Summary
Bariatric surgery is currently the most effective procedure for the treatment of obesity. Given the role of the gut microbiota in regulating host metabolism and adiposity, we investigated the long-term effects of bariatric surgery on the microbiome of patients randomized to Roux-en-Y gastric bypass or vertical banded gastroplasty and matched for weight and fat mass loss. The two surgical procedures induced similar and durable changes on the gut microbiome that were not dependent on body mass index and resulted in altered levels of fecal and circulating metabolites compared with obese controls. By colonizing germ-free mice with stools from the patients, we demonstrated that the surgically altered microbiota promoted reduced fat deposition in recipient mice. These mice also had a lower respiratory quotient, indicating decreased utilization of carbohydrates as fuel. Our results suggest that the gut microbiota may play a direct role in the reduction of adiposity observed after bariatric surgery.
Graphical Abstract
Highlights
-
•
RYGB and VBG induce long-term alterations in the human gut microbiome
-
•
The changes in the microbiome do not depend on BMI
-
•
RYGB and VBG have different effects on bile acid and TMAO metabolism
-
•
The surgically altered microbiome contributes to fat mass regulation
Bariatric surgery durably alters the human gut microbiome. Here, Tremaroli et al. demonstrate that two types of bariatric surgery, Roux-en-Y gastric bypass and vertical banded gastroplasty, produce long-term alterations of the gut microbiome independently of BMI and that these alterations modulate host metabolism and fat mass deposition.
Introduction
The prevalence of obesity and its metabolic comorbidities, such as type 2 diabetes and cardiovascular disease, is on the rise worldwide. Although a number of genetic and environmental factors have been identified, the mechanisms responsible for excessive fat mass accumulation and development of obesity are not fully understood. Recent research shows that our intestinal microbes contribute to the regulation of energy homeostasis and fat storage and may play a role in obesity and its complications (Karlsson et al., 2013; Qin et al., 2012; Smith et al., 2013; Tremaroli and Bäckhed, 2012). Indeed, obesity has been associated with altered gut microbiota composition, reduced microbial diversity, and reduced gene richness (Le Chatelier et al., 2013; Ley et al., 2005; Turnbaugh et al., 2009). In addition, dietary weight loss interventions have been shown to increase gene richness of the microbiota and shift its composition toward that of lean individuals (Cotillard et al., 2013; Ley et al., 2006).
Bariatric surgery is currently the best treatment for sustained weight loss and reduction of obesity-related comorbidities (Sjöström et al., 2007, 2012). There are a number of bariatric surgery procedures, of which Roux-en-Y gastric bypass (RYGB) is considered one of the most effective, promoting both weight reduction and improvement of metabolism (Sjöström et al., 2007; Werling et al., 2013a). The underlying mechanisms have not been fully elucidated, but reduced food ingestion, changes in food preferences, increased satiety, release of satiety-promoting gut hormones (e.g., glucagon-like peptide 1 [GLP-1] and peptide YY [PYY]), increased gastric emptying, and a shift in bile acid metabolism together with an increased signaling through the bile acid nuclear receptor farnesoid X receptor (FXR) have all been suggested to play a role (le Roux and Bloom, 2005; Miras and le Roux, 2013; Ryan et al., 2014).
Recent work has also indicated that the gut microbiota may mediate some of the beneficial effects of bariatric surgery, and changes in the composition and diversity of the gut microbiota have been observed in the short term after RYGB in humans (Furet et al., 2010; Graessler et al., 2013; Kong et al., 2013; Zhang et al., 2009), as well as after vertical sleeve gastrectomy and RYGB in mice (Liou et al., 2013; Ryan et al., 2014). However, there are no long-term investigations of the stability of these changes or of the effects of different procedures on the human gut microbiota. Additionally, the causal link between the alterations to the microbiota and the reduction in adiposity observed after bariatric surgery has not been previously analyzed in humans.
Here we used shotgun sequencing of the human fecal metagenome to analyze the gut microbiota of weight-stable women 9 years after randomization to either RYGB or vertical banded gastroplasty (VBG) (Olbers et al., 2006) and matched for weight and fat mass loss. As a reference, we also analyzed the gut microbiota of two groups of non-operated women with body mass index (BMI) matched to the patients’ pre-surgical BMI (OBS) and post-surgical BMI (Ob). We aimed to identify whether alterations in the gut microbiota observed previously in the short term remained stable over time and if RYGB and VBG induced specific alterations in the gut microbiome. Moreover, we investigated the causal link between the surgically altered human gut microbiota and adiposity phenotype by performing microbiota transplantations of human stools in germ-free (GF) mice.
Results and Discussion
Bariatric Surgery has Long-Term Effects on Gut Microbiota Composition
To investigate the long-term effects of bariatric surgery on the gut microbiota, we obtained fecal samples from 14 women 9.4 years after they were randomized to RYGB (n = 7) or VBG (n = 7) (Olbers et al., 2006). These women were included in a recent study of the mechanisms underlying the maintenance of long-term weight loss (Werling et al., 2013b). RYGB and VBG patients were matched for pre- and post-operative weight, BMI, and body composition, and had thus experienced similar weight and fat mass loss. We also included fecal samples from seven non-operated women with severe obesity (OBS) and a BMI similar to the mean pre-surgical BMI of RYGB and VBG patients. Clinical and biometric measurements of these three groups of women are summarized in Table S1.
Genomic DNA was extracted from fecal samples using a repeated bead beating procedure (Salonen et al., 2010) and sequenced on Illumina HiSeq 2000. In total, we obtained 63 Gbp of paired-end reads, with an average of 3.0 ± 3.0 (SD) Gbp for each sample (Table S2). To determine the composition of the gut microbiota, we aligned high-quality Illumina reads devoid of human DNA to 2,382 reference genomes obtained from the National Center for Biological Information (NCBI) and the Human Microbiome Project (HMP) databases (http://hmpdacc.org) (Table S2) using methods from our MEDUSA platform (Karlsson et al., 2012; Karlsson et al., 2014).
