Abstract
The interest in the role of metabotropic glutamate receptor 4 (mGlu4) in CNS related disorders has increased the need for methods to investigate the binding of allosteric drug candidates. Our aim is to present the first fully characterized in vitro binding assay of mGlu4 positive allosteric modulators (PAMs). Results suggest that mGlu4 PAMs have characteristic co-operative binding with orthosteric glutamate, which offers a notable insight to the further development of mGlu4 targeted therapies.
Keywords: Metabotropic glutamate receptor, Parkinson disease, mGlu4, Binding affinity
1. Introduction
Glutamate (L-Glutamate acid, 1 in Fig. 1) is a classical neurotransmitter, but it has also impact on non-neuronal systems (Nedergaard et al., 2002). During the last decade there has been an increasing interest in the role of metabotropic glutamate receptors (mGlus) in central nervous system (CNS) related disorders, like Parkinson disease (PD) (Marino et al., 2003;Niswender and Conn, 2010). The metabotropic glutamate receptor 4 (mGlu4) is a member of the Family C Seven Transmembrane Spanning/G-protein coupled receptors (7TMRs/GPCRs) Group III, which is primarily expressed in presynaptic region and plays an important role especially in basal ganglia, which is one of the most essential pathological area in PD (Lindsley and Hopkins, 2012). Moreover, the endogenous glutamate binding activates the mGlu4 and reduces transmission at both striatopallidal synapses and synapses between subthalamic nucleus and the substantia nigra compacta, both of which are overacting in PD (Marino et al., 2003). Current treatment of PD is primarily aimed at restoring dopaminergic function, which has been associated with complications such as dyskinesias and fluctual motor responses (Rascol et al., 2000). By passing the classical dopamine system in the treatment of CNS related disorders should produce fewer side effects, which has activated investigations to use mGlu4 as a drug target (Lindsley et al., 2009;Lindsley and Hopkins, 2012).
Figure 1. Structures of mGlu4 targeting compounds.
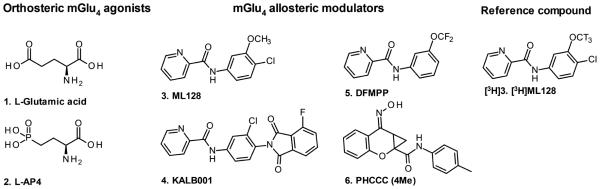
The orthosteric agonists L-Glutamate (1), L-AP4 (2), and positive allosteric modulators of mGlu4, ML128 (3) and KALB001 (4), DFMPP (5) and PHCCC (4Me) (6) are known structures to bind to mGlu4.
The basic function of mGlu4 requires endogenous glutamate binding to induce dynamic change in the receptor conformation, which ultimately leads to interaction with a heterotrimeric G protein and further intracellular signaling (Kenakin and Miller, 2010). The localization and function on GABAenergic neurons suggest that selective activation of mGlu4 decreases GABA release in the synapse, which leads to the reduction or even elimination of PD symptoms (Gregory et al., 2011). The glutamate binding site targeted drugs (such as compound 2 in Fig. 1) suffer from receptor specificity and the search for molecules that behave as allosteric modulators has led to finding of small molecular scaffolds targeting to other distinct binding cavities of the mGlu4 (Christopoulos, 2002). The targeting of distinct allosteric binding site of mGlus with small molecular scaffolds made it possible to non-invasively image these receptors using nuclear medicine technologies, which have increased the knowledge of mGlus and mGlu targeting drugs in multiple diseases (Black et al., 2010; Drouin-Ouellet et al., 2011; Kil et al., 2013, 2014a, 2014b; Wang et al., 2012). During the last decades there has been a substantial progress in identifying positive allosteric modulators (PAM) for mGlu4 (Conn et al., 2009). New mGlu4 PAMs are usually discovered with functional in vitro assays although the allosteric modulators induce functional change to stimulus cascades and at the same time increase the complexity of pharmacological effects, mechanism of action, and ligand binding characteristics, which could explain why promising compounds fail in vivo and the difficulties to develop new imaging tracers (Kenakin and Miller, 2010). Functional methods are needed to evaluate the binding characteristics of new mGlu4 targeted drugs and imaging agents in order to develop drug treatments and imaging tracers of the receptor.
2. Materials and Methods
2.1 Synthesis protocols
[3H]Iodomethane (1 Ci/ml in DMF) was purchased from American Radiolabeled Chemicals Inc., and used without further purification. Otherwise specified, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). The products were identified by LC-MS and 1H-NMR and 13C-NMR using Varian 500 MHz spectrometer. All NMR samples were dissolved in chloroform-d (CDCl3) containing tetramethylsilane as a reference standard. HRMS was obtained from High Resolution Mass Spectrometry Facility at University California, Riverside using electrospray ionization (ESI)/atmospheric pressure chemical ionization (APCI) technique (Agilent Time of Flight (TOF) LC-MS). Melting point (mp) was measured using Mettler MP50 melting point system. The purities of all compounds were over 95% determined by HPLC.
