Abstract
Birth defects are a major cause of morbidity and mortality worldwide. There has been much progress in understanding the genetic basis of familial and syndromic forms of birth defects. However, the etiology of nonsydromic birth defects is not well-understood. Although there is still much work to be done, we now have the tools to accomplish the task. Advances in next-generation sequencing have introduced a sea of possibilities, from disease-gene discovery to clinical screening and diagnosis. These advances have been fruitful in identifying a host of candidate disease genes, spanning the spectrum of birth defects. With the advent of CRISPR-Cas9 gene editing, researchers now have a precise tool for characterizing this genetic variation in model systems. Work in model organisms has also illustrated the importance of epigenetics in human development and birth defects etiology. Here we review past and current knowledge in birth defects genetics. We describe genotyping and sequencing methods for the detection and analysis of rare and common variants. We remark on the utility of model organisms and explore epigenetics in the context of structural malformation. We conclude by highlighting approaches that may provide insight into the complex genetics of birth defects.
Introduction
Nearly 8 million children are born in the world each year with a serious birth defect (Christianson et al., 2005). In the United States, birth defects affect at least 1 in every 33 newborns and result in considerable mortality and long-term disability (Centers for Disease Control and Prevention, 2008). Progress has been made in identifying environmental risk factors in nonsyndromic birth defects;1 however, ample work remains in terms of characterizing the genetic basis for most of these conditions. Here we review the genetic basis of nonsyndromic structural birth defects, with a focus on the four most common structural birth defects: congenital heart defects (CHD), neural tube defects (NTD), clefts of the lip and/or palate (CLP), and hypospadias. We provide a historical perspective and describe current microarray- and sequencing-based approaches for identifying common and rare variants underlying structural birth defects. We discuss the strengths and limitations of each technique and provide examples of the successful implementation of each approach to identify genetic factors influencing the risk of nonsyndromic birth defects.
CHD, NTD, CLP, and hypospadias account for nearly half of the birth defects that occur in the United States (Parker et al., 2010; Porter et al., 2005). CHDs are abnormalities of the heart or great vessels that are present at birth. They are the most common type of birth defect. These malformations occur in about 8 of every 1000 live births, and approximately 40% of babies born with the most serious CHDs die in infancy (Gilboa et al., 2010; Hoffman and Kaplan, 2002; Mathews et al., 2013; Moller et al., 1993; Pierpont et al., 2007; Yoon et al., 2001). Affected infants who survive often require repeated surgeries and lengthy hospitalization. Similarly, neural tube defects are often severe and debilitating. These malformations result from improper closure of the skull or vertebrae, leaving the brain or spinal cord exposed. In the United States, NTDs affect 0.6 in every 1000 births (Parker et al., 2010). Rates of NTDs are even higher in some developing countries (Castilla et al., 2003). CLP is a congenital malformation in which facial development is disrupted. It affects 2 of every 1000 births in the United States. Although it is not a major cause of infant mortality, children with craniofacial malformations often require surgery to repair the cleft lip or cleft palate and may encounter problems with feeding, speaking, hearing, or social stigmatism. Hypospadias is a structural malformation in which the opening of the urethra is located on the underside of the penis rather than on the tip. It affects approximately 3 per 1000 births (Dolk et al., 2004; Fisch et al., 2009; Porter et al., 2005).
Genetic Landscape: Past
Several lines of evidence, in both animal and human studies, indicate that most nonsyndromic defects have a genetic component. Existing evidence from human studies includes increased concordance among monozygotic twins compared to dizygotic twins, among full siblings compared to half siblings, and among first-degree relative compared to second- and third-degree relatives. Such studies point to a genetic basis for CHD (Oyen et al., 2009), NTD (Janerich and Piper, 1978), CLP (Christensen and Mitchell, 1996), and hypospadias (Schnack et al., 2008).
Candidate Gene Studies
Early genetic studies of nonsyndromic birth defects focused on testing the association of a small number of candidate single-nucleotide polymorphisms (SNPs) with common birth defects, including CLP, NTDs, and CHDs. For many of these studies, candidate genes were selected based upon mouse models of normal and abnormal development. For example, animal studies initially highlighted growth factors involved in development of the palate. Several of these developmental genes were later included in a small case-control study of nonsyndromic cleft lip with or without cleft palate (Ardinger et al., 1989). This work revealed an association between transforming growth factor-alpha (TGFA) and CLP – an association that has been replicated in subsequent studies (Lu et al., 2014). Characterization of NTDs in two mouse models (spin cycle and crash) led to the discovery of CELSR1 (Curtin et al., 2003) and associated proteins within the planar cell polarity pathway. Genes within this pathway (e.g. CELSR1, FUZ, VANGL1, VANGL2, and SCRIB) have since been linked to NTDs among humans. In the case of nonsyndromic CHDs, many of the critical cardiac transcription factors (e.g. NKX2-5, GATA4) were first characterized in the mouse then included as targets in candidate gene studies (Lyons et al., 1995; Molkentin et al., 1994). Discovery of mutations in the transcription factors, ZIC3, GATA4, and NKX2-5 in CHD has since highlighted the critical role of these proteins in cardiac development (McCulley and Black, 2012).
Candidate gene studies of birth defects not only built upon findings from developmental biology but also gained insight from epidemiologic studies. It was clear by 19922 that periconceptional folate intake reduced the risk of NTD. Subsequent work established a link between maternal periconceptional multivitamin use and reduced risk of conotruncal heart defects, limb deficiencies, and CLP (Shaw et al., 1995a; Shaw et al., 1995b). Frosst et al. (1995) described a polymorphism in methylenetetrahydrofolate reductase (MTHFR 677C>T) that encodes a thermolabile enzyme with diminished activity. Individuals with this form of MTHFR have a decreased concentration of serum folate and an increased concentration of homocysteine. Recognizing the potential implications, researchers soon tested for associations between MTHFR 677C>T and common birth defects. By the end of the decade, MTHFR 677C>T was established as an important risk factor in NTD (van der Put, Nathalie MJ et al., 1998) and conotruncal heart defects (Junker et al., 2001; van Beynum et al., 2006; Yin et al., 2012).
As custom genotyping microarrays became more available in the mid-2000s, researchers began to genotype entire pathways rather than individual genes. Despite changes in technology, folate-related genes continued to be an important focus of study (Hobbs et al., 2014; Shaw et al., 2009; Zhu et al., 2012). This approach cast a broader net in search of common variants affecting disease risk. It did so by examining tens to hundreds of polymorphisms, within the context of gene-environment (Hobbs et al., 2014; Zhu et al., 2012) and maternal-fetal interactions (Li et al., 2014). An increasingly large number of study participants were necessary in order to account for multiple testing. Therefore, common birth defects (CHD, NTD, and CLP) received much of the initial focus.