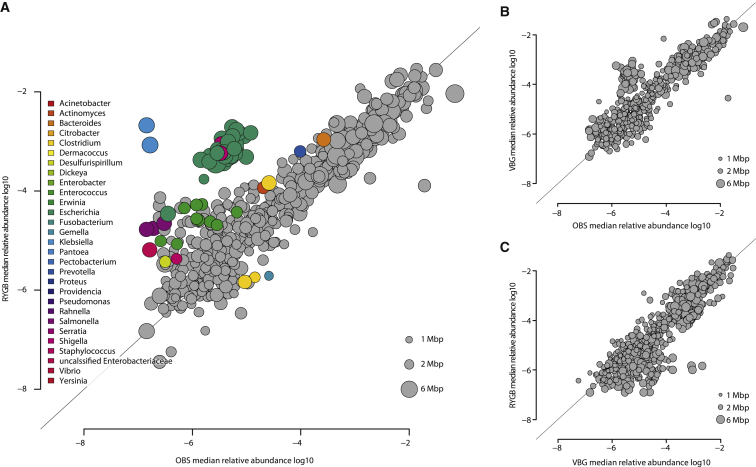
We observed significant differences in microbiota composition for RYGB versus OBS samples, but not for VBG versus OBS or RYGB versus VBG (Figure 1; Table S3). The abundance of species belonging to 29 microbial genera differed significantly between RYGB and OBS samples (Figure 1A; Table S3); in particular, the levels of several species in the Gammaproteobacteria class were higher while the levels of three species in the Firmicutes phylum (Clostridium difficile, Clostridium hiranonis, and Gemella sanguinis) were lower in RYGB versus OBS samples (Table S3). At the genus level, several facultative anaerobes in the Proteobacteria (e.g., Escherichia, Klebsiella, and Pseudomonas) were present at increased relative abundance in RYGB (Table S3). We observed a different relative abundance for several species when comparing VBG with OBS samples, and RYGB with VBG samples, although the differences were not significant after correction for multiple testing (Table S3). E. coli tended to increase, while Eubacterium rectale and Roseburia intestinalis tended to decrease in VBG compared to OBS (Table S3). Klebsiella species tended to increase, while Bifidobacterium species tended to decrease in RYGB in comparison to VBG (Table S3).
Figure 1.
Long-term Effects of Bariatric Surgery on Gut Microbiota Composition
(A) Scatter plot of median species abundance in RYGB patients and severely obese subjects (OBS).
(B) Scatter plot of median species abundance in VBG patients and OBS subjects.
(C) Scatter plot of median species abundance in RYGB and VBG patients.
The genus affiliations of differentially abundant species are indicated by color (Adj. p < 0.05, Wilcoxon rank-sum test). See also Table S3.
These analyses demonstrated that women who have undergone RYGB had increased abundance of Proteobacteria in comparison to non-operated severely obese women. Proteobacteria are generally not considered to be beneficial due to their proinflammatory properties (Amar et al., 2011; Cani et al., 2007; Mukhopadhya et al., 2012). However, the RYGB and VBG patients in our study were not under inflammatory stress as determined by plasma CRP levels (Table S1). Moreover, our results are in agreement with previous findings showing an expansion of Proteobacteria shortly after RYGB, both in humans (Furet et al., 2010; Graessler et al., 2013; Kong et al., 2013; Zhang et al., 2009) and mice (Liou et al., 2013). Additionally, despite the obvious differences between the two surgical procedures (Olbers et al., 2006), we did not observe distinct microbiota profiles for RYGB and VBG patients. Although this result could be due to the small sample size of our cohort, it is also possible that other mechanisms common to the two surgical procedures might affect the composition of the gut microbiota. For example, increases in pH have been shown to promote the growth of E. coli in vitro (Duncan et al., 2009), and both RYGB and VBG are associated with reduced luminal exposure to gastric acidity.
Bariatric Surgery Has Long-Term Effects on the Genetic Content of the Gut Microbiota
To explore the genetic content of the gut microbiome after bariatric surgery, we aligned our reads to the MetaHIT human gut microbial gene catalog (Qin et al., 2010). We calculated the abundance of KEGG orthologs (KOs), and analyzed the abundance of KEGG pathways using the reporter features algorithm to obtain a global view of the changes (Oliveira et al., 2008; Patil and Nielsen, 2005).
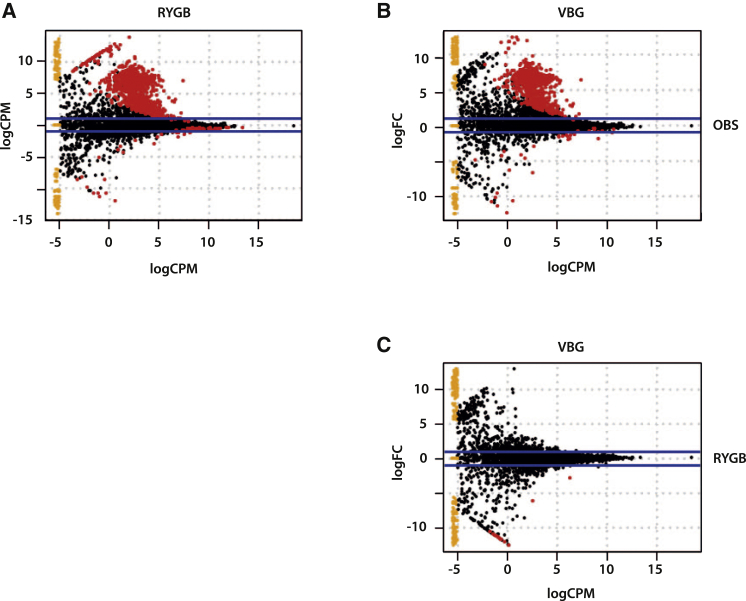
We observed differences in the microbiome of operated and non-operated severely obese women (Figure 2; Table S4): 928 KOs were enriched and 60 were depleted in RYGB versus OBS samples (Figure 2A; Table S4), while 682 KOs were enriched and 33 were depleted in VBG versus OBS samples (Figure 2B; Table S4). However, only 17 KOs were differentially abundant between RYGB and VBG samples, all enriched in VBG (Figure 2C; Table S4).
Figure 2.
Functional Differences in the Microbiomes of RYGB, VBG and OBS Subjects as Measured by the Abundance of KOs
(A) Comparison of RYGB versus OBS samples.
