Abstract
Cyclophilins are ubiquitously expressed proteins that bind to prolines and can catalyse cis/trans isomerization of proline residues. There are 17 annotated members of the cyclophilin family in humans, ubiquitously expressed and localized variously to the cytoplasm, nucleus or mitochondria. Surprisingly, all eight of the nuclear localized cyclophilins are found associated with spliceosomal complexes. However, their particular functions within this context are unknown. We have therefore adapted three established assays for in vitro pre-mRNA splicing to probe the functional roles of nuclear cyclophilins in the context of the human spliceosome. We find that four of the eight spliceosom-associated cyclophilins exert strong effects on splicing in vitro. These effects are dose-dependent and, remarkably, uniquely characteristic of each cyclophilin. Using both qualitative and quantitative means, we show that at least half of the nuclear cyclophilins can act as regulatory factors of spliceosome function in vitro. The present work provides the first quantifiable evidence that nuclear cyclophilins are splicing factors and provides a novel approach for future work into small molecule-based modulation of pre-mRNA splicing.
Keywords: nuclear cyclophilins, pre-messenger RNA (mRNA) splicing, spliceosome
INTRODUCTION
Splicing, the process by which introns are removed from pre-mRNA, is accomplished by the complex macromolecular machinery called the spliceosome. The spliceosome promotes the two trans-esterification reactions necessary to release intron sequence from the pre-mRNA substrate and join together flanking exons. The spliceosome is composed of five U-rich small nuclear RNAs, each in complex with several protein partners (U snRNPs) that join with an evolutionarily conserved set of core proteins to recognize splice sites and position the pre-mRNA correctly for splicing [1–3]. The assembly of these components on a pre-mRNA substrate proceeds through ordered addition and dissociation of unique sub-sets of snRNPs and core proteins, which creates a series of intermediate splicing complexes.
Multiple proteomic studies of purified spliceosomal complexes from humans show that, in addition to these core snRNPs and associated core proteins, there are additional ‘accessory’ protein components of the spliceosome that also dynamically rearrange during spliceosome assembly and catalysis [2–4]. The definition of what is a ‘core’ compared with an ‘accessory’ protein is somewhat unclear, because both sets of proteins associate tightly with spliceosomal complexes and associate with specific complexes at each stage of the splicing complex. More importantly, most ‘core’ and ‘accessory’ proteins found associated with the spliceosome have no annotated function within the context of pre-mRNA splicing and their individual roles in cells have been difficult to characterize. A conservative accounting of the proteins that tightly associate with isolated and purified spliceosomal complexes would include approximately 30 U1 and U2 snRNP proteins and ~25 non-snRNP proteins that associate stably with the mammalian spliceosome during the earliest A complex stage [5–7]. In the conversion from the A complex to the pre-catalytic B complex stage, many of these early proteins leave or become much less stably associated, whereas 30 or so U4/U5/U6 snRNP-associated proteins join the spliceosome. Simultaneously, there is an influx of ~40 additional proteins, including those of the highly conserved hPRP19–Cdc5L (human pre-mRNA processing factor 19–cell division cycle 5-like complex) [5,8]. As the spliceosome is activated to form catalytically competent Bact- and C-complexes, at least 50 proteins dissociate or become much less-stably associated at the same time as the U1 and U4 snRNPs leave, whereas a new cohort of more than 40 new proteins join or become more stably associated with the complex [4,9–11]. In summary, each of the early, intermediate and catalytic splicing complexes that can currently be purified from human cells have protein components that numbers ~100 proteins; and there are ~170 unique proteins in total that are reproducibly and stably associated at some stage of the splicing cycle. Of those 170, a little more than 80 can be assigned an orthologue in Saccharomyces cerevisiae; many of the rest are found in spliceosomes purified from plants and animals, implying roles in splicing regulation [5]. The question now becomes one of functional characterization. Although some spliceosomal proteins have DEAD/DEAH RNA-dependent ATPase or RNA-recognition motif (RRM) domains that suggest direct interaction with RNA, it is not clear what roles the many other proteins found tightly associated with the spliceosome may play in spliceosome formation or catalytic function.
One intriguing class of proteins identified in human spliceosomes by proteomics is the cyclophilins. These proteins are widely expressed in mammalian cells and highly conserved members can be found in prokaryotes and eukaryotes and at least one virus [12,13]. Seventeen unique cyclophilins are expressed in human; eight localize to the cytoplasm or are secreted, one associates with mitochondria and eight are found in the nucleus [13,14]. Most cyclophilins are peptidyl-prolyl isomerases (PPIs), with the characteristic enzymatic activity of interconverting cis- and trans-isomers of proline on model peptide substrates in vitro [15–17]. The proteins are well characterized in terms of the structure of their PPI domains, their isomerase activity and high-affinity interaction with the drug cyclosporine, which directly blocks proline binding to the active site [15,18–21]. Point mutations that abrogate both cyclosporine binding and isomerase activity have been identified [15,22,23], but even with this knowledge it has not been a straightforward process to identify endogenous targets or to unambiguously identify their cellular functions.
All eight of the human nuclear cyclophilins have consistently been found to be associated with mammalian spliceosomes [4–6,8–11,24–29]. Various proteomics studies have shown that the nuclear cyclophilins join spliceosomal complexes at different stages of assembly. PPIH joins at B-complex with the tri-snRNP and leaves at the same time as U4 snRNP [5,8]. PPIE and PPIL1 join B-complex along with the PRP19 complex and remain through C-complex [1,9,16]. PPIL2 and CWC27 are strongly detected in activated spliceosomes prior to first step chemistry (Bact-complex) [1,3,9,16]. PPIL3, PPWD1 and PPIG are found in spliceosomes following first step chemistry (C-complex) [1,15,16]. These results suggest that the nuclear cyclophilins are distributed throughout the splicing cycle in order to play some regulatory role, although the isomerase domain has never been implicated in RNA–protein interactions. Furthermore, multiple experimental approaches, including yeast two-hybrid screens, co-immunoprecipitations and pull-down assays and other studies with purified proteins have shown that the nuclear cyclophilins interact directly with known spliceosome-associated splicing factors including PRPF4 (pre-mRNA processing factor 4), Aquarius, PCBP1 (poly-c binding protein 1) and Slu7 [19,30,31]. It is assumed that these interactions may somehow be mediated through, or have an impact on, proline isomerization activity of the cyclophilin involved in the interaction. It is not clear, however, what affect prolyl-isomerase activity would have on pre-mRNA catalysis, although many spliceosome components are unusually proline rich (e.g. U2 snRNP proteins SF3A1 (splicing factor 3A, subunit 1) and SF3A2 (splicing factor 3A, subunit 2), which encode for 15% and 25% proline respectively) [32,33]. Finally, several of the nuclear cyclophilins also have additional domains, including RRM (PPIE), U-box (PPIL2) and WD40 (PPWD1) motifs, which indicate other possible interaction mechanisms with components of the spliceosome. Taken together, it seems that the nuclear cyclophilins are probably playing some regulatory role within the spliceosome, perhaps mediated through the unique set of protein–protein interactions with other splicing factors found throughout spliceosome assembly and catalysis.
