Abstract
Immune cells function in an interacting hierarchy that coordinates activities of various cell types according to genetic and environmental contexts. We developed graphical approaches to construct an extensible immune reference map from mass cytometry data of cells from different organs, incorporating landmark cell populations as flags on the map to compare cells from distinct samples. The maps recapitulated canonical cellular phenotypes and revealed reproducible, tissue-specific deviations. The approach revealed influences of genetic variation and circadian rhythms on immune system structure, enabled direct comparisons of murine and human blood cell phenotypes, and even enabled archival fluorescence-based flow cytometry data to be mapped onto the reference framework. This foundational reference map provides a working definition of systemic immune organization to which new data can be integrated to reveal deviations driven by genetics, environment, or pathology.
The immune system is a systemically mobile network of cells with emergent properties derived from dynamic cellular interactions. Unlike many solid tissues, where cells of given functions are localized into substructures that can be readily defined, the distribution of phenotypically similar immune cells into various organs complicates discerning differences between them. Much research has necessarily focused on understanding the individual cell types within the immune system, and, more recently, towards identifying interacting cells and the messengers they use to communicate. Methods of single cell analysis, such as flow cytometry, have been at the heart of this effort to enumerate and quantitatively characterize immune cell populations (1-3). As research has accelerated, the number of markers required to identify cell types and explain detailed mechanisms has surpassed the technical limitations of fluorescence-based flow cytometry (1-4). Consequently, insights have often been limited because only a few cell subsets could be examined, independent of the immune system as a whole (5, 6).
Although individual immune cell populations have been examined extensively, no comprehensive or standardized reference map of the immune system has been developed, primarily because of the difficulty of data normalization and lack of co-expression measurements that would enable “merging” of results. In other analysis modalities, such as transcript profiling of cell populations, reference standards and minable databases have shown extraordinary utility (7-14). A comprehensive reference map defining the organization of the immune system at the single cell level would similarly offer new opportunities for organized data analysis. For example, macrophages exhibit tissue-specific phenotypes (15), and adaptive immune responses are influenced by genetics (16), but discerning these properties of immune organization required integrating the results of many disparate studies. Even current analytical tools that do provide a systems-level view do not compare new samples to an existing reference framework, making them unsuitable for this objective (17, 18). In contrast, a reference map that is extensible could provide a biomedical foundation for a systematized, dynamic, community-collated resource to guide future analyses and mechanistic studies.
We leveraged mass cytometry, a platform that allows measurement of multiple parameters simultaneously at the single-cell level, to initiate a reference map of the immune system (19-21). By combining the throughput of flow cytometry with the resolution of mass spectrometry, this hybrid technology enables the simultaneous quantification of 40 parameters in single cells. Use of mass cytometry allows fluorophore reporters to be replaced with isotopically-pure, stable heavy metal ions conjugated to antibodies or affinity reagents (22). These reporter ions are then quantified by time-of-flight mass spectrometry to provide single-cell measurements, enabling a more detailed characterization of complex cellular systems for a robust reference map.
An Analytical Framework for a Reference Map
A useful reference map should enable a data-driven organization of cells and should be flexible enough to accommodate different types of measurements. This would result in a map with underlying consistency but also robust enough to allow overlay of new data (or even of archival data from different measurement modalities) according to cell similarities. The approach is meant to provide templates for representing the system as a whole to enable systems-level comparisons, similar to other efforts to compare biological networks (23-28). Although we provide one template here, the framework is built to enable users to construct individualized or community-organized versions.
Building a reference map requires the ability to overlay data from multiple samples onto a foundational reference sample(s), which is not accommodated by algorithms like SPADE and viSNE, which necessitate incorporating data from all samples at the onset (17, 18). Without this feature, the reference map would not be an extensible solution. Moreover, the reference map ought to incorporate information on millions of individual cells to comprehensively represent the numerous cell types within complex samples, which remains beyond the capacity of other approaches (18). The mapping procedure should also enable users to implement one of the many available clustering algorithms or their own subjective definitions to determine cell groupings (29). Perhaps most importantly, positions of landmark cell populations are marked as flags on the map to allow users to compare cells in new samples to cells described in existing literature (30).
Force-directed graphs are a type of graphical model commonly used to spatially organize complex data in an intuitive and flexible manner (31). Force-directed graphs might also enable a method for grouping cells with similar features in a space that is defined by the molecular features of the individual cells (32). Force-directed approaches are based on a set of “forces” that guide data organization into, usually, a 2-dimensional plane (33, 34). Nodes (in this case groups of cells) that are similar are connected by edges with a length proportional to their resemblance (in our implementation, cosine similarity). These nodes are then spatialized into a graph: All nodes repel one another as if they were the same poles of magnets, but edges pull similar nodes together, acting like springs. We adapted this concept to build a new method to visualize complex cellular samples termed Scaffold (Single-Cell Analysis by Fixed Force- and Landmark-Directed) maps.
Scaffold maps enable a model to be built that incorporates prior knowledge from the literature but also allows the discovery and analysis of unanticipated cell types or behavioral states. Such an extensible map can allow for new datasets to be incorporated and linked to their mechanistic conclusions with references—as do transcriptomics or genomics databases (7, 11, 13, 14).
Systematic Analysis for an Immune Reference Map
We initiated a prototype high-resolution reference map of the murine immune system by characterizing the expression of 39 cell-surface proteins and transcription factors (selected to delineate immune cell types) on over 3*107 single cells from ten different anatomical locations (Fig. S1A, Table S1 and Materials and Methods). Single-cell suspensions from the bone marrow, blood, spleen, skin-draining (inguinal) lymph node (SLN), mesenteric lymph node (MLN), thymus, lungs, liver, small intestine, and colon of 12-week old male C57BL/6, Balb/c, and 129S1/Sv mice were simultaneously processed in replicate. Measurements were done under conditions that limited measuring error (35, 36), and all antibodies were validated to bind target proteins by standard protocols. As such, one antibody cocktail was used for all samples, and cells were barcoded and pooled by tissue before cell staining to minimize technical variability (Materials and Methods). Single-cell protein expression was quantified using a CyTOF mass cytometer. The data for these samples were normalized to account for variability in instrument sensitivity over time (36). Cells from each condition were subsequently identified by their barcode and written into a unique flow cytometry standard file for each sample (see data distribution instructions).
Defining Immune Organization in the Bone Marrow
Because the bone marrow contains most developing and mature immune cell types, we used the cells therein to build a foundational map as a point of comparison (Fig. 1A-i). “Landmark” populations of immune cells commonly recognized in the literature were identified in the bone marrow data of all C57BL/6 replicates by conventional criteria (Fig. 1A-ii, S1B). These populations ranged from hematopoietic stem cells to terminally-differentiated lymphocytes and myeloid cells and served as landmarks within the map (visualized by red nodes) to demarcate the location of cell populations of interest (Fig. 1A-iii).