Candidate gene studies are especially well-suited to situations where there is strong evidence for involvement of a pathway in disease. However, this method may be vulnerable to inadvertent bias in the selection of candidate genes. It is also possible that a significant association is detected by chance alone. Therefore, it is important that results be replicated. In the past, there have been a number of questionable genotype-phenotype associations (Hirschhorn et al., 2002), which might have been clarified by rigorous validation efforts. To address this issue, the National Human Genome Research Institute (NHGRI) working group has outlined several best practices for replicating genotype-phenotype associations (Chanock et al., 2007). The guidelines have become requisite standards for GWAS, but they are equally applicable to candidate gene studies. The report recommends that a comparable phenotype and population should be analyzed in both the initial study and the replication. It also stresses that the replication study should be large enough to identify the initial association. Ideally, the replication sample should be at least as large in number as the discovery sample.
Genetic Landscape: Present
Birth defects sometimes cluster within families and have a higher recurrence rate among full-siblings compared to half-siblings. This is especially true of syndromic birth defects, which often segregate as autosomal dominant, autosomal recessive, or X-linked traits (Fahed et al., 2013). In contrast, genotype may play only a minor role in birth defects caused by maternal exposure to a teratogen such as isotretinoin (Rosa, 1983).
Genetic variation among humans is often classified as either rare or common. Common variants are arbitrarily defined as those with a minor allele frequency (MAF) of at least 5% (1000 Genomes Project Consortium, 2012); whereas, rare variants are often characterized as having a MAF less than 1%. There is a longstanding debate about the respective contributions of common and rare variants to complex diseases (Bodmer and Bonilla, 2008; Gibson, 2012). In reality, both common and rare variants are thought to contribute to risk of nonsyndromic birth defects. In the following pages, we will describe both categories and will review methods for variant detection and analysis.
Common Variants
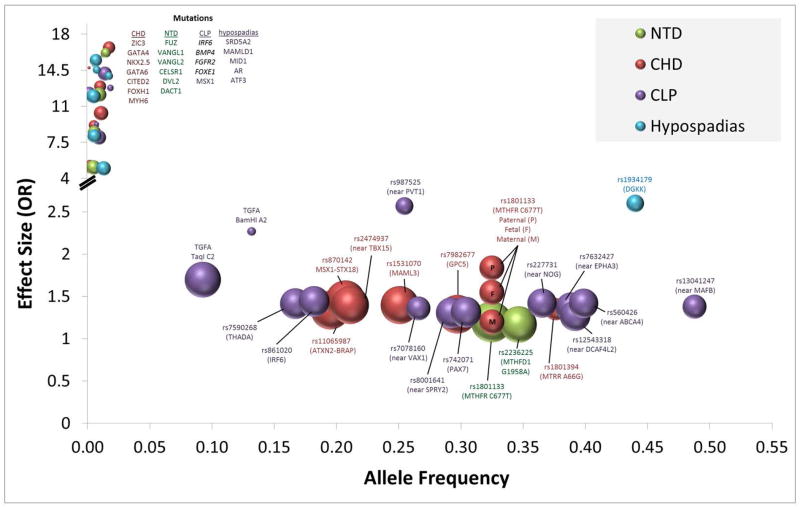
Genome wide association studies (GWAS) interrogate hundreds of thousands to millions of SNPs in order to identify associations between a genotype and complex disease. Nearly 2000 GWAS have been reported since the initial publication of Ozaki et al. (2002). These studies have identified numerous common variants that are risk factors for disease (Welter et al., 2014). The majority of GWAS of birth defects have focused on CLP, CHD, and hypospadias (Table 1). To our knowledge, none have include NTDs. GWAS of CHDs have identified 5 SNPs reaching genome-wide significance (p < 5 × 10−8). The CHD-associated SNPs have modest odds ratios (OR) ranging from 1.2 to 1.5 and have global minor allele frequency (GMAF) of 0.2–0.3 (Table 1). Meanwhile, GWAS on CLP have reported SNPs with greater effects size (OR = 1.4–2.6). Two GWAS has been conducted on hypospadias. The first identified a common variant (GMAF=.44) in the gene DGKK that is strongly associated with risk of hypospadias (van der Zanden, Loes FM et al., 2011). The second, much larger study identified 18 SNPs with genome-wide significance, accounting for a total of 8.7% of disease liability (Geller et al., 2014). A total of 56.9% disease liability was explained when all SNPs in the study were considered. This lends support to the infinitesimal model, in which hundreds or thousands of loci contribute to disease risk. In this model, statistically significant GWAS results represent only the largest of effects drawn from a normal distribution (Gibson, 2012).
Table 1.
Genome wide association studies of congenital heart defects, clefts of the lip and/or palate, and hypospadias.
| Phenotype | SNP | Allelea | GMAF | Chr. | Gene | P | OR het (95% CI) | OR hom (95% CI) | Author |
|---|---|---|---|---|---|---|---|---|---|
| Atrial septal defect | rs870142 | T/C | 0.207 | 4p16.2 | MSX1 - STX18 (between) | 2.60 × 10−10 | 1.46 (NR) | Cordell HJ et al. | |
| CHD | rs1531070 | A/G | 0.251 | 4q31.1 | MAML3 | 5.0 × 10−12 | 1.40 (1.27 – 1.54) | Hu Z et al. | |
| CHD | rs2474937 | C/T | 0.211 | 1p12 | TBX15 (closest) | 8.0 × 10−10 | 1.40 (1.26 – 1.56) | Hu Z et al. | |
| TOF | rs11065987 | G/A | 0.195 | 12q24.12 | ATXN2 - BRAP (between) | 7.7 × 10−11 | 1.34 (1.21 – 1.50) | Cordell HJ et al. | |
| TOF | rs7982677 | G/A | 0.297 | 13q31.3 | GPC5 | 3.03 × 10−11 | 1.29 (1.15 – 1.44) | Cordell HJ et al. | |
| CLP | rs560426 | G/A | 0.399 | 1p22.1 | ABCA4 (closest) | 5.01 × 10−12 | 1.42 (1.29 – 1.59) | Beaty TH et al. | |
| CLP | rs987525 | A/C | 0.254 | 8q24 | PVT1 - GSDMC between) | 3.34 × 10−24 | 2.57 (2.02 – 3.26) | 6.05 (3.88 – 9.43) | Birnbaum S et al. |
| CLP | rs7078160 | A/G | 0.266 | 10q25 | KIAA1598 - VAX1 | 1.92 × 10−8 | 1.36 (1.21 – 1.53) | 2.50 (1.95 – 3.21) | Mangold E et al |
| CLP | rs227731 | C/A | 0.365 | 17q22 | NOG (closest) | 1.07 × 10−8 | 1.38 (1.21 – 1.56) | 1.91 (1.63 – 2.24) | Mangold E et al |
| CLP | rs13041247 | C/T | 0.488 | 20q12 | MAFB (closest) | 1.44 × 10−11 | .70 (0.64 – 0.78) | Beaty TH et al. | |
| CLP | rs861020 | A/G | 0.182 | 1q32.2 | IRF6 | 3.24 × 10−12 | 1.44 (1.27 – 1.64) | 2.04 (1.60 –2.60) | Ludwig KU et al. |
| CLP | rs742071 | T/G | 0.304 | 1p36 | PAX7 | 7.02 × 10−9 | 1.32 (1.13 – 1.54) | 1.88 (1.52 – 2.32) | Ludwig KU et al. |
| CLP | rs7590268 | G/T | 0.167 | 2p21 | THADA | 1.25 × 10−8 | 1.42 (1.23 – 1.64) | 1.98 (1.47 – 2.66) | Ludwig KU et al. |
| CLP | rs7632427 | C/T | 0.388 | 3p11.1 | EPHA3 (closest) | 3.90 × 10−8 | 0.73 (0.64 – 0.83) | 0.61 (0.49 – 0.76) | Ludwig KU et al. |
| CLP | rs12543318 | C/A | 0.392 | 8q21.3 | DCAF4L2 (closest) | 1.90 × 10−8 | 1.27 (1.11 – 1.46) | 1.68 (1.40 – 2.01) | Ludwig KU et al. |
| CLP | rs8001641 | A/G | 0.292 | 13q31.1 | SPRY2 (closest) | 2.62 × 10−10 | 1.31 (1.13 – 1.51) | 1.86 (1.54 – 2.26) | Ludwig KU et al. |
| Hypospadias | rs1934179 | T/C | 0.438 | Xp11.22 | DGKK | 2.80 × 10−21 | 2.60 (2.10 – 3.10) | Van der Zanden LFM et al. | |
| Hypospadias | 18 SNPsb | Geller F et al. |
Odds Ratios (ORs) are given with the major allele set as baseline. Chr., chromosome; het, heterozygous; hom, homozygous; CHD, congenital heart defect; TOF, Tetralogy of Fallot, CLP, clefts of the lip and/or palate;
The minor allele is given first. The risk allele is shown in bold.
