mutations were detected in the 1000 Genomes database. The mutated residues are all highly conserved in rat, mouse, chicken and dog (data not shown).
Structural and Genotype–Phenotype Correlations
The identification of homozygous carriers of mutations p.Arg207His and p.Val136Met is helpful to establish a correlation between the biochemical and structural changes caused by these mutations and the phenotypes of the patients in whom these mutations were found.
Both mutations, p.Arg207His and p.Val136Met, are associated with the most severe phenotypes and fatal outcome, yet the kinetics and the structural changes are remarkably different. The p.Arg207His is located close to His442 in the active catalytic tunnel and reduces catalytic and overall efficiencies by 100-fold; even though Arg207 polar residue is located on the surface, it still donates multiple hydrogen-bonds with neighboring residues that were changed with the mutation, and consequently the highest reduction in enzymatic activity. In contrast, p.Val136Met is located at the beginning of the NH2 terminal domain, far from the substrate binding and catalytic sites, and yet still produces a negative effect on enzyme activity by reducing 29% of the overall catalytic efficiency. The negative effect is probably a result of an energy interaction with residues contributing to the acetyl-CoA binding site, like Trp193 (Shen et al., 2011). While both mutations cause a decrease in the protein expression, the reduction is more severe for p.Val136Met than for p.Arg207His. It is evident that patients with homozygous mutations produce a very damaging phenotype because of the lack of normal alleles to express a functional enzyme.
The other two mutations causing large structural and kinetic changes are p.Arg566Cys and p.Arg186Trp. On the one hand, p.Arg566Cys is on the surface and very close to His442 at the active-site tunnel, which not only causes major disruption of hydrogen-bond interactions, because Arg566 has multiple hydrogen-bonding with surrounding neighbors, but also results in more than 50% reduction of the overall catalytic efficiency. Because of the mutation, four hydrogen-bond interactions are changed, including the most important one with catalytic residue His442. Notably, p.Arg566Cys is close to Ser558 and Arg560, which are both important for the P-loop, and Ser558 is important for acetyl-CoA binding. Arg560 acts as an active site residue, whose function is to stabilize coenzyme binding by interacting with the 3′-phosphate of the acetyl-CoA (Wu and Hersh, 1995).
Arg186 is partially surface exposed in the NH2 terminal domain, far from the substrate binding or catalytic sites. However, the p.Arg186Trp affects a key hydrogen-bond interaction with Trp193, and the mutated residue Trp186 has a large hydrophobic aromatic ring, which may contribute to the enzyme structural instability by making clash interactions with neighbor residues. The mutation p.Arg186Trp reduces the overall enzymatic activity by 90%. In comparison with p.Val136Met, p.Arg186Trp is closer to Trp193 than Val136, and consequently has a more damaging effect on the enzymatic activity.
The phenotype of the patient in whom we found these mutations is moderate to severe, and associated with both motor and cognitive deficits. However, since the patient is compound heterozygous for both p.Arg566Cys and p.Arg186Trp mutations, it may not be possible to state to which extent each of the mutations contributed to the severity of the phenotype.
The mutation p.Ser694Cys is buried and far from the catalytic site, but close to the substrate binding site, and involve a hydrogen-bond interaction with Tyr670, which is important for choline binding. The mild structural changes are correlated with 27% reduction of the overall catalytic efficiency. The phenotype of the patient carrying heterozygous mutations, p.Ser694Cys and p.Ser548Ter, were moderate and defined by p.Ser694Cys, because p.Ser548Ter, a null allele, produced a truncated enzyme.
Although p.Val194Leu is on a surface loop and closer to the substrate binding site than p.Arg186Trp, it does not cause the severe catalytic impairment caused by p.Arg186Trp. In fact, p.Val194Leu reduces the overall catalytic efficiency by only 21%, in part because the mutation changes from an aliphatic residue to a larger aliphatic residue. Furthermore, p.Val194Leu is predicted to be tolerated by SIFT analysis. The relatively mild phenotype of the two siblings who are compound heterozygous for p.Val194Leu and p.Val507Leu correlates with the results.
Finally, the mutation p.Pro211Ala is located in the catalytic domain and produces a 56% reduction of the overall catalytic efficiency. Proline to alanine mutations tends to be slightly destabilizing of stability and function. The mutation produces not only the least structural changes, but also no hydrogen-bond changes, which correlates well with the SIFT score prediction as tolerated, indicating that this mutated residue may not affect ChAT enzymatic activity. Consistent with these findings was the mild phenotype of the two siblings, who were biallelic for p.Pro211Ala and the adjacent mutation p.Leu210Pro, previously identified and biochemically characterized by another group (Ohno et al., 2001). These two patients did not experience episodes of apnea, and the CMAP amplitudes in response to RNS showed no decrement at rest, but only after 5 minutes of nerve stimulation at 10 Hz (Maselli et al., 2003).