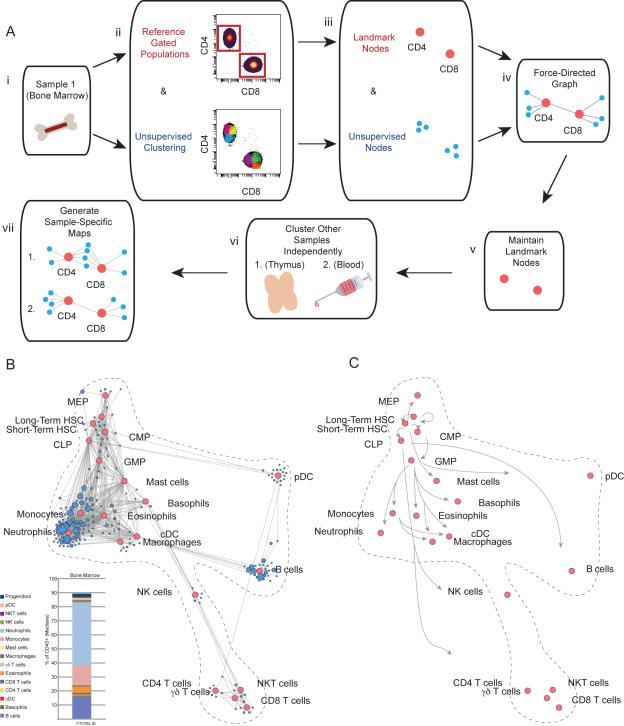
Figure 1. Scaffold Maps Reveal Immune Organization of the Bone Marrow.
(A) Schematic of the Scaffold map algorithm. (i) Bone marrow from C57BL/6 mice was chosen as the reference sample. (ii) Leukocytes were grouped according to prior knowledge to define landmark cell populations as reference points on the map. The same leukocytes were subjected to unsupervised clustering to provide an objective view of the tissue composition and organization. An illustration is provided with the two major lineages of mature T cells, which express either the cell-surface co-receptor cluster of differentiation 4 (CD4) or cluster of differentiation 8 (CD8). (iii-iv) Both landmark populations (red nodes) and unsupervised clusters (blue nodes) were utilized to generate a force-directed graph in which similar nodes are located close together according to the similarity of their protein expression. Thus, similar nodes fall in proximity to one another while disparate nodes segregate apart from one another. Size of unsupervised clusters denotes the relative number of cells in that grouping. (v) Landmark populations from the bone marrow were fixed in place for subsequent maps to provide points of reference for rapid human interpretation. (vi) Additional samples were each subjected to unsupervised clustering via the same clustering algorithm. (vii) The resulting clusters for each sample were overlaid onto the original landmark nodes to generate tissue-specific Scaffold maps.
(B) Bone marrow Scaffold map for C57BL/6 mice. Red nodes denote landmark manually-gated cell populations; blue nodes represent unsupervised cell clusters from the same data. Insert: median frequencies of cell populations defined by conventional criteria from the bone marrow of C57BL/C mice, n = 14.
(C) Scaffold map showing only the position of the landmark nodes with arrows annotating established maturation relationships in hematopoietic development.
We also took a data-driven approach to group similar cells into “clusters” according to their expression of the measured proteins. Grouping similar cells by clustering allows all of the data to be visualized at once. We therefore performed an unsupervised clustering of the C57BL/6 bone marrow leukocytes from all biological replicates with a modified Partitioning Around Medioids (PAM) algorithm (adapted for larger datasets) (Fig. 1A-ii, Materials and Methods)(37). We chose a number of clusters (200) that we expect exceeds the number of “true” cell populations present in the data. Therefore, we do not expect each cluster to represent a recognized functional cell subset, but rather to over-partition the data to ensure that two populations of distinct natures are not merged through under-clustering. We believe this to be an appropriate tradeoff, as the proximity of clusters immediately reveals groups of highly similar cells and thereby provides clarity during visualization. This enables an intuitive browsing of the data rather than relying on clustering to define the “true” number of cell populations, which depends on evolving semantic conventions and understandings of cellular functions. Manual analysis of cell populations by traditional criteria, which we visualized by landmark nodes, remains the standard to which automated clustering algorithms are routinely compared (29).
The reference map was built by combining these unsupervised cell clusters (blue nodes) with the manually identified cell populations (red nodes) (Fig. 1A-iii). Cluster sizes are scaled to reflect the relative cell frequencies in these initial maps, though this option can be modified. A force-directed algorithm was applied to the data, attracting cell clusters with similar phenotypes while separating those with dissimilar phenotypes (Fig. 1A-iv). When mapping C57BL/6 bone marrow cells (Fig. 1B), the landmark and unsupervised nodes were arranged (with no manual intervention or organization) into a structure that recapitulated most known developmental relations between these populations (Fig. 1C)(17, 20). For instance, the hematopoietic stem cell (HSC) landmark was situated at the top of the map and linked to progenitors and more mature populations below. Different granulocytes (including neutrophils, eosinophils, basophils, and mast cells) occupied nearby portions of the map. Macrophages and conventional dendritic cells (cDC) fell adjacent, and the various T cell populations (CD4+, CD8+, NKT and γδ) grouped together.
Because clusters serve as a means of partitioning the data in this map, the density of clusters also reflected the relative frequencies of immune cells in the bone marrow that correspond to cell types as defined by established criteria (Fig. 1B, insert). For instance, the map exhibited the densest concentration of unsupervised clusters (blue nodes) surrounding the neutrophil, monocyte and B cell landmarks. Rarer populations, such as dendritic cells, eosinophils and basophils, were more sparsely represented. The progenitor zone contained cell clusters proximal to every multipotent population identified by established criteria with cell clusters also falling in between them, revealing the transition states between classically-defined progenitors. This graph represents the data from all C57BL/6 biological replicates combined, though the data from individual mice consistently demonstrated these trends (Fig. S2).
The Scaffold map of the bone marrow thus reflected the expected biological relations between immune cell populations and enabled an unsupervised visualization of its composition and complexity. The profiles of cells in any cluster, or group of clusters, can also be visualized by conventional histograms. We used this as the initiating reference template and mapped other organs onto this map for comparison.
Mapping Immune Organization across the Body
After determining that Scaffold maps effectively convey the organization of the immune cells present in the bone marrow, we determined how immune cells from other lymphoid organs or the blood might map into this space. By fixing the identity and position of the landmark (red) nodes that represent canonical populations in the bone marrow, we retained a common reference across all samples (Fig. 1A-v). We performed unsupervised clustering of total leukocytes from each tissue independently (Fig. 1A-vi). We then overlaid these cell clusters (blue nodes) onto the reference map by allowing them to find their location according to the attractive and repulsive forces described above (Fig. 1A-vii and 2).