2.1.1 Radiosynthesis of N-(4-Chloro-3-methoxyphenyl)-2-picolinamide, [3H]ML128, ([3H]3)
[3H]Iodomethane (25 mCi, 1 Ci/ml in DMF, American Radiolabeled Chemicals Inc.) was added to a mixture solution of 9 (1.5 mg) and 5 M potassium hydroxide solution (3.0 μl) in DMF (0.3 ml) containing. The reaction mixture was heated at 90 °C for 10 min, and diluted with 1.0 ml of HPLC solvents. Then, the aliquot was injected into HPLC equipped with Gemini-NX C18 semi-preparative column (250 mm × 10 mm, 5 μm, Phenomenex Inc.), flow scintillation detector, and internal UV detector eluting with a solution of 55% acetonitrile and 45% 0.1 M ammonium formate at a flow rate of 4 ml/min. The fractions containing compound ([3H]3) were collected between 11–13 min. The radioactive product was diluted with 30 ml of water and passed through C18 Sep-Pak Plus followed by additional wash with 5 ml of sterile water. Finally total 6.34 mCi of ([3H]3) was eluted from cartridge with ethanol (25% yield). The radioactivity was measured and the final concentration was set as 10 μM in EtOH and stored in class vial at −20 °C during the experiments.
2.1.2 Preparation of N-(4-Chloro-3-hydroxyphenyl)-2-picolinamide (9): (Scheme 1.)
Scheme 1.
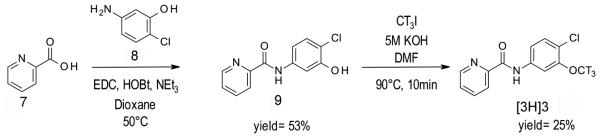
Syntheses of [3H]ML128 ([3H]3) starting from commercial reagents (Kil et al., 2014a).
The preparation of 9 was prepared by the method published previously by (Kil et al., 2014a). 5-Amino-2-chlorophenol (8, 1.20 g, 8.36 mmol) was added to a mixture solution of 2-picolinic acid (7, 0.857 g, 6.97 mmol), N-hydroxybenzotriazole hydrate (HOBt·H2O, 1.066 g, 6.97 mmol), diisopropylethylamine (DIPEA, 1.80 g, 13.93 mmol) and N-(3-methylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC·HCl, 2.0 g, 10.45 mmol) in dry 1,4-dioxane (80 mL) at room temperature. The reaction solution was heated at 50°C overnight. The reaction mixture was diluted with 300 mL of dichloromethane (DCM), and sequentially washed three times with 100 ml of water. After the organic layer was separated, dried over anhydrous sodium sulfate, filtered, and concentrated, the residue was purified by silica-gel chromatography using gradient hexane/ethyl acetate from 95/5 to 30 /70 to give the product 9 as a pale-yellow solid (0.482 g, 1.94 mmol, 27.8% yield). mp 140-142°C. 1H NMR (500 MHz, CDCl3) δ ppm 10.02 (s, 1H), 8.62 (d, J = 5.0 Hz, 1H), 8.30 (d, J = 8.0 Hz, 1H), 7.92 (ddd, J = 7.6, 7.6, 1.6 Hz, 1H), 7.64 (d, J = 1.5 Hz, 1H), 7.50 (dd, J = 7.0, 5.0 Hz, 1H), 7.31 (d, J = 8.5 Hz, 1H), 7.27 (d, J = 2.5 Hz, 1H), 5.68 (bs, 1H); 13C NMR (125 MHz, DMSO-d6) δ ppm 163.02, 154.39, 150.91, 149.32, 139.37, 138.96, 130.71, 127.82, 123.15, 116.13, 112.88, 109.11. LC-MS, calculated for C12H9N2O2Cl: 248.04; observed: 249.0 [MH]+. HRMS m/z calcd for C12H9ClN2O2 (MH+), 249.0425; found 249.0437.
2.1.3 Preparation of N-(4-Chloro-3-methoxyphenyl)-2-picolinamide, ML128 (3)
The preparation of 3 was prepared by the method published previously by (Kil et al., 2013)
2.1.4 N-(3-Chloro-4-(4-fluoro-1,3-dioxo-isoindolin-2-yl)phenyl)-2-picolinamide, KALB001 (4)
The preparation of 4 was performed by the method published previously by (Kil et al., 2014a).
2.1.5 N-(3-(Difluoromethoxy)phenyl)-2-picolinamide, DFMPP (5)
The preparation of 5 was performed by the method published previously by (Kil et al., 2013).
2.1.6 N-(p-tolyl)-7-(hydroxyimino)cyclopropa[b]chromen-1a-carboxamide, PHCCC (4Me) (6)
Was purchased from Tocris Inc.
2.1.7 Quality control (QC) analysis
The chemical identity of [3H]3 was confirmed by injection with the cold reference compound into radio-HPLC.