The importance of ChAT phosphorylation in cholinergic neurons has been demonstrated by two groups (Bruce and Hersh, 1989; Schmidt and Rylett, 1993); furthermore, it has been shown that ChAT phosphorylation can alter its enzymatic activity (Dobransky et al., 2000). We found that the wild-type ChAT purified from mammalian cells showed a higher turnover number (kcat) than the wild-type ChAT isolated from bacteria. Our results are in agreement with the results reported by another group (Dobransky et al. 2000), demonstrating that phosphorylated ChAT has a higher enzymatic activity than nonphosphorylated.
Summary
There is a striking variability in the severity of phenotypes resulting from mutations in CHAT, which is the only gene so far known to be linked with congenital deficiency of ACh synthesis. This variability is remarkable, even in members of the same family carrying similar genotypes. The identified ChAT mutations have different degrees of decreasing the synthesis of ACh in cholinergic neurons, which is correlated with the variability of the severity of phenotypes resulting from these mutations. The most damaging mutations have a trend in common: they affect residues that are important for the binding of acetyl-CoA and choline substrates at the active tunnel, so the substrates can properly bind to the catalytic residue His442. Thus, both substrates need to be properly bound at the enzyme for the synthesis of ACh neurotransmitter to occur at a normal rate. However, when the mutations modify the binding of any of the substrates, the ACh synthesis is diminished at varied degrees, depending on the position and the significance of the mutated residues in ChAT tertiary structure.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Irwin H. Segel for his technical assistance in enzyme kinetics and Dr. David W. Rodgers for providing the human ChAT cDNA clone. Dr. Robert Faircough and Dr. Enoch Baldwin provided editorial help. Flora Rees-Arredondo for her help on the figures, and Sandra A. Arredondo for her technical help. This work was supported by the National Institutes of Health, Grant R01NS049117-01 to RAM, and the Myasthenia Gravis Foundation of California. Conflict of Interest statement. None declared.
Funding:
This work was supported by the National Institutes of Health, Grant R01NS049117-01 to RAM, and the Myasthenia Gravis Foundation of California.
REFERENCES
- Arredondo J, Chernyavsky AI, Webber RJ, Grando SA. Biological effects of SLURP-1 on human keratinocytes. J Invest Dermatol. 2005;125:1236–1241. doi: 10.1111/j.0022-202X.2005.23973.x. [DOI] [PubMed] [Google Scholar]
- Arredondo J, Chernyavsky AI, Karaouni A, Jolkosky DL, Pinkerton KE, Grando SA. Receptor-mediated tobacco toxicity: cooperation of the Ras/Raf-1/MEK1/ERK and JAK-2/STAT-3 pathways downstream of alpha7 nicotinic receptor in oral keratinocytes. FASEB J. 2006;20:2093–2101. doi: 10.1096/fj.06-6191com. [DOI] [PubMed] [Google Scholar]
- Arredondo J, Chernyavsky AI, Jolkovsky DL, Pinkerton KE, Grando SA. Receptor-mediated tobacco toxicity: acceleration of sequential expression of α5 and α7 nicotinic receptor subunits in oral keratinocytes exposed to cigarette smoke. FASEB J. 2008;22:1356–1368. doi: 10.1096/fj.07-9965.com. [DOI] [PubMed] [Google Scholar]
- Barisic N, Muller JS, Paucic-Kirincic E, Gazdik M, Lah-Tomulic K, Pertl A, Sertic J, Zurak N, Lochmuller H, Abicht A. Clinical variability of CMS-EA (congenital myasthenic syndrome with episodic apnea) due to identical CHAT mutations in two infants. Eur J Pediatr Neurol. 2005;9:7–12. doi: 10.1016/j.ejpn.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Bruce G, Hersh LB. The phosphorylation of choline acetyltransferase. Neurochem Res. 1989;14:613–620. doi: 10.1007/BF00964869. [DOI] [PubMed] [Google Scholar]
- Bruce G, Wainer BH, Hersh LB. Immunoaffinity purification of human choline acetyltransferase: comparison of the brain and placental enzymes. J Neurochem. 1985;45:611–620. doi: 10.1111/j.1471-4159.1985.tb04030.x. [DOI] [PubMed] [Google Scholar]
- Cai Y, Cronin CN, Engel AG, Ohno K, Hersh LB, Rodgers D. Choline acetyltransferase structure reveals distribution of mutations that cause motor disorders. EMBO J. 2004;23:2047–2058. doi: 10.1038/sj.emboj.7600221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbini LA, Hersh LB. Functional analysis of conserved histidines in choline acetyltransferase by site-directed mutagenesis. J Neurochem. 1993;61:247–253. doi: 10.1111/j.1471-4159.1993.tb03561.x. [DOI] [PubMed] [Google Scholar]