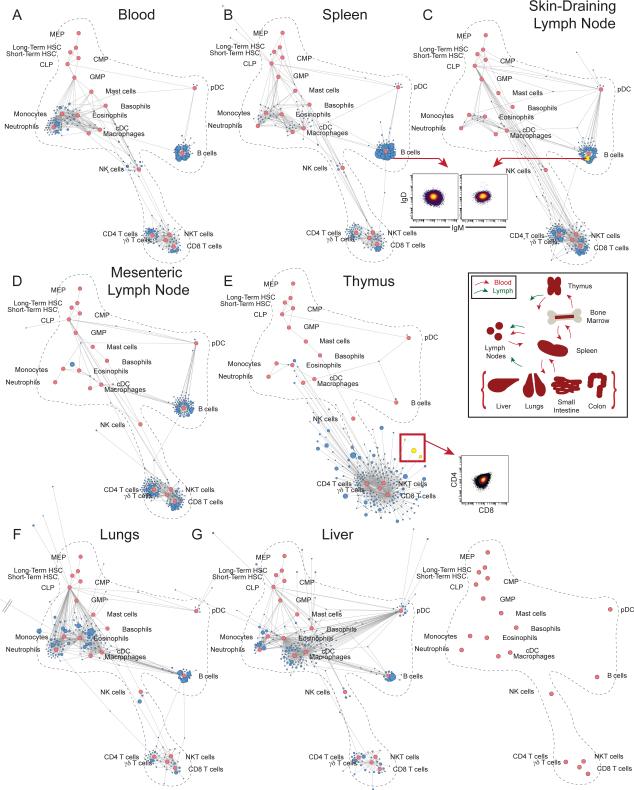
Figure 2. Mapping Systemic Immune Organization by Tissue.
Scaffold maps for lymphoid organs and peripheral solid organs from C57BL/6 mice using bone marrow as the reference sample to define landmark nodes (red). (A) Blood (B) Spleen (C) Skin- Draining (Inguinal) Lymph Node (SLN) (D) Mesenteric Lymph Node (MLN) (E) Thymus (F) Lungs (G) Liver, n = 14. Grey double bars denote a cluster extending behind another map for visualization purposes. Inserts, from top to bottom: Cells comprising B cell clusters from the spleen and SLN are visualized by 2D scatter plot. Schematic of immune cell circulation through and within the tissues characterized by mass cytometry. Cells comprising a deviant thymic T cell population cluster are visualized by 2D scatter plot.
By inspecting the composition of the peripheral blood on the map, it was apparent that the cell populations overlapped with those found in the bone marrow—as evident by the proximity of unsupervised clusters to the landmarks. (Fig. 2A). As expected, the blood did not contain cells localized to the HSC/progenitor portion of the map. Rather, cell clusters associated with landmark nodes of mature cell populations known to predominate in circulating blood at steady state, including granulocytes, monocytes, B cells, T cells, and NK cells (Fig. S3-S4, Table S2). Because unsupervised cell clusters from the blood positioned close to landmark populations, there were no substantial unanticipated populations present in the circulation.
In comparison, maps for the secondary lymphoid organs (spleen, skin-draining lymph node (SLN), mesenteric lymph node (MLN)) all exhibited an immune landscape dominated by mature lymphoid cells of the T and B cell lineages (Fig. 2B-D). Indeed, these populations were also comparable when viewed by conventional 2D dot plots (Fig. 2B and C, insert). Many of the myeloid cells in these tissues mapped more closely to the macrophage and dendritic cell zones and expressed major histocompatibility complex (MHC) class II used to present antigens, consistent with the presence of mature antigen-presenting cells (APCs) in these organs (Fig. 2B-D)(38). The clusters from the secondary lymphoid organs also largely mapped near a landmark population, indicating that most cells found in these tissues belong to well-characterized populations. The subtle differences in the cellular organization of these organs become evident thorough investigation of their maps, revealing enrichment in the NK cell, monocyte, macrophage, and conventional dendritic cells (cDC) in the spleen compared to frequencies of those cells in lymph nodes (p < 0.0001 for each by ANOVA). A higher frequency of macrophages (p = 0.0006 by two-sided t-test) and lower frequency of cDC (p = 0.013 by two-sided t-test) were present in the skin-draining lymph node than in the mesenteric lymph node. An appreciation for the distinct cellular composition of different secondary lymphoid organs provides an opportunity to examine how each cellular environment shapes the immune responses initiated in these locations.
Many of the cell clusters in the thymus radiated far away from the landmarks on the map. Inspection of these clusters indicated that many comprised CD4+CD8+ double-positive (DP) T cells that were absent from the bone marrow (Fig. 2E, red arrow). As the thymus largely contains developmental T cells, this was expected. However, the increased length of the lines connecting these ubiquitous DP T cell clusters to their nearest landmarks denotes cells that deviate from the characterized reference. We also observed these trends when cell populations from the spleen were used to define landmarks (Fig. S5).
Immune cell subsets in peripheral solid organs were compared to the reference map of the bone marrow (Fig. 2F and G, Fig. S6). The region of the maps representing myeloid cells was, in general, more densely filled (Fig. S3 and S4). For instance, cells from the lungs exhibited many clusters distributed between the macrophage, cDC, and eosinophil landmarks, indicating cells in this tissue were phenotypically distinct from those in bone marrow and even spleen. Alveolar macrophages in the lung expressed the proteins CD11c and Siglec-F, which are canonically markers of cDC and eosinophils, respectively (Fig. 2F)(39). Similarly, the liver map exhibited many clusters connected to the macrophage landmark, though the length of the lines connecting them was longer than those for the macrophages in the bone marrow (p = 0.0004 by one-sided Wilcoxon rank sum test, see Materials and Methods), consistent with the unique characteristics of liver macrophages (Kupffer cells) (Fig. 2G)(40). Overall, these maps of peripheral solid organs, including the gut (Fig. S6), exhibited less fidelity than those of lymphoid organs to the bone marrow reference, indicating that immune cells in these sites are likely distinct in their phenotypes and functions. Several previously uncharacterized cellular phenotypes are listed in Table S3. For future studies cell populations present in any tissue could also be used to define landmarks for organ-specific maps. Moreover, a comparative analysis of immune organization within the gut revealed site-specific characteristics, with significantly lower frequencies of CD4 and CD8 T cells and higher frequencies of macrophages and cDC in the colon than in the small intestine (p = 2.8*10−15, 0.001, 9.4*10−7, 1.0*10−5 respectively by one-sided t-test; Fig. S6). This understanding will inform further investigations of immune responses and pathologies within regions of the gut.