2.2 Biological assays
2.2.1 mGlu4 Transfection vector
The vector of mGlu4 from rat was obtained as a gift from Drs. Tanabe and Shigetada Nakanishi’s laboratory (Osaka Bioscience Institute, Osaka, Japan), and its structure is described in the previous literature (Tanabe et al., 1992). To extract mGlu4 cDNA insert out of its backbone, pBluescript II KS(+), EcoR1 (New England Biolabs) was treated, and incubated for 4 h at +37 °C. The electrophoresis was performed, and mGlu4 insert (3704 bp) was isolated from its backbone (2954 bp) using gel purification kit (Qiagen Inc.) following the manufacturers protocol. The target backbone, pcDNA, was also digested by the same procedure and concentrations were analyzed using NanoDrop (Thermo Scientific). The pcDNA (30 fmol) and mGlu4 insert (90 fmol) were mixed and incubated at room temperature for 2 h mediated by T4 ligase (Invitrogen® Life Technology). After the ligase was deactivated by heating, the combined vectors were incorporated into DH5E competent cells (Invitrogen® Life Technology) by electroporation. The aliquot was transferred to B agar dish containing ampicillin, and grown at +37 °C for 24 hours. Clones were amplified in LB media containing 0.1% ampicillin and screened by electrophoresis and DNA sequencing. Successful clones were further cultured in LB broth (1 L) containing 0.1% ampicillin stock solution. The resultant vectors containing mGlu4 cDNA was obtained using Maxi-Prep Column kit (Qiagen Inc.) by the standard procedure.
2.2.2 CHO cell transfection with mGlu4
The mGlu4 DNA and Plus™ reagent (Invitrogen® Life Technology) were mixed in reduced serum media (Opti-MEM® Life Technology), and added to the reduced serum media containing Lipofectamine® (Invitrogen® Life Technology). The resultant aliquot was incubated for 15 min at room temperature. The CHO cells were seeded onto six-wall plate, and cultured overnight until confluence at +37 °C in the culture media (Corning cellgro F12K Medium containing 10% FBS and 1% Penicillin-Streptomycin solution). The medium was replaced with OPTI-MEM in prior of transfection using the Lipofectamine method according the manufacturer’s protocol. Transfected cells were incubated at +37 °C for 3 h. The media was replaced with culture media containing G418 (3000 μg/ml) as selection reagent. The selection media was replaced every three days during the culturing. The selected cell clone was further analyzed by western blot using anti-mGlu4 antibody (Millipore) as a primary antibody.
2.2.3 Western blot
The SDS PAGE was done using 20 μg of protein sample and the proteins were transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer’s protocols (Bio-Rad). After incubation with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Tween 20) for 60 min, the membrane was washed once with TBST and incubated with antibodies against mGlu4 (1:2000) at 4 °C for 12 h. Membranes were washed three times for 10 min and incubated with a secondary antibody (1:5000 dilution) ECL Anti-rabbit IgG (horseradish peroxidase linked whole antibody from donkey NA934V (GE Healthcare UK)) for 1 h. Blots were washed with TBST three times and developed (#34080 Thermo Supersignal West Pico Chemiluminescent substrate) on HyBlot CL Autoradiography Film (Cat No.: E3012 Denvill Scientific Inc.).
2.2.4 Cell handling
CHO cells expressing mGlu4 were used for all binding assay protocols. Each individual test tube contained 50000 freshly harvested CHO cells expressing mGlu4 in HAMs cell culture medium (Life technologies, Ham's F-12 Nutrient Mix, GlutaMAX™). The cells were washed twice with ice cold PBS in prior of manual removal from culturing flask (CELLSTAR® Cell Culture Flasks with Filter Cap) to falcon tube. The cells were gently centrifuged with 800 rpm at +4 °C for 5 min and the pellet was resuspended to DMEM Glutamate free culturing media. The cells were counted and the final concentration was set as 50000 cells 98 μl of buffer. The cells were kept at +4 °C until the incubation was started. The final concentration of organic solvent (EtOH) was kept under 2% in all experiments. All experiments were done in 0.65 ml VWR® Micro Centrifuge tubes (polypropylene).
2.2.5 Saturation assay
Tested compounds and radioligand ([3H]3) were dissolved in ethanol and the incubation was started by addition of 1μl of each solution to the test tube. After 30 min of incubation at room temperature, the tubes were centrifuged at 1500 rpm for 4min at 4°C. The supernatant was gently removed from the tubes and the pellets were washed 5 times with 200 μl of ice-cold glutamate free DMEM cell culture medium. After the final wash the supernatant was removed and samples were mixed with the scintillation liquid (Perkin Elmer, Optima Gold) in prior to counting in a scintillation counter (Packard TriCarb Model, 1 min/vial) using Solvent-Saver™ scintillation vials (VWR International LLC.).
2.2.6 Co-operative assay:
Cold compound and radioligand [3H]3 (10 nM as final concentration) were dissolved in ethanol and the incubation was started by addition of 1μl of each solution to the test tube. After 30 min of incubation at room temperature with varied concentrations of orthosteric ligands (Glutamate 1 and L-AP4 2), the tubes were centrifuged at 1500 rpm for 4min at 4°C. The supernatant was gently removed from the tubes and the pellets were washed 5 times with 200 μl of ice-cold DMEM cell culture medium. After the final wash the supernatant was removed and samples were mixed with the scintillation liquid (Perkin Elmer, Optima Gold) in prior to counting in a scintillation counter (Packard TriCarb Model, 1 min/vial) using Solvent-Saver™ scintillation vials (VWR International LLC.).