The association of nuclear cyclophilins with distinct stages of human spliceosome assembly points to potential functions for these proteins in splicing regulation. To begin characterizing their roles in splicing, we examined the influence of human nuclear cyclophilins on splicing catalysis and spliceosome assembly in an in vitro assay system. We show that altering the levels of several cyclophilin proteins inhibits splicing chemistry and interferes with spliceosomal complex formation in vitro. These studies open the door to understanding the cellular functions of nuclear cyclophilins and to identifying their biological substrates.
EXPERIMENTAL
Cloning, expression and purification of cyclophilins
All of the cyclophilin expression constructs used in the current study have been previously published, along with detailed expression and purification protocols [15,18]. To summarize, expression constructs for all cyclophilins used in the current study were cloned using cDNA from the Mammalian Gene Collection (accession numbers follow: PPIA – BC003026, PPIE – BC008451, PPIG – BC001555, PPIH – BC003412, PPIL1 – BC003048, PPIL2 – BC000022, PPIL3 – BC007693, PPWD1 – BC015385, CWC27 – BC012117). All clones were generated using the In-Fusion ligation-independent-cloning system (BD Biosciences) and with the exception of PPIH and PPIL3 the expression vector was a ligation independent cloning (LIC)-compatible version of pET28a which encodes an N-terminal His6-affinity tag and a thrombin protease cleavage site. PPIH was cloned into a LIC-compatible version of pET28a in which the solubility enhancer GST is expressed N-terminally to the His6-tag used for purification. PPIL3 was cloned into pTYB2 (NEB), which encodes a self-cleaving intein sequence upstream of a C-terminal chitin-binding protein-affinity tag. Expression constructs were transformed into BL21(DE3) cells (Life Technologies). All cyclophilin proteins were induced at a final concentration of 100 μM IPTG when at D600 = 0.8–1.0 and incubated at 37°C overnight. Purification protocols for all cyclophilins except PPIL3 were as in [15]; briefly, proteins were lysed by sonication in the base buffer containing 20 mM Tris, pH 7.5, 500 mM NaCl and a protease inhibitor cocktail (Sigma). Cyclophilins were initially purified by nickel affinity (His-Link, Promega), except for PPIL3 which was purified using chitin resin (NEB) and eluted with the base buffer plus 10 mM 2-mercaptoethanol. In all cases, affinity chromatography was followed by size exclusion chromatography on a Superdex 200 column (GE Healthcare). Final protein buffer consisted of 20 mM Tris, pH 7.6–7.9, 10 mM 2-mercaptoethanol and potassium chloride at 125–250 mM depending on the solubility of the individual cyclophilin. Point mutations were generated using primer mutagenesis and verified by DNA sequencing; expression and purification protocols were as for wild-type in all cases.
Pre-mRNA substrates and in vitro transcription
For the reverse transcriptase (RT)-quantitative PCR (qPCR) assay and for some of the denaturing gel splicing assays, the adenovirus major (AdML)-derived splicing substrate HMS388 was utilized (containing 311 nucleotides; exon1-intron-exon2 sizes are 133, 123 and 56 respectively). For most of the denaturing gel splicing assays and for all of the native gel assays, the related substrate HMS389 was used. Both HMS388 and 389 contain AdML exons 1 and 2 with a truncated intron sequence and both have branch point/3′ splice site sequences subcloned from the β-globin transcript. HMS389 has a longer polypyrimidine tract and importantly has an AG-GG mutation at the 3′ splice site that allows for the first step of splicing, but not the second step. Both templates have been described in detail elsewhere [4,34,35]. In other experiments, both the fushi tarazu (ftz) and the β-globin splicing substrates were used. The ftz splicing substrate is derived from the ftz gene from Drosophila melanogaster, specifically the SalI/BglII fragment (555 nucleotides; exon1-intron-exon2 sizes are 271, 147 and 171 respectively) [35]. The β-globin substrate used in the current studies contains the first and second exons of the human β-globin gene with a truncated exon (493 nucleotides; exon1-intron-exon2 sizes are 153, 130 and 210 respectively) [35]. For all splicing substrates, in vitro transcription was accomplished using T7 runoff transcription and the 32P-labelled G(5′)ppp(5′)G capped pre-mRNA was gel purified after synthesis.
In vitro splicing
Reactions consisted of 20%–40% HeLa cell nuclear extract, 2–6 mM magnesium acetate, 120 mM potassium glutamate, 3 mM ATP, 5 mM creatine phosphate, 0.05 mg/ml tRNA and 5–10 nM G(5′)ppp(5′)G capped pre-mRNA substrate. Protein was added to final concentrations of 1–200 μM as described in the text prior to incubation at 30°C.
RT-qPCR-based analysis of in vitro splicing
Based on the protocol outlined in [36], aliquots of in vitro splicing reactions were immediately diluted 1:65 in water, vortexed and kept on ice until all samples were ready for analysis. Using the TaqMan® One-Step RT-PCR kit (Applied Biosystems), 2 μl of the diluted splicing reactions were added in triplicate samples as template to 10 μl of reactions that included primers to ends of the exons of the pre-mRNA substrate and TaqMan probe to the exon junction of spliced mRNA. Experiments were performed and analysed using an iCycler iQ (BioRad). iCycler software (BioRad) was used to generate threshold cycles (Ct) for each well; triplicate samples were averaged and samples with S.D. more than one cycle were tagged for manual inspection. If an outlier was found, the average of duplicate samples was used for downstream analysis. To determine ΔCt, triplicate-averaged Ct for each protein-added sample was compared with triplicate-averaged buffer-only splicing from the same experiment. The S.D.s of triplicate measurements of Ct were used for error propagation. When a given protein concentration was measured in more than two experiments, ΔCt was averaged across experiments and the average errors calculated.
Denaturing gel analysis
Aliquots of in vitro splicing reaction were incubated with splicing dilution buffer (100 mM Tris, pH 7.5, 10 mM EDTA, 1% SDS, 150 mM NaCl, 0.3 M NaAc, pH 5.2) for 5 min at room temperature. RNA was isolated by phenol–chloroform–isoamyl extraction and ethanol precipitation. RNA pellets were resuspended in sample buffer (95% formamide, 20 mM EDTA, Bromophenol Blue, Cyan Blue) and loaded on to 15% acrylamide gels. Gels were run for 1.5 h at 35 W. Gels were immobilized and exposed to phosphorimaging screens, which were digitized with a Typhoon Scanner (Molecular Dynamics) and quantified with ImageQuant software (Molecular Dynamics).
Native gel analysis
Based on the protocol outlined in [37], before splicing, nuclear extract was pre-incubated at 30°C for 15 min to deplete endogenous ATP. Samples from different time points from in vitro splicing reactions were kept on ice until all samples were ready for analysis. Ten microlitres of splicing reaction samples were mixed with 5 μl of native gel loading buffer (20 mM Trizma base, 20 mM glycine, 25% glycerol, 0.05% Cyan Blue, 0.05% Bromophenol Blue, 2.5 mg/ml heparin sulfate) and incubated at room temperature for 5 min before loading on to a 2.1% agarose gel. Gels were run at 72 V for 3.5 h, dried on to Whatman paper and exposed to phosphorimaging screens, which were digitized with a Typhoon Scanner (Molecular Dynamics).