Genetic Variation Impacts Immune Cell Composition and Phenotype
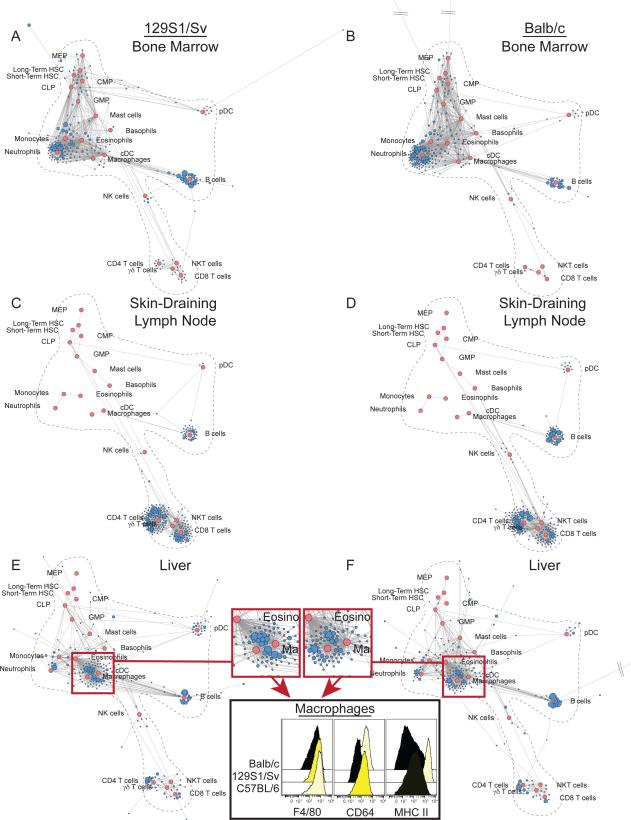
We used the reference maps to reveal the impact of genetic diversity on immune cell phenotypes and organization. We generated Scaffold maps of immune cells from two common inbred mouse strains, 129S1/Sv and Balb/c (Fig. 3). Mapping cells from the bone marrow from these animals onto the C57BL/6 reference map revealed that the vast majority of clusters fell close the C57BL/6 landmarks (Fig. 3A and B). However, certain cell clusters were distinct from those in the C57BL/6 reference. This likely reflects genetic variability, such as the relative lack of T cells in Balb/c mice, which we confirmed by conventional analysis of T cell populations (CD4 T cells, p = 0.0007; CD8 T cells, p = 0.001; γδ T cells, p = 2.2*10−7; NKT cells, p = 6.2*10−8 by ANOVA).
Figure 3. Immune Organization Across Inbred Mouse Strains.
Scaffold maps for several tissues from 129S1/Sv and Balb/c mice using C57BL/6 bone marrow as the reference sample to define landmark nodes (red). (A) Bone Marrow (B) Skin-Draining (Inguinal) Lymph Node (C) Liver, n = 3. Histograms of Fcγ receptor I (CD64) and major histocompatibility complex class II (MHC II) expression on liver macrophages from representative mice of each strain.
Similarly, analysis of the maps for lymphoid organs from these strains demonstrated high fidelity between unsupervised clusters and landmarks, with enrichment for mature lymphocytes. Other cell types in these organs also reflected the underlying genetics, such as pDC and NK cells, which were over-represented in the SLN of Balb/c mice (p = 1.2*10−6; p = 7.5*10−8 respectively by ANOVA) (Fig. 3C and D and S2, Table S2). In contrast, the SLN in C57BL/6 mice contained significantly more cDC and NKT cells but fewer CD4 T cells than SLN from the other strains (p = 5.0*10−5; p = 2.9*10−7; p = 5.5*10−10 respectively by ANOVA). Analysis of peripheral solid organs revealed other apparent impacts of genetic variation. In the liver, an unexpected shift in cell density from the macrophage to the cDC landmark was observed only in 129S1/Sv mice. Further investigation of these cells demonstrated differential expression of Fc gamma receptor I (CD64) and MHC II in liver macrophages from these inbred strains, causing these cells to adopt a phenotype more similar to that of cDC (Fig. 3E and F, red arrows). The difference in CD64 staining could be attributable to a polymorphism in the gene expressed by 129S1/Sv mice (41). However, this difference in MHC II expression was not observed when comparing macrophages in other solid organs, suggesting that this disparity is specific to the liver.
These results illustrate the ability of Scaffold maps to highlight sample-specific differences in immune cell characteristics. These maps convey a common global structure of immune cell populations along with specific influences of genetic variance.
Circadian Influences on Immune Organization
To investigate circadian immune fluctuations, which can powerfully regulate immune system behavior (42, 43), we obtained organs from C57BL/6 mice in 4 batches, either in the morning (8-9am; Zeitgeber time 1-2) or afternoon (1-2pm; Zeitgeber time 6-7) of two consecutive days.
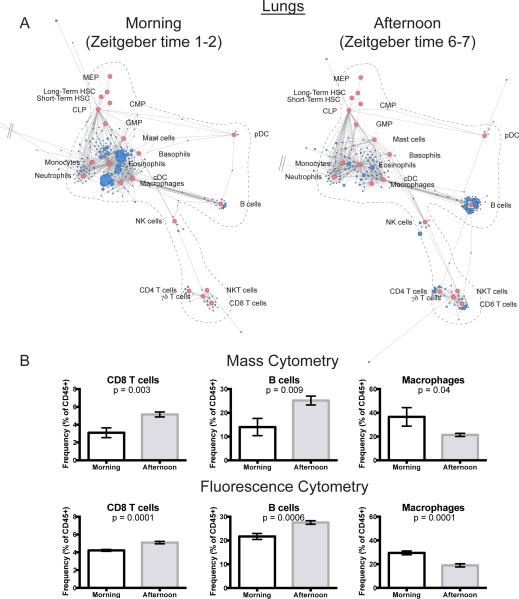
Analysis of the maps revealed a number of cell populations that fluctuated according to the time of day. Unexpectedly, these were significantly more pronounced in the peripheral solid organs than in the lymphoid tissues. The lungs displayed clear circadian patterns with remodeling of the ratios for several immune cell populations (Fig. 4A). To validate these findings, we used fluorescence-based flow cytometry to investigate the composition of the lungs in a new cohort of animals. In both analyses the frequencies of CD8 T cells and B cells were significantly higher in the afternoon than in the morning (Fig. 4B). In contrast, the frequency of macrophages increased in the morning, revealing a compensatory shift in composition from myeloid to lymphoid cells (Fig. 4B). Scaffold maps in which cell populations from the lungs were used as the Landmarks additionally recapitulated these results (Fig. S7). Further investigation of the macrophage compartment by generating a population-specific force-directed map revealed differential remodeling of alveolar and interstitial macrophages in a circadian manner (Fig. S8A). Validation by conventional criteria corroborated that alveolar macrophages were more prevalent in the morning, whereas interstitial macrophages were increased in frequency in the afternoon (Fig. S8B and C). Thus, reference map analysis revealed previously undetected influence of circadian rhythms on immune organization of peripheral organs that was particularly prominent in pulmonary lymphocytes and macrophages. The symptom severity of patients diagnosed with infectious or atopic lung pathologies (i.e. allergies, asthma, and viral pneumonias) fluctuate in a circadian manner (44, 45). These results provide a potential explanation for these trends, as the lung-resident immune compartment undergoes circadian reorganization. This suggests that certain modes of antigen presentation could become exacerbated during different times of the day, or could indicate that nasally applied vaccines or therapeutics might have differing influences on immune function depending on the time of application.
