Abstract
Background and Purpose
This study investigates 36 subjects aged 55 to 65 from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database to expand our knowledge of early-onset (EO) Alzheimer’s Disease (EO-AD) using neuroimaging biomarkers.
Methods
Nine of the subjects had EO-AD, and 27 had EO mild cognitive impairment (EO-MCI). The structural ADNI data were parcellated using BrainParser, and the 15 most discriminating neuroimaging markers between the two cohorts were extracted using the Global Shape Analysis (GSA) Pipeline workflow. Then the Local Shape Analysis (LSA) Pipeline workflow was used to conduct local (per-vertex) post-hoc statistical analyses of the shape differences based on the participants’ diagnoses (EO-MCI+EO-AD). Tensor-based Morphometry (TBM) and multivariate regression models were used to identify the significance of the structural brain differences based on the participants’ diagnoses.
Results
The significant between-group regional differences using GSA were found in 15 neuroimaging markers. The results of the LSA analysis workflow were based on the subject diagnosis, age, years of education, APOE(ε4), MMSE, visiting times, and logical memory as regressors. All the variables had significant effects on the regional shape measures. Some of these effects survived the False Discovery Rate (FDR) correction. Similarly, the TBM analysis showed significant effects on the Jacobian displacement vector fields, but these effects were reduced after FDR correction.
Conclusions
These results may explain some of the differences between EO-AD and EO-MCI, and some of the characteristics of the EO cognitive impairment subjects.
Keywords: Alzheimer’s Disease, early-onset, ADNI, neuroimaging, brain mapping
Introduction
Alzheimer’s disease (AD) is the most common cause of neurodegenerative dementia. It leads to irreversible neuronal loss and progressive cognitive decline, and spreads from the memory to all other cognitive domains, eventually causing death1. Advancing age is the single most important risk factor of AD; and with life expectancy and population increases, its incidence is expected to double in the next two decades.2 AD prevalence among individuals especially between 65 and 85 years of age is exponentially increasing.2 AD has a strong genetic component, with up to 80% heritability, as estimated from twin-concordance studies.3,4
AD is divided into two main subtypes: early-onset AD (EO-AD) and sporadic AD. Early-onset dementia diagnosis is determined when the disease presentation begins before the age of 65. Researchers have discovered three genes associated with EO-AD. These genes are APP,5 which encodes amyloid precursor protein on chromosome 21; PS-1,6 encoding presenilin 1 on chromosome 14; and PS-2,7,8 which encodes presenilin 2 on chromosome 1. However, these known genetic mutations account for only 2% of all cases of EO-AD.9 Sporadic AD is the other group which is most commonly termed late-onset AD (LO-AD). It is defined by the disease presentation after the age of 65, and it is well-known that Apolipoprotein E (APOE) ε4 can influence it. However, according to the current viewpoint, classifying AD into EO and LO is probably not useful from a mechanistic point of view because mutations in APP, PS-1, and PS-2 can be found in both EO and LO. Similarly, APOE ε4 increases the risk of AD in both EO-AD and LO-AD.10
Recent proposed consensus criteria for AD have underlined the role played by neuroimaging phenotypes for the disease diagnosis.11 Accordingly, MRI-based measures of atrophy in several structures, including the hippocampus,12–17 entorhinal cortex,18 and temporal lobe volumes,19 as well as of ventricular enlargement12,20 have been claimed as the fingerprint of preclinical AD. Of all the MRI markers of AD, hippocampal atrophy assessed on high-resolution T1-weighted MRIs is perhaps the most significantly established and validated.
However, despite the wide body of literature on the accuracy of neuroimaging markers in identifying subjects at risk of developing AD, much less attention has been devoted to EO-AD. There may be different reasons for this. First, EO-AD is rarer disorder than LO-AD, and thus, it is difficult to have a reasonable number of study subjects for it to achieve reliable results. Second, in most cases, researchers, considering EO-AD to be genetically based, give too much attention to genetics determinants and less attention to biomarkers, including neuroimaging.21
It has been widely demonstrated that the cognitive pattern of EO-AD differs from that of LO-AD, in that in the former, the neocortical functions are more affected, which shows that EO-AD and LO-AD differ in their typical topographic patterns of brain atrophy.22–25 Yet there have been few studies on the differences between cohorts, including EO-AD and EO-MCI (mild cognitive impairment) cohorts, or in some characteristics of EO cognitive impairment subjects (EO-AD+EO-MCI), in terms of neuroimaging, especially using volume- and shape-based morphometrics.
This article investigates subjects aged 55 to 65 to broaden our understanding the EO cognitive impairment, including EO-AD and EO-MCI, in terms of neuroimaging. We employ the Global Shape Analysis (GSA), Local Shape Analysis (LSA), and Tensor-based Morphometry (TBM) workflows via the Pipeline workflow environment. Neuropsychology data can be integrated in the imaging analysis of volume- and shape-based measurements. We expect this neuroimaging study to explain some of the differences between the two cohorts, and some of the characteristics of EO cognitive impairment subjects (EO-AD+EO-MCI) in terms of volume- and shape-based morphometrics using GSA, LSA, and TBM under the LONI Pipeline environment.
Methods
ADNI Data
The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. Its primary goal to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early-stage AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principle Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California San Francisco. ADNI is the result of the efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, aged 55–90, to participate in the research—approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years. For up-to-date information see http://www.adni-info.org. Baseline and longitudinal structural MRI scans are collected from the full sample every 6–12 months.
Study Participants
Data (ADNI study design)
The participants were screened, enrolled, and followed up prospectively according to the ADNI study protocol described in detail elsewhere.26 The degree of clinical severity for each participant was evaluated with an annual semi-structured interview. This interview generated an overall Clinical Dementia Rating (CDR) score and the CDR Sum of Boxes.27 The Mini-Mental State Examination (MMSE) for each structural MRI scan was also conducted. The Logical Memory (LM) Test is a modified version of the episodic memory assessment from the Wechsler Memory Scale-Revised (WMS-R).28 The subjects were asked to recall a short story with 25 pieces of information, both immediately after the story was read to the subject, and after a 30-minute delay. The maximum score was 25, with each recalled piece of information given 1 point. The LM test was done repeatedly with 12-month intervals. APOE genotyping was determined using DNA obtained from the subjects’ blood samples and was performed at the University of Pennsylvania.
The participants were selected if they were classified at the baseline as (1) patients with MCI with an MMSE score between 24 and 29, a subjective memory complaint verified by an informant, objective memory loss as measured by the Wechsler Memory Scale-Revised, a CDR score of 0.5, absence of significant levels of impairment in other cognitive domains, essentially preserved activities of daily living, absence of dementia at the time of the baseline MRI scan, and with amnestic-type MCI based on the revised MCI criteria; and (2) patients with AD who met criteria for probable AD (CDR score of 1). The MCI subjects were scanned at 0, 6, 12, 18, 24, and 36 months, and the MRI scans for the AD subjects were collected at 0, 6, 12, and 24 months.