RESULTS
Spliceosome-associated cyclophilins exhibit differential effects on pre-mRNA splicing in vitro
In order to investigate whether cyclophilins found associated with spliceosomal complexes play a role in splicing, we examined their affect in an in vitro splicing system, which consists of incubating a synthetic pre-mRNA substrate in the presence of nuclear extracts prepared from HeLa cells. Nuclear extract contains the components of the spliceosome, which recognize the intron in the substrate pre-mRNA, assemble into splicing complexes and catalyse the two steps of splicing chemistry. We hypothesized that if cyclophilins have a role in the spliceosome, addition of an excess of these proteins could affect normal spliceosome function and result in a change in efficiency of the splicing of the pre-mRNA substrate. In order to test this hypothesis, we expressed and purified the eight nuclear cyclophilins to homogeneity and added increasing concentrations of the individual proteins to in vitro splicing reactions. We also expressed, purified and tested the cytoplasmic cyclophilin PPIA; this canonical family member is very similar in primary sequence and atomic structure to the nuclear cyclophilins, but does not stably associate with the spliceosome in cells and does not interact with spliceosomal proteins [5,15,38]. Table 1 illustrates the identity, sequence similarity to the control protein PPIA and the spliceosomal association patterns of proteins used in the current study.
Table 1.
Cyclophilins used in the current study and their association with spliceosome complexes
| Gene Name | Domain Organization | Identity/Similarity to PPIA | Spliceosome Association |
|---|---|---|---|
| PPIH | 54/69 | A, B, Bact | |
|
|
|||
| PPIE | 68/81 | B, Bact, C | |
|
|
|||
| PPIL1 | 51/67 | B, Bact, C | |
|
|
|||
| PPIL2 | 49/61 | B, Bact, C | |
|
|
|||
| CWC27 | 43/61 | B, Bact, C | |
|
|
|||
| PPIL3 | 39/54 | Bact, C | |
|
|
|||
| PPIG | 52/63 | C | |
|
|
|||
| PPWD1 | 49/59 | C | |
|
|
|||
| PPIA | 100/100 | none | |
|
|
|||
Cyclophilin constructs and their domain organization are schematized. As described in the text, expression constructs of PPI domains alone or of RRM or U-box motifs were also used in splicing reactions; these domain boundaries are also noted where applicable. The third column notes the amino acid identity and similarity for the PPI domain of each nuclear cyclophilin to that of the control cyclophilin PPIA as calculated by BLAST. Cyclophilin association with splicing complexes is indicated in the fourth column with highest abundance noted in bold as per [5].
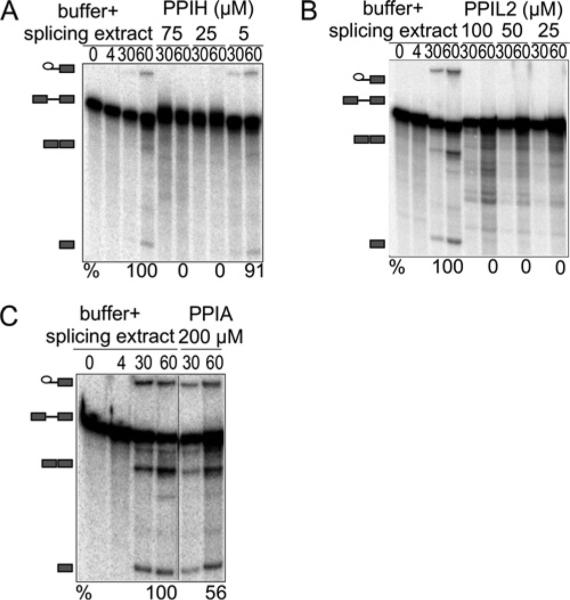
Figure 1 shows denaturing acrylamide gel analysis of radiolabelled RNA extracted from a series of in vitro splicing reactions using a modified AdML intron [4,34,35]. The reactions included different concentrations of individual cyclophilin proteins or protein buffer alone, added to nuclear extract derived from HeLa cells. We quantified splicing efficiency as the amount of mRNA product compared with the sum of pre-mRNA, first step intermediates and mRNA product. With increasing amounts of protein, we found that PPIH and PPIL2 reduced splicing efficiency in a dose-dependent manner relative to adding buffer alone. As expected, the addition of PPIA to the in vitro splicing reaction did not affect splicing efficiency, even at concentrations double that of the nuclear cyclophilins tested.
Figure 1. PPIH and PPIL2 inhibit splicing in vitro as measured by denaturing gel electrophoresis.
The panels show denaturing gel analysis of radiolabelled HMS388 splicing substrate, isolated from in vitro splicing reactions supplemented with increasing concentrations of the indicated cyclophilin proteins or with buffer alone. RNA species are schematized at the left as (from top to bottom) lariat intron intermediate, pre-mRNA, mRNA, free 5′-exon. Percentage splicing efficiency (amount of mRNA compared with total RNA species) is indicated at the bottom of each gel image. As indicated, relatively low concentrations of PPIH (A) or PPIL2 (B) efficiently inhibit the splicing reaction; in contrast, high concentrations of PPIA (C) do not block splicing activity. Note that unless indicated by a black line, lanes are contiguous and from a single gel, although experiments were performed at least three times. Also note that identical splicing substrate (HMS388) is loaded in each lane; slight differences in mobility are probably due to effects from overloading samples in order to detect low populations of splicing intermediates.