Figure 4. Mapping Circadian Changes in the Lungs.
(A) Scaffold maps of lungs of representative animals collected in the morning (8-9am; left) or the afternoon (1-2 pm; right).
(B) Population frequencies in the lungs between morning and afternoon as defined by traditional criteria from both the original mass cytometry dataset (n=7 morning and afternoon) and a follow-up fluorescence experiment (n=7 morning; n=8 afternoon). Bars represent mean ± SEM, and p-values result from one-sided t-test.
Integrating Human Data into the Reference Map
Because immune cell types are well conserved between mice and humans, we analyzed human data overlaid onto the murine reference map (46). Mass cytometry data from whole peripheral blood from four healthy human donors was passed through the Scaffold map algorithm. We calculated distance between clusters on the basis of 15 cell-surface markers that have similar cell subset expression patterns between humans and mice (Fig. 5A-C). Differences between the species were apparent, such as the increased frequency of neutrophils and relative scarcity of B cells in human peripheral blood (47). However, the similar overlay pattern confirmed a common global structure of immunity. We also generated a map of murine blood using only the same 15 proteins to measure distance from the established landmarks (Fig. 5C). This similarity is not surprising. Gene expression networks from humans to mice have strong similarities—even to the point of enabling drug screening based on gene network similarities (48). The human data were not normalized or differentially transformed in any manner, underscoring the robustness of the mapping approach. Efforts to generate a human-centric reference map may enable more detailed mapping of human immune organization, but these results demonstrate the feasibility of comparing cellular features across the species barrier.
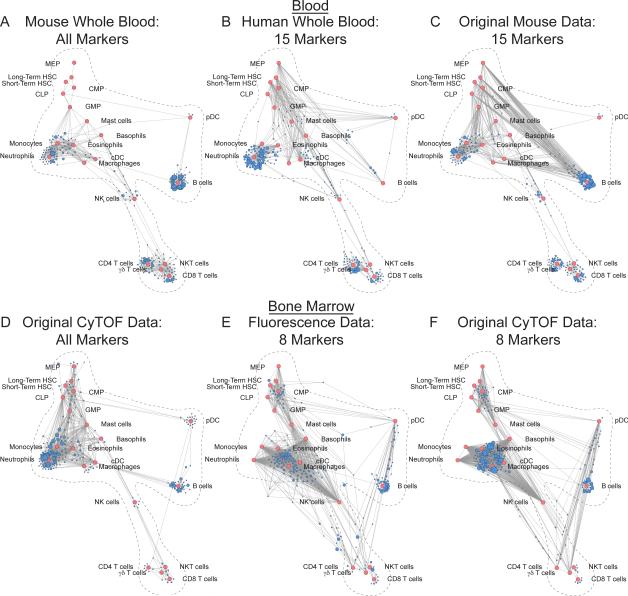
Figure 5. Mapping Human and Archival Data onto the Reference Map.
(A) Original mass cytometry whole blood Scaffold map from C57BL/6 mice, n = 14.
(B) Scaffold map of human whole blood interrogated by 15-parameter mass cytometry with distance measured using only those 15 dimensions for layout of unsupervised clusters onto the reference. Human parameters were assigned to murine correlate markers with similar cellular distribution, including canonical surface markers used for identification of cell populations by conventional criteria as well as several orthologous proteins, n = 4.
(C) Scaffold map of original murine blood mass cytometry data with distance measured using only the same 15 dimensions for layout of unsupervised clusters onto the reference.
(D) Original mass cytometry bone marrow Scaffold map from C57BL/6 mice.
(E) Scaffold map of C57BL/6 bone marrow interrogated by 8-color fluorescence-based flow cytometry from a previously published dataset (Qiu et al., 2011) with distance measured using only those 8 dimensions (B cell isoform of CD45 (B220), integrin αM (CD11b), T cell receptor β chain (TCRβ), cluster of differentiation 4 (CD4), cluster of differentiation 8 (CD8), stem cell growth factor receptor (c-Kit), stem cell antigen 1 (Sca-1), signaling lymphocytic activation molecule 1 (CD150)) for layout of unsupervised clusters onto the reference.
(F) Scaffold map of original mass cytometry data with distance measured using only the same 8 dimensions for layout of unsupervised clusters onto the reference.
Mapping Archival Data
The ability to map data from independent experiments would increase the utility of a reference map, creating a dynamic resource in which knowledge could accrue over time. Therefore, we mapped archival fluorescence-based flow cytometry data onto the reference map (Fig. 5D-F). We used a previously published dataset of bone marrow cells from C57BL/6 mice obtained with 8-color flow cytometry including lineage-specific markers (B cell isoform of CD45 (B220) for B cells, integrin αM (CD11b) for myeloid cells, T cell receptor β chain (TCRβ) for T cells, cluster of differentiation 4 (CD4) and 8 (CD8) to distinguish the major types of mature T cells) as well as stem cell/progenitor markers (stem cell growth factor receptor (c-Kit), stem cell antigen 1 (Sca-1), signaling lymphocytic activation molecule 1 (CD150))(17). We used only the information contained in these 8 dimensions to calculate similarity (Fig. 5E). As a point of reference, we also generated a Scaffold map from the original mass cytometry data of the C57BL/6 bone marrow using these same 8 dimensions (Fig. 5F).
Cells from the fluorescence data occupied the major regions of the Scaffold map with frequencies similar to those in the original reference. Moreover, the maps generated from both fluorescence and mass cytometry data using the same 8 dimensions exhibited strong similarity, suggesting that the underlying structure of the system remained the primary driver of the layout organization. Cell populations for which no unique markers exist and for which complex combinations of markers define cell types (such as the different myeloid cell subsets) exhibited lower resolution on the map and as such grouped in the center of several landmark nodes. Thus, although the specific selection of measured features impacts the ability to discriminate between similar cell populations, even a few key parameters can drive cell clusters toward cognate known reference cell subsets within the map.