A total of 18 loci were significantly associated with hypospadias
Genotyping Technology
Genotyping methods have evolved rapidly since the first GWAS was completed in 2002. The number of variants assayed by GWAS has increased from around 10,000 SNPs in the early 2000s to 5 million SNPs at present (Hopper et al., 2012). The additional SNPs provide increased resolution of haplotypes, increased coverage of low frequency variants, and improved ability to infer genomic structural variation (Alkan et al., 2011). Structural variation, including insertions, deletions, and inversions, is broadly associated with nonsyndromic birth defects (Southard et al., 2012). SNP microarray and array comparative genomic hybridization (array CGH) have historically been the workhorses for detecting insertions and deletions, referred to as copy number variation (CNV). However, SNP microarrays may be gaining an edge because of their versatility. Increased density of SNP microarrays has made it possible to detect smaller structural variation and accurately resolve their breakpoints. Although the high density SNP microarray is a powerful tool, it is biased by a lower sensitivity in detecting single copy gains compared to single copy deletions. Even with high density chips, it can be challenging to consistently detect small events. Next-generation sequencing (NGS), a high-throughput method for sequencing millions or billions of DNA strands in parallel, overcomes many of these limitations and is arguably better suited at detecting small structural variation. There are multiple bioinformatics and statistical considerations in CV detection (Alkan et al., 2011) that are beyond the scope of this paper.
GWAS of CHD, CLP, and hypospadias have been successful in identifying common variants that influence disease risk (Beaty et al., 2010; Birnbaum et al., 2009; Cordell et al., 2013a; Cordell et al., 2013b; Geller et al., 2014; Hu et al., 2013; Ludwig et al., 2012; van der Zanden, Loes FM et al., 2011). These SNPs (Table 1) occur frequently in the population (af = 0.17-.49) and have modest effects size (OR = 1.3–2.6) (Figure 1). They may be directly involved in the disease etiology (i.e. functional or causal variant) or may “tag” a nearby functional variant that is involved in development of the disease. In most cases, the SNP being “tagged” is common within the population and has an effect size that is similar, if not slightly larger than the original SNP. In some instances, the GWAS points to genomic regions that are not only susceptible to perturbations caused by common variants but are also vulnerable to rare variants. Ventral anterior homeobox 1 (VAX1) proves an example of a gene that contains both common SNPs and rare functional variants. A GWAS first identified SNPs within VAX1 that were strongly associated with CLP. Sequencing of the gene among affected family trios later replicated the risk markers from the GWAS and identified both common and rare variants associated within CLP (Butali et al., 2013). Interferon regulatory factor 6 (IRF6) provides a similar example. Mutations in this gene are known to cause Van der Woude syndrome, an autosomal dominant form of cleft lip and palate. Not only do rare variants in IRF6 cause syndromic cleft lip and palate, but a common variant within an IRF6 enhancer influences disease risk by disrupting transcription factor binding (Rahimov et al., 2008). Genomic variants that have a major impact on health or reproductive fitness are more likely to be under selective pressures, limiting allele transmission and reducing allele frequencies within the population, as shown in the upper left-hand quadrant of Figure 1. These mutations often have large effects but occur at such low frequency that even in large GWAS they fail to meet genome-wide significance.
Figure 1. Allele frequency, effect size, and select birth defects.
Common single nucleotide polymorphisms (SNPs) with modest effect size (middle-center) have been identified by association studies of sporadic cases of congenital heart defects (CHD), neural tube defects (NTD), clefts of the lip and/or palate (CLP), and hypospadias. In contrast, certain rare mutations appear to have high penetrance/effect size for the above-listed birth defects. The genes listed in the upper left are rare causal mutations that have been substantiated among patient groups and characterized in model systems. There are few if any adequately powered studies for these mutations; therefore, the OR for mutations is estimated based upon published literature and is intended to illustrate the general trend. Disease-associated SNPs are identified in the lower middle part of the figure by spheres, which are scaled in size based upon the total number of cases and controls in combined discovery and validation studies, from 479 individuals in the smallest study to 10,091 individuals in the largest study. Allele frequency represents global minor allele frequency (GMAF) from dbSNP. Effect sizes in the figure are based upon GWAS in Table 1 and meta-analyses of candidate-gene association studies (Cai et al., 2014; Feng et al., 2014; Jiang et al., 2014; Lu et al., 2014; Yadav et al., 2014; Yin et al., 2012).
Risk loci identified by GWAS account for only a small fraction of the observed heritability of any particular birth defect (Gibson, 2012). Rare variants are seen to play an increasingly important role in the etiology of birth defects, and it is likely that rare alleles explain some of the missing heritability of complex traits. NGS provides an effective means of identifying rare variants. Whole-exome sequencing (WES) is an especially efficient approach for functional variant discovery. WES uses probe hybridization enrichment to capture 50–60 Mb of genomic DNA including, protein coding sequences, micro RNA, and in some cases untranslated regions flanking each gene (UTR). The exome comprises only 2% of the human genome; however, it contains the majority of known, disease-causing mutations. WES has been widely successful in identifying mutations responsible for inherited Mendelian diseases (Rabbani et al., 2012) and familial forms of birth defects (Arrington et al., 2012; Yu et al., 2013). As the cost of WES continues to fall, it becomes a more attractive tool for studying complex disorders, including birth defects. For example, exome sequencing of 362 cases-parent trios with CHD and 264 control-parent trios recently identified histone-modifying genes that are involved in CHD (Zaidi et al., 2013). Because CHD is under strong selective pressure, the investigators in this example focused on de novo mutations that might account for the sporadic pattern of occurrence among their cases. They found a very similar number of de novo protein-altering mutations among cases and controls, but interestingly the mutations among cases were more likely to occur in genes required for heart development. Based on the number de novo mutations in heart developmental genes, the paper estimated that such mutations have a role in 10% of severe nonsyndromic CHD. The results suggest that risk for isolated CHD is influenced by mutations that affect any one of a broad range of developmental genes. This mirrors a study among trios with autism, which showed that predicted damaging, de novo mutations among cases were more likely to affect genes expressed in the developing brain (Sanders et al., 2012).