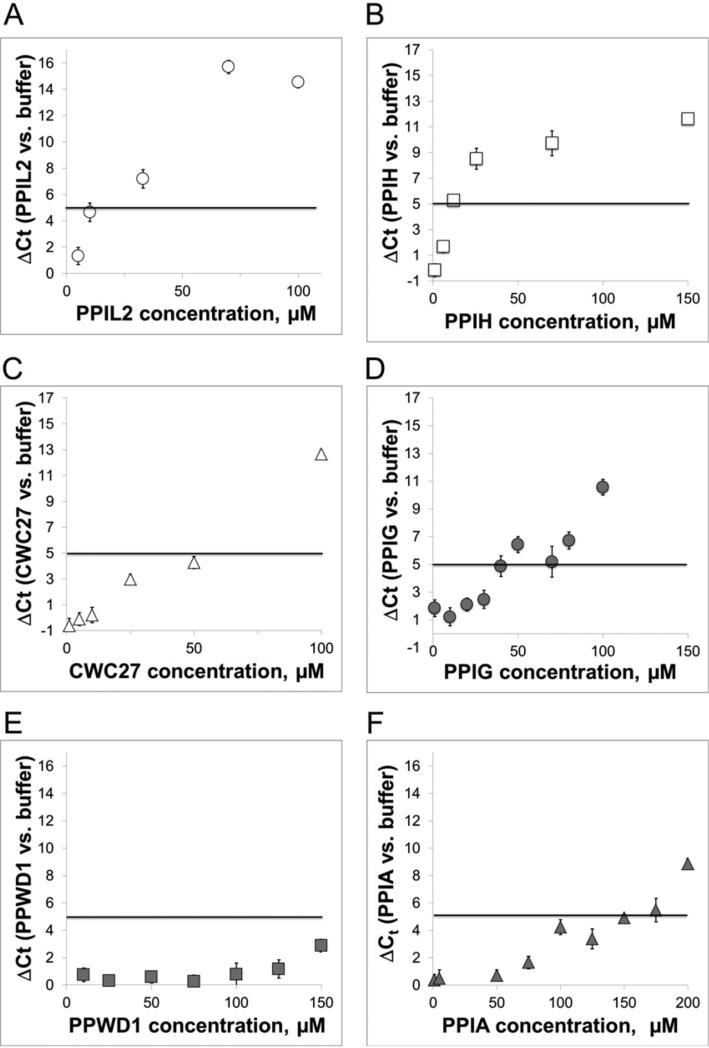
To examine the dose dependence of protein concentration on the inhibitory splicing effect and to more easily expand our analysis to the entire set of nuclear cyclophilins, we next employed an RT-qPCR based assay. This assay is built on the use of a TaqMan® probe to the exon–exon junction that forms upon completion of splicing of our AdML-based substrate [36]. As configured, the assay has much higher throughput than standard gel-based splicing detection and allowed us to test multiple concentrations of all the nuclear cyclophilins, along with the control PPIA protein. To analyse the RT-qPCR results, we determined the change in threshold cycle relative to a protein buffer control (ΔCt), which we plotted against protein concentration (Figure 2A). A shift to higher ΔCt indicates less mRNA in the originating sample, which we attribute to lower splicing efficiency. We see variability of less than one ΔCt between triplicate RT-PCR reactions within a given experiment and 1–2 threshold cycle variation between replicate experiments. Our probe set provides 90% + PCR efficiency upon our substrate. We are therefore confident that a significant amount of inhibition has occurred when the ΔCt exceeds five cycles, which would correspond to more than a 20-fold drop in mRNA production, although under optimized splicing conditions we can detect between 10 and 15 cycles difference between buffer plus splicing extract and protein-added splicing reactions.
Figure 2. PPIL2, PPIH, CWC27 and PPIG inhibit splicing as measured by an exon-junction specific probe.
The amount of AdML mRNA produced in the splicing reaction is detected via RT-qPCR by a TaqMan probe to the spliced exon junction sequence. Plotted is the average difference in cycles required to reach threshold (ΔCt) between reactions supplemented with cyclophilins compared with buffer alone. The five-cycle ΔCt threshold, as mentioned in the text, corresponds to significant inhibition in the production of mRNA and is shown in all RT-qPCR plots as a solid black line. We find that PPIL2 (A) and PPIH (B) are strong inhibitors of our AdML splicing substrate; CWC27 (C) and PPIG (D) slightly less so; and the other four nuclear cyclophilins are not inhibitory in this experimental setup [see PPWD1 in panel (E) and also Supplementary Figure S1]. The control PPIA (F), as in Figure 1, shows only slight inhibition of splicing at high protein concentrations.
Our results segregate the nuclear cyclophilins into three classes: (1) cyclophilins that yield a five-cycle increase over buffer control at low concentrations (10–50 μM); (2) cyclophilins that reach ΔCt > 5 at concentrations higher than 50 μM; and (3) cyclophilins that require higher concentrations to observe significant effects on ΔCt. As expected from the previous assays analysed by denaturing gel, PPIL2 and PPIH fall into the more inhibitory class of nuclear cyclophilins (Figures 2A and 2B). We also find that CWC27 and PPIG significantly block mRNA production in the 10–50 μM range (Figures 2C and 2D). The remaining cyclophilins tested, including PPWD1 and the control PPIA (Figures 2E and 2F), only exhibited inhibitory effects at much higher protein concentrations if at all (Supplementary Figure S1).
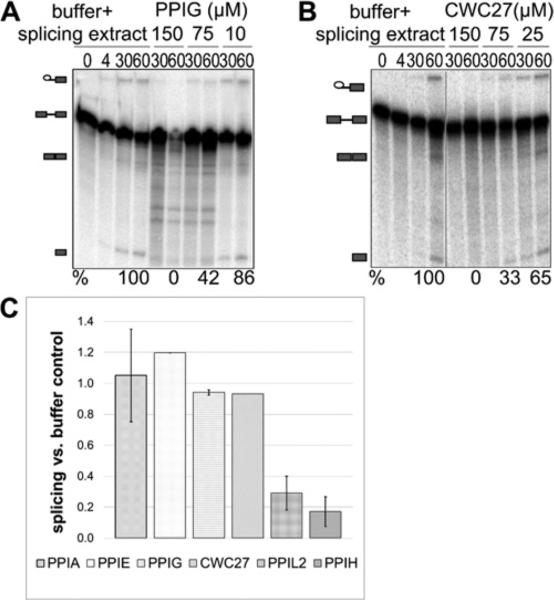
We confirmed effects on AdML substrate splicing by CWC27 and PPIG and then quantified the degree of inhibition by the nuclear cyclophilins, using denaturing gels to measure splicing efficiency. Those results confirmed a similar trend to the RT-qPCR data (Figure 3). In summary, four of the eight nuclear cyclophilins have an inhibitory effect on in vitro splicing of AdML substrate, indicating that they are regulating splicing in vitro.
Figure 3. Inhibition of splicing as seen in the RT-qPCR assay is replicated using denaturing gel analysis.
(A) PPIG is shown to inhibit splicing of HMS388 splicing substrate. (B) CWC27 is also shown to be inhibitory. Panels are labelled as in Figure 1. Panel (C) shows quantification of splicing efficiency for PPIA, PPIE, PPIG, CWC27, PPIL2 and PPIH using denaturing gel analysis. Values and associated error are calculated by averaging the data from multiple lanes on acrylamide gels.
Nuclear cyclophilins affect in vitro splicing efficiency on a variety of pre-mRNA substrates
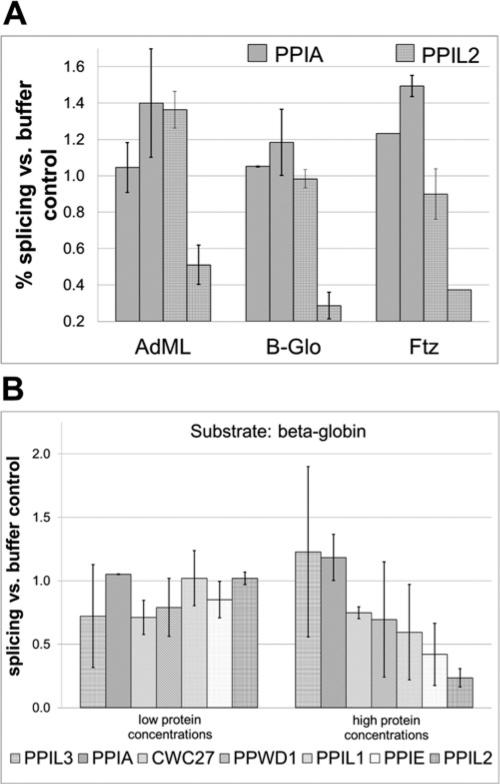
We do not know what cellular pre-mRNA substrates are the actual targets of nuclear cyclophilin regulation in vivo. It is possible that the AdML splicing substrate used in our in vitro experiments does not reflect a general function of these proteins in pre-mRNA splicing. In an effort to examine this possibility and to see if the effects on splicing that we observe are substrate specific, we examined the effect of cyclophilins on splicing of substrates derived from the human β-globin and ftz sequences [35]. Based on denaturing gel analysis of in vitro splicing, we quantified the effects of PPIL2 and PPIA on splicing efficiency using AdML, β-globin and ftz substrates (Figure 4A). We find that PPIL2 is strongly inhibitory on all three substrates and that PPIA is not (and may even stimulate splicing at higher concentrations of protein, perhaps via mass action). We note that other modulators of in vitro splicing using the β-globin substrate show a range of behaviours. For instance, lower concentrations of added protein may actually improve splicing efficiency; this is seen, for instance, for serine-rich (SR) proteins, although at higher concentrations squelching mechanisms cause inhibition (Melissa Jurica, personal communication). By contrast, we did not see statistically significant enhancement of splicing efficiency upon the β-globin substrate by our nuclear cyclophilins. On the other hand, we observe appreciable inhibition of the splicing efficiency on the β-globin substrate even for cyclophilins that do not effect splicing on AdML (PPWD1, PPIL1 and PPIE; Figure 4B). From these data, we can say that modulation of splicing efficiency by nuclear cyclophilins is not isolated to one substrate and that pre-mRNA sequence can influence sensitivity to different cyclophilins.