We included only the EO-MCI and EO-AD cohorts in the GSA, LSA, and TBM analyses. Nine had EO-AD (male: 4 and female: 5) and twenty-seven had EO-MCI (male: 15 and female: 12). None of the EO-AD subjects carried mutations of APP or PS-1 and PS-2 genes. These 36 EO-MCI and EO-AD subjects were chosen from among all subjects in the ADNI-1 database (DB) that met the criterion of 55 ≤ age ≤ 65 as of September 2010.
In this study, 106 MRI scans for 27 EO-MCI subjects and 28 MRI scans for EO-AD subjects were included in the GSA, LSA, and TBM analyses. Some EO-MCI and EO-AD subjects were scanned fully at all intervals, but other subjects dropped out of the follow-up scans (Table 1). Our statistical analyses used linear mixed effect models29,30 where the random effect of the scan-time (e.g., baseline, 12 month follow up, etc.) were accounted for to ensure that within subject dependences were modeled appropriately and the model residuals contain only random white noise.
Table 1.
Demographic data and scanning/testing frequency
| N | Age µ ± σ |
Gender (M/F) |
Education (years, mean±SD) |
MMSE | ApoE(ε4) (+/−) | Imaging Scans |
|
|---|---|---|---|---|---|---|---|
| MCI | 27 | 61.2±2.87 | 15/12 | 16.226 ± 2.764 | 26.745 ± 2.342 | 14/13 | 106 |
| AD | 9 | 60.4±3.34 | 4/5 | 16.142 ± 2.304 | 21.571 ± 3.795 | 5/4 | 28 |
| p-value | - | 0.0810 | 0.5630 | 0.8834 | <0.0001 | 0.8471 |
MRI Data (ADNI Data)
We used the data obtained from the ADNI database (http://www.loni.usc.edu/ADNI). The ADNI MRIs analyzed here were the baseline, screening, or follow-up scans at the entry to the study; they were acquired at multiple sites using the GE Health Care (Buckinghamshire, England), Siemens Medical Solutions USA (Atlanta, Georgia), or Philips Electronics 1.5 T (Philips Electronics North America; Sunnyvale, California) system.31 Two high-resolution T1-weighted volumetric magnetization-prepared 180° radiofrequency pulses and rapid gradient-echo scans were collected from each subject and was normalized for intensity in homogeneities, non-brain tissue was removed, and subcortical white matter and deep gray matter volumetric structures were segmented.32,33 The raw DICOM (Digital Imaging and Communications in Medicine) images were downloaded from the public ADNI site (http://ADNI.loni.usc.edu). All the MRIs were processed according to previously published methods34 using the FreeSurfer V4 software package (http://surfer.nmr.mgh.harvard.edu).35 The intensity gradients were followed outward from the white matter surface to find the gray matter surface (gray–cerebrospinal fluid boundary).36,37
LONI Pipeline Environment
LONI Pipeline38,39 is a graphical workflow environment that allows the design, execution, validation, and provision of complex heterogeneous computational protocols. In this manuscript, the Pipeline environment was used to investigate the interrelations between the subjects’ phenotypes, genotypes, and biomedical imaging markers, including the volumetric and shape-based measures of their brain structure. For the EO ADNI subjects, our analysis protocols included automatic imaging feature extraction, geometric modeling, and statistical analysis of various global and regional anatomical measures.
The GSA Pipeline workflow39 provides an automated protocol for high-throughput data preprocessing [e.g., skull-stripping,40 volumetric registration41)], brain anatomical parcellation into 56 regions of interest (ROIs)42,43), and extraction of shape and volume measures (average mean curvature, surface area, volume, shape index, and curvedness), between the group statistical analyses of the shape regional differences, as well as generation of 3D scene files that illustrate the statistically significant regional anatomical differences between the EO-MCI and EO-AD cohorts.
Table 2 shows the definitions of the 5 intrinsic geometric cortical measures used in this study, and the formulas used to compute them. The principal curvatures (k1, k2) were computed using triangulated surface models that represented the boundaries of different brain areas.44 ID(x,y,z) represents the indicator function of the ROI (D),45 and SΩ:r = r(u, v), (u, v) ∈ Ω is the parametric surface representation of the region boundary.46 In the LSA protocol, the structural attributes and cortical measures are calculated per-vertex in the specific shape regions that are first co-registered across subjects to establish homologous anatomical features before statistically analyzing them against various subject demographic, clinical or phenotypic data.38