(B) Comparison of VBG versus OBS samples.
(C) Comparison of RYGB versus VBG samples.
Each spot represents a KO, and red spots represent KOs whose abundance is significantly different. FC, fold change; CPM, counts per million. See also Table S4 and Figures S1 and S2.
The reporter pathways for phosphotransferase systems and fluorobenzoate degradation were enriched in RYGB versus OBS microbiomes, and the reporter pathways for flagellar assembly, sulfur relay system, glutathione metabolism, glyoxylate and dicarboxylate metabolism, ABC transporters (in particular for lysine/arginine/ornithine, histidine, and putrescine), and phenylalanine metabolism were enriched in VBG versus OBS samples (Table 1; Table S4). In addition, the reporter pathways for nitrogen metabolism, fatty acid metabolism, and two-component systems were enriched both in RYGB versus OBS and VBG versus OBS microbiomes (Tables 1 and S4).
Table 1.
KEGG Pathways Enriched in the Gut Microbiomes of RYGB and VBG Patients in Comparison to OBS Women
| RYGB Enriched Compared to OBS | VBG Enriched Compared to OBS |
|---|---|
| – | Flagellar assembly |
| – | Sulfur relay system |
| – | Glutathione metabolism |
| – | Glyoxylate and dicarboxylate metabolism |
| – | ABC transporters |
| – | Phenylalanine metabolism |
| Nitrogen metabolism | Nitrogen metabolism |
| Fatty acid metabolism | Fatty acid metabolism |
| Two-component system | Two-component system |
| Phosphotransferase system | – |
| Fluorobenzoate degradation | – |
Two-component systems are sensory systems that enable bacteria to sense and respond to changes in the surrounding environment (Mascher et al., 2006). We found that the two-component systems responding to nitrogen availability (nifA), phosphoglycerate transport (pgtCAP), and short-chain fatty acid (SCFA) metabolism (acetoacetate, atoCE) were specifically enriched in RYGB compared to OBS, while salt stress response and twitching motility were specifically enriched in VBG compared to OBS (Table S4). Two-component system genes for response to antimicrobial peptides, outer membrane stress, and trimethylamine N-oxide (TMAO) were enriched in both RYGB and VBG compared to OBS microbiomes. Furthermore, the gene coding for TMAO reductase (torA, K07811), which is involved in the respiration of TMAO to TMA, was also enriched in both RYGB and VBG compared to OBS microbiomes (Table S4).
The increase in tor genes likely reflects the high levels of E. coli in both RYGB and VBG samples, as E. coli can use TMAO as a terminal electron acceptor (Barrett and Kwan, 1985). However, we observed increased plasma concentrations of TMAO only in RYGB and not in VBG women (Figure S1A); the levels of choline and betaine, known dietary precursors for TMA production by the gut microbiota, did not differ between the groups (Figures S1B and S1C). The increased levels of TMAO in RYGB samples might be the consequence of a less anaerobic metabolism in the RYGB intestine, which is supported by the broad increase of facultative anaerobes in RYGB microbiomes, and could occur due to the shortening of the small bowel. We speculate that TMAO could be produced in the intestine by oxidation of TMA catalyzed by TMA monooxygenase; this enzyme is present in bacteria such as Pseudomonas (Barrett and Kwan, 1985), which showed increased abundance in RYGB microbiomes.
These observations are of interest because high TMAO levels linked to TMA oxidation by the human hepatic flavin monooxygenase FMO3 have been associated with cardiovascular diseases (CVDs) (Tang et al., 2013; Wang et al., 2011), while it is known that RYGB is associated with a lower CVD risk (Sjöström et al., 2012).
Bariatric Surgery Has Long-Term Effects on Microbial Fermentation
The enrichment of KOs related to phosphoglycerate, acetoacetate, and SCFAs transporters and to phosphotransferase systems pointed to increased energy flux into sugar metabolism and glycolysis in RYGB microbiomes. By contrast, the enrichment of pathways for amino acid uptake and metabolism and for glyoxylate metabolism pointed to the utilization of amino acids and acetate for energy production by the VBG microbiome. As the fermentation activities of the gut microbiota contribute to host nutrition and health (Flint et al., 2012), we functionally tested whether the differences in the microbiome resulted in altered microbial fermentation of carbohydrate and protein by measuring the levels of fecal SCFAs and branched-chain fatty acids (BCFAs). The SCFAs acetate, propionate, and butyrate were present in comparable proportions, but their concentrations tended to decrease in RYGB and VBG samples (Figures S2A–S2D), while the BCFAs isobutyrate and isovalerate, produced from the amino acids valine and leucine, respectively, either showed no change or a slight tendency to increase in RYGB and VBG samples (Figures S2E–S2G). As a result, we observed a decrease in the SCFA/BCFA ratio for bariatric surgery samples, which was significant for RYGB (Figure S2H). Importantly, total fiber intake did not differ, while protein intake decreased for RYGB and VBG women (Table S1), indicating that the lower SCFA/BCFA ratio was not a consequence of dietary consumption.
The similar SCFA/BCFA ratio and the analysis of pathways enriched in RYGB and VBG compared to OBS subjects reveal some common effects of the two bariatric surgery procedures and indicate altered intestinal fermentation shifted toward reduced production of SCFAs and increased amino acid fermentation after bariatric surgery. As the total fiber intake was similar for RYGB, VBG, and OBS patients, our results point to a diet-independent adaptation of the gut microbiota to bariatric surgery. In particular the decreased SCFAs levels in RYGB and VBG fecal samples also point to reduced energy harvest from the diet, as previously proposed (Turnbaugh et al., 2006).
Bariatric Surgery Has Long-Term Effects on Bile Acid Metabolism
Bile acids are cholesterol metabolites that facilitate the absorption of dietary fat and fat-soluble molecules and are now also recognized as regulators of energy metabolism through activation of the receptors TGR5 and FXR (Hylemon et al., 2009; Lefebvre et al., 2009). Increased circulating levels of primary and secondary bile acids have been observed after RYGB (Ahmad et al., 2013; Glicksman et al., 2010; Kohli et al., 2013; Patti et al., 2009) and have been linked to increased GLP-1 and reduced glucose and triglyceride levels (Nakatani et al., 2009; Patti et al., 2009; Pournaras et al., 2012; Simonen et al., 2012). The postprandial bile acid response is blunted in obesity and enhanced after RYGB, indicating that this response might be important for inducing satiety, reduced food intake, and lower body weight (Ahmad et al., 2013; Glicksman et al., 2010).