Figure 4. The nuclear cyclophilins inhibit splicing of multiple pre-mRNA substrates.
(A) A low and a high concentration of PPIA and PPIL2 are tested against the AdML, β-globin and ftz substrates. Results are quantified from denaturing gel analysis and error calculated as the S.D. between lanes on the gel. (B) Results for six nuclear cyclophilins and the control PPIA are quantified for the β-globin substrate. In general, the nuclear cyclophilins characterized as inhibitory using the AdML substrate are also inhibitory against β-globin. PPIL3 and PPIA are slightly stimulatory and PPWD1 and PPIL1 are inhibitory against β-globin.
The in vitro effect of the nuclear cyclophilins, while mediated through the isomerase domain, is largely unaffected by isomerization activity
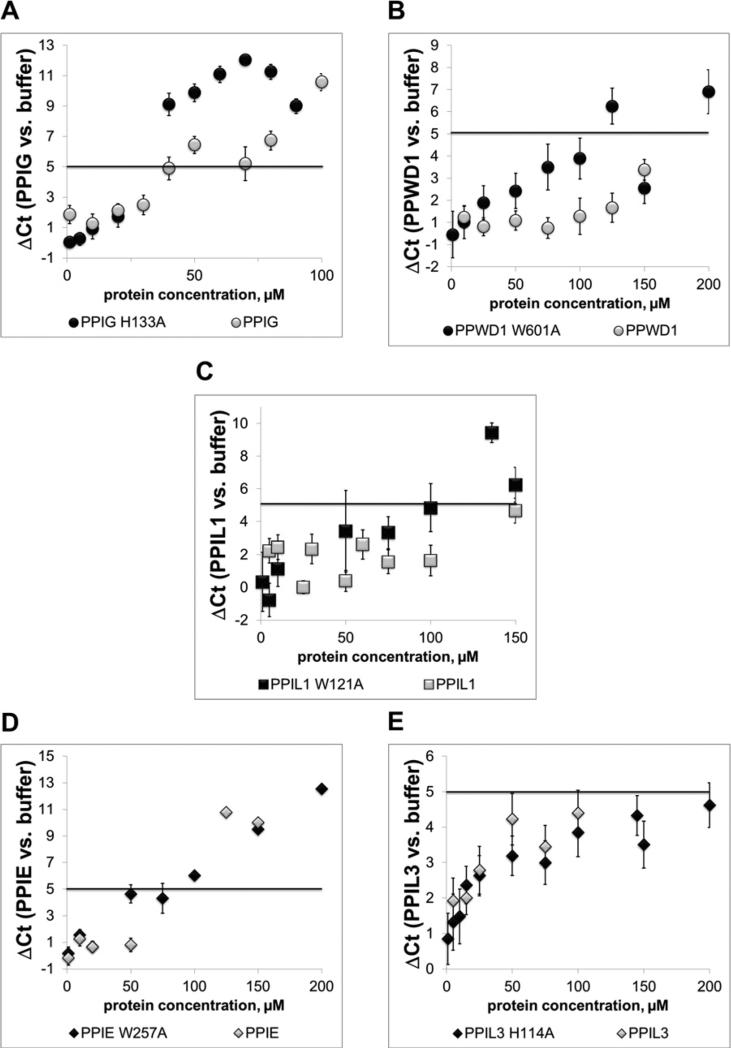
For the multi-domain nuclear cyclophilins PPIE and PPIL2 we used the RT-qPCR assay to compare the activity of full-length protein with isolated domains. For PPIL2, we found that both the isolated PPI and the U-box motifs had a smaller effect than full-length PPIL2, although both isolated domains altered splicing efficiency (Supplementary Figure S2A). In contrast, the isolated PPI domain of PPIE had slightly stronger effects than either full-length or isolated RRM motif (Supplementary Figure S2B). For all other nuclear cyclophilins, the results we measure in our assays are tied to the presence of only the isomerase domain, either because that is the only domain present in the gene (PPIH, PPIL1, PPIL3) or because the isomerase domain is the only soluble version of the cyclophilin we have expressed in vitro (PPWD1, PPIG, CWC27). The simplest explanation of the function of the nuclear cyclophilins in splicing would invoke their ability to isomerize proline. To test this, we altered a subset of cyclophilin proteins by mutating the isomerase active site and examined the impact of these changes on in vitro splicing. Six of the eight nuclear cyclophilins are capable of catalysing the cis/trans isomerization of proline residues on any number of synthetic peptide sequences; the exceptions being PPIL2 and CWC27, both of which have naturally-occurring substitutions in their proline-binding pockets that abrogate this activity [15–17]. For isomerase-active cyclophilins, mutation of any one of several residues in the proline-binding pocket inactivates the enzyme [23,39]. The two cyclophilins that most potently inhibit splicing efficiency in our experiments, PPIL2 and CWC27, cannot be tested with these mutations. For the remaining five spliceosomal cyclophilins (PPIG, PPIL3, PPIE, PPIL1 and PPWD1) we mutated a key active site residue (a tryptophan or histidine analogous to Trp121 in PPIA) and tested its effect on AdML splicing, using RT-qPCR to measure splicing efficiency. Mutation of the active site of PPIG, which was the most potent wild-type protein we could test in this way, increased the inhibitory effect on AdML substrate (Figure 5A). Mutation of the previously non-inhibitory PPWD1 also significantly decreases splicing catalytic efficiency (Figure 5B). Mutation of PPIL1 led to a modest decrease in splicing efficiency (Figure 5C). For PPIE and PPIL3 we see no gain of function by mutation of the active site residue (Figures 5C–5E). These results suggest a limited role for proline isomerization in in vitro splicing, although once again we see a variety of phenotypes indicating that the role of isomerization in the context of each nuclear cyclophilin may be different.
Figure 5. The isomerase activity of nuclear cyclophilins is not necessary to detect splicing inhibition.