Table 2.
Intrinsic geometric cortical features and their definitions.
| Geometric Measure | Mathematical formulas | |
|---|---|---|
| Volume |
|
|
| Surface Area |
|
|
| Mean Curvature |
|
|
| Shape Index |
|
|
| Curvedness |
|
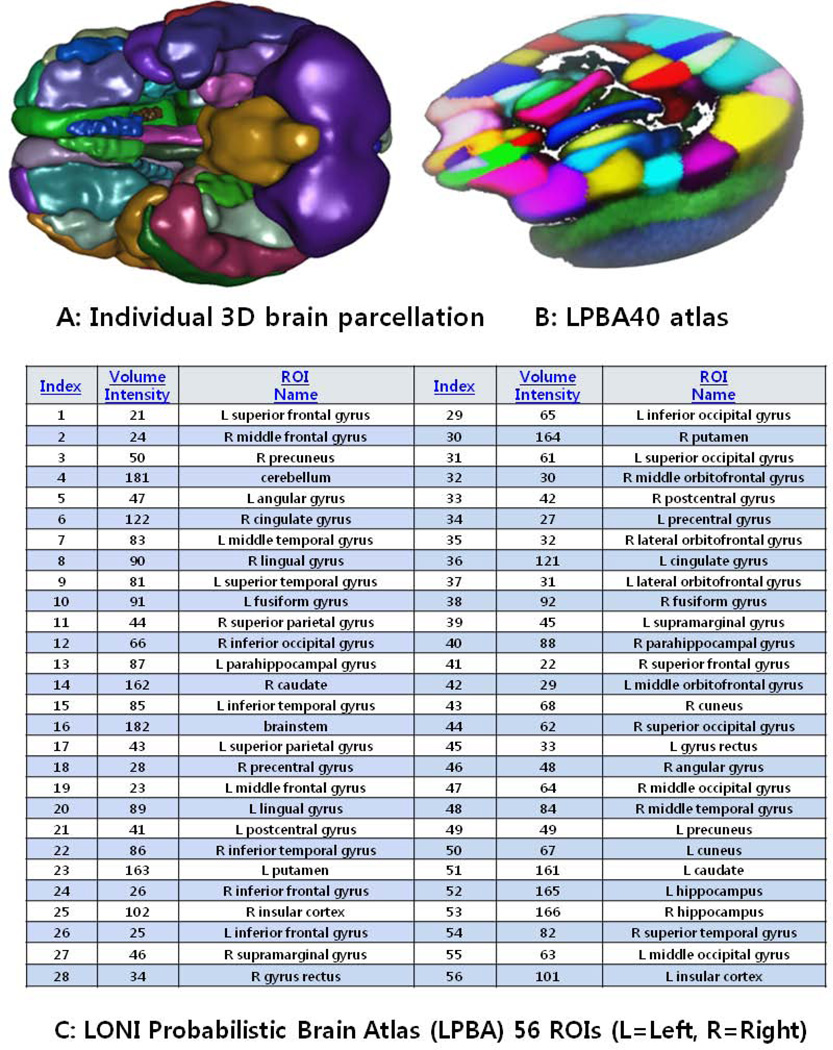
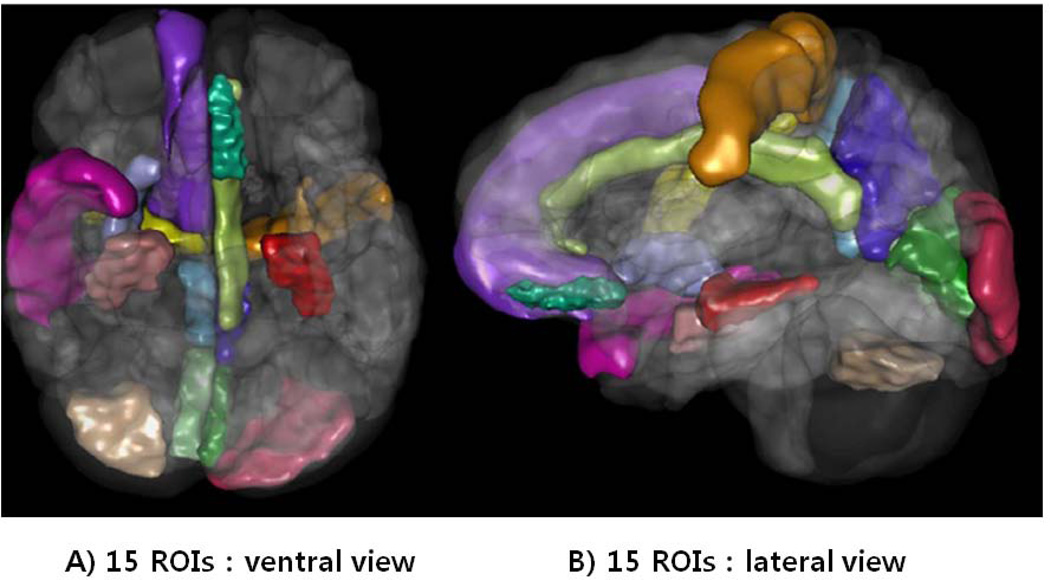
From the collection of 280 imaging markers (56 ROIs × 5 shape measures), we chose the 15 most significant neuroimaging biomarkers that provided the highest discrimination between the EO-MCI and EO-AD groups using t-tests that compared the EO-MCI and EO-AD groups (with an a priori false-positive rate of 0.05, p < 0.05) (These biomarkers are described in Table 3). The 15 neuroimaging biomarkers were derived from the structural imaging data (between 106 scans for the 27 EO-MCI subjects and 28 scans for the 9 EO-AD subjects) using the GSA workflow and were based on the automated ROI extractions generated by BrainParser.42,43 Fig 1 illustrates the LPBA40 atlas, an example of the 3D reconstruction of the BrainParser output for one subject, and the names of the 56 ROIs. We used this GSA Pipeline workflow to obtain a set of 15 neuroimaging biomarkers. Fig 2 shows one 3D scene file that corresponded to the 15 ROI volume metric.
Table 3.
Summary of the most significant imaging phenotypes – 15 derived-bioimaging markers (p< 0.05)
| Neuroimaging phenotypes | Shape & Volume Measures | P-value |
|---|---|---|
| L_hippocampus (Volume) | Volume | 0.00067 |
| R_hippocampus (Volume) | Volume | 0.00539 |
| L_gyrus_rectus (Surface area) | Surface area | 0.01728 |
| L_middle_occipital_gyrus (Volume) | Volume | 0.01805 |
| R_precuneus (Shape index) | Shape index | 0.03186 |
| L_cingulate_gyrus (Average mean curvature) | Average mean curvature | 0.03350 |
| R_superior_temporal_gyrus (Volume) | Volume | 0.03353 |
| R_precentral_gyrus (Shape index) | Shape index | 0.03411 |
| R_putamen (Curviness) | Curvedness | 0.03504 |
| R_superior_frontal_gyrus (Volume) | Volume | 0.03706 |
| L_precentral_gyrus (Volume) | Volume | 0.04125 |
| R_cuneus (Surface area) | Surface area | 0.04203 |
| L_cuneus (Shape index) | Shape index | 0.04952 |
| R_inferior_occipital_gyrus (Curviness) | Curvedness | 0.05037 |
| L_precuneus (Volume) | Volume | 0.05080 |
Fig 1.
Summary of the 56 regions of interest (ROIs) (A,C) extracted by the BrainParser software using the LPBA40 atlas (B).
Fig 2.
One example of a 3D scene output file indicating statistically significant (p-value < 0.05) volumetric differences in between the EO-AD and EO-MCI.
Fifteen ROIs in the 3D scene : “L_hippocampus, R_hippocampus, R_precuneus, L_precuneus, L_cingulate_gyrus, R_superior_temporal_gyrus, L_gyrus_rectus, L_middle_occipital_gyrus, R_precentral_gyrus, R_putamen, R_superior_frontal_gyrus, L_precentral_gyrus, R_cuneus, L_cuneus, and R_inferior_occipital_gyrus”
We used the Local Shape Analysis (LSA) Pipeline workflow to conduct local (per-vertex) post-hoc statistical analyses of the shape differences between the two cohorts in the left and right hippocampus, left and right middle frontal gyri, and left and right middle temporal gyri. It is already known that the hippocampal brain volume is reduced early in dementia patients.47–52 The temporal and frontal lobes were also chosen because there is significant evidence that the associative areas are involved in dementia.49,53–55 Thus, we chose two middle gyri as representative samples of these two lobes. After generating the localized neuroimaging measures of the anatomy, we focused our attention bilaterally on 3 regions. We used a design matrix that included diagnosis, age, APOE(ε4), MMSE, years of education, number of scanning repetition, and LM (immediate and delayed Recall) as regressors. We used the statistical method of the multivariate linear regression (MLR) model to conduct the LSA analysis using 106 EO-MCI scans and 28 EO-AD scans (for a total of total 134 scans), by doing eight independent analyses for the regressors. Thus, we compared the cohorts of EO patients based on the participant’s diagnosis (EO-MCI+EO-AD), with anatomical morphometric measures as predictors of the diagnosis as response variables.