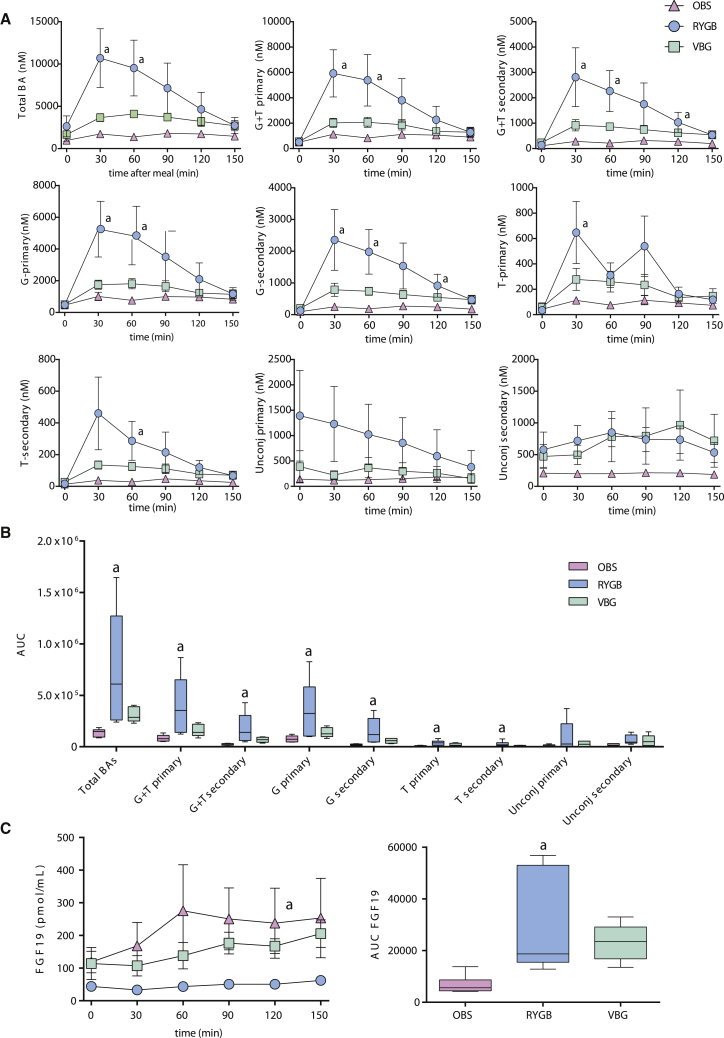
Because the composition of the bile acid pool is regulated by the gut microbiota (Sayin et al., 2013), we examined the relationship between the altered gut microbiota and bile acid pool composition after bariatric surgery. We quantified putative microbial genes for bile acid metabolism and showed that levels of bsh genes (for bile salt hydrolases) did not differ, while baiB, baiCD, baiE, baiF, and baiG (genes for the 7α-dehydroxylation of primary bile acids) increased in RYGB compared to OBS, although not significantly (Figure S3A). Fasting bile acid concentrations did not differ between RYGB, VBG, and OBS patients, but we confirmed a blunted postprandial bile acid response for OBS women and observed a slight increase in postprandial bile acid levels for VBG patients (Figures 3A and 3B), which was significant only for the secondary bile acid glycine-conjugated lithocholic acid (Figure S3B). However, we observed a pronounced postprandial bile acid response in RYGB compared to OBS patients, with a significant increase in total and glycine- and taurine-conjugated bile acids, but not unconjugated bile acids (Figures 3A and 3B). The increased bile acid levels in RYGB women were attributable to increases in both primary and secondary conjugated bile acids (Figure S3B, ANOVA p < 0.05). Finally, we measured the circulating levels of FGF19, an intestinal factor that regulates bile acid, carbohydrate, lipid, and energy metabolism through bile acid-mediated activation of FXR (Beenken and Mohammadi, 2009). We observed an increased FGF19 response in RYGB women compared to OBS (Figure 3C), indicating increased FXR signaling.
Figure 3.
Postprandial Bile Acid and FGF19 Responses in RYGB, VBG, and OBS Women
(A) Circulating concentrations of total bile acids (Total BA), total conjugated primary (G+T primary), total conjugated secondary (G+T secondary), glycine-conjugated primary (G-primary), glycine-conjugated secondary (G-secondary), taurine-conjugated primary (T-primary), taurine-conjugated secondary (T-secondary), unconjugated primary, and unconjugated secondary bile acids.
(B) Tukey box plots showing the area under the curve (AUC) as a measure of the bile acid postprandial responses.
(C) Circulating concentrations of FGF19 and AUC showing FGF19 postprandial response in RYGB, VBG, and OBS women.
Plasma samples were collected during fasting and every 30 min for 2.5 hr after a standard meal. Samples from colecystectomized patients (one in the RYGB and one in the VBG group) were excluded from the analysis, and plasma from one VBG woman could not be obtained, so the results represent the mean ± SEM for six RYGB, five VBG, and seven OBS women. a p < 0.05 according to one-way ANOVA with Tukey’s correction for multiple comparisons for RYGB compared to OBS. See also Figure S3.
Our results provide evidence for an increased abundance in RYGB microbiomes of microbial genes involved in the dehydroxylation of primary to secondary bile acids, which is in line with the increased response of secondary bile acids in RYGB patients. However, these results need to be further validated in larger cohorts and at shorter time points after bariatric surgery.