For cyclophilins that are active isomerases against proline-containing substrates, point mutations were made at the position corresponding to Trp121 in PPIA. This mutation should abrogate proline turnover and cyclosporine binding [39]. In panels (A) and (B), it can be seen that mutation of PPIG or PPWD1 increases splicing inhibition. In panel (C), the modest effect of mutating PPIL1 is seen. In (D and E), mutation of the inhibitory PPIE PPI domain or of PPIL3 has no effect on splicing inhibition.
Spliceosome-associated cyclophilins differentially affect spliceosome assembly in vitro
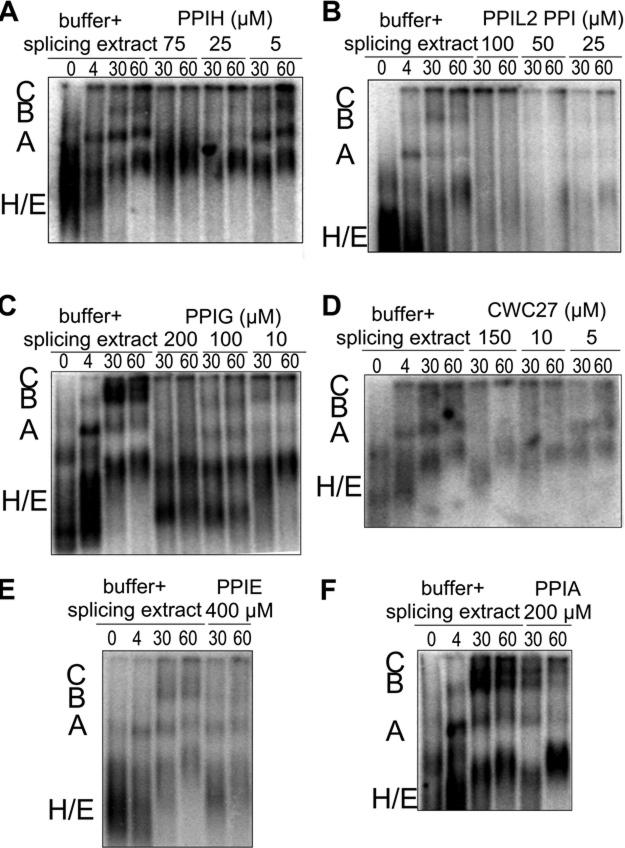
We have focused thus far only on the effect of adding exogenous protein on splicing catalysis in vitro. It is possible that the proteins are not directly affecting catalysis, but instead interfere with spliceosome complex assembly on the pre-mRNA substrate. To investigate this possibility, we carried out native gel analysis of in vitro splicing reactions in the presence or absence of PPIH, PPIL2, PPIG or CWC27 (Figure 6). We also examined higher concentrations of PPIE or PPIA as controls.
Figure 6. The effect of cyclophilins on spliceosome assembly visualized by native agarose gel analysis of in vitro splicing reactions.
In each panel, the first four lanes show a time course of splicing reactions with buffer and nuclear extract, along with the radiolabelled AdML substrate. The relative positions of E/H, A, B and C complexes are indicated to the left of each gel. Subsequent lanes show 30 and 60 min time-points of splicing reactions containing the indicated concentrations of PPIH (A), PPIL2 (B), PPIG (C), CWC27 (D), PPIE (E) or PPIA (F).
When buffer alone is added to the reactions, we see the characteristic appearance of higher mobility bands over time, which have been previously annotated as A, B and C intermediate splicing complexes [40]. We find that adding cyclophilin protein to assembling spliceosomes has an effect consistent with their impact on splicing chemistry. PPIL2, PPIG, PPIH and CWC27 all appear to interfere with spliceosome assembly at concentrations where they inhibit splicing chemistry (Figures 6A–6D) and neither PPIA nor PPIE significantly affected high-order complex formation (Figures 6E and 6F). These effects are dose-dependent, since adding lower amounts of inhibitory protein allow for higher-order complex formation to occur over time. The effects on assembly appear different for the different cyclophilins. For instance, PPIG addition seems to exert stronger effects on C complex, which does not form in any of our test cases as compared with buffer control. By comparison, PPIH and PPIL2 results could be interpreted as slowing assembly at an earlier step, as the formation of both A and B complexes are strongly inhibited at all concentrations tested. However, as this is a strictly qualitative measure of spliceosomal assembly in vitro, caution must be used in interpreting these gels with regard to mechanism. However, we do conclude that there is a correlation between inhibition of splicing catalytic efficiency in vitro and the ability of the spliceosome to assemble efficiently upon the AdML pre-mRNA substrate.
DISCUSSION
In the present study, we provide the first comprehensive picture of the eight nuclear cyclophilins and their impact on spliceosome assembly and function in vitro. Until recently, the roles of cyclophilins in the cell have been largely uncharacterized. For the nuclear cyclophilins (PPIE, PPIG, PPIH, PPIL1, PPIL2, PPIL3, PPWD1 and CWC27) almost nothing has been reported as to their cellular functions beyond their association with mammalian spliceosomes in multiple proteomics studies [4,5,8–11,24,26–29,41,42]. Our data indicate that cyclophilins can differentially affect the ability of spliceosomes to assemble and function properly in vitro, indicating that they are capable of affecting these processes in vivo as well.
For the eight nuclear cyclophilins, we found that four had significant effects on splicing chemistry and assembly in multiple assay systems. Addition of PPIL2, CWC27, PPIG and PPIH all clearly blocked splicing chemistry in a concentration-dependent manner, especially in comparison with the other four nuclear cyclophilins and the cytoplasmic cyclophilin PPIA (Figures 1–3). These effects were dependent on the concentration of protein used, indicating a specific effect for each cyclophilin within the context of the spliceosomal machinery. Inhibitory effects were seen on multiple splicing substrates, implying a general mechanism of action (Figure 4). We note that, as in the case of other splicing factors, the inhibition of splicing in vitro does not imply a particular mechanism in vivo. It is quite possible that the nuclear cyclophilins are acting to enhance splicing efficiency in cells and experiments are underway in our laboratory to investigate that possibility. However, the fact that we see effects on splicing efficiency, whether positive or negative in vitro, indicate a role for these proteins in regulating splicing efficiency in vivo. Four of the nuclear cyclophilins also exerted specific effects on spliceosomal assembly in a native gel experiment, linking their inhibition of catalytic efficiency to the ability of the assembling spliceosome to form higher-order complexes (Figure 6).
It is interesting that our most potent regulators are so dissimilar in their domain structure and in their presumed functional role within the spliceosome. For instance, their integration into the spliceosome follows unique patterns. PPIH joins spliceosomes as part of tri-snRNP and is the earliest cyclophilin to integrate into the complex. PPIL2 and CWC27 integrate into complexes later in the splicing cycle and are most highly detected in the late intermediate Bact-complex [5]. PPIL2 is strongly detected only in Bact and is not appreciably present in C-complex, a very specific pattern that implies a particular role in the transition from intermediate to catalytically-active complex. Finally, PPIG is found only as part of catalytic spliceosome complexes. These integration patterns are replicated to some degree by the results of our native gel analysis, which indicate that adding purified PPIH or PPIL2 seems to affect spliceosomal assembly at an earlier stage than PPIG (Figure 6).