In the Local shape analysis (LSA) Pipeline workflow, the 3D structural MIR data are first preprocessed (skull-stripped, spatially normalized, parcellated)39,42, then shape models of 56 brain regions are generated as genus-zero 2D-manifolds56,57. By traversing the triangulated boundary manifolds (vertex-by-vertex), statistical significance maps are obtained that represent the group differences (EO-MCI vs. EO-AD) in two complementary shape metrics. The radial distance and displacement vector field measures at each vertex encode the magnitude and direction of local shape morphometry which quantify the discrepancy between each subject that the “mean shape” (boundary) for each of the 56 regions of interest. Probability-values corresponding to the test-statistics are overlaid on the mean boundary shape for each region to illustrate the group differences.
Tensor Based Morphometry (TBM) is a volumetric image analysis technique58–61 that produces 3D volumetric maps of change. For instance, when applied to structural brain data, TBM uses the deformation of one brain to match another (typically a reference atlas like ICBM62 or a cohort-derived phantom atlas) to generate individual maps of brain changes (Jacobian maps). These Jacobian maps represent the magnitude of the localized displacement vector field required to co-register the data into a template, e.g., an average group minimum distance template (MDT).63 In our case, we used the MDT atlas that is derived as a canonical phantom atlas representing a point of gravitational balance for all scans in the study. TBM identifies regional structural differences from the gradients of the non-linear deformation fields that align or warp images to a common anatomical template. At each voxel, a color-coded Jacobian determinant value indicates the local volume excess or deficit relative to the corresponding anatomical structures in the template.64–66
Similar to our prior LSA analysis, the TBM study used a design matrix that included the diagnosis, age, APOE(ε4), MMSE, years of education, number of scanning repetitions, and LM (immediate and delayed Recall) as regressors. However, TBM analysis provided a wide range of regional assessments through whole brain analysis in this study, unlike the LSA analysis. We used the MLR model to perform whole brain TBM analytics using 106 EO-MCI scans and 28 EO-AD scans (for a total of 134 scans), in the process performing eight independent analyses for the regressors. The two cohorts of EO patients were compared based on the participants’ diagnoses (EO-MCI+EO-AD).
Results
1. Demographic characteristics
The demographic and clinical data of the subjects at the baseline are summarized in Table 1 (using Chi-square and t-test analyses). In this study, we chose EO subjects (aged between 55 and 65 years) from the ADNI datasets. There was no statistically significant difference in age between the EO-AD and EO-MCI subjects. The age distribution of the EO subjects is shown in Table 4.
Table 4.
Age distribution for the EO-AD and EO-MCI.
| Cohort | Gender | N | Total | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCI | M | 15 | 27 | 2 | 1 | 1 | 3 | 4 | 3 | 1 | 15 | |||||
| F | 12 | 2 | 1 | 2 | 2 | 1 | 3 | 1 | 12 | |||||||
| AD | M | 4 | 9 | 1 | 1 | 2 | 4 | |||||||||
| F | 5 | 1 | 2 | 1 | 1 | 5 |
2. Neuroimaging biomarker selection
The most significant 15 neuroimaging biomarkers were selected (p < 0.05) from 56 ROIs and the 5 different volume- and shape-based metrics, based on how well they discriminated between the 2 cohorts. They are shown in Table 3. The ROIs for the 15 neuroimaging biomarkers were the L_hippocampus, R_hippocampus, R_precuneus, L_precuneus, L_cingulate_gyrus, R_superior_temporal_gyrus, L_gyrus_rectus, L_middle_occipital_gyrus, R_precentral_gyrus, R_putamen, R_superior_frontal_gyrus, L_precentral_gyrus, R_cuneus, L_cuneus, and R_inferior_occipital_gyrus.
3. Local Shape Analysis (LSA)
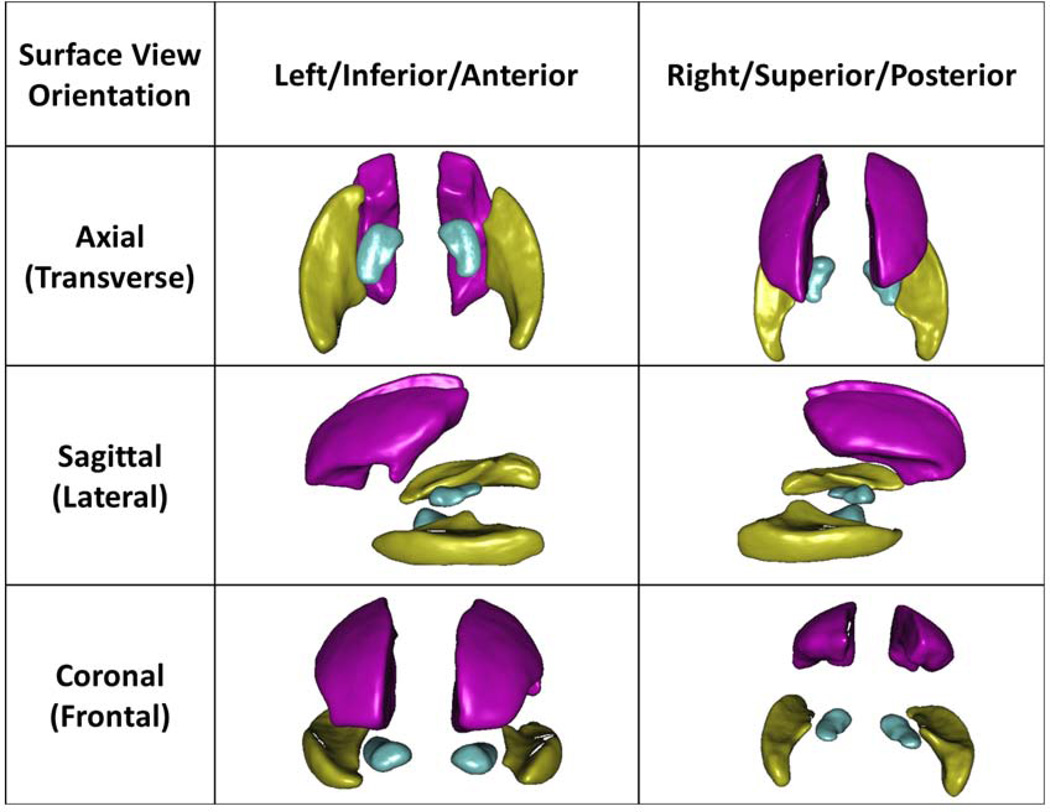
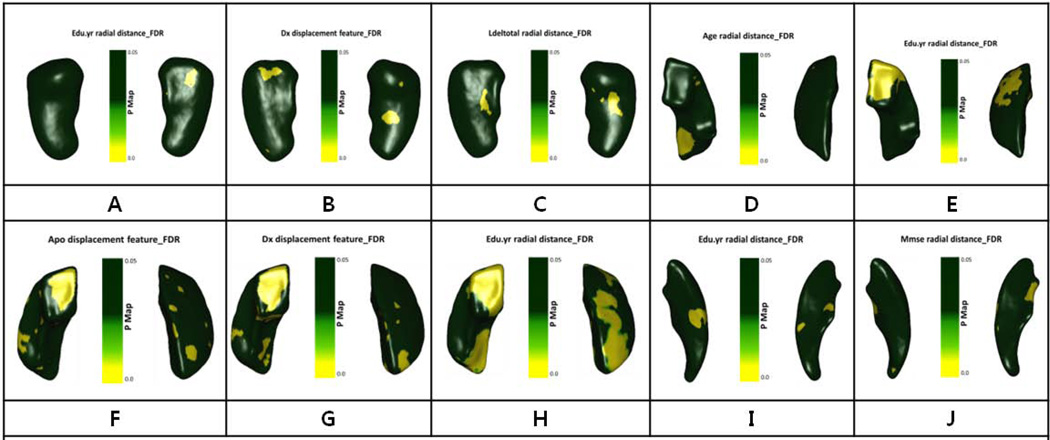
In the LSA analysis, we looked for regional effects (bilaterally in the hippocampus, middle frontal gyrus, and temporal lobe) of different phenotypic variables, such as the diagnosis, age, years of education, APOE(ε4), MMSE, and LM (immediate and delayed recall) as regressors for the MLR model on the local shape-based registrations across subjects. Some results survived the FDR correction67,68. Fig 3 illustrates the mean shapes, their poses, and 3D spatial interrelations of the 3 regions that we studied in detail bilaterally – the left and right hippocampi (labels 165–166), middle frontal gyri (labels 23–24), and middle temporal gyri (labels 83–84). All the results that survived the FDR correction are shown as p-value color maps in Fig 4 (A–T). In particular, the LM, including immediate and delayed recall should be very sensitive to the decline of the cognitive function.69
Fig 3.