WES has recently been used alongside SNP arrays in order to identify de novo CNV in CHD (Glessner et al., 2014). In a study conducted by the Pediatric Cardiac Genomics Consortium, two complementary technologies, WES and microarray, were used to detect de novo mutations among 538 CHD case-parent trios and 1301 healthy controls. SNP microarray is very effective at identifying large CNV that reside throughout the genome; however, it is unable to identify small CNV, has a bias towards detecting single copy losses, and cannot map the location of copy number gains. In contrast, exome sequencing is often inaccurate at identifying large structural variation, but is well-suited at detecting small CNV, which take the form of insertions or deletions (indels). By pairing WES with microarray, the investigators enhanced the effectiveness of both technologies and were able to identify recurrent de novo CNV at 15q11.2 and detect CNVs in genes that interact with key CHD proteins, NKX2-5 and GATA4 (Glessner et al., 2014).
WES has certain limitations. First, it covers only a small fraction of the genome. Regulatory elements play a critical role in development and disease risk (Wamstad et al., 2014); however, only a fraction of these elements are assessed by WES. A second limitation of WES is that it does not provide uniform coverage. Some regions of the exome have high coverage; whereas others receive limited coverage and suffer from low sensitivity in SNP detection. Uniformity of coverage for WES has improved in recent years, but there is still has much room for improvement (Meynert et al., 2014). WES also has limited accuracy in identifying structural variation greater than 30 bp. Synthetic long-read approaches may improve accuracy of indel detection from WES datasets (McCoy et al., 2014), but microarray and whole genome sequencing are currently more accurate alternatives for detecting long indels (Fang et al., 2014).
Whole-genome sequencing overcomes many of the limitations of WES. This method uses NGS technology to determine the DNA sequence of the human genome. Unlike WES, whole-genome sequencing does not depend upon targeted enrichment; thus, it provides fairly uniform coverage across much of the genome and has less bias in detecting non-reference alleles (Meynert et al., 2014). Because of improved coverage, whole-genome sequencing requires a mean depth of approximately 14 reads to achieve 95% sensitivity; whereas, WES requires a mean on-target depth of 40 reads to reach this threshold (Meynert et al., 2014). Whole-genome sequencing has been used to study autism, CHD, and other complex diseases (Chaiyasap et al., 2014; Michaelson et al., 2012). It is able to identify potential, disease-causing variants both within and between genes and is more accurate than WES in detecting structural variation, such as insertions or deletions (Fang et al., 2014). With the release of the Illumina HiSeq X Ten system in 2014, the cost of sequencing the whole genome at 30X depth has dropped to approximately $1000 per sample (Meynert et al., 2014). An economic analysis by Meynert et al. (2014) recently demonstrated that the cost of sequencing the whole exome and the whole genome are roughly equivalent for institutions with access to a HiSeq X Ten system. Because access to this sequencing platform is limited, WES still remains in most cases a more affordable option for sequencing protein coding regions. A major consideration with whole genome sequencing is that it generates an enormous amount of data, which must be stored and analyzed. Analysis requires bioinformatics and statistical expertise that is not broadly available. There are also challenges in predicting the functional consequences of variants, especially those within the intragenic region (Kircher et al., 2014).
Identifying de novo mutations is another strategy for discovering rare causal variants. Samocha et al. (2014) recently described a framework for interpreting de novo mutations, in which the number of de novo mutations within a gene or gene-set is compared to the expected number of mutations. In this framework, expected mutation rates are estimated based on local sequence context and selective constraint. Evolutionary constraint is not only informative within this context, but is also an important factor in prioritizing rare sequence variants. Several algorithms are available for estimating sequencing conservation among species: GERP++, PhyloP, SiPhy (Liu et al., 2013). Sequence variants may also be prioritized using functional prediction algorithms, such as PolyPhen2, SIFT, MutationTaster, MutationAssessor, FATHMM, or LRT (Liu et al., 2013). Functional prediction methods are individually prone to false positives; therefore, it is prudent to evaluate the consensus among models (Tennessen et al., 2012). Software is also available to help predict the effects of genomic variation upon RNA splicing; Human Splicing Finder represents one such tool (Desmet et al., 2009). Conservation- and functional-prediction tools are useful for prioritizing single nucleotide variation (SNV) within the exon. However, there are still major challenges to interpreting SNV within noncoding regions (Khurana et al., 2013). Prediction models are also not well suited to evaluating the consequences of insertions and deletions.
Functional Validation
Despite limitations, functional prediction models can be very helpful for population-based studies. Studies often identify multiple disease-associated variants. Functional predictions can help prioritize variants for further study. Likely causal variants may then be characterized in cell culture or in animal models. In studies where there is appropriate consent, lymphoblastoid cell lines can be generated from peripheral B lymphocytes. Cell lines possessing the candidate variant can be screened to detect phenotypic changes at a cellular level. In cases where a cell line from the proband is unavailable, gene editing may be used to introduce the candidate variant into an appropriate cell type. Several options are available for gene editing, including clustered regularly interspaced short palindromic repeats (CRISPRs), zinc-finger nucleases (ZFNs) and transcription activator–like effector nucleases (TALENs) (Gaj et al., 2013). The most recent addition to this group, CRISPR/Cas, has proven to be a powerful tool for sequence specific gene editing. CRISPR/Cas systems can be used to efficiently introduce a putative causal variant into model systems (Cong et al., 2013). CRISPR/Cas systems have been used generate mice (Heckl et al., 2014), zebrafish (Hwang et al., 2013), and Xenopus tropicalis (Nakayama et al., 2013) models with targeted mutations.
Gene knockdown provides an alternative method of studying a candidate disease gene. Fakhro et al. (2011) used a Xenopus tropicalis gene knockdown system to identify human gene orthologs that are responsible for heterotaxy, which is a type of congenital heart disease caused by defects in left-right body patterning. A large excess of copy number variants in 61 genes had been discovered in human heterotaxy subjects compared to unaffected subjects. Twenty-two of these genes had Xenopus orthologs, and 7 of these were found to be expressed in the ciliated left-right organizer. Gene knockdown experiments with 5 of the genes resulted in left-right heart morphological anomalies, thereby validating their function in cardiac left-right patterning. Animal models provide much of the foundation for what we know about developmental processes. However, these models are not without their shortcomings. Animals and humans often have differences in the rate and production of birth defects related to environment, teratogens, and modifying factors. In addition, genes are not uniformly conserved between humans and models organisms. These differences can hinder translation of findings between animals to humans. Nonetheless, model systems are invaluable in the study of birth defects and will play an increasingly vital role in characterizing candidate disease-genes related to human birth defects.