PPIH is a minimal cyclophilin, coding only for the core PPI domain [15,20,43,44]. On the other hand, PPIL2, CWC27 and PPIG are all multi-domain cyclophilins, with an additional U-box motif, region of low complexity and SR-motif respectively [44–46]. The roles of these additional sequences is not known; we find for PPIL2 that the inhibitory effect we see is due both to the isolated U-box and to the PPI domains, suggesting a role for both domains in the function of PPIL2 in splicing and certainly the SR-motif is well known as a modulator of splicing efficiency. For CWC27 and PPIG we have soluble expression constructs only for the isomerase domain, so we can say that the isomerase domain is minimally capable of interfering with splicing in vitro. PPIL2 and CWC27 have substitutions in the isomerase domain that render them incapable of isomerizing proline residues; and we find that mutation of the PPIG-active site actually makes the isomerase domain a slightly more effective inhibitor in our in vitro assays (Figure 5). The inhibitory activity of wild-type PPIH and the modest effects of active-site mutation on the inhibition measured using PPIG, PPWD1 and PPIL1, imply that the function of the isomerase-active site is neither necessary nor sufficient for regulating in vitro splicing catalysis. Indeed, previously published data and unpublished results from our own laboratory indicate that the binding of several nuclear cyclophilins to their specific protein-binding partners within the spliceosome is due to residues outside the isomerase active site (Tara L. Davis and S. RaElle Jackson, unpublished data) [19,47]. Previously it has been shown that addition of the macrocyclic inhibitor cyclosporine A to in vitro reactions has a surprisingly limited effect on splicing activity, considering that six spliceosome-associated cyclophilins ligate this inhibitor with nanomolar affinity [48]. Cyclosporine A binds to the nuclear cyclophilins precisely within the proline-binding pocket of the active site, so it is a reasonable assumption that if isomerase activity were the main functional role of the nuclear cyclophilins, stronger effects on splicing would have been observed when adding this inhibitor to cells. Taken together, these data imply that many members of the nuclear cyclophilin family play cyclophilin-specific roles and/or have regulatory functions that minimally involve the proline-binding surface of the protein.
It is interesting, based on these data, to speculate on the potential mode of action of the cyclophilins within the spliceosome. There are structures of complexes between PPIH and a peptide derived from the spliceosomal factor PRPF4 and between PPIL1 and the spliceosomal protein SNW1 [19,21,47]; in both cases, surfaces outside the cyclophilin-active site are engaged in the interaction. Tight interaction between spliceosomal proteins and nuclear cyclophilins may help to anchor proteins to each other in the midst of the dynamic conformational rearrangements that occur at the RNA and protein level during the splice cycle, in order to keep them oriented productively within the ever-changing splicing machinery. Alternatively, changing the relative amounts of proteins available to form spliceosomal complexes can effect catalytic efficiency (and, indirectly, alternative splice choice); perhaps cyclophilins and other highly-ordered protein–protein interaction domains within the spliceosome act to ensure proper stoichiometry and/or prevent off-target interactions of the large numbers of disordered proteins of the spliceosome [33]. Either model could explain why proline isomerase activity does not affect splicing assembly and catalytic efficiency in vitro. It is also possible that the regulation of protein conformation by proline isomerization regulates splicing outside of the processes of spliceosome assembly and catalysis, but that this involves transient interactions with proteins not intimately associated with the spliceosome. For instance, it is possible that isomerization of an SR-containing protein or their regulatory kinases (SRPKs), might greatly affect splicing efficiency. These effects would be subtle at best in vitro and would perhaps not be seen at all in our isomerase inactive assays. Efforts are underway in our laboratory to characterize physiologically-relevant splicing phenotypes induced by cyclophilin knockdown in human cells, in order to capture potential isomerase-dependent effects of the nuclear cyclophilins on splicing.
In summary, we see in the current study that effects of the cyclophilins on splicing and spliceosome assembly are not general, but vary significantly depending upon the identity of the cyclophilin examined. These data support the hypothesis that the nuclear cyclophilins perform important and unique functions in the spliceosome. The next steps will be to specify those functions and their molecular mechanisms and ultimately place them into a cellular context. Given the complexity of multi-intron pre-mRNAs and alternative splicing in higher eukaryotes, this will not be an easy task. One path for experimentation may be found by exploiting small molecule inhibitors for the cyclophilins. Six of the nuclear cyclophilins bind cyclosporine A with nanomolar affinity and there are additional tight-binding inhibitors with alternate scaffolds available [15,22,49]. Furthermore, the availability of high-quality crystal structures for the majority of spliceosomal cyclophilins allows for structural-based screening of new ligands. Such molecules may serve to specifically modulate the function of the cyclophilins and thereby the spliceosome. Our work in the present study indicates that this conserved enzyme family plays an intriguing role in the regulation of pre-mRNA splicing.
Supplementary Material
ACKNOWLEDGEMENTS
Most of the original expression constructs utilized in this manuscript originated from the Structural Genomics Consortium in Toronto, Ontario, Canada (www.thesgc.org). Walter Bray and Kerstin Effenberger, both of UCSC, developed and optimized the RT-qPCR assay for splicing studies. Zane Barrett, of UCSC, assisted Melissa Jurica with denaturing gel analysis of splicing reactions not included in the current manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of General Medical Sciences, National Cancer Institute or the National Institutes of Health.
FUNDING
This was supported by the National Institutes of General Medical Sciences [grant numbers K99GM094293] (to T.L.D.) and R01GM72649](to M.S.J.)]; and the National Cancer Institute [grant number R01CA136762 (to M.S.J.)].
Abbreviations
- AdML
adenovirus major-late
- ftz
fushi tarazu
- LIC
ligation-independent cloning
- PPI
peptidyl-prolyl isomerase
- PRPF4
pre-mRNA processing factor 4
- qPCR
quantitative PCR
- RRM
RNA-recognition motif
- RT
reverse transcriptase
- snRNP
small nuclear RNA–protein complex
Footnotes
AUTHOR CONTRIBUTION
Melissa Jurica and Tara Davis conceived of the concepts and designed the flow of experiments. B. Adams, Miranda Coates, RaElle Jackson and Tara Davis expressed and purified cyclophilin proteins and performed RT-qPCR splicing experiments and B. Adams and Tara Davis analysed the data from these experiments. Melissa Jurica and Tara Davis performed denaturing gel analysis of splicing reactions. Tara Davis performed native gel analysis of splicing reactions. Melissa Jurica and Tara Davis wrote the paper with input and editing from the other authors.