Mean geometric models of the left and right hippocampi (turquoise), middle frontal gyri (pink), and the middle temporal gyri (yellow). The surface-based statistical maps are computed on each vertex of these atlas shapes.
Fig 4.
Statistical maps generated by the LSA workflow. Each insert image illustrates on the 3D shapes the statistically significant p-values, which are generated by fitting linear models at each vertex on the triangulated shapes. The dependent variable was the radial distance morphometry measure (deviation of individual shape model from mean shape atlas) and independent regressors including diagnosis, age, education years, APOE(ε4), MMSE, Visiting times, and Logical memory (Immediate and Delayed recall).
P-maps (A) for Lt. hippocampus, P-maps (B,C) for Rt. Hippocampus, P-maps (D,E) for Lt. middle frontal gyrus, P-maps (F,G,H) for Rt. middle frontal gyrus, P-maps (I, J) for Lt. middle temporal gyrus.
4. Tensor Based Morphometry (TBM)
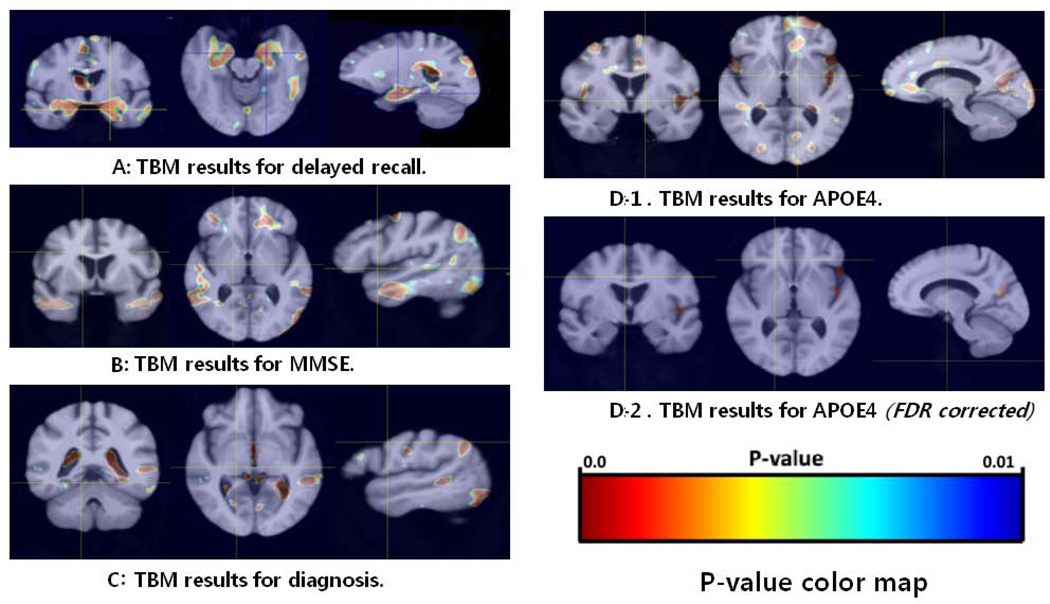
The TBM pipeline workflow and the corresponding TBM results [using the diagnosis, APOE(ε4) including the FDR corrected results, MMSE, and delayed recall as predictors] are shown in Fig 5. These results indicate an association between the phenotypes (delayed recall) and the structural neuroimaging data (the TBM anatomical maps) in the medial temporal area, including the hippocampi, parahippocampal gyri, and amygdala. Additional correlations between the TBM and MMSE scores were observed in the frontal area and the middle and medial temporal gyri, and a correlation between TBM and the diagnosis was detected in the medial temporal area, occipital area, visual cortex, insula, and ventricles. A correlation between TBM and ApoE(ε4) was further observed in the frontal area, cingulated gyri, insular lobe, lateral sulci, occipital area, and procuneus. Especially, the correlation between TBM and ApoE(ε4) survived the FDR correction in the left lateral sulcus, insula, and precuneus.
Fig 5.
TBM Results: Cross-sectional statistical maps delayed recall (A), MMSE (B), diagnosis (C), and APOE (D). The p-value maps are overlaid on the MDT brain atlas constructed using the structural imaging data for all (N=36) subjects.
Discussion
Global Shape Analysis (GSA)
The left precuneus volume was significantly reduced in the EO group, which supports the results of some previously mentioned studies and may explain why precuneal atrophy is more prominent in patients with EO relative to LO.70,71 Both the left and right hippocampal volumes were some of the most significant neuroimaging biomarkers, as we initially hypothesized. We identified the ‘shape index’ of the right precuneus and the ‘average mean curvature’ of the left cingulated gyrus and the left superior temporal gyrus as significant covariates associated with the dementia phenotypes (EO vs. LO Alzheimer’s). This suggests that EO-AD differences may be related to atrophy of the posterior cingulated cortex and temporal lobe.72 Other ROI-based shape measurements (e.g., of the ‘surface area’ and ‘curviness’) were also associated with the group phenotype, but we were not able to find previously published reports consistent with this finding. Albeit, most prior studies relied on volumetric measures, whereas we employ shape-morphometry metrics.