A Cross-Sectional View of Cellular Compartments
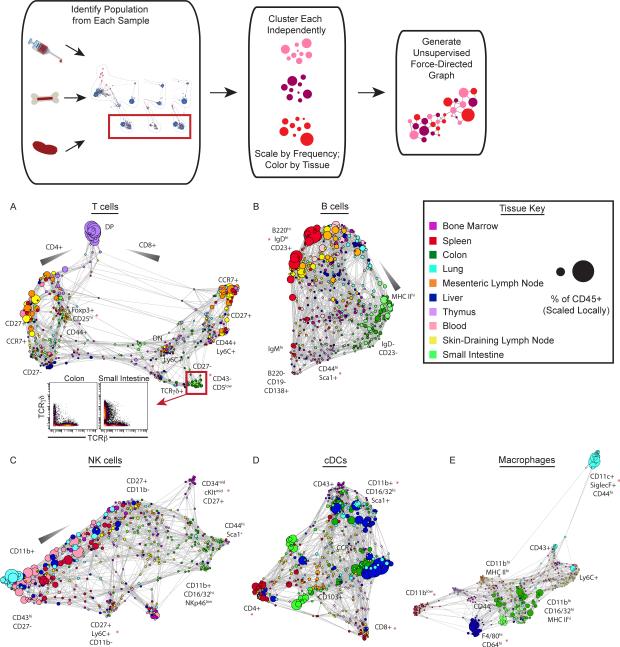
It would be useful to reveal detailed local structure of cell subsets that lack pre-existing landmarks to enable characterization of similarities and deviations. Having identified distinctions within given cell subsets across anatomical locations, we used unsupervised force-directed graphs (lacking landmark populations) to organize cells of a given cell type (T cells or dendritic cells, for instance) defined by traditional criteria such that differences between them would become apparent (Fig. 6). Each major cell population from every tissue was clustered and mapped together into force-directed graphs, resulting in a phenotypic landscape for that given cell type. As noted, manually-defined landmarks were omitted, though they could be defined in subsequent analyses as desired by the user. Cell clusters were colored according to their tissue of origin to reveal how each tissue is represented within the global similarity map for each cell type. Scaling each cluster proportionally to the percent of total leukocytes represented the relative frequency of cells in each cluster.
Figure 6. Defining the Landscape of Immune Cell Populations.
Population-specific landscapes were generated as follows: Cell populations were manually gated, subjected to unsupervised clustering and laid out in an unsupervised force-directed graph. Clusters are colored according to tissue of origin and sized by the number of cells in each cluster as a percent of the total number of leukocytes in the tissue of origin. Each plot is scaled independently.
(A) T cell landscape including Lineage marker (Lin)− cluster of differentiation 3 (CD3)+ cells. Cells comprising T cell clusters from the colon and small intestine falling within the red box are visualized by 2D scatter plot, n = 14.
(B) B cell landscape including Lin− B cell isoform of CD45 (B220)+ and Lin− syndecan-1 (CD138)+ cells, n = 14.
(C) NK cell landscape including Lin− cluster of differentiation 49b (CD49b)+ cells, n = 14.
(D) cDC landscape including Lin− integrin αx (CD11c)hi major histocompatibility complex class II (MHC II)hi cells, n = 14.
(E)Macrophage cell landscape including Lin− Fcγ receptor 1 (CD64)+ EGF-like module-containing mucin-like hormone receptor-like 1 (F4/80)+ cell, n = 14.
Lineage markers (Lin) defined in Materials and Methods.
We began by examining the landscape of T cells across the body, as T cells are well known to exhibit organ-specific properties. The mapping shows that a large group of cell clusters was exclusively located in the thymus and expressed both CD4 and CD8, characteristic of developmental double-positive (DP) T cells (Fig. 6A, S9, Table S4). The T cell map then showed two predominant branches characterized by CD4 (left) or CD8 expression (right), which were bridged by smaller clusters lacking high expression of either. Some of these cell clusters expressed the γδ T cell receptor (TCR)(Fig. 6A, insert). Others expressing TCRβ were localized to the gut and lungs, likely representing recently described mucosa-associated invariant T (MAIT) cells (Fig. S9, Table S4)(49). Among the CD4+ and CD8+ T cells expressing the αβ TCR, further divisions were driven by C-C chemokine receptor type 7 (CCR7), CD27 and CD44, which are common markers that distinguish differentiation states (Fig. S9, Table S4)(50). The tissue distribution of these subsets appeared skewed, with enrichment of effector and memory T cells in the peripheral solid organs. A group of CD4+ αβ T cell clusters expressed CD25 and forkhead box P3 (Foxp3), characteristic of regulatory T cells, and were overrepresented in the gut (Fig. S9, Table S4).
Whereas T cells demonstrate a largely bifurcated set of phenotypes with “bridging” cell subsets, the B cell landscape was markedly different, exhibiting a continuum of phenotypes in tissues distributed across the body (Fig. 6B). Although B cells in the bone marrow exhibited a wide range of phenotypes reflecting developmental stages, those in the secondary lymphoid organs expressed higher amounts of B220 and CD19 (a cell-surface co-receptor expressed by most mature B cells) with variable expression of the B cell receptor isotypes IgM and IgD and CD23 (the low-affinity IgE receptor)(Fig. S10, Table S4). The majority in peripheral solid organs exhibited reduced amounts of IgD and CD23 with increased MHC II (Fig. S10, Table S4)(51). Many thymic B cells exhibited a unique phenotype, characterized by the extracellular matrix receptor CD44 and Sca-1, and mapped near the plasma cells, which express syndecan 1 (CD138)(Fig. S10, Table S4). Thus, the B cell landscape was characterized by a phenotypic continuum with enrichment of specific phenotypes according to tissue of residence.
The NK cell landscape was predominantly organized by expression of CD11b and CD27, which delineate NK cell maturation stages (Fig. 6C, Fig. S11, Table S4)(52). A discrete population of NK cells expressing higher levels of developmental markers CD34 and cKit (CD117) was found in the bone marrow (Fig. S11, Table S4). In the peripheral solid organs, large populations of NK cells were present in the liver and lung with fewer in the gut. A group of NK cells with broad tissue distribution expressed Ly6C, which has been associated with NK cell memory (Fig. S11, Table S4)(53). These results recapitulated the known landscape of lymphoid cell biology and provided new insights regarding immune organization across the body according to the tissues in which the immune cells resided (Table S4).
Definitive statements regarding myeloid phenotypes and their functions remains a matter of interest (54, 55) and occasional contention (56). For instance, examining the cDC landscape revealed several sub-groups, some of which expressed CD4 or CD8 (the expression of which we noted were mutually exclusive and overrepresented in the secondary lymphoid organs (Fig. 6D). Several of the thymic cDC clusters expressed CD8, a feature characteristic of cross-presenting DC, which may reflect their need to present intracellular antigens in the context of both MHC I and II to promote T cell tolerance (Fig. S12, Table S4)(57). Many cDC in peripheral solid organs and the bone marrow were CD11b+ and expressed higher levels of Fcγ receptors (CD16/CD32), suggesting they may be more sensitive to antibody-mediated activation (Fig. S12, Table S4).