REFERENCES
- 1.Valadkhan S, Jaladat Y. The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics. 2010;10:4128–4141. doi: 10.1002/pmic.201000354. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 3.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a0037074. doi: 10.1101/cshperspect.a003707. CrossRef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agafonov DE, Deckert J, Wolf E, Odenwalder P, Bessonov S, Will CL, Urlaub H, Luhrmann R. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell. Biol. 2011;13:2667–2682. doi: 10.1128/MCB.05266-11. CrossRef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, Lührmann R. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Lührmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16719–16719. doi: 10.1073/pnas.262483899. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Luhrmann R. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA. 2010;16:2384–2403. doi: 10.1261/rna.2456210. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 11.Makarov EM. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 12.Thai V, Renesto P, Fowler CA, Brown DJ, Davis T, Gu W, Pollock DD, Kern D, Raoult D, Eisenmesser EZ. Structural, biochemical, and in vivo characterization of the first virally encoded cyclophilin from the mimivirus. J. Mol. Biol. 2008;378:71–86. doi: 10.1016/j.jmb.2007.08.051. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6:226–226. doi: 10.1186/gb-2005-6-7-226. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galat A. Peptidylproline cis-trans-isomerases: immunophilins. Eur. J. Biochem. 1993;216:689–707. doi: 10.1111/j.1432-1033.1993.tb18189.x. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 15.Davis TL, Walker JR, Campagna-Slater V, Finerty PJ, Paramanathan R, Bernstein G, MacKenzie F, Tempel W, Ouyang H, Lee WH, et al. Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol. 2010;8:e1000439. doi: 10.1371/journal.pbio.1000439. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison RK, Stein RL. Substrate specificities of the peptidyl prolyl cis-trans isomerase activities of cyclophilin and FK-506 binding protein: evidence for the existence of a family of distinct enzymes. Biochemistry. 1990;29:3813–3816. doi: 10.1021/bi00468a001. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 17.Piotukh K. Cyclophilin a binds to linear peptide motifs containing a consensus that is present in many human proteins. J. Biol. Chem. 2005;280:23668–23674. doi: 10.1074/jbc.M503405200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 18.Davis TL, Walker JR, Ouyang H, MacKenzie F, Butler-Cole C, Newman EM, Eisenmesser EZ, Dhe-Paganon S. The crystal structure of human WD40 repeat-containing peptidylprolyl isomerase (PPWD1). FEBS J. 2008;275:2283–2295. doi: 10.1111/j.1742-4658.2008.06381.x. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 19.Reidt U, Wahl MC, Fasshauer D, Horowitz DS, Lührmann R, Ficner R. Crystal structure of a complex between human spliceosomal cyclophilin H and a U4/U6 snRNP-60K peptide. J. Mol. Biol. 2003;331:45–56. doi: 10.1016/s0022-2836(03)00684-3. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 20.Teigelkamp S, Achsel T, Mundt C, Göthel SF, Cronshagen U, Lane WS, Marahiel M, Lührmann R. The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA. 1998;4:127–141. PubMed. [PMC free article] [PubMed] [Google Scholar]
- 21.Stegmann CM, Luhrmann R, Wahl MC. The crystal structure of PPIL1 bound to cyclosporine A suggests a binding mode for a linear epitope of the SKIP protein. PLoS One. 2010;5:e10013. doi: 10.1371/journal.pone.0010013. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daum S, Schumann M, Mathea S, Aumüller T, Balsley MA, Constant SL, de Lacroix BFA, Kruska F, Braun M, Schiene-Fischer C. Isoform-specific inhibition of cyclophilins. Biochemistry. 2009;48:6268–6277. doi: 10.1021/bi9007287. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, Walsh CT. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y-IG, Moore RE, Ge HY, Young MK, Lee TD, Stevens SW. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilagan J, Yuh P, Chalkley RJ, Burlingame AL, Jurica MS. The role of exon sequences in C complex spliceosome structure. J. Mol. Biol. 2009;394:363–375. doi: 10.1016/j.jmb.2009.09.019. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng R, Hawkins I, Link AJ, Patton JG. The splicing factor PSF is part of a large complex that assembles in the absence of pre-mRNA and contains all five snRNPs. RNA Biol. 2006;3:69–76. doi: 10.4161/rna.3.2.3017. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 28.Rappsilber J. Large-Scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 30.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Luhrmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 31.Kuraoka I, Ito S, Wada T, Hayashida M, Lee L, Saijo M, Nakatsu Y, Matsumoto M, Matsunaga T, Handa H, et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J. Biol. Chem. 2007;283:940–950. doi: 10.1074/jbc.M706647200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 32.Kofler M, Schuemann M, Merz C, Kosslick D, Schlundt A, Tannert A, Schaefer M, Lührmann R, Krause E, Freund C. Proline-rich sequence recognition. Mol. Cell. Proteomics. 2009;8:2461–2473. doi: 10.1074/mcp.M900191-MCP200. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korneta I, Bujnicki JM. Intrinsic disorder in the human spliceosomal proteome. PLoS Comput. Biol. 2012;8:e1002641. doi: 10.1371/journal.pcbi.1002641. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–2778. doi: 10.1101/gad.1030602. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 36.Effenberger KA, Perriman RJ, Bray WM, Lokey RS, Ares M, Jr, Jurica MS. A high-throughput splicing assay identifies new classes of inhibitors of human and yeast spliceosomes. J. Biomol. Screen. 2013;18:1110–1120. doi: 10.1177/1087057113493117. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou D, Mei Q, Li J, He H. Cyclophilin A and viral infections. Biochem. Biophys. Res. Commun. 2012;424:647–650. doi: 10.1016/j.bbrc.2012.07.024. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Chen CM, Walsh CT. Human and Escherichia coli cyclophilins: sensitivity to inhibition by the immunosuppressant cyclosporin A correlates with a specific tryptophan residue. Biochemistry. 1991;30:2306–2310. doi: 10.1021/bi00223a003. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 40.Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 2010;38:6664–6672. doi: 10.1093/nar/gkq494. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behzadnia N. Functional spliceosomal A complexes can be assembled in vitro in the absence of a penta-snRNP. RNA. 2006;12:1738–1746. doi: 10.1261/rna.120606. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 2008;29:281–301. doi: 10.1128/MCB.01415-08. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingelfinger D. Two protein-protein interaction sites on the spliceosome-associated human cyclophilin CypH. Nucleic Acids Res. 2003;31:4791–4796. doi: 10.1093/nar/gkg660. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 45.Cavarec L, Kamphausen T, Dubourg B, Callebaut I, Lemeunier F, Metivier D, Feunteun J, Fischer G, Modjtahedi N. Identification and characterization of Moca-cyp. A Drosophila melanogaster nuclear cyclophilin. J. Biol. Chem. 2002;277:41171–41182. doi: 10.1074/jbc.M203757200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 46.Scanlan MJ, Welt S, Gordon CM, Chen YT, Gure AO, Stockert E, Jungbluth AA, Ritter G, Jager D, Jager E, et al. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62:4041–4047. PubMed. [PubMed] [Google Scholar]
- 47.Xu C. Solution structure of human peptidyl prolyl isomerase-like Protein 1 and insights into its interaction with SKIP. J. Biol. Chem. 2006;281:15900–15908. doi: 10.1074/jbc.M511155200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 48.Horowitz DS, Lee EJ, Mabon SA, Misteli T. A cyclophilin functions in pre-mRNA splicing. EMBO J. 2002;21:470–480. doi: 10.1093/emboj/21.3.470. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zenke G, Strittmatter U, Fuchs S, Quesniaux VFJ, Brinkmann V, Schuler W, Zurini M, Enz A, Billich A, Sanglier JJ, et al. Sanglifehrin A, a novel cyclophilin-binding compound showing immunosuppressive activity with a new mechanism of action. J. Immunol. 2001;166:7165–7165. doi: 10.4049/jimmunol.166.12.7165. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.