Local Shape Analysis (LSA)
In this study, we investigated the sensitivity in the left and right hippocampi, the middle frontal gyri, and the middle temporal gyri. We selected these 3 bilateral ROIs because we expected these ROIs to show us some of the differences between the 2 cohorts, and some of the characteristics of the EO cognitive impairment subjects (EO-MCI+EO-AD). We chose both hippocampi to find out if their shape-measures are also associated with changes in various phenotypic variables (regressors). The hippocampus is not a homogeneous structure but consists of several subfields with distinct histological characteristics: the subiculum, the three cornu ammonis sectors (CA1,CA2 and CA3), and the dentate gyrus73. The results shown in Fig 4 illustrate the hippocampal radial distance association with the years of education at the bottom of the left subiculum. Hippocampal displacement feature atrophy associated with the diagnosis was observed in the right bottom subiculum and the top CA2 and CA3, and hippocampal radial distance atrophy correlated with delayed recall was seen in the right bottom subiculum and the top CA2 and CA3.
We found some significance with delayed recall but not for immediate recall in both hippocampi. The lack of a significant relationship between immediate recall and the hippocampal volume is consistent with the results of some studies that the hippocampal volume was more strongly related to delayed recall than immediate recall.74–76 The shape-based LSA analysis suggests that the left and right hippocampi are closely associated with delayed recall of LM. This finding implies that the shape morphometry may be coupled with diminishing logically associated episodic memory performance, and is correlated more with delayed recall than immediate recall. The middle frontal gyrus radial distance atrophy in the left posterior inferior area was found to be associated with age. A strong correlation between the middle frontal gyrus radial distance and the years of education was observed in the left bottom anterior inferior and top superior anterior areas. An association between the middle frontal gyrus displacement feature and APOE(ε4) was detected in the right bottom anterior inferior area. The middle frontal gyrus radial distance atrophy in the right inferior area and superior posterior area was related to the years of education. Finally, the years of education and the MMSE score were correlated to the radial distance atrophy in the left middle temporal gyrus. These results reflect significant effects that survived post-hoc false discovery rate (FDR) correction for multiple testing. Despite the small sample size, some significant genotype effects (after the FDR correction) on the shape metrics were observed, as shown in Fig 4(A–T).
Tensor Based Morphometry (TBM)
The TBM analysis used the MLR to study the multivariate relations between anatomical morphometric measures (response variables) and a diverse array of regressors (the diagnosis, age, APOE(ε4), MMSE, number of scanning repetitions, and LM, including the immediate and delayed recall). The medial temporal area (MTA), including the hippocampus, parahippocampus, and amygdale, was significantly associated with the LM test scores for delayed recall. Thus, MTA degeneration may be considered to be associated with context-related episodic memory performance, especially with delayed recall. One possible explanation is that in these areas, the brain atrophied over time and affected the LM delayed recall for all the EO-AD and EO-MCI subjects. There were some diagnosis effects such as ventricles expansion, which indirectly implies gray matter atrophy in the surrounding tissue (representing differences between the two cohorts). A correlation between TBM and ApoE(ε4) was observed in the frontal area, cingulated gyri, insular lobe, lateral sulci, and procuneus. However, these did not survive the rigorous post-hoc FDR correction, except for the correlation between TBM and ApoE(ε4) in the left lateral sulcus, insula, and precuneus. The left lateral sulcus and insula may be associated with the atrophy of the left medial temporal area, and precuneal atrophy can reflect the view in some reports that disproportionate precuneus atrophy is more prominent in patients with a younger age of onset as we discussed earlier in this paper.25,70,71 The precuneus is located in the medial aspect of the posterior parietal lobe, and its borders are the parietooccipital sulcus posteriorly. Previous studies have also shown that the precuneus might be functionally impaired in younger patients with AD.77,78 All the results are shown as 3D images that represent the raw p-value maps in Fig 5.
Limitations
The sample size for this early onset Alzheimer’s disease study was rather small, due to significant data stratification and lack of available data. EO-AD is less prevalent than LO-MCI, which may have contributed to the relative weakness of the statistical results. We did not do the statistical analyses of individual APOE ε2, ε3, and ε4 alleles because the sample size was insufficient. This prevented us from analyzing the effects of the APOE ε4 genotypes (ε3/ε4 and ε4/ε4). This study did not include neuroimaging data from asymptomatic normal controls (NC). Future studies may explore the underlying differences between NC and MCI or AD and NC subjects, provided appropriate data is available.
Conclusions
In summary, even with the small number of EO subjects, we were able to make several neuroimaging observations regarding the EO-AD and EO-MCI cohorts using our GSA, LSA, and TBM workflows. We developed a graphical pipeline protocol for performing neuroimaging analysis using the LONI Pipeline environment. The methodology presented here can be used as a basis for future large-scale neuroimaging studies with hundreds and thousands of subjects of varying phenotypes
Acknowledgments
The study design, data analyses and writing of this manuscript were supported in part by NIA P 50 AG16570, NIBIB EB01651, NLM LM05639, NCRR RR019771, NIMH R01 MH071940, NIBIB 9P41EB015922-15, NCRR 2-P41-RR-013642-15, P20 NR015331, U54 EB020406, P50 NS091856, P30 DK089503, and NSF grants 0716055 and 1023115.
This work was partially supported by a grant from the Korean Health Technology R&D Project, Ministry for Health, Welfare, & Family Affairs, Republic of Korea (Grant No. A092077) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2012R1A1A4A01013120).
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is Rev March 26, 2012 coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Readers who are interested in having the full workflow details of the image analysis pipeline used for this study, should contact the authors.