Epigenetics and non-syndromic birth defects
Epigenetic modifications include DNA methylation, modification of histones, and DNA interactions with non-coding RNA. DNA methylation, the most studied epigenetic modification, constitutes an epigenetic mechanism whereby a methyl group is covalently bound to a cytosine base in the context of CpG dinucleotides. Methylation of cytosines in DNA has been implicated in the mechanism of silencing of gene expression, genomic imprinting, chromosomal stability and protection against repetitive element expression.
During pregnancy, folate-dependent nucleotide synthesis and DNA methylation are increased (Oommen et al., 2005), and altered DNA methylation may be an underlying mechanism in the development of birth defects (Blom et al., 2006; Li et al., 2005; Okano et al., 1999). Indeed, recent studies showed that altered DNA methylation is associated with NTD and CHD (Chen et al., 2010; Chowdhury et al., 2011a; Chowdhury et al., 2011b; Wang et al., 2010). DNA methylation is an attractive therapeutic target for congenital defects because maternal dietary supplementation may restore DNA methylation patterns and negate the hypomethylating effects of harmful maternal exposures such as bisphenol-A (Dolinoy et al., 2007). Various maternal factors (Waterland and Jirtle, 2003) implicated in abnormal fetal development have been shown to affect DNA methylation patterns (Baccarelli et al., 2009; Candiloro and Dobrovic, 2009; Cooney et al., 2002). Alterations in epigenetic phenomena, such as DNA methylation, are likely to play a crucial role in determining the fetal phenotype (Wolff et al., 1998). The combined effects of genetics and epigenetics in the intrauterine environment and subsequent fetal development are not well understood and warrant further investigation.
Animal studies have confirmed the importance of folic acid and folate metabolism in normal fetal growth and development. A homomorphic mutation in the mouse MTRR gene, which is necessary for utilization of methyl groups from folate metabolism, resulted in developmental delay and congenital malformations, including neural tube and heart defects (Padmanabhan et al., 2013). Transgenerational effects of MTRR deficiency were also observed. When maternal grandparents were MTRR deficient, wild type female grand progeny exhibited wide-spread genomic instability and altered placental gene expression, as well as an increased level of congenital malformations. These malformations persisted in wild type progeny for five generations and were independent of maternal environment, suggesting transgenerational epigenetic inheritance, triggered by defects in MTRR.
In human studies, McKay et al. (2012) found that interindividual differences in DNA methylation patterns at birth are influenced by environmental factors, such as maternal vitamin B12 levels, genetic factors such as infant MTRR and maternal MTHFR genotypes, as well as length of gestation. These factors influence folate metabolism and affect the pool of methyl groups available for DNA methylation. Differences in methylation at the IGF2 locus, important in intrauterine growth (Börzsönyi et al., 2012), were associated with maternal MTHFR 677C>T polymorphism. They conclude that both global and gene-specific DNA methylation patterns in the developing fetus are dependent on genetic factors in both fetus and mother that influence folate metabolism, as well as the intrauterine environment (vitamin B12 levels). In a study of one pair of monozygotic twins who were discordant for renal agenesis, Jin et al. (2014) found no differences in SNPs, CNVs or indels between the twins. They did, however, find 514 differentially methylated regions that were localized to 10 signaling pathways and 25 genes, including 6 genes that are known to be involved in organ development. These data implicate DNA methylation in the mechanism of organogenesis as well as in congenital malformations.
Recent work by Zaidi et al. (2013) suggests that aberrant histone modification may play a role in certain birth defects. This study compared the frequency of harmful de novo mutations among infants affected by CHD compared to healthy controls. Infants with CHD had a 7.5-fold excess of protein-altering de novo mutations (premature termination, frameshift or splice site) in genes that are important in cardiac development. Notably, cases also had a significant excess of mutations in genes involved in histone modifications. Histone modifications influence gene transcription throughout life and are especially important in regulating developmental genes.
Most research concerning congenital malformations has focused on maternal or fetal genetics and/or epigenetics. The paternal genetic component of human birth defects has not been sufficiently studied. Some recent evidence has emerged from experimental work to suggest paternal genetics contribute to risk of birth defects in offspring. For example, Lambrot et al. (2013) determined that paternal dietary folate deficiency increases birth defects in offspring. It was demonstrated that a folate deficient diet alters the sperm DNA methylation at loci associated with genes responsible for normal development and disease. Also, sperm histone methylation was altered in folate deficient males, suggesting that dietary insufficiency of folate could alter gene expression. Folate deficient males had a significantly reduced pregnancy rate when mated to control females due to increased post-implantation resorption. An increase in the frequency of malformations was observed in offspring of folate deficient males, including hydrocephalus, limb and muscle or skeletal defects. These data suggest that male folate levels may also be important in the prevention of structural birth defects. Therefore, it may be useful to consider both maternal and paternal folate deficiency in future studies of birth defects.
Genetic Landscape: Future
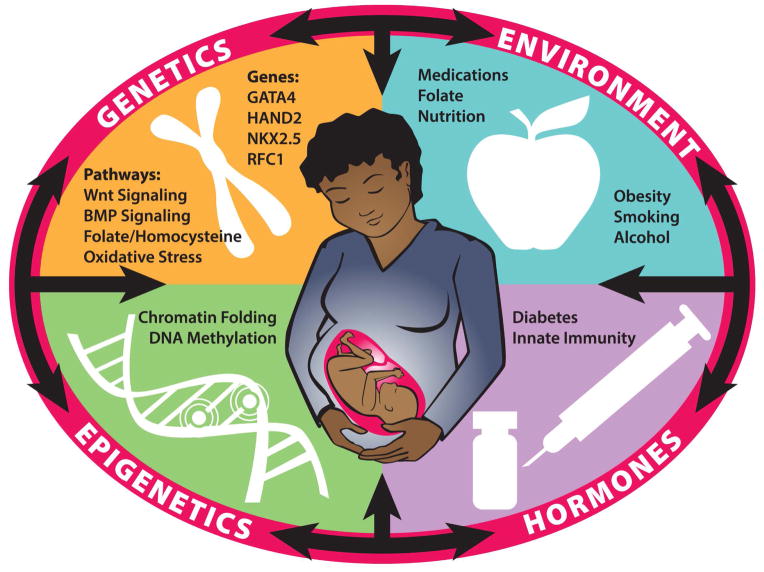
As Figure 2 illustrates, most birth defects are multifactorial in origin. Specifically, maternal environment (e.g. medications, folate, nutrition, obesity, smoking, alcohol) interacts with maternal genetics, epigenetics, and hormones to influence various metabolic processes and signaling pathways. These factors shape the intrauterine environment and can interact with fetal genetics and epigenetics to either facilitate or disrupt embryogenesis. Recent developments have helped to better understand such risk factors. In short, epidemiology has identify environmental risk factors; genomics has provided an unprecedented tool for identifying genetic variants within an affected population; model systems and gene editing have been invaluable in studying development and disease; and new analytical approaches have provided a means of interpreting complex datasets. Aided by these tools, research has shifted from a gene-centered analysis to a systems-based analysis that examines the interactions among metabolic pathways and gene networks within the context of maternal and intrauterine environments. Much progress has been made; however, there is still much work to be done.