Analysis of BMI as a Possible Confounding Factor
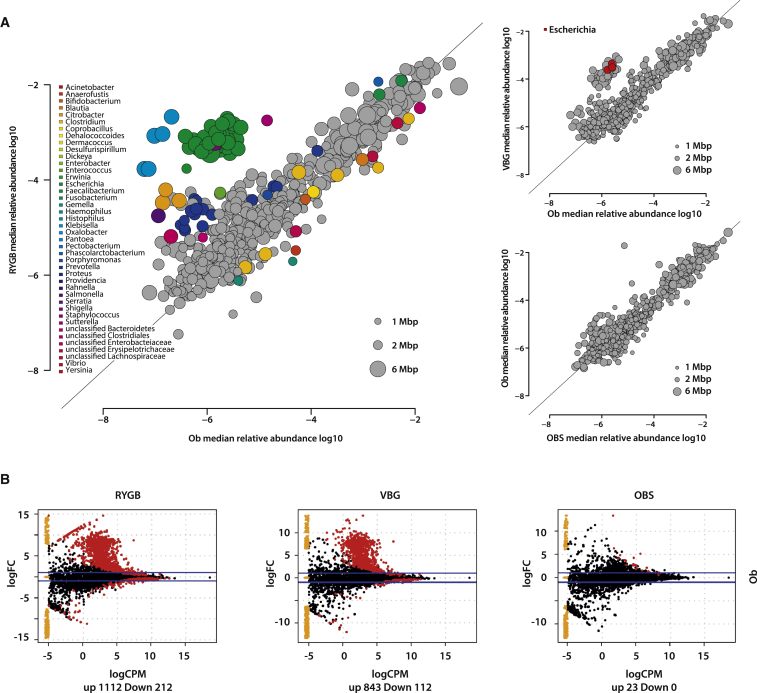
To control for differences in BMI, we compared the gut microbiome of RYGB and VBG patients with that of non-operated women (Ob, n = 9) with BMI matched to the post-surgical BMI of the RYGB and VBG patients (BMI = 31.9 ± 2.7 [SD]) (Table S1 and S5). The Ob group was selected from a larger cohort recently analyzed with methods comparable to those used here (Karlsson et al., 2013). We found again an expansion in Proteobacteria and a reduction in several clostridial species after RYGB, and also a significant increase in the relative abundance of three E. coli species in VBG (Figure 4A; Table S5).
Figure 4.
Gut Microbiome of RYGB and VBG Women in Comparison to Non-Operated Women (Ob) with BMI Matched to the Post-Surgical BMI of the Bariatric Surgery Patients
(A) Gut microbiota composition. Scatter plot of median species abundance in RYGB and Ob, in VBG and Ob, and in Ob and OBS women. The genus affiliations of differentially abundant species are indicated by color (Adj. p < 0.05, Wilcoxon rank-sum test).
(B) Functional differences in the microbiomes measured by the abundance of KOs. Each spot represents a KO, and red spots represent KOs whose abundance is significantly different. The number of KOs significantly increased (up) and depleted (down) is shown under each plot. The direction of change is defined as up being the title above and down being the title to the right of each plot. FC, fold change; CPM, counts per million. See also Table S5 and Figure S4.
We observed differences in the genetic content of RYGB, VBG, and Ob microbiomes (Figure 4B; Table S5). Several pathways (e.g., two-component systems, fluorobenzoate degradation, and glyoxylate and dicarboxylate metabolism) and KOs (e.g., nifA, pgtCAP, ato, and deg genes) enriched in RYGB or VBG compared to OBS were also enriched in comparison to Ob microbiomes (Table S5). In particular, torA (K07811) was enriched in RYGB and VBG compared to both OBS and Ob (Figure S4).
These results indicate that the differences in gut microbiota composition and genetic content result from bariatric surgery and not BMI per se, since the OBS and Ob microbiomes were similar (Figure 4; Table S5). Indeed, previous studies of the gut microbiota during dietary interventions for weight loss have not reported an increase in Proteobacteria but a shift in taxa such as Bacteroidetes (Ley et al., 2006), Eubacterium/Roseburia (Duncan et al., 2008), and Eubacterium rectale (Cotillard et al., 2013). Together with these observations, our results suggest that bariatric surgery produces a specific shift in the microbiota that persists for up to a decade after surgery and is different from shifts related to dietary interventions for weight loss.
Transmission of the Human Adiposity Phenotype by Transplantation of the Gut Microbiota
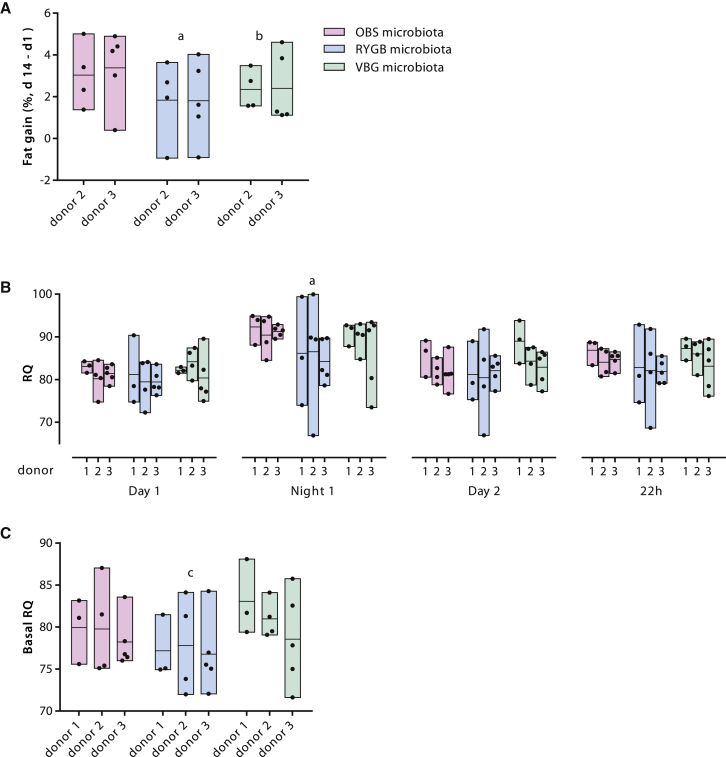
To investigate whether the altered gut microbiota of bariatric surgery patients directly affects adiposity and metabolism, we transplanted the fecal microbiota of RYGB, VBG, and OBS patients into female GF mice. We observed that mice colonized with RYGB and VBG microbiota for 2 weeks accumulated 43% and 26% less body fat, respectively, than mice colonized with OBS microbiota (Figure 5A), while mice colonized with RYGB microbiota had the highest average increase in lean mass (Figures S5A–S5C); body weight gain and food intake did not differ between the groups during the 2-week colonization period (Figures S5D and S5E). In addition, we measured oxygen (O2) consumption and carbon dioxide (CO2) production in the mice colonized with RYGB, VGB, and OBS microbiota. Although O2 consumption, body weight gain, and food intake did not differ during the measurements (Figures S5F–S5I), GF mice colonized with RYGB microbiota had a lower respiratory quotient ([RQ]; ratio between CO2 produced and O2 consumed) than mice colonized with OBS microbiota (active phase, Night1, Figure 5B) or with VBG microbiota (resting phase, Figure 5C), thus indicating decreased utilization of carbohydrates and increased utilization of lipids as fuel in recipients of RYGB microbiota.