References
- 1.Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiological reviews. 2001 Apr;81(2):741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 2.Mayeux R. Alzheimer's disease: epidemiology. Handbook of clinical neurology. 2008;89:195–205. doi: 10.1016/S0072-9752(07)01218-3. [DOI] [PubMed] [Google Scholar]
- 3.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiat. 2006 Feb;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 4.Ertekin-Taner N. Genetics of Alzheimer's Disease: A centennial review. Neurol. Clin. 2007 Aug;25(3):611–667. doi: 10.1016/j.ncl.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991 Feb 21;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 6.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995 Jun 29;375(6534):754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 7.Levy-Lahad E, Wasco W, Poorkaj P, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995 Aug 18;269(5226):973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 8.Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995 Aug 31;376(6543):775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 9.Pericak-Vance MA, Grubber J, Bailey LR, et al. Identification of novel genes in late-onset Alzheimer's disease. Experimental gerontology. 2000 Dec;35(9–10):1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 10.Ertekin-Taner N. Genetics of Alzheimer disease in the pre- and post-GWAS era. Alzheimer's research & therapy. 2010;2(1):3. doi: 10.1186/alzrt26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois B, Feldman HH, Jacova C, et al. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet neurology. 2007 Aug;6(8):734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 12.Thompson PM, Hayashi KM, De Zubicaray GI, et al. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004 Aug;22(4):1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 13.Jack CR, Jr, Slomkowski M, Gracon S, et al. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003 Jan 28;60(2):253–260. doi: 10.1212/01.wnl.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolz R, Heckemann RA, Aljabar P, et al. Measurement of hippocampal atrophy using 4D graph-cut segmentation: application to ADNI. NeuroImage. 2010 Aug 1;52(1):109–118. doi: 10.1016/j.neuroimage.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Morra JH, Tu Z, Apostolova LG, et al. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Human brain mapping. 2009 Sep;30(9):2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu A, Fennema-Notestine C, Dale AM, Miller MI Alzheimer's Disease Neuroimaging I. Regional shape abnormalities in mild cognitive impairment and Alzheimer's disease. NeuroImage. 2009 Apr 15;45(3):656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apostolova LG, Thompson PM, Green AE, et al. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Human brain mapping. 2010 May;31(5):786–797. doi: 10.1002/hbm.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardenas VA, Chao LL, Studholme C, et al. Brain atrophy associated with baseline and longitudinal measures of cognition. Neurobiology of aging. 2011 Apr;32(4):572–580. doi: 10.1016/j.neurobiolaging.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X, Lee S, Hibar DP, et al. Mapping Alzheimer's disease progression in 1309 MRI scans: power estimates for different inter-scan intervals. NeuroImage. 2010 May 15;51(1):63–75. doi: 10.1016/j.neuroimage.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou YY, Lepore N, Saharan P, et al. Ventricular maps in 804 ADNI subjects: correlations with CSF biomarkers and clinical decline. Neurobiology of aging. 2010 Aug;31(8):1386–1400. doi: 10.1016/j.neurobiolaging.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padovani A, Gilberti N, Borroni B. The usefulness of biological and neuroimaging markers for the diagnosis of early-onset Alzheimer's disease. International journal of Alzheimer's disease. 2011;2011:296374. doi: 10.4061/2011/296374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frisoni GB, Pievani M, Testa C, et al. The topography of grey matter involvement in early and late onset Alzheimer's disease. Brain : a journal of neurology. 2007 Mar;130(Pt 3):720–730. doi: 10.1093/brain/awl377. [DOI] [PubMed] [Google Scholar]
- 23.Hanyu H, Nakano S, Abe S, Arai H, Iwamoto T, Takasaki M. Differences of neuroimaging between early-onset and late-onset Alzheimer-type dementia. Rinsho shinkeigaku = Clinical neurology. 1995 Oct;35(10):1104–1109. [PubMed] [Google Scholar]
- 24.Pievani M, Bocchetta M, Boccardi M, et al. Striatal morphology in early-onset and late-onset Alzheimer's disease: a preliminary study. Neurobiology of aging. 2013 Jul;34(7):1728–1739. doi: 10.1016/j.neurobiolaging.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Frisoni GB, Testa C, Sabattoli F, Beltramello A, Soininen H, Laakso MP. Structural correlates of early and late onset Alzheimer's disease: voxel based morphometric study. J Neurol Neurosurg Psychiatry. 2005 Jan;76(1):112–114. doi: 10.1136/jnnp.2003.029876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010 Jan 19;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993 Nov;43(11):2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 28.Wechsler D. WMS-R Wechsler Memory Scale-Revised Mannual. New York: The Psychological Corporation. Harcourt Brace Jovanovich, Inc.; 1987. [Google Scholar]
- 29.McCulloch CE. Generalized linear mixed models. Wiley Online Library. 2006 [Google Scholar]
- 30.Verbeke G, Molenberghs G. Linear mixed models for longitudinal data. Springer; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jack CR, Jr, Bernstein MA, Fox NC, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008 Apr;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 1999 Feb;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 33.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999 Feb;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 34.Desikan RS, Cabral HJ, Fischl B, et al. Temporoparietal MR imaging measures of atrophy in subjects with mild cognitive impairment that predict subsequent diagnosis of Alzheimer disease. AJNR Am J Neuroradiol. 2009 Mar;30(3):532–538. doi: 10.3174/ajnr.A1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. [September 26, 2000];Proceedings of the National Academy of Sciences of the United States of America. 2000 97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002 Jan 31;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 37.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000 Sep 26;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinov I, Torri F, Macciardi F, et al. Applications of the Pipeline Environment for Visual Informatics and Genomics Computations. BMC Bioinformatics. 2011;12(1):304. doi: 10.1186/1471-2105-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinov I, Lozev K, Petrosyan P, et al. Neuroimaging Study Designs, Computational Analyses and Data Provenance Using the LONI Pipeline. PLoS ONE. 2010;5(9):e13070. doi: 10.1371/journal.pone.0013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pieper S, Lorensen B, Schroeder W, Kikinis R. The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D slicer as an open platform for the medical image computing community. Paper presented at: Biomedical Imaging: Nano to Macro, 2006. 3rd IEEE International Symposium on; 6–9 April 2006.2006. [Google Scholar]
- 42.Tu Z, Narr KL, Dinov I, Dollar P, Thompson PM, Toga AW. Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models. IEEE Transactions on Medical Imaging. 2008;27(4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shattuck DW, Mirza M, Adisetiyo V, et al. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39(3):1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terzopoulos D. The computation of visible-surface representations. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1988:417–438. [Google Scholar]
- 45.Larson R, Edwards BH. Calculus. Brooks/Cole Pub Co; 2009. [Google Scholar]