2.2.7 Association assay
Cold compound and radioligand [3H]3 (10 nM as final concentration) were dissolved in ethanol and the incubation was started by addition of 1μl of each solution to the test tube. After 1, 3, 5, 10, 15, 20, 30, 45, and 60 min of incubation at room temperature with 100 nM of Glu, the tubes were centrifuged at 1500 rpm for 4min at 4°C. The supernatant was gently removed from the tubes and the pellets were washed 5 times with 200 μl of ice-cold DMEM cell culture medium. After the final wash the supernatant was removed and samples were mixed with the scintillation liquid (Perkin Elmer, Optim Gold) in prior to counting in a scintillation counter (Packard TriCarb Model, 1 min/vial) using Solvent-Saver™ scintillation vials (VWR International LLC.).
2.2.8 Dissociation assay
The dissociation assay was started after 1 hour of incubation of mGlu4 expressing cells with 10 nM of [3H]ML128 ([3H]3) by adding (time = 60 min) 10 μM excess of ML128 (3). Aliquots were removed at 61, 63, 65, 70, 75, 80, 90, 105 and 120 min time points, the tubes were centrifuged at 1500 rpm for 4 min at 4°C. The supernatant was gently removed from the tubes and the pellets were washed 5 times with 200 μl of ice-cold DMEM cell culture medium and the specific binding was determined. The K+1 and K−1 were analyzed using GraphPad Prism software and the estimated Kd was calculated as Kd= K−1/ K+1.
2.2.9 mGlu4 competitive binding assay
For the competitive binding 10 nM of [3H]ML128 ([3H]3) was used together with increased concentrations of the test compound ranging from 0.01 nM to 10 μM. The DMEM glutamate free F12 medium was used as incubation buffer with 100 nM of Glu. Compound ([3H]3) (10 nM) was added to the cell with or without test compounds on ice and incubated for 30 min at room temperature. Samples were centrifuged at 1200 rpm for 4 min at +4 °C and washed five times with cold DMEM cell culture medium (with the addition of glutamate) as washing buffer in prior of liquid scintillation counting.
2.2.10 Temperature dependency
The binding of 10 nM ([3H]3) was measured at +4°C, 25°C and 45°C. The DMEM glutamate free F12 medium was used as incubation buffer with 100 nM of Glu. Compound ([3H]3) (10 nM) was added to the harvested cells on ice and incubated for 30 min at varied temperatures. Samples were centrifuged at 1200 rpm for 4 min at +4 °C and washed five times with cold DMEM cell culture medium (with the addition of glutamate) as washing buffer in prior of liquid scintillation counting.
2.2.11 Ion dependency
Binding buffer contained 30mM of Hepes with or without NaCl 100 mM, MgCl2 1 mM, KCl 5 mM, and CaCl2 2.5 mM, pH = 7.4 adjusted with NaOH. The cells were washed three times with using different compositions of ions binding buffer at +4 °C over a 6h period. The binding buffer included also10 nM [3H]3 and 30 μM of L-AP4. The non-specific binding was defined by the presence of 10 μM of 3. Samples were centrifuged at 1200 rpm for 4 min at +4 °C and washed five times with cold DMEM cell culture medium (with the addition of glutamate) as washing buffer before liquid scintillation counting.
2.2.12 Liquid Scintillation counting (LSC)
After washing the samples were kept at +4 °C and the scintillation liquid (Perkin Elmer, Optima Gold, 0.5 ml) was added into the cells in reaction vials. The tubes were transferred to Solvent-Saver™ scintillation vials (VWR International LLC.) to be counted in a scintillation counter (Packard TriCarb Model, 1 min/vial).
2.2.13 Analyses
Nonspecific binding was determined using 10 μM nonradioactive ML128 (3) and specific binding was determined by extracting the nonspecific binding from the total binding. All measurements were done with three parallels. The statistical significance was tested using Students t-test. All the analyses were done with GraphPad Prism software (GraphPad Software Inc.).
3. Results and Discussion
The mechanism of action for mGlu4 allosteric modulators can involve multiple pharmacological properties like district conformational chances that modulate the binding of endogenous ligand, modulation of intracellular responses leading to altered signal capacity and/or the modulator can behave as an agonist or inverse agonist (Conn et al., 2009; Urwyler, 2011). Despite the important role of binding characteristics for drug development there is no fully characterized cell based binding assay method available for mGlu4 allosteric modulators (Kil et al., 2014a, 2013; Le Poul et al., 2012; Motlagh et al., 2014; Patent US20120245153). Moreover, the binding affinity of specific compound can be considered as an intrinsic property of the molecule, the co-operative effect is unique and dependent on the orthosteric/allosteric ligand pair in a specific microenvironment (Langmead, 2011).