Figure 5.
The Gut Microbiota Influences Fat Accumulation and Metabolism in Colonized Mice
(A) Fat gain in mice colonized with human stools from OBS, RYGB, and VBG women. Fat gain was calculated as the difference between fat content normalized over body weight at the end of the experiment (d14) and fat content normalized over body weight 1 day after colonization (d1). The results presented were obtained from colonization of GF mice with fecal microbiota from two patients in each of the RYGB, VBG, and OBS groups (four to five mice per donor microbiota).
(B) RQ (ratio between CO2 produced and O2 consumed) calculated for the light and dark phases and for the overall 22 hr period in the Somedic chamber.
(C) RQ in the resting phase (basal RQ, calculated as the average RQ for the lowest 10 min average of O2 consumption).
The results presented in (B) and (C) were obtained from colonization of GF mice with fecal microbiota from three patients in each of the RYGB, VBG, and OBS groups (3–5 mice per donor microbiota).
Bars show the results of colonization with the same donor microbiota, with black dots showing individual mice and lines indicating the mean. Statistical significance of differences between the means was tested by one-way ANOVA with Tukey’s correction for multiple comparisons (a p < 0.05 for RYGB compared to OBS, b p < 0.05 for VBG compared to OBS, and c p < 0.05 for RYGB compared to VBG). See also Figure S5.
The reduced fat mass gain in GF mice transplanted with RYGB and VBG microbiota is in agreement with our results on the composition of the gut microbiome and with a short-term human study showing an inverse correlation between E. coli abundance and fat mass after RYGB (Furet et al., 2010). These results are also consistent with a mouse study showing a reduced ability of the RYGB microbiota to promote adiposity (Liou et al., 2013). As we observed decreased SCFAs in RYGB and VBG samples, reduced energy harvest from the diet might be a mechanism for the reduced fat mass gain in the mice transplanted with bariatric surgery microbiota.
We also observed that mice receiving RYGB microbiota gained the least fat mass and had a reduced RQ, suggesting reduced utilization of carbohydrates and increased utilization of fat for metabolic processes. Reduced RQ has been observed in humans after RYGB and associated to reduced glucose oxidation, increased lipid oxidation, and increased plasma levels of conjugated bile acids (Simonen et al., 2012). As we found increased levels of secondary bile acids in the plasma of RYGB patients, microbial regulation of bile acid metabolism could contribute to the improved metabolic phenotype of RYGB patients by promoting increased utilization of fat, and to the superior long-term weight loss and metabolic improvements observed after RYGB compared to VBG (Sjöström et al., 2007; Werling et al., 2013a). However, as we compared the microbiomes of RYGB and VBG patients matched for weight loss, future studies of the microbiome from patients achieving different levels of weight and fat loss are required to validate our findings.
Perspective
Our study shows that bariatric surgery has long-term effects on the composition and functional capacity of the gut microbiota and that these changes have the potential to modulate host metabolic regulation, thus adding evidence for the transmissibility of the human adiposity phenotype through the gut microbiota.
Our results also show that two different bariatric surgery procedures, namely RYGB and VBG, have similar long-term effects on the gut microbiome in women matched for BMI and fat mass loss. However, the two bariatric surgery procedures might result in different functionality due to different intestinal environmental conditions, as we exemplified for TMA/TMAO metabolism. Importantly, the changes in the microbiome were not dependent on BMI or degree of weight and fat mass loss, thus revealing shifts in the gut microbiota that were specific to bariatric surgery.
Experimental Procedures
Subjects and Sample Collection
The operated women included in this study participated in a previous clinical trial for the measurement of energy expenditure after RYGB (n = 7) and VBG (n = 7) (Werling et al., 2013b). The clinical trial and the enrollment of subjects were described previously (Werling et al., 2013b). We also included samples from severely obese women with a BMI similar to the mean pre-surgical BMI of the operated women (OBS, n = 7), and these women underwent measurements of energy expenditure as the RYGB and VBG patients. None of the women included in our study had diabetes. However, the RYGB patients had significantly reduced fasting insulin compared with OBS, whereas there was a trend toward reduction in the VBG patients (Table S1).
Blood samples were collected during the measurements of energy expenditure in the indirect calorimetry chamber; 17 out of 21 stool samples were also collected during the measurements, while the remaining four samples were collected at home 2 to 8 days after. Stool samples collected in the calorimetry chamber were immediately frozen at −80°C. Samples collected at home were kept at room temperature and mailed to the hospital, where they arrived within 24 hr from collection and were immediately frozen at −80°C. The 21 women in our study did no use antibiotics for at least 6 weeks before inclusion in the study. The studies were performed in accordance with the Declaration of Helsinki and were approved by the Regional Ethical Review Board in Gothenburg, Sweden (no: 359-09 and 740-10). All study participants were informed verbally and in writing and signed a consent form.
Quantification of Organic Acids, TMAO, and Bile Acids
Fecal SCFAs and BCFAs were determined by gas chromatography-mass spectrometry (GC-MS) as previously described (Ryan et al., 2014). TMAO, choline, betaine, and bile acids were extracted from plasma samples and measured by ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). Detailed methods are provided in the Supplemental Experimental Procedures.