- 46.Santal LA. Integral geometry and geometric probability. Cambridge Univ Pr; 2004. [Google Scholar]
- 47.Frisoni GB, Sabattoli F, Lee AD, Dutton RA, Toga AW, Thompson PM. In vivo neuropathology of the hippocampal formation in AD: A radial mapping MR-based study. Neuroimage. 2006;32(1):104–110. doi: 10.1016/j.neuroimage.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 48.Apostolova LG, Dinov ID, Dutton RA, et al. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease. Brain. 2006 Nov;129(Pt 11):2867–2873. doi: 10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- 49.Hua X, Leow AD, Parikshak N, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer's disease: an MRI study of 676 AD, MCI, and normal subjects. NeuroImage. 2008 Nov 15;43(3):458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf H, Grunwald M, Kruggel F, et al. Hippocampal volume discriminates between normal cognition; questionable and mild dementia in the elderly. Neurobiology of aging. 2001 Mar-Apr;22(2):177–186. doi: 10.1016/s0197-4580(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 51.Luders E, Thompson PM, Kurth F, et al. Global and regional alterations of hippocampal anatomy in long-term meditation practitioners. Human brain mapping. 2012 Jul 19; doi: 10.1002/hbm.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morra JH, Tu Z, Apostolova LG, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. NeuroImage. 2009 Mar;45(1 Suppl):S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho AJ, Hua X, Lee S, et al. Comparing 3 T and 1.5 T MRI for tracking Alzheimer's disease progression with tensor-based morphometry. Human brain mapping. 2010 Apr;31(4):499–514. doi: 10.1002/hbm.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen L, Kim S, Risacher SL, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. NeuroImage. 2010 Nov 15;53(3):1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Risacher SL, Shen L, West JD, et al. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiology of aging. 2010 Aug;31(8):1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutman BA, Hua X, Rajagopalan P, et al. Maximizing power to track Alzheimer's disease and MCI progression by LDA-based weighting of longitudinal ventricular surface features. Neuroimage. 2013;70:386–401. doi: 10.1016/j.neuroimage.2012.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Song Y, Rajagopalan P, et al. Surface-based TBM boosts power to detect disease effects on the brain: An N= 804 ADNI study. Neuroimage. 2011;56(4):1993–2010. doi: 10.1016/j.neuroimage.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hua X, Gutman B, Boyle CP, et al. Accurate measurement of brain changes in longitudinal MRI scans using tensor-based morphometry. Neuroimage. 2011;57(1):5–14. doi: 10.1016/j.neuroimage.2011.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho AJ, Hua X, Lee S, et al. Comparing 3 T and 1.5 T MRI for tracking Alzheimer's disease progression with tensor-based morphometry. Human Brain Mapping. 2010;31(4):499–514. doi: 10.1002/hbm.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lepore N, Brun C, Chou YY, Chiang MCA, Dutton RA, Hayashi KM, Luders EA, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. Generalized Tensor-Based Morphometry of HIV/AIDS Using Multivariate Statistics on Deformation Tensors. Medical Imaging, IEEE Transactions on. 2008;27(1):129–141. doi: 10.1109/TMI.2007.906091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang M, Leow A, Klunder A, et al. Fluid Registration of Diffusion Tensor Images Using Information Theory. Medical Imaging, IEEE Transactions on. 2008;27(4):442–456. doi: 10.1109/TMI.2007.907326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazziotta JTA, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001 Aug;356(1412):1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature. 2000 Mar 9;404(6774):190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- 64.Freeborough PA, Fox NC. Modeling brain deformations in Alzheimer disease by fluid registration of serial 3D MR images. Journal of computer assisted tomography. 1998 Sep-Oct;22(5):838–843. doi: 10.1097/00004728-199809000-00031. [DOI] [PubMed] [Google Scholar]
- 65.Chung MK, Worsley KJ, Paus T, et al. A unified statistical approach to deformation-based morphometry. NeuroImage. 2001 Sep;14(3):595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- 66.Riddle WR, Li R, Fitzpatrick JM, DonLevy SC, Dawant BM, Price RR. Characterizing changes in MR images with color-coded Jacobians. Magnetic resonance imaging. 2004 Jul;22(6):769–777. doi: 10.1016/j.mri.2004.01.078. [DOI] [PubMed] [Google Scholar]
- 67.Chumbley JR, Friston KJ. False discovery rate revisited: FDR and topological inference using Gaussian random fields. Neuroimage. 2009;44(1):62–70. doi: 10.1016/j.neuroimage.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 68.Che A, Cui J, Dinov I. SOCR Analyses: Implementation and Demonstration of a New Graphical Statistics Educational Toolkit. JSS. 2009 Apr;30(3):1–19. doi: 10.18637/jss.v030.i03. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lange KL, Bondi MW, Salmon DP, et al. Decline in verbal memory during preclinical Alzheimer's disease: examination of the effect of APOE genotype. J Int Neuropsychol Soc. 2002 Nov;8(7):943–955. doi: 10.1017/s1355617702870096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karas G, Scheltens P, Rombouts S, et al. Precuneus atrophy in early-onset Alzheimer's disease: a morphometric structural MRI study. Neuroradiology. 2007 Dec;49(12):967–976. doi: 10.1007/s00234-007-0269-2. [DOI] [PubMed] [Google Scholar]
- 71.Moller C, Vrenken H, Jiskoot L, et al. Different patterns of gray matter atrophy in early- and late-onset Alzheimer's disease. Neurobiology of aging. 2013 Aug;34(8):2014–2022. doi: 10.1016/j.neurobiolaging.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 72.Shiino A, Watanabe T, Kitagawa T, et al. Different atrophic patterns in early- and late-onset Alzheimer's disease and evaluation of clinical utility of a method of regional z-score analysis using voxel-based morphometry. Dementia and geriatric cognitive disorders. 2008;26(2):175–186. doi: 10.1159/000151241. [DOI] [PubMed] [Google Scholar]
- 73.Mueller SG, Schuff N, Raptentsetsang S, Elman J, Weiner MW. Selective effect of Apo e4 on CA3 and dentate in normal aging and Alzheimer's disease using high resolution MRI at 4 T. NeuroImage. 2008 Aug 1;42(1):42–48. doi: 10.1016/j.neuroimage.2008.04.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kohler S, Black SE, Sinden M, et al. Memory impairments associated with hippocampal versus parahippocampal-gyrus atrophy: an MR volumetry study in Alzheimer's disease. Neuropsychologia. 1998 Sep;36(9):901–914. doi: 10.1016/s0028-3932(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 75.O'Driscoll GA, Florencio PS, Gagnon D, et al. Amygdala-hippocampal volume and verbal memory in first-degree relatives of schizophrenic patients. Psychiatry research. 2001 Aug 25;107(2):75–85. doi: 10.1016/s0925-4927(01)00095-6. [DOI] [PubMed] [Google Scholar]
- 76.Kramer JH, Schuff N, Reed BR, et al. Hippocampal volume and retention in Alzheimer's disease. Journal of the International Neuropsychological Society : JINS. 2004 Jul;10(4):639–643. doi: 10.1017/S1355617704104050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sakamoto S, Ishii K, Sasaki M, et al. Differences in cerebral metabolic impairment between early and late onset types of Alzheimer's disease. Journal of the neurological sciences. 2002 Aug 15;200(1–2):27–32. doi: 10.1016/s0022-510x(02)00114-4. [DOI] [PubMed] [Google Scholar]
- 78.Scarmeas N, Habeck C, Anderson KE, et al. Altered PET functional brain responses in cognitively intact elderly persons at risk for Alzheimer disease (carriers of the epsilon4 allele) The American journal of geriatric psychiatry : official journal of the American Association for Geriatric Psychiatry. 2004 Nov-Dec;12(6):596–605. doi: 10.1176/appi.ajgp.12.6.596. [DOI] [PMC free article] [PubMed] [Google Scholar]