- Cossins J, Belaya K, Hicks D, Salih MA, Finlayson S, Carboni N, Liu WW, Maxwell S, Zoltowska K, Farsani GT, Laval S, Seidhamed MZ, et al. Congenital myasthenic syndromes due to mutations in ALG2 and ALG14. Brain. 2013;136:944–956. doi: 10.1093/brain/awt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbini L, Rodriguez G, Hersh LB. Kinetic and inactivation studies of recombinant Drosophila choline acetyltransferase. Brain Res Bull. 1990;24:119–124. doi: 10.1016/0361-9230(90)90295-b. [DOI] [PubMed] [Google Scholar]
- Dobransky T, Rylett RJ. A model for dynamic regulation of choline acetyltransferase by phosphorylation. J Neurochem. 2005;95:305–313. doi: 10.1111/j.1471-4159.2005.03367.x. [DOI] [PubMed] [Google Scholar]
- Dobransky T, Davis WL, Xiao GH, Rylett RJ. Expression, purification and characterization of recombinant human choline acetyltransferase: phosphorylation of the enzyme regulates catalytic activity. Biochem J. 2000;349:141–151. doi: 10.1042/0264-6021:3490141. pt 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AG, Shen XM, Selcen D, Sine S. New horizons for congenital myasthenic syndromes. Ann N Y Acad Sci. 2012;1275:54–62. doi: 10.1111/j.1749-6632.2012.06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AG. Current status of the congenital myasthenic syndromes. Neuromuscul Disord. 2012;22:99–111. doi: 10.1016/j.nmd.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AG, Lambert EH. Congenital myasthenic syndromes. Electroencephalogr Clin Neurophysiol Suppl. 1987;39:9. [PubMed] [Google Scholar]
- Fonnum F. Radiochemical micro assays for the determination of choline acetyltransferase and acetylcholinesterase activities. Biochem J. 1969;115:465–472. doi: 10.1042/bj1150465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hersh LB, Nair RV, Smith DJ. The reaction of choline acetyltransferase with sulfhydryl reagents. Methoxycarbonyl-CoA disulfide as an active site-directed reagent. J Biol Chem. 1979;254:11988–11992. [PubMed] [Google Scholar]
- Hersh LB. The lack of specificity towards salts in the activation of choline acetyltransferase from human placenta. J Neurochem. 1979;32:991–996. doi: 10.1111/j.1471-4159.1979.tb04585.x. [DOI] [PubMed] [Google Scholar]
- Hersh LB, Barker LA, Rush B. Effect of sodium chloride on changing the rate-limiting step in the human placental choline acetyltransferase reaction. J Biol Chem. 1978;253:4966–4970. [PubMed] [Google Scholar]
- Hersh LB. Kinetic studies of the choline acetyltransferase reaction using isotope exchange at equilibrium. J Biol Chem. 1982;257:12820–12825. [PubMed] [Google Scholar]
- Hersh LB, Peet M. Re-evaluation of the kinetic mechanism of the choline acetyltransferase reaction. J Biol Chem. 1977;252:4796–4802. [PubMed] [Google Scholar]
- Holt DJ, Bachus SE, Hyde TM, Wittie M, Herman MM, Vangel M, Saper CB, Kleinman JE. Reduced density of cholinergic interneurons in the ventral striatum in schizophrenia: an in situ hybridization study. Biol Psychiatry. 2005;58:408–416. doi: 10.1016/j.biopsych.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Jogl G, Tong L. Crystal structure of carnitine acetyltransferase and implications for the catalytic mechanism and fatty acid transport. Cell. 2003;112:113–122. doi: 10.1016/s0092-8674(02)01228-x. [DOI] [PubMed] [Google Scholar]
- Kim AR, Rylett RJ, Shilton BH. Substrate binding and catalytic mechanism of human acetylcholine transferase. Biochemistry. 2006;45:14621–14631. doi: 10.1021/bi061536l. [DOI] [PubMed] [Google Scholar]
- Mallory LA, Shaw JG, Burgess SL, Estrella E, Nurko S, Burpee TM, Agus MS, Darras BT, Kunkel LM, Kang PB. Congenital myasthenic syndrome with episodic apnea. Pediatr Neurol. 2009;41:42–45. doi: 10.1016/j.pediatrneurol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek KW, Vijay IK, Marth JD. A recessive deletion in the GlcNAc-1-phosphotransferase gene results in peri-implantation embryonic lethality. Glycobiology. 1999;9:1263–1271. doi: 10.1093/glycob/9.11.1263. [DOI] [PubMed] [Google Scholar]