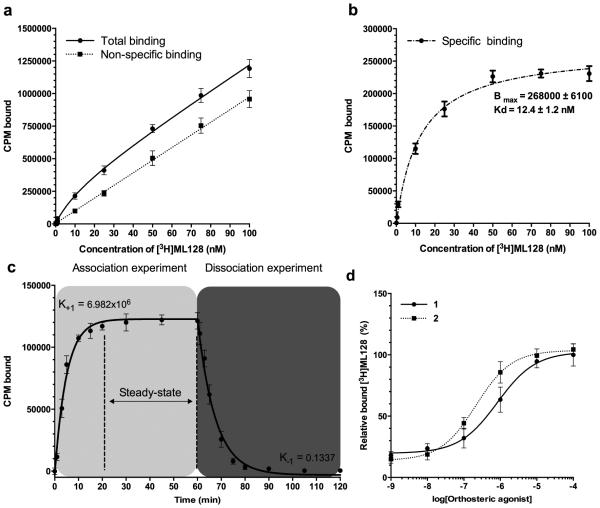
Well characterized mGlu4 PAM 3 (see Fig. 1) was selected as a candidate for tritium labeling and reference compound ([3H]3) based on published in vitro and in vivo activity data, good aqueous solubility, biodistribution, and drug properties (Kil et al., 2013). ML128 represents the most potent and selective mGlu4 probe, which is also developed as an mGlu4 positron emission tomography (PET) tracer (Hopkins et al., 2010; Kil et al., 2013). Furthermore, ML128 is centrally penetrating upon systemic dosing, displays excellent pharmacokinetics and has strong anti-Parkinsonian activity in preclinical models (Hopkins et al., 2010). During the experiments the orthosteric-allosteric co-operative binding mode was detected in the association assays between tritium labeled ([3H]3) and mGlu4, where the interaction and the binding site density of ([3H]3) were enhanced with increased amount of orthosteric agonists (see Fig. 2). Results indicate strong positive co-operativity function (α > 1) between these co-binders. The logEC50 (n=3) values were −6.08 (SE 0.07) and −6.67 (SE 0.05) for 1 and 2 respectively. Despite the results with both tested agonists (see Compounds 1 and 2 in Fig. 1) are quite similar it is essential to remind that the probe dependence may lead e.g. to significant changes in receptor activities or binding potentials depending on the used concentration and orhosteric-allosteric ligand pair. The equilibrium analysis, using constant amount of glutamate (100 μM), indicates that ([3H]3), at concentrations ranging from 0.2 to 200 nM, bound in a saturable manner with an apparent Kd of 12.4 nM following a one site-binding model (see Fig. 2)
Figure 2.
Equilibrium analysis of compound 5 binding to mGlu4 in CHO cells revealed co-operative binding with orthosteric agonist. (a) Total and nonspecific binding were analyzed using 0.1, 0.5, 1.0, 10, 25, 50, 75, and 100 nM of ([3H]3). Method was based on whole cell binding assay, where incubations (30 min at room temperature) and washings (at +4 °C) were made in constant 100 nM concentration of glutamate. Sensitive centrifugation method was used to extract the unbound and bound fraction from each other. Nonradioactive 3 (10 μM, 1000x excess to hot ligand) was used to block specific binding site. (b) The specific binding was calculated based on the difference between analyzed total and nonspecific binding and presented as an average together with the standard deviation. The data was analyzed using GraphPad Prism software to estimate the presented Bmax and Kd values. (c) The association of the ([3H]3) was measured using 10 nM of hot ligand with or without 1000 fold excess of unlabeled compound to assess the nonspecific binding. Specific binding data are plotted at 0, 1, 3, 5, 10, 15, 20, 30, 45, and 60 min time points. The dissociation study was started 1 hour after the labeling of mGlu4 expressing cells with 10 nM of [3H]3 by adding (time = 60 min) 1000 fold excess of ([3H]3). Aliquots were removed at 61, 63, 65, 70, 75, 80, 90, 105 and 120 min time points, and the specific binding was determined. The K+1 and K−1 were analyzed using GraphPad Prism software and the estimated Kd was calculated as Kd= K−1/ K+1. (d) Co-operative binding between 1or 2 and ([3H]3) was studied by addition of orthosteric agonists. The specific binding is presented together with standard deviation from three experiments (n=3).
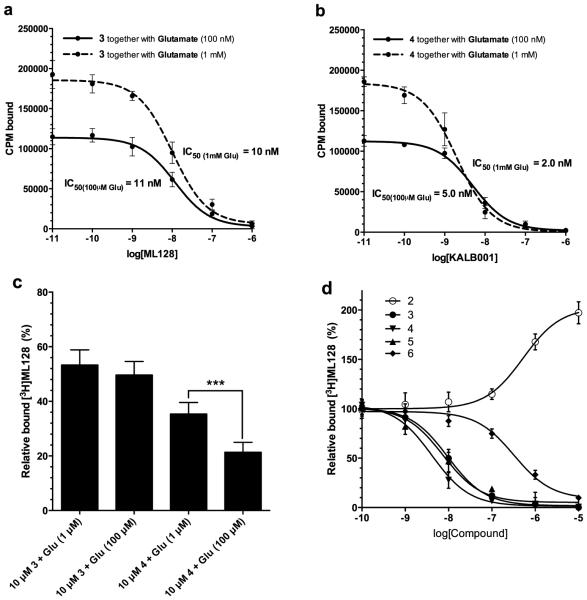
In the competitive assay, the compound ([3H]3) was displaced in a concentration-dependent manner in the mGlu4 expressing cells by reference PAMs 3, 4 and 5 (see Fig. 1). The IC50 value for unlabeled 3 was similar in the binding inhibition assays (11 nM with 100 nM of 1 and 10 nM with 1 mM of 1, (see Fig. 3) compared to what was measured in the direct saturation (12.3 nM, see Fig. 2) as well as from the association and dissociation assays (19 nM, see Fig. 2). However, the competitive binding assay gave significantly (p<0.05) better binding inhibition activity (IC50 value) for experimental PAM 4 when higher concentration of glutamate was used (logIC50 values −8.30 and −8.70 respectively with corresponding SE values of 0.05 and 0.06 n=3). The single concentration assay (10nM) (Fig. 3c) gave the similar trend and the inhibition potency of 4 was increased when higher concentration of glutamate was used (bound fractions of [3H]3 35.4% and 21.4% with SE 1.4 and 1.2 respectively (n=9)). Results suggest that the binding of an individual PAM to allosteric binding site have unique characteristics depending on the orthosteric-allosteric ligand pair in specific microenvironment. The competitive binding assay was further analyzed with known mGlu4 targeting compounds. Results (n=3) indicated logIC50 values of −8.02 (SE 0.05), −8,13 (SE 0.10), −8,38 (SE 0.08), −6,49 (SE 0.10) and EC50 value of −6,28 (SE 0.11) for compounds 3, 4, 5, 6 and 2 respectively. The total amount of specific binding (See Fig. 4a) of [3H]3 is quite similar at +4 °C and +25 °C, however the higher temperature (+45 °C) drops the specific binding apparently due to denaturation of the proteins. The ion dependency assay (See Fig. 4b) indicated that the specific binding of [3H]3 was related to chloride ion. However, this might be due to the binding of orthosteric ligand, which demands the existent of chloride ion in binding buffer (Kuang and Hampson, 2006).