DNA Extraction and Metagenome Data Analysis
Genomic DNA was extracted from fecal samples using repeated bead beating (Salonen et al., 2010). DNA fragments of approximately 300 bp were sequenced on an Illumina HiSeq 2000 instrument. Metagenomic sequence data was analyzed using methods from the MEDUSA platform (Karlsson et al., 2012, 2014). The statistical analysis was performed in the software R (R Core Team, 2012). All p values were corrected for multiple testing with the method from Benjamini and Hochberg (1995). Detailed methods are provided in the Supplemental Experimental Procedures.
Mouse Experiments
For microbiota transplantations, we selected stool samples from three RYGB, three VBG, and three OBS patients who did not use medications and did not receive cholecystectomy and used these samples to colonize GF Swiss Webster female mice in three independent experiments. Mice were colonized by oral gavage of fecal slurry at 8–10 weeks of age, for a period of 2 weeks; more details are provided in the Supplemental Experimental Procedures. Mice were fed autoclaved chow and sterile water; food consumption and body weight were recorded 1, 7, and 14 days after colonization. In two of the experiments, body composition was determined with an EchoMRI instrument (EchoMRI) 1 and 14 days after colonization. All mice were analyzed by indirect calorimetry in the Somedic INCA system (Somedic, Hörby) for measurements of O2 consumption and CO2 production. All animal experiments were approved by the Gothenburg Animal Ethics Committee.
Author Contributions
M.W., T.O., L.F., C.W.l.R., and F.B. conceived and designed the human study. V.T. and F.B. designed and analyzed microbiota transplantation experiments in GF mice. M.S. developed the methods and quantified the levels of TMAO, choline, betaine, and bile acids. P.K.-D. quantified the levels of SCFAs and BCFAs. V.T., F.K., and M.W. performed the experiments. F.K. and J.N. analyzed the gut microbiota sequence data. V.T., F.K., and F.B. wrote the manuscript. All authors were involved in the discussion of the results and commented on the manuscript.
Conflicts of Interest
F.K. is employed by Metabogen AB; J.N. and F.B. are founders and shareholders of Metabogen AB.
Acknowledgments
We thank Rosie Perkins for editing the manuscript and Anna Hallén for assistance with figures and artwork. We also thank Carina Arvidsson, Sara Nordin-Larsson, Ulrica Enquist, and Ying Shiuan Lee for technical assistance on the mice and the Centre for Physiology and Bio-Imaging (CPI) at the University of Gothenburg for assistance with the Somedic INCA system. This study was supported by the Swedish Research Council, the NovoNordisk foundation, Torsten Söderberg’s foundation, Ragnar Söderberg’s foundation, Swedish Diabetes foundation, Swedish Heart Lung Foundation, Göran Gustafsson’s foundation, IngaBritt och Arne Lundbergs foundation, Knut and Alice Wallenberg foundation, the Swedish Foundation for Strategic Research, the FP7 sponsored program METACARDIS, the regional agreement on medical training and clinical research (ALF) between Region Västra Götaland and Sahlgrenska University Hospital, and Science Foundation Ireland (SFI) ref 12/YI/B2480. Computations were performed at Chalmers Centre for Computational Science and Engineering (C3SE) provided by the Swedish National Infrastructure for Computing (SNIC). F.B. is a recipient of an ERC Consolidator Grant (European Research Council, Consolidator grant 615362 - METABASE).
Published: August 4, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes five figures, five tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2015.07.009.
Accession Numbers
Metagenomic sequence data has been deposited at the European Nucleotide Archive under the accession number ENA: ERP009310.
Supplemental Information
References
- Ahmad N.N., Pfalzer A., Kaplan L.M. Roux-en-Y gastric bypass normalizes the blunted postprandial bile acid excursion associated with obesity. Int. J. Obes. 2013;37:1553–1559. doi: 10.1038/ijo.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar J., Chabo C., Waget A., Klopp P., Vachoux C., Bermúdez-Humarán L.G., Smirnova N., Bergé M., Sulpice T., Lahtinen S. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol. Med. 2011;3:559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E.L., Kwan H.S. Bacterial reduction of trimethylamine oxide. Annu. Rev. Microbiol. 1985;39:131–149. doi: 10.1146/annurev.mi.39.100185.001023. [DOI] [PubMed] [Google Scholar]
- Beenken A., Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc. B. Met. 1995;57:289–300. [Google Scholar]
- Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., Neyrinck A.M., Fava F., Tuohy K.M., Chabo C. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Cotillard A., Kennedy S.P., Kong L.C., Prifti E., Pons N., Le Chatelier E., Almeida M., Quinquis B., Levenez F., Galleron N., ANR MicroObes consortium Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Duncan S.H., Lobley G.E., Holtrop G., Ince J., Johnstone A.M., Louis P., Flint H.J. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Duncan S.H., Louis P., Thomson J.M., Flint H.J. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 2009;11:2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- Flint H.J., Scott K.P., Louis P., Duncan S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- Furet J.P., Kong L.C., Tap J., Poitou C., Basdevant A., Bouillot J.L., Mariat D., Corthier G., Doré J., Henegar C. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glicksman C., Pournaras D.J., Wright M., Roberts R., Mahon D., Welbourn R., Sherwood R., Alaghband-Zadeh J., le Roux C.W. Postprandial plasma bile acid responses in normal weight and obese subjects. Ann. Clin. Biochem. 2010;47:482–484. doi: 10.1258/acb.2010.010040. [DOI] [PubMed] [Google Scholar]