- Maselli RA, Chen D, Mo D, Bowe C, Fenton G, Wollman RL. Choline acetyltransferase mutations in myasthenic syndrome due to deficient acetylcholine synthesis. Muscle Nerve. 2003;27:180–187. doi: 10.1002/mus.10300. [DOI] [PubMed] [Google Scholar]
- Maselli RA, Arredondo J, Cagney O, Anderson JA, Williams C, Soliven B. MUSK tyrosine kinase domain mutations causing congenital myasthenia syndrome. Hum Mol Genet. 2010;19:2370–2379. doi: 10.1093/hmg/ddq110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Lambert EH, Engel AG. Synaptic vesicle abnormality in familial infantile myasthenia. Neurology. 1987;37:206–214. doi: 10.1212/wnl.37.2.206. [DOI] [PubMed] [Google Scholar]
- Mubumbila V, Sutter A, Ptok U, Heun R, Quirin-Stricker C. Identification of a single nucleotide polymorphism in the choline acetyltransferase gene associated with Alzheimer's disease. Neurosci Lett. 2002;333(1):9–12. doi: 10.1016/s0304-3940(02)00955-2. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nachmansohn D, Machado AL. The formation of acetylcholine. A new enzyme choline acetylase. J Neurophysiol. 1943;6:397–403. [Google Scholar]
- Ohno K, Tsujino A, Brengman JM, Harper CM, Bajzer Z, Udd B, Beyring R, Robb S, Kirkham FJ, Engel AG. Choline acetyltransferase mutations cause myasthenic syndrome associated with episodic apnea in humans. Proc Natl Acad Sci USA. 2001;98:2017–2022. doi: 10.1073/pnas.98.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JH, McGeer PL, McGeer EG. Anti-human choline acetyltransferase fragments antigen binding (FAB)–sepharose chromatography for enzyme purification. Neurochem Res. 1983;8:1481–1486. doi: 10.1007/BF00965003. [DOI] [PubMed] [Google Scholar]
- Ramsay RR, Gandour RD, Van der Leij FR. Molecular enzymology of carnitine transfer and transport. Biochim Biophys Acta. 2001;1546:21–24. doi: 10.1016/s0167-4838(01)00147-9. [DOI] [PubMed] [Google Scholar]
- Ray B, Simon JR, Lahiri DK. Determination of high-affinity choline uptake (HACU) and choline acetyltransferase (ChAT) activity in the same population of cultured cells. Brain Res. 2009;1297:160–168. doi: 10.1016/j.brainres.2009.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régal L, Shen XM, Selcen D, Verhille C, Meulemans S, Creemers JW, Engel AG. PREPL deficiency with or without cystinuria causes a novel myasthenic syndrome. Neurology. 2014;82(14):1254–1260. doi: 10.1212/WNL.0000000000000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr Choline acetyltransferase. Inhibition by thiol reagents. J Biol Chem. 1974;249:2156–2159. [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senderek J, Müller JS, Dusl M, Strom TM, Guergueltcheva V, Diepolder I, Laval SH, Maxwell S, Cossins J, Krause S, Muelas N, Vilchez JJ, et al. Hexosamine biosynthetic pathway mutations cause neuromuscular transmission defect. Am J Hum Genet. 2011;88:162–172. doi: 10.1016/j.ajhg.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segel IH. Steady-state kinetics of multireactant enzymes. In: Segel IH, editor. Enzyme Kinetics. Wiley; New York: 1975. pp. 505–845. [Google Scholar]
- Shen XM, Crawford TO, Brengman J, Acsadi G, Iannaconne S, Karaca E, Khoury C, Mah JK, Edvardson S, Bajzer Z, Rodgers D, Engel AG. Functional consequences and structural interpretation of mutations of human choline acetyltransferase. Hum Mutat. 2011;32:1259–1267. doi: 10.1002/humu.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt BM, Rylett RJ. Phosphorylation of rat brain choline acetyltransferase and its relationship to enzyme activity. J Neurochem. 1993;61:1774–1781. doi: 10.1111/j.1471-4159.1993.tb09815.x. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Kulkarni S, Dharmadhikari AV, Sampath S, Bhatt SS, Shaikh TH, Xia Z, Pursley AN, Cooper ML, Shinawi M, Paciorkowski AR, Grange DK, et al. Recurrent deletions and reciprocal duplications of 10q11.21q11.23 including CHAT and SLC18A3 are likely mediated by complex low-copy repeats. Hum Mutat. 2012;33(1):165–79. doi: 10.1002/humu.21614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse PJ. The cholinergic deficit in Alzheimer’s disease. J Clin Psychiatry. 1998;59:19–22. [PubMed] [Google Scholar]
- Wu D, Hersh LB. Identification of an active site arginine in rat choline acetyltransferase by alanine scanning mutagenesis. J Biol Chem. 1995;270:29111–29116. doi: 10.1074/jbc.270.49.29111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.