Figure 3.
Competitive binding assays indicate the importance of probe dependency phenomenon of mGlu4 targeting compounds. Competitive binding curves for (a) 3 and (b) 4 (see Fig. 1) were analyzed using varied concentrations (0.01, 0.1, 1.0, 10, 100, 1000 and 10000 nM) of cold compound and constant amount (10 nM) of labeled [3H]3. The specific binding was analyzed using GraphPad Prism software and averages from at least three parallels are presented together with standard deviation for each concentration. Incubations were done in room temperature for 30min using two different concentrations (100 nM and 1 mM) of orthosteric glutamate. (c) Single dose analysis of the binding potency with different concentrations of glutamate. The binding of 4 was significantly increased compared to reference compound [3H]3 with higher concentration of glutamate. (d) Relative inhibition curve analysis of known mGlu4 targeting compounds (Engers et al., 2009)(Zhang and Brownell, 2012).
Figure 4.
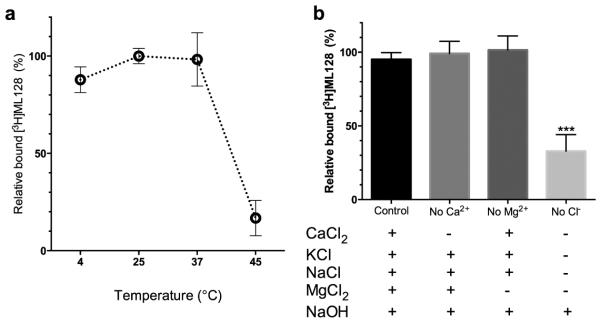
Temperature and ion dependency. (a) The relative binding of 10 nM [3H]3 in different temperatures were analyzed. (b) Ion dependency of 10 nM [3H]3 was analyzed using varied salt formulations. The binding of [3H]3 to mGlu4 was significantly reduced by removal of chloride ion from the binding buffer.
The interest in the role of metabotropic glutamate receptor 4 (mGlu4) in CNS related disorders has increased a need for methods to investigate the binding of positive allosteric modulators (PAMs). The developed binding assay allows an easy and low cost centrifugation based method to detect the binding mechanism of mGlu4 allosteric modulators to the orthosteric binding site. The results suggest that the individual mGlu4 PAMs have a unique co-operative binding with the specific orhosteric ligand, which should be carefully considered when binding assays are developed and the data is analyzed and especially when the functional or imaging data is used for drug discovery, quantification of receptor density or evaluating the possible drug dose for individual patients. The probe dependence may lead to different activities and the allosteric binding sites have unique characteristics depending on the orthosteric-allosteric ligand pair in specific microenvironment (Langmead, C.J., 2011). Moreover, new mGlu4 allosteric ligands, deeper knowledge of the mGlu4 during the disease progression and the mechanism of action of mGlu4 allosteric modulators are needed to resolve issues seen with drug candidates: low functionality, poor bioavailability and/or metabolic stability as well as patenting issues (Flor and Acher, 2012;Gubellini et al., 2014). The obtained results highlight the importance of knowing the basics for the individual drug binding mechanism when potent imaging tracers and drug candidates are tested and used for evaluating different pharmacological effects or receptor densities. The resent findings of the interactions between different mGlus have increased the knowledge of the complex pharmacological nature of mGlu4 (Yin et al., 2014). The concentrations of glutamate and the mGlu4 allosteric modulators have characteristic co-operative binding. However, the mechanisms of the mGlu4 modulation need further clarification and the presented basic radioligand-binding assay offers an invaluable insight to investigate the characteristics of mGlu4 targeted allosteric ligands. Based on the results, it could be assumed that the mGlu4 density in specific microenvironment does not unambiguously correlate with the allosteric modulator binding.