Figure 2. Interactions between genetics, epigenetics, maternal hormonal levels, and environmental exposures during embryonic development.
Disruptions or anomalies in these interrelated systems may perturb the precise developmental program, leading to an increased risk of structural birth defects. Environmental and lifestyle factors, such as obesity, cigarette smoke, alcohol, nutrient intake, and folate supplementation influence folate and homocysteine metabolism and may impact oxidative stress. Oxidative stress and increased homocysteine can affect signaling pathways that include critical developmental genes, such as GATA4, HAND2, NKX2.5 and RFC1. Aberrant folate or homocysteine metabolism can also drain the pool of methyl groups that is critical to maintaining gene expression levels through DNA- and histone-methylation. This reflects the multifactorial origin of most nonsyndromic birth defects.
There have been successful reductions in some birth defects through research and population-based initiatives, for example, folate supplementation and fortification; yet, birth defects continue to be a global public health problem. Developing countries may not have the resources to follow through with nutritional or environmental preventive measures when malnutrition remains all too common. Birth defects persist in developed countries because nutritional, genetic, epigenetic environmental and unknown factors have not been fully investigated. Continued research in the areas of epidemiology, genetics, and epigenetics will lessen the global burden of birth defects through personalized and population-based preventative strategies.
Acknowledgments
The authors wish to thank Ashley S. Block for assistance in the preparation of this manuscript.
Funding Source: Support provided by the National Institute of Child Health and Human Development (5R01HD039054-12) and the National Center on Birth Defects and Developmental Disabilities (U01DD000491-05).
Footnotes
Nonsyndromic birth defects are defined in the context of this paper as congenital malformations that are not associated with a known or identifiable syndrome.
In 1992, the United States Public Health Service made the recommendation that women of childbearing age consume 0.4 mg of folic acid per day to reduce the risk of having a pregnancy affected by a neural tube defect (Houk et al., 1992).
References
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardinger HH, Buetow KH, Bell GI, Bardach J, VanDemark DR, Murray JC. Association of genetic variation of the transforming growth factor-alpha gene with cleft lip and palate. Am J Hum Genet. 1989;45:348–353. [PMC free article] [PubMed] [Google Scholar]
- Arrington CB, Bleyl SB, Matsunami N, Bonnell GD, Otterud BE, Nielsen DC, Stevens J, Levy S, Leppert MF, Bowles NE. Exome analysis of a family with pleiotropic congenital heart disease. Circ Cardiovasc Genet. 2012;5:175–182. doi: 10.1161/CIRCGENETICS.111.961797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, de Assis NA, Alblas MA. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Blom HJ, Shaw GM, den Heijer M, Finnell RH. Neural tube defects and folate: case far from closed. Nat Rev Neurosci. 2006;7:724–731. doi: 10.1038/nrn1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börzsönyi B, Demendi C, Pajor A, Rigo J, Jr, Marosi K, Ágota A, Nagy ZB, Joó JG. Gene expression patterns of the 11β-hydroxysteroid dehydrogenase 2 enzyme in human placenta from intrauterine growth restriction: the role of impaired feto-maternal glucocorticoid metabolism. Eur J Obstet Gynecol Reprod Biol. 2012;161:12–17. doi: 10.1016/j.ejogrb.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Butali A, Suzuki S, Cooper ME, Mansilla AM, Cuenco K, Leslie EJ, Suzuki Y, Niimi T, Yamamoto M, Ayanga G. Replication of genome wide association identified candidate genes confirm the role of common and rare variants in PAX7 and VAX1 in the etiology of nonsyndromic CL (P) Am J Med Genet A. 2013;161:965–972. doi: 10.1002/ajmg.a.35749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Zhang T, Zhong R, Zou L, Zhu B, Chen W, Shen N, Ke J, Lou J, Wang Z. Genetic Variant in MTRR, but Not MTR, Is Associated with Risk of Congenital Heart Disease: An Integrated Meta-Analysis. PloS one. 2014;9:e89609. doi: 10.1371/journal.pone.0089609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev Res (Phila) 2009;2:862–867. doi: 10.1158/1940-6207.CAPR-09-0056. [DOI] [PubMed] [Google Scholar]
- Castilla EE, Orioli IM, Lopez-Camelo JS, Dutra MdG, Nazer-Herrera J. Preliminary data on changes in neural tube defect prevalence rates after folic acid fortification in South America. Am J Med Genet A. 2003;123:123–128. doi: 10.1002/ajmg.a.20230. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Update on overall prevalence of major birth defects--Atlanta, Georgia, 1978–2005. MMWR Morb Mortal Wkly Rep. 2008;57:1–5. [PubMed] [Google Scholar]
- Chaiyasap P, Kulawonganunchai S, Srichomthong C, Tongsima S, Suphapeetiporn K, Shotelersuk V. Whole Genome and Exome Sequencing of Monozygotic Twins with Trisomy 21, Discordant for a Congenital Heart Defect and Epilepsy. PloS one. 2014;9:e100191. doi: 10.1371/journal.pone.0100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chen X, Guo J, Lei Y, Zou J, Lu X, Bao Y, Wu L, Wu J, Zheng X, Shen Y. Global DNA hypomethylation is associated with NTD-affected pregnancy: A case-control study. Birth Defects Res A Clin Mol Teratol. 2010;88:575–581. doi: 10.1002/bdra.20670. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Cleves MA, MacLeod SL, James SJ, Zhao W, Hobbs CA. Maternal DNA hypomethylation and congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011a;91:69–76. doi: 10.1002/bdra.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Erickson SW, MacLeod SL, Cleves MA, Hu P, Karim MA, Hobbs CA. Maternal genome-wide DNA methylation patterns and congenital heart defects. PloS one. 2011b;6:e16506. doi: 10.1371/journal.pone.0016506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Mitchell LE. Familial recurrence-pattern analysis of nonsyndromic isolated cleft palate--a Danish Registry study. Am J Hum Genet. 1996;58:182–190. [PMC free article] [PubMed] [Google Scholar]