The macrophage landscape exhibited distinct segregation by location, consistent with their tissue- specific homeostatic functions and self-renewal (Fig. 6E)(15). Compared to macrophages present in the SLN and MLN, which exhibited high expression of the CD11b integrin and MHC II, red-pulp macrophages in the spleen expressed significantly less CD11b (Fig. S13, Table S4). The macrophages in the gut exhibited the highest expression of MHC II and Fcγ receptors (CD16/CD32), which might reflect a greater capacity to present antigen to CD4 T cells or sensitivity to activation via antibodies (Fig. S13, Table S4). Macrophages in the liver (Kupffer cells) expressed the highest levels of F4/80 and CD64 (Fcγ receptor I), whereas alveolar macrophages in the lung segregated far away based on their high expression of the CD11c integrin, the Siglec-F lectin and CD44 (Fig. S13, Table S4).
Thus, the force-directed graphical landscapes enabled rapid identification of the features that distinguish each population across the samples of interest, providing a model for characterizing the predominant differences between multiple conditions.
Summary and Conclusions
We exploited the increased parameterization afforded by mass cytometry to generate a consolidated, extensible reference map of the murine immune system with single-cell resolution. By assessing the composition and characteristics of immune populations throughout the body, this provides the basis for a systematic model of immune organization.
This objective necessitated new analytical methods for comparing groups of complex cellular samples. Our visualization algorithm combines unsupervised clustering with cellular landmarks defined by prior knowledge. The resulting Scaffold maps enabled global characterization of the steady-state immune structure from different anatomical locations, genetic backgrounds, circadian time points, and species barriers. When compared to an unsupervised graph across the organismal immune system (Fig. S14), the advantages of such a framework become apparent. The incorporation of landmarks assists in the interpretation of the graphical organization. They also importantly provide the reference points for comparing data, enabling the unique features of new, uncharacterized samples to stand out by comparison to a characterized baseline sample. A reference map of this nature will be useful in additional iterations when merged with immunological perturbations such as infection, autoimmune disease, or cancer to identify how altered immune states deviate from the steady state.
Beyond providing an analytical framework to understand immune organization from the unified dataset generated here, the approaches we describe can serve as a data repository for collating experimental data from the research community (Fig. S15). This would provide several distinct benefits. First, users could mine the data included in these studies to investigate the characteristics and distribution of cell types of interest in a dynamic way. Second, user modification of defined parameters (such as the definition of landmark populations) could provide analyses of immune structure not biased by prior strictures.
Perhaps more urgent to the community at large, mapping of newly created datasets onto a reference structure will assist in global comparisons of archival animal experiments with clinical human data. Investigators can merge newly mapped data to compare cellular features across previously mapped features in the reference landscape. With the implementation of standard regression analysis, the presence or absence of given clinical outcomes due to certain immune configurations might be discerned—much as has been the case with accessible archival gene expression datasets (9). In one analysis, the expression of a newly discovered regulatory molecule from ongoing forward genetics efforts (58, 59) could be defined in all immune cell types during health and disease. This could be achieved by measuring such a molecular feature by mass cytometry in addition to the proteins included here and mapping the resulting data. Alternatively, changes in metabolism or cell death programs within the global immune system during chronic inflammation or aging would be revealed, providing knowledge to inform the design of precise therapeutic strategies. Moreover, as the number of measurable parameters on a single-cell basis increases, the framework could easily be updated to reflect more detailed datasets.
Scaffold maps demonstrate the capacity to align data from distinct analysis platforms, including fluorescence-based flow cytometry, or across species of interest, such as the demonstration of mapping human immune data onto a murine framework. As the throughput of other single-cell analysis modalities, such as single-cell RNA-Seq (60, 61), continues to develop, these data could also be incorporated into the map along with other metadata types such as publication records, clinical phenotypes, and other relevant assays analogous to other strategies for data integration (62, 63). Therefore, this core infrastructure forms the basis for a centralized repository in which single-cell data can accrue over time, providing a unified reference map for understanding the organization and behavior of complex cellular systems. Efforts that characterize cellular behavior in this open-source approach will continue to improve upon the initiating reference presented here to reveal the inherent structure in biological networks of immunity for clinical benefit.
Supplementary Material
Acknowledgments
We thank J. Kenkel, B. Burt, D-H. Wang and M.Ch'ng for their assistance in tissue processing, A.Trejo and A. Jager for mass cytometry quality control and maintenance, B. Gaudilliere and M. Angst for access to human whole blood data, and M. Angelo, C. Loh, N. Reticker-Flynn and L. Sanman for constructive feedback.
M.H.S. was supported by a George D. Smith Stanford Graduate Fellowship and currently by NIH F31CA189331. P.F.G. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. G.F. is supported by a Stanford Bio-X Graduate Fellowship. E.R.Z. was supported by CIRM Basic Biology II RB2-01592 and NIH NRSA F32 GM093508-01. S.C.B. is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-2017-09) and K99GM104148-01. This work was supported by NIH 1 R01 DK096038; NIH 5 U01 CA141468; NIH 5 R01 DK082537; NIH 5 P01 HL075462; NIH 1 R01 CA163441 to E.G.E. and 0158 G KB065; 1R01CA130826; 5U54CA143907NIH; CIRM: DR1-01477; HEALTH.2010.1.2-1; HHSF223201210194C - FDA: BAA-12-00118; HHSN272200700038C; N01-HV-00242; NIH 41000411217; NIH 5-24927; P01 CA034233-22A1; PN2EY018228; RB2-01592; RFA CA 09-009; RFA CA 09-011; U19 AI057229; U54CA149145; W81XWH-12-1-0591 OCRP-TIA NWC, NIH S10 SIG S10RR027582-01 and the Rachford and Carlota A. Harris Endowed Professorship to G.P.N.