- Graessler J., Qin Y., Zhong H., Zhang J., Licinio J., Wong M.L., Xu A., Chavakis T., Bornstein A.B., Ehrhart-Bornstein M. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2013;13:514–522. doi: 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- Hylemon P.B., Zhou H., Pandak W.M., Ren S., Gil G., Dent P. Bile acids as regulatory molecules. J. Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F.H., Fåk F., Nookaew I., Tremaroli V., Fagerberg B., Petranovic D., Bäckhed F., Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F.H., Tremaroli V., Nookaew I., Bergström G., Behre C.J., Fagerberg B., Nielsen J., Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Karlsson F.H., Nookaew I., Nielsen J. Metagenomic data utilization and analysis (MEDUSA) and construction of a global gut microbial gene catalogue. PLoS Comput. Biol. 2014;10:e1003706. doi: 10.1371/journal.pcbi.1003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli R., Bradley D., Setchell K.D., Eagon J.C., Abumrad N., Klein S. Weight loss induced by Roux-en-Y gastric bypass but not laparoscopic adjustable gastric banding increases circulating bile acids. J. Clin. Endocrinol. Metab. 2013;98:E708–E712. doi: 10.1210/jc.2012-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L.C., Tap J., Aron-Wisnewsky J., Pelloux V., Basdevant A., Bouillot J.L., Zucker J.D., Doré J., Clément K. Gut microbiota after gastric bypass in human obesity: increased richness and associations of bacterial genera with adipose tissue genes. Am. J. Clin. Nutr. 2013;98:16–24. doi: 10.3945/ajcn.113.058743. [DOI] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J.M., Kennedy S., MetaHIT consortium Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- le Roux C.W., Bloom S.R. Why do patients lose weight after Roux-en-Y gastric bypass? J. Clin. Endocrinol. Metab. 2005;90:591–592. doi: 10.1210/jc.2004-2211. [DOI] [PubMed] [Google Scholar]
- Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Liou A.P., Paziuk M., Luevano J.M., Jr., Machineni S., Turnbaugh P.J., Kaplan L.M. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci. Transl. Med. 2013;5:178ra41. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T., Helmann J.D., Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras A.D., le Roux C.W. Mechanisms underlying weight loss after bariatric surgery. Nat. Rev. Gastroenterol. Hepatol. 2013;10:575–584. doi: 10.1038/nrgastro.2013.119. [DOI] [PubMed] [Google Scholar]
- Mukhopadhya I., Hansen R., El-Omar E.M., Hold G.L. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- Nakatani H., Kasama K., Oshiro T., Watanabe M., Hirose H., Itoh H. Serum bile acid along with plasma incretins and serum high-molecular weight adiponectin levels are increased after bariatric surgery. Metabolism. 2009;58:1400–1407. doi: 10.1016/j.metabol.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Olbers T., Björkman S., Lindroos A., Maleckas A., Lönn L., Sjöström L., Lönroth H. Body composition, dietary intake, and energy expenditure after laparoscopic Roux-en-Y gastric bypass and laparoscopic vertical banded gastroplasty: a randomized clinical trial. Ann. Surg. 2006;244:715–722. doi: 10.1097/01.sla.0000218085.25902.f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A.P., Patil K.R., Nielsen J. Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst. Biol. 2008;2:17. doi: 10.1186/1752-0509-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K.R., Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. USA. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti M.E., Houten S.M., Bianco A.C., Bernier R., Larsen P.R., Holst J.J., Badman M.K., Maratos-Flier E., Mun E.C., Pihlajamaki J. Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring) 2009;17:1671–1677. doi: 10.1038/oby.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras D.J., Glicksman C., Vincent R.P., Kuganolipava S., Alaghband-Zadeh J., Mahon D., Bekker J.H., Ghatei M.A., Bloom S.R., Walters J.R. The role of bile after Roux-en-Y gastric bypass in promoting weight loss and improving glycaemic control. Endocrinology. 2012;153:3613–3619. doi: 10.1210/en.2011-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- R Core Team (2012). R: A Language and Environment for Statistical Computing (Vienna).
- Ryan K.K., Tremaroli V., Clemmensen C., Kovatcheva-Datchary P., Myronovych A., Karns R., Wilson-Pérez H.E., Sandoval D.A., Kohli R., Bäckhed F., Seeley R.J. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., Nikkilä J., Jalanka-Tuovinen J., Immonen O., Rajilić-Stojanović M., Kekkonen R.A., Palva A., de Vos W.M. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Sayin S.I., Wahlström A., Felin J., Jäntti S., Marschall H.U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Simonen M., Dali-Youcef N., Kaminska D., Venesmaa S., Käkelä P., Pääkkönen M., Hallikainen M., Kolehmainen M., Uusitupa M., Moilanen L. Conjugated bile acids associate with altered rates of glucose and lipid oxidation after Roux-en-Y gastric bypass. Obes. Surg. 2012;22:1473–1480. doi: 10.1007/s11695-012-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöström L., Narbro K., Sjöström C.D., Karason K., Larsson B., Wedel H., Lystig T., Sullivan M., Bouchard C., Carlsson B., Swedish Obese Subjects Study Effects of bariatric surgery on mortality in Swedish obese subjects. N. Engl. J. Med. 2007;357:741–752. doi: 10.1056/NEJMoa066254. [DOI] [PubMed] [Google Scholar]
- Sjöström L., Peltonen M., Jacobson P., Sjöström C.D., Karason K., Wedel H., Ahlin S., Anveden Å., Bengtsson C., Bergmark G. Bariatric surgery and long-term cardiovascular events. JAMA. 2012;307:56–65. doi: 10.1001/jama.2011.1914. [DOI] [PubMed] [Google Scholar]
- Smith M.I., Yatsunenko T., Manary M.J., Trehan I., Mkakosya R., Cheng J., Kau A.L., Rich S.S., Concannon P., Mychaleckyj J.C. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.H., Wang Z., Levison B.S., Koeth R.A., Britt E.B., Fu X., Wu Y., Hazen S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B., Feldstein A.E., Britt E.B., Fu X., Chung Y.-M. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling M., Fändriks L., Björklund P., Maleckas A., Brandberg J., Lönroth H., le Roux C.W., Olbers T. Long-term results of a randomized clinical trial comparing Roux-en-Y gastric bypass with vertical banded gastroplasty. Br. J. Surg. 2013;100:222–230. doi: 10.1002/bjs.8975. [DOI] [PubMed] [Google Scholar]
- Werling M., Olbers T., Fändriks L., Bueter M., Lönroth H., Stenlöf K., le Roux C.W. Increased postprandial energy expenditure may explain superior long term weight loss after Roux-en-Y gastric bypass compared to vertical banded gastroplasty. PLoS ONE. 2013;8:e60280. doi: 10.1371/journal.pone.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., DiBaise J.K., Zuccolo A., Kudrna D., Braidotti M., Yu Y., Parameswaran P., Crowell M.D., Wing R., Rittmann B.E., Krajmalnik-Brown R. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.