- Christianson A, Howson CP, Modell B. March of Dimes: global report on birth defects, the hidden toll of dying and disabled children 2005 [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Bentham J, Topf A, Zelenika D, Heath S, Mamasoula C, Cosgrove C, Blue G, Granados-Riveron J, Setchfield K. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet. 2013a;45:822–824. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Topf A, Mamasoula C, Postma AV, Bentham J, Zelenika D, Heath S, Blue G, Cosgrove C, Granados Riveron J, Darlay R, Soemedi R, Wilson IJ, Ayers KL, Rahman TJ, Hall D, Mulder BJ, Zwinderman AH, van Engelen K, Brook JD, Setchfield K, Bu’Lock FA, Thornborough C, O’Sullivan J, Stuart AG, Parsons J, Bhattacharya S, Winlaw D, Mital S, Gewillig M, Breckpot J, Devriendt K, Moorman AF, Rauch A, Lathrop GM, Keavney BD, Goodship JA. Genome-wide association study identifies loci on 12q24 and 13q32 associated with tetralogy of Fallot. Hum Mol Genet. 2013b;22:1473–1481. doi: 10.1093/hmg/dds552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, Henderson DJ, Spurr N, Stanier P, Fisher EM. Mutation of Celsr1 Disrupts Planar Polarity of Inner Ear Hair Cells and Causes Severe Neural Tube Defects in the Mouse. Curr Biol. 2003;13:1129–1133. doi: 10.1016/s0960-9822(03)00374-9. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolk H, Vrijheid M, Scott JE, Addor MC, Botting B, de Vigan C, de Walle H, Garne E, Loane M, Pierini A, Garcia-Minaur S, Physick N, Tenconi R, Wiesel A, Calzolari E, Stone D. Toward the effective surveillance of hypospadias. Environ Health Perspect. 2004;112:398–402. doi: 10.1289/ehp.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112:707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci U S A. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Wu Y, Narzisi G, O’Rawe JA, Barrón LTJ, Rosenbaum J, Ronemus M, Iossifov I, Schatz MC, Lyon GJ. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med. 2014;6:89. doi: 10.1186/s13073-014-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Zhang E, Duan W, Xu Z, Zhang Y, Lu L. Association between polymorphism of TGFA Taq I and cleft Lip and/or palate: a meta-analysis. BMC oral health. 2014;14:88. doi: 10.1186/1472-6831-14-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch H, Lambert SM, Hensle TW, Hyun G. Hypospadias rates in New York State are not increasing. J Urol. 2009;181:2291–2294. doi: 10.1016/j.juro.2009.01.059. [DOI] [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, Van Den Heuvel L. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. 1995 doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller F, Feenstra B, Carstensen L, Pers TH, van Rooij IA, Körberg IB, Choudhry S, Karjalainen JM, Schnack TH, Hollegaard MV. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat Genet. 2014 doi: 10.1038/ng.3063. [DOI] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa SM, Salemi JL, Nembhard WN, Fixler DE, Correa A. Mortality Resulting From Congenital Heart Disease Among Children and Adults in the United States, 1999 to 2006Clinical Perspective. Circulation. 2010;122:2254–2263. doi: 10.1161/CIRCULATIONAHA.110.947002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Bick AG, Ito K, Homsy JG, Rodriguez-Murillo L, Fromer M, Mazaika E, Vardarajan B, Italia M, Leipzig J, DePalma SR, Golhar R, Sanders SJ, Yamrom B, Ronemus M, Iossifov I, Willsey AJ, State MW, Kaltman JR, White PS, Shen Y, Warburton D, Brueckner M, Seidman C, Goldmuntz E, Gelb BD, Lifton R, Seidman J, Hakonarson H, Chung WK. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115:884–896. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Hobbs CA, Cleves MA, MacLeod SL, Erickson SW, Tang X, Li J, Li M, Nick T, Malik S. Conotruncal heart defects and common variants in maternal and fetal genes in folate, homocysteine, and transsulfuration pathways. Birth Defects Res A Clin Mol Teratol. 2014;100:116–126. doi: 10.1002/bdra.23225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JIE, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Hopper J, Makalic E, Schmidt D, Bui M, Stone J, Kapuscinski M, Park D, Jenkins M, Southey M. ‘Next-generation’genome wide association studies. Hered Cancer Clin Pract. 2012;10:A48. [Google Scholar]
- Houk V, Oakley G, Jr, Erickson JD, Mulinare J, James LM. Recommendations for the use of folic acid to reduce the number of cases of spina bifida and other neural tube defects. Morb Mortal Weekly Rep. 1992;41:1–7. [PubMed] [Google Scholar]
- Hu Z, Shi Y, Mo X, Xu J, Zhao B, Lin Y, Yang S, Xu Z, Dai J, Pan S. A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations. Nat Genet. 2013;45:818–821. doi: 10.1038/ng.2636. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janerich DT, Piper J. Shifting genetic patterns in anencephaly and spina bifida. J Med Genet. 1978;15:101–105. doi: 10.1136/jmg.15.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhang Y, Wei L, Sun Z, Liu Z. Association between MTHFD1 G1958A Polymorphism and Neural Tube Defects Susceptibility: A Meta-Analysis. PloS one. 2014;9:e101169. doi: 10.1371/journal.pone.0101169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Zhu S, Hu P, Liu D, Li Q, Li Z, Zhang X, Xie Y, Chen X. Genomic and Epigenomic Analyses of Monozygotic Twins Discordant for Congenital Renal Agenesis. Am J Kidney Dis. 2014 doi: 10.1053/j.ajkd.2014.01.423. [DOI] [PubMed] [Google Scholar]
- Junker R, Kotthoff S, Vielhaber H, Halimeh S, Kosch A, Koch HG, Kassenbohmer R, Heineking B, Nowak-Gottl U. Infant methylenetetrahydrofolate reductase 677TT genotype is a risk factor for congenital heart disease. Cardiovasc Res. 2001;51:251–254. doi: 10.1016/s0008-6363(01)00286-3. [DOI] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Colonna V, Mu XJ, Kang HM, Lappalainen T, Sboner A, Lochovsky L, Chen J, Harmanci A, Das J, Abyzov A, Balasubramanian S, Beal K, Chakravarty D, Challis D, Chen Y, Clarke D, Clarke L, Cunningham F, Evani US, Flicek P, Fragoza R, Garrison E, Gibbs R, Gumus ZH, Herrero J, Kitabayashi N, Kong Y, Lage K, Liluashvili V, Lipkin SM, MacArthur DG, Marth G, Muzny D, Pers TH, Ritchie GR, Rosenfeld JA, Sisu C, Wei X, Wilson M, Xue Y, Yu F, Dermitzakis ET, Yu H, Rubin MA, Tyler-Smith C, Gerstein M. 1000 Genomes Project Consortium. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrot R, Xu C, Saint-Phar S, Chountalos G, Cohen T, Paquet M, Suderman M, Hallett M, Kimmins S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat Commun. 2013;4 doi: 10.1038/ncomms3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Erickson SW, Hobbs CA, Li J, Tang X, Nick TG, Macleod SL, Cleves MA. Detecting Maternal-Fetal Genotype Interactions Associated With Conotruncal Heart Defects: A Haplotype-Based Analysis With Penalized Logistic Regression. Genet Epidemiol. 2014;38:198–208. doi: 10.1002/gepi.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Pickell L, Liu Y, Wu Q, Cohn JS, Rozen R. Maternal methylenetetrahydrofolate reductase deficiency and low dietary folate lead to adverse reproductive outcomes and congenital heart defects in mice. Am J Clin Nutr. 2005;82:188–195. doi: 10.1093/ajcn.82.1.188. [DOI] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP v2. 0: A Database of Human Non-synonymous SNVs and Their Functional Predictions and Annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XC, Yu W, Tao Y, Zhao PL, Li K, Tang LJ, Zheng JY, Li LX. Contribution of transforming growth factor alpha polymorphisms to nonsyndromic orofacial clefts: a HuGE review and meta-analysis. Am J Epidemiol. 2014;179:267–281. doi: 10.1093/aje/kwt262. [DOI] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Mathews TJ, MacDorman MF Division of Vital Statistics. Infant Mortality Statistics from the 2010 Period Linked Birth/Infant Death Data Set. 2013;62(8) [PubMed] [Google Scholar]