4. Conclusions
Our results suggest that the characteristics of mGlu4 allosteric modulators should be interpreted with the co-operative orthosteric-allosteric factor depending on the concentrations of the drugs and the glutamate in the specific microenvironment. The mGlu4 targeting drugs and imaging agents have a high potential to revolutionize the PD treatment in the near future. The basic in vitro methods needed to estimate the structure binding relationships and the effects of the environment should be taken account when the binding data is analyzed or used to design novel drug compounds. All this ultimately aims to increase the knowledge of the drug-protein interactions, helping to interpret the functional and imaging data, and selection of the proper drug dosing for individual patient for clinical trials.
Highlights.
Binding methods are essential when new mGlu4 allosteric modulators are developed.
Glutamate and allosteric compounds have co-operative binding to mGlu4.
The microenvironment affects the binding of PAMs.
Acknowledgements
Studies were supported by the grants NIBIB-R01EB012864 and NIMH-R01MH091684 to A-LB. PP was supported by the Orion Farmos Research Foundation and Kuopio University Foundation. We would like also to thank Drs. Tanabe and Shigetada Nakanishi’s laboratory (Osaka Bioscience Institute, Osaka, Japan) for providing us the vector of mGlu4 from rat as a gift. In addition, Dr. Shilpa Prabhakar and Dr. Xandra Breakefield from Molecular Neurogenetics Unit, Massachusetts General Hospital (Boston, MA, USA) are greatly acknowledged for helping with the transfection of the cell line.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
PP, ZZ, ALB wrote the manuscript and developed the binding assays; KK did radiolabeling and contributed to transfection studies; ZZ synthetized the compounds; DK and BT contributed to development of transfection vector and transfection of the cells; ALB designed the study.
Competing financial interest statement
The authors declare no competing financial interest.
References
- Black YD, Xiao D, Pellegrino D, Kachroo A, Brownell A-L, Schwarzschild MA. Protective effect of metabotropic glutamate mGluR5 receptor elimination in a 6-hydroxydopamine model of Parkinson’s disease. Neurosci. Lett. 2010;486:161–165. doi: 10.1016/j.neulet.2010.09.043. doi:10.1016/j.neulet.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nat. Rev. Drug Discov. 2002;1:198–210. doi: 10.1038/nrd746. doi:10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. doi:10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin-Ouellet J, Brownell A-L, Saint-Pierre M, Fasano C, Emond V, Trudeau L-E, Lévesque D, Cicchetti F. Neuroinflammation is associated with changes in glial mGluR5 expression and the development of neonatal excitotoxic lesions. Glia. 2011;59:188–199. doi: 10.1002/glia.21086. doi:10.1002/glia.21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engers DW, Niswender CM, Weaver CD, Jadhav S, Menon UN, Zamorano R, Conn PJ, Lindsley CW, Hopkins CR. Synthesis and Evaluation of a Series of Heterobiarylamides That Are Centrally Penetrant Metabotropic Glutamate Receptor 4 (mGluR4) Positive Allosteric Modulators (PAMs) J. Med. Chem. 2009;52:4115–4118. doi: 10.1021/jm9005065. doi:10.1021/jm9005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor PJ, Acher FC. Orthosteric versus allosteric GPCR activation: the great challenge of group-III mGluRs. Biochem. Pharmacol. 2012;84:414–424. doi: 10.1016/j.bcp.2012.04.013. doi:10.1016/j.bcp.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Gregory KJ, Dong EN, Meiler J, Conn PJ. Allosteric modulation of metabotropic glutamate receptors: structural insights and therapeutic potential. Neuropharmacology. 2011;60:66–81. doi: 10.1016/j.neuropharm.2010.07.007. doi:10.1016/j.neuropharm.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubellini P, Melon C, Dale E, Doller D, Kerkerian-Le Goff L. Distinct effects of mGlu4 receptor positive allosteric modulators at corticostriatal vs. striatopallidal synapses may differentially contribute to their antiparkinsonian action. Neuropharmacology. 2014;85:166–177. doi: 10.1016/j.neuropharm.2014.05.025. doi:10.1016/j.neuropharm.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Hopkins CR, Niswender CM, Lewis LM, Weaver CD, Lindsley CW. Discovery of a potent, selective and in vivo active mGluR4 positive allosteric modulator, in: Probe Reports from the NIH Molecular Libraries Program. National Center for Biotechnology Information (US), Bethesda (MD) 2010 [PubMed] [Google Scholar]