Footnotes
Supplementary material contains the following:
Materials and Methods
Supplementary Text
Figs. S1 to S20
Tables S1 to S4
References (64-68)
Author Contributions
References
- 1.Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747–749. doi: 10.1126/science.166.3906.747. [DOI] [PubMed] [Google Scholar]
- 2.Herzenberg LA, Herzenberg LA. Toward a layered immune system. Cell. 1989;59:953–954. doi: 10.1016/0092-8674(89)90748-4. [DOI] [PubMed] [Google Scholar]
- 3.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler's guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahnke Y, Chattopadhyay P, Roederer M. Publication of optimized multicolor immunofluorescence panels. Cytometry. 2010;77A:814–818. doi: 10.1002/cyto.a.20916. [DOI] [PubMed] [Google Scholar]
- 5.Aderem A, Hood L. Immunology in the post-genomic era. Nature Immunology. 2001;2:373–375. doi: 10.1038/87665. [DOI] [PubMed] [Google Scholar]
- 6.Zak DE, Tam VC, Aderem A. Systems-Level Analysis of Innate Immunity. Annu. Rev. Immunol. 2014;32:547–577. doi: 10.1146/annurev-immunol-032713-120254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heng TSP, Painter MW. Immunological Genome Project Consortium, The Immunological Genome Project: networks of gene expression in immune cells. Nature Immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 8.Jojic V, et al. Identification of transcriptional regulators in the mouse immune system. Nature Immunology. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sirota M, Butte AJ. The role of bioinformatics in studying rheumatic and autoimmune disorders. Nat Rev Rheumatol. 2011;7:489–494. doi: 10.1038/nrrheum.2011.87. [DOI] [PubMed] [Google Scholar]
- 10.Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M-S, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium TEP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2013;488:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutkowski J, et al. NeXO Web: the NeXO ontology database and visualization platform. Nucleic Acids Research. 2013;42:D1269–D1274. doi: 10.1093/nar/gkt1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto D, et al. Tissue-Resident Macrophages Self-Maintain Locally throughout Adult Life with Minimal Contribution from Circulating Monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinzel FP, Sadick MD, Holaday BJ, Coffman RL, Locksley RM. Reciprocal expression of interferon gamma or interleukin 4 during the resolution or progression of murine leishmaniasis. Evidence for expansion of distinct helper T cell subsets. J Exp Med. 1989;169:59–72. doi: 10.1084/jem.169.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nature Biotechnology. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amir E-AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature Biotechnology. 2013 doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendall SC, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:677–678. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendall SC, et al. Single-Cell Trajectory Detection Uncovers Progression and Regulatory Coordination in Human B Cell Development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ornatsky O, et al. Highly multiparametric analysis by mass cytometry. J. Immunol. Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Lou X, et al. Polymer-Based Elemental Tags for Sensitive Bioassays. Angew. Chem. Int. Ed. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidal M, Cusick ME, Barabási A-L. Interactome Networks and Human Disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozenblatt-Rosen O, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2013;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, et al. nbt.1563. Nature Biotechnology. 2009;27:829–837. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter H, Hofree M, Ideker T. Genotype to phenotype via network analysis. Current Opinion in Genetics & Development. 2013;23:611–621. doi: 10.1016/j.gde.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litvak V, et al. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 2013;490:421–425. doi: 10.1038/nature11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aghaeepour N, et al. Critical assessment of automated flow cytometry data analysis techniques. Nature Methods. 2013;10:228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ideker T, Dutkowski J, Hood L. Boosting Signal-to-Noise in Complex Biology: Prior Knowledge Is Power. Cell. 2011;144:860–863. doi: 10.1016/j.cell.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutkowski J, et al. A gene ontology inferred from molecular networks. Nature Biotechnology. 2012;31:38–45. doi: 10.1038/nbt.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP. A Continuous Molecular Roadmap to iPSC Reprogramming through Progression Analysis of Single-Cell Mass Cytometry. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eades P, Meek DS, Rees VGHJ. A Heuristic for Graph Drawing. Congressus Numerantium. 1984;42:149–160. [Google Scholar]
- 34.Fruchterman TM, Reingold EM. Graph drawing by force-directed placement. Software: Practice and experience. 1991;21:1129–1164. [Google Scholar]
- 35.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nature Biotechnology. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman L, Rousseeuw PJ. Finding Groups in Data. John Wiley & Sons; 1990. [Google Scholar]
- 38.Itano AA, Jenkins MK. Antigen presentation to naive CD4 T cells in the lymph node. Nature Immunology. 2003;4:733–739. doi: 10.1038/ni957. [DOI] [PubMed] [Google Scholar]
- 39.Kirby AC, Coles MC, Kaye PM. Alveolar Macrophages Transport Pathogens to Lung Draining Lymph Nodes. The Journal of Immunology. 2009;183:1983–1989. doi: 10.4049/jimmunol.0901089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Movita D, et al. Kupffer cells express a unique combination of phenotypic and functional characteristics compared with splenic and peritoneal macrophages. J. Leukoc. Biol. 2012;92:723–733. doi: 10.1189/jlb.1111566. [DOI] [PubMed] [Google Scholar]
- 41.Gavin AL, Leiter EH, Hogarth PM. Mouse FcgammaRI: identification and functional characterization of five new alleles. Immunogenetics. 2000;51:206–211. doi: 10.1007/s002510050033. [DOI] [PubMed] [Google Scholar]
- 42.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nature Reviews Immunology. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen KD, et al. Circadian Gene Bmal1 Regulates Diurnal Oscillations of Ly6Chi Inflammatory Monocytes. Science. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smolensky MH, Reinberg A, Labrecque G. Twenty-four hour pattern in symptom intensity of viral and allergic rhinitis: treatment implications. J. Allergy Clin. Immunol. 1995;95:1084–1096. doi: 10.1016/S0091-6749(95)70212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panzer SE, Dodge AM, Kelly EAB, Jarjour NN. Circadian variation of sputum inflammatory cells in mild asthma. J. Allergy Clin. Immunol. 2003;111:308–312. doi: 10.1067/mai.2003.65. [DOI] [PubMed] [Google Scholar]
- 46.Shay T, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 48.Usary J, et al. Predicting Drug Responsiveness in Human Cancers Using Genetically Engineered Mice. Clinical Cancer Research. 2013;19:4889–4899. doi: 10.1158/1078-0432.CCR-13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le Bourhis L, et al. Mucosal-associated invariant T cells: unconventional development and function. Trends Immunol. 2011;32:212–218. doi: 10.1016/j.it.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 51.Shimomura Y, et al. A unique B2 B cell subset in the intestine. Journal of Experimental Medicine. 2008;205:1343–1355. doi: 10.1084/jem.20071572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiossone L, et al. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 53.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8+ T cells. Nature Reviews Immunology. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature Immunology. 2012 doi: 10.1038/ni.2419. doi:10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller JC, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nature Immunology. 2012 doi: 10.1038/ni.2370. doi:10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unravelling mononuclear phagocyte heterogeneity. Nature Reviews Immunology. 2010;10:453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hadeiba H, Butcher EC. Thymus-homing dendritic cells in central tolerance. Eur. J. Immunol. 2013;43:1425–1429. doi: 10.1002/eji.201243192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoebe K, et al. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 59.Beutler B, Goodnow CC. How host defense is encoded in the mammalian genome. Mamm. Genome. 2011;22:1–5. doi: 10.1007/s00335-010-9312-4. [DOI] [PubMed] [Google Scholar]
- 60.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ideker T. Integrated Genomic and Proteomic Analyses of a Systematically Perturbed Metabolic Network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 63.Hwang D, et al. A data integration methodology for systems biology: experimental verification. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17302–17307. doi: 10.1073/pnas.0508649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Behbehani GK, et al. Transient partial permeabilization with saponin enables cellular barcoding prior to surface marker staining. Cytometry A. 2014;85:1011–1019. doi: 10.1002/cyto.a.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez A, Laio A. Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 68.Jacomy M, Venturini T, Heymann S, Bastian M, Muldoon MR. ForceAtlas2, a Continuous Graph Layout Algorithm for Handy Network Visualization Designed for the Gephi Software. PLoS ONE. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.All mass cytometry data are accessible at. http://54.215.233.207:8080/. R scripts are available at https://github.com/nolanlab.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.