- McCoy RC, Taylor RW, Blauwkamp TA, Kelley JL, Kertesz M, Pushkarev D, Petrov DA, Fiston-Lavier A. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly repetitive transposable elements. 2014 doi: 10.1371/journal.pone.0106689. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JA, Groom A, Potter C, Coneyworth LJ, Ford D, Mathers JC, Relton CL. Genetic and non-genetic influences during pregnancy on infant global and site specific DNA methylation: role for folate gene variants and vitamin B12. PLoS One. 2012;7:e33290. doi: 10.1371/journal.pone.0033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics. 2014;15:247-2105-15-247. doi: 10.1186/1471-2105-15-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Kalvakolanu DV, Markham BE. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the alpha-myosin heavy-chain gene. Mol Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller J, Allen H, Clark E, Dajani A, Golden A, Hayman L, Lauer R, Marmer E, McAnulty J, Oparil S. Report of the task force on children and youth. Circulation. 1993;88:2479–2486. doi: 10.1161/01.cir.88.5.2479. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oommen AM, Griffin JB, Sarath G, Zempleni J. Roles for nutrients in epigenetic events. J Nutr Biochem. 2005;16:74–77. doi: 10.1016/j.jnutbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. doi: 10.1161/CIRCULATIONAHA.109.857987. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- Padmanabhan N, Jia D, Geary-Joo C, Wu X, Ferguson-Smith AC, Fung E, Bieda MC, Snyder FF, Gravel RA, Cross JC. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell. 2013;155:81–93. doi: 10.1016/j.cell.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, Anderson P, Mason CA, Collins JS, Kirby RS. Updated national birth prevalence estimates for selected birth defects in the United States, 2004–2006. Am J Med Genet A. 2010;88:1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- Pierpont ME, Basson CT, Benson DW, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL. Genetic basis for congenital heart defects: current knowledge. Circulation. 2007;115:3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- Porter MP, Faizan MK, Grady RW, Mueller BA. Hypospadias in Washington State: maternal risk factors and prevalence trends. Pediatrics. 2005;115:e495–9. doi: 10.1542/peds.2004-1552. [DOI] [PubMed] [Google Scholar]
- Rabbani B, Mahdieh N, Hosomichi K, Nakaoka H, Inoue I. Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J Hum Genet. 2012;57:621–632. doi: 10.1038/jhg.2012.91. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C. Disruption of an AP-2α binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa F. Teratogenicity of isotretinoin. Lancet. 1983;322:513. doi: 10.1016/s0140-6736(83)90538-x. [DOI] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack TH, Zdravkovic S, Myrup C, Westergaard T, Christensen K, Wohlfahrt J, Melbye M. Familial aggregation of hypospadias: a cohort study. Am J Epidemiol. 2008;167:251–256. doi: 10.1093/aje/kwm317. [DOI] [PubMed] [Google Scholar]
- Shaw GM, Lu W, Zhu H, Yang W, Briggs FBS, Carmichael SL, Barcellos LF, Lammer EJ, Finnell RH. 118 SNPs of folate-related genes and risks of spina bifida and conotruncal heart defects. BMC Medical Genetics. 2009;10:49. doi: 10.1186/1471-2350-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, O’Malley CD, Wasserman CR, Tolarova MM, Lammer EJ. Maternal periconceptional use of multivitamins and reduced risk for conotruncal heart defects and limb deficiencies among offspring. Am J Med Genet. 1995a;59:536–545. doi: 10.1002/ajmg.1320590428. [DOI] [PubMed] [Google Scholar]
- Shaw GM, Wasserman C, O’Malley C, Tolarova M, Lammer E. Risks of orofacial clefts in children born to women using multivitamins containing folic acid periconceptionally. Lancet. 1995b;346:393–396. doi: 10.1016/s0140-6736(95)92778-6. [DOI] [PubMed] [Google Scholar]
- Southard AE, Edelmann LJ, Gelb BD. Role of copy number variants in structural birth defects. Pediatrics. 2012;129:755–763. doi: 10.1542/peds.2011-2337. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beynum IM, Kouwenberg M, Kapusta L, den Heijer M, van der Linden IJM, Daniels O, Blom HJ. MTRR 66A> G polymorphism in relation to congenital heart defects. Clin Chem Lab Med. 2006;44:1317–1323. doi: 10.1515/CCLM.2006.254. [DOI] [PubMed] [Google Scholar]
- van der Put, Nathalie MJ, Gabreëls F, Stevens E, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel, Lambert P, Blom HJ. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zanden, Loes FM, van Rooij IA, Feitz WF, Knight J, Donders ART, Renkema KY, Bongers EM, Vermeulen SH, Kiemeney LA, Veltman JA. Common variants in DGKK are strongly associated with risk of hypospadias. Nat Genet. 2011;43:48–50. doi: 10.1038/ng.721. [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Wang X, Demuren OO, Boyer LA. Distal enhancers: new insights into heart development and disease. Trends Cell Biol. 2014;24:294–302. doi: 10.1016/j.tcb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang F, Guan J, Le J, Wu L, Zou J, Zhao H, Pei L, Zheng X, Zhang T. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am J Clin Nutr. 2010;91:1359. doi: 10.3945/ajcn.2009.28858. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- Yadav U, Kumar P, Yadav SK, Mishra OP, Rai V. Polymorphisms in folate metabolism genes as maternal risk factor for Neural Tube Defects: an updated meta-analysis. Metab Brain Dis. 2014:1–18. doi: 10.1007/s11011-014-9575-7. [DOI] [PubMed] [Google Scholar]
- Yin M, Dong L, Zheng J, Zhang H, Liu J, Xu Z. Meta analysis of the association between MTHFR C677T polymorphism and the risk of congenital heart defects. Ann Hum Genet. 2012;76:9–16. doi: 10.1111/j.1469-1809.2011.00687.x. [DOI] [PubMed] [Google Scholar]
- Yoon PW, Rasmussen SA, Lynberg MC, Moore CA, Anderka M, Carmichael SL, Costa P, Druschel C, Hobbs CA, Romitti PA. The National Birth Defects Prevention Study. Public Health Rep. 2001;116:32. doi: 10.1093/phr/116.S1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Wynn J, Cheung YH, Shen Y, Mychaliska GB, Crombleholme TM, Azarow KS, Lim FY, Chung DH, Potoka D. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum Genet. 2013;132:285–292. doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Yang W, Lu W, Etheredge AJ, Lammer EJ, Finnell RH, Carmichael SL, Shaw GM. Gene variants in the folate-mediated one-carbon metabolism (FOCM) pathway as risk factors for conotruncal heart defects. Am J Hum Genet. 2012;158:1124–1134. doi: 10.1002/ajmg.a.35313. [DOI] [PMC free article] [PubMed] [Google Scholar]