- Jeffrey Conn P, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. doi:10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol. Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. doi:10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil K-E, Poutiainen P, Zhang Z, Zhu A, Choi J-K, Jokivarsi K, Brownell A-L. Radiosynthesis and Evaluation of an (18)F-Labeled Positron Emission Tomography (PET) Radioligand for Metabotropic Glutamate Receptor Subtype 4 (mGlu4) J. Med. Chem. 2014a doi: 10.1021/jm501245b. doi:10.1021/jm501245b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil K-E, Zhang Z, Jokivarsi K, Gong C, Choi J-K, Kura S, Brownell A-L. Radiosynthesis of N-(4-chloro-3-[(11)C]methoxyphenyl)-2-picolinamide ([(11)C]ML128) as a PET radiotracer for metabotropic glutamate receptor subtype 4 (mGlu4) Bioorg. Med. Chem. 2013;21:5955–5962. doi: 10.1016/j.bmc.2013.07.046. doi:10.1016/j.bmc.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil K-E, Zhu A, Zhang Z, Choi J-K, Kura S, Gong C, Brownell A-L. Development of [(123)I]IPEB and [(123)I]IMPEB as SPECT Radioligands for Metabotropic Glutamate Receptor Subtype 5. ACS Med. Chem. Lett. 2014b;5:652–656. doi: 10.1021/ml500007z. doi:10.1021/ml500007z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang D, Hampson DR. Ion dependence of ligand binding to metabotropic glutamate receptors. Biochem. Biophys. Res. Commun. 2006;345:1–6. doi: 10.1016/j.bbrc.2006.04.064. doi:10.1016/j.bbrc.2006.04.064. [DOI] [PubMed] [Google Scholar]
- Langmead CJ. Determining allosteric modulator mechanism of action: integration of radioligand binding and functional assay data. Methods Mol. Biol. Clifton NJ. 2011;746:195–209. doi: 10.1007/978-1-61779-126-0_10. doi:10.1007/978-1-61779-126-0_10. [DOI] [PubMed] [Google Scholar]
- Le Poul E, Boléa C, Girard F, Poli S, Charvin D, Campo B, Bortoli J, Bessif A, Luo B, Koser AJ, Hodge LM, Smith KM, DiLella AG, Liverton N, Hess F, Browne SE, Reynolds IJ. A potent and selective metabotropic glutamate receptor 4 positive allosteric modulator improves movement in rodent models of Parkinson’s disease. J. Pharmacol. Exp. Ther. 2012;343:167–177. doi: 10.1124/jpet.112.196063. doi:10.1124/jpet.112.196063. [DOI] [PubMed] [Google Scholar]
- Lindsley CW, Hopkins CR. Metabotropic glutamate receptor 4 (mGlu4)-positive allosteric modulators for the treatment of Parkinson’s disease: historical perspective and review of the patent literature. Expert Opin. Ther. Pat. 2012;22:461–481. doi: 10.1517/13543776.2012.679437. doi:10.1517/13543776.2012.679437. [DOI] [PubMed] [Google Scholar]
- Lindsley CW, Niswender CM, Engers DW, Hopkins CR. Recent progress in the development of mGluR4 positive allosteric modulators for the treatment of Parkinson’s disease. Curr. Top. Med. Chem. 2009;9:949–963. [PubMed] [Google Scholar]
- Marino MJ, Williams DL, Jr, O’Brien JA, Valenti O, McDonald TP, Clements MK, Wang R, DiLella AG, Hess JF, Kinney GG, Conn PJ. Allosteric modulation of group III metabotropic glutamate receptor 4: a potential approach to Parkinson’s disease treatment. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13668–13673. doi: 10.1073/pnas.1835724100. doi:10.1073/pnas.1835724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. doi:10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard M, Takano T, Hansen AJ. Beyond the role of glutamate as a neurotransmitter. Nat. Rev. Neurosci. 2002;3:748–755. doi: 10.1038/nrn916. doi:10.1038/nrn916. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. doi:10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patent US20120245153 - Mglur4 allosteric potentiators, compositions, and methods of treating … - Google Patents [WWW Document], n.d. URL http://www.google.com/patents/US20120245153 (accessed 10.9.14)
- Rascol O, Brooks DJ, Korczyn AD, De Deyn PP, Clarke CE, Lang AE. A five-year study of the incidence of dyskinesia in patients with early Parkinson’s disease who were treated with ropinirole or levodopa. 056 Study Group. N. Engl. J. Med. 2000;342:1484–1491. doi: 10.1056/NEJM200005183422004. doi:10.1056/NEJM200005183422004. [DOI] [PubMed] [Google Scholar]
- Stocchi F. The levodopa wearing-off phenomenon in Parkinson’s disease: pharmacokinetic considerations. Expert Opin. Pharmacother. 2006;7:1399–1407. doi: 10.1517/14656566.7.10.1399. doi:10.1517/14656566.7.10.1399. [DOI] [PubMed] [Google Scholar]
- Urwyler S. Allosteric modulation of family C G-protein-coupled receptors: from molecular insights to therapeutic perspectives. Pharmacol. Rev. 2011;63:59–126. doi: 10.1124/pr.109.002501. doi:10.1124/pr.109.002501. [DOI] [PubMed] [Google Scholar]
- Wang J-Q, Zhang Z, Kuruppu D, Brownell A-L. Radiosynthesis of PET radiotracer as a prodrug for imaging group II metabotropic glutamate receptors in vivo. Bioorg. Med. Chem. Lett. 2012;22:1958–1962. doi: 10.1016/j.bmcl.2012.01.039. doi:10.1016/j.bmcl.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Noetzel MJ, Johnson KA, Zamorano R, Jalan-Sakrikar N, Gregory KJ, Conn PJ, Niswender CM. Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J. Neurosci. Off. J. Soc. Neurosci. 2014;34:79–94. doi: 10.1523/JNEUROSCI.1129-13.2014. doi:10.1523/JNEUROSCI.1129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Brownell A-L. Imaging of Metabotropic Glutamate Receptors (mGluRs) In: Bright P, editor. Neuroimaging - Clinical Applications. InTech; 2012. doi: 10.5772/23714. [Google Scholar]