Abstract
Serum is a common sample of convenience for metabolomics studies. Its processing time can be lengthy and may result in the loss of metabolites including those of red blood cells (RBC). Unlike serum, whole blood (WB) is quickly processed, minimizing the influence of variable hemolysis while including RBC metabolites. To determine differences between serum and WB metabolomes, both sample types, collected from healthy volunteers, were assayed by 1H-NMR spectroscopy. A total of 34 and 50 aqueous metabolites were quantified from serum and WB, respectively. Free hemoglobin (Hgb) levels in serum were measured and the correlation between Hgb and metabolite concentrations was determined. All metabolites detected in serum were at higher concentrations in WB with the exception of acetoacetate and propylene glycol. The 18 unique metabolites of WB included adenosine, AMP, ADP and ATP, which are associated with RBC metabolism. The use of serum results in the underrepresentation of a number of metabolic pathways including branched chain amino acid degradation and glycolysis and gluconeogenesis. The range of free Hgb in serum was 0.03-0.01 g/dL and 8 metabolites were associated (p ≤ 0.05) with free Hgb. The range of free Hgb in serum samples from 18 sepsis patients was 0.02-0.46 g/dL. WB and serum have unique aqueous metabolite profiles but the use of serum may introduce potential pathway bias. Use of WB for metabolomics may be particularly important for studies in diseases like sepsis in which RBC metabolism is altered and mechanical and sepsis-induced hemolysis contributes to variance in the metabolome.
Keywords: metabolomics, hemolysis, sepsis, critical care
Introduction
Metabolomics is a rapidly advancing field of discovery science aimed at furthering knowledge of the biological consequences of metabolic changes in an organism or cell (1). In human studies, blood samples are used to capture a physiological average of the host's metabolic status and serum is routinely collected, which makes it a frequent sample of convenience for metabolomics studies. However, the processing time for serum is lengthy (30-60 min) (2) and can be variable, which may lead to changes in metabolite concentrations. In addition, the analysis of the serum metabolome does not take into account the contribution of erythrocytes (RBC). The reliability of serum as a biofluid for metabolomics can be further compromised by ex vivo mechanical RBC hemolysis that can occur due to suboptimal blood collection technique or technical difficulties and centrifugation (3-5). The extent of hemolysis can vary tremendously leading to a broad range of free hemoglobin (Hgb) levels in serum samples (6). This phenomenon is known to impact the reliability of a number of clinical tests including the measurement of lactate dehydrogenase and liver function tests such as ALT and AST (4). Until recently, RBC were considered relatively unimportant in critical illnesses such as sepsis with their oxygen carrying capacity being viewed as their primary function. New knowledge and understanding of RBC function shows that their metabolic activity extends beyond glycolysis (7-9) and they play an essential role in transporting amino acids (10). In sepsis, RBC have gained recognition because they modulate the microcirculation, the RBC membrane participates in the regulation of blood rheology, and RBC width distribution is associated with survivorship (11, 12). In addition, pathogen (6, 13) and complement-induced (14) hemolysis as well as sepsis- induced RBC fragility, enhances the likelihood of in vivo hemolysis. The influence of RBC in sepsis is furthered by the recommendation that patients with septic shock receive RBC transfusion (15), which can lead to hemolysis.
Red blood cells are metabolically active, producing millimolar amounts of ATP and transporting a number of amino acids (10). As such, it is reasonable to expect that RBC make a significant contribution to the blood metabolome that could provide additional insight into sepsis-induced changes. In addition, the quicker processing of WB likely minimizes changes in metabolite levels, including those contributed by the RBC. In this study, we hypothesized that whole blood (WB), which can be quickly and consistently processed, is an alternative metabolomics test material to serum that will provide more metabolic detail and less variation related to processing than serum. To test this hypothesis, we aimed to determine the extent of the differences between the serum and WB metabolomes by comparing the quantified metabolic profiles using 1H-NMR spectroscopy. We also assessed the extent of the association between free hemoglobin (Hgb) levels (as a surrogate of hemolysis) and serum metabolites from samples collected from healthy volunteers and conducted pathway analysis of the two datasets. Finally, in order to assess whether these findings had real world relevance and the potential to affect clinical research conclusions, we measured the range of hemolysis present in representative serum samples from patients with sepsis.
Materials and Methods
Healthy Subjects
Normal, healthy volunteers were identified and recruited for study participation via the Claude D. Pepper Older Americans Independence Center (OAIC) Research Participant Program at the University of Michigan's Geriatric Center and the Michigan Institute of Clinical and Health Research (MICHR) clinical studies website (UMClinicalStudies.org) (16). The study and its associated informed consent form were approved by the University of Michigan's Institutional Review Board approved study (UM IRB; protocol number, HUM00038122).
For study eligibility, subjects had to be non-smoking, and non-obese with no known medical conditions that required chronic drug therapy. On the day of sample collection, volunteers presented to MICHR's clinical research unit (http://www.michr.umich.edu). Following the acquisition of written informed consent, fasting (12 h) blood samples (serum and WB) were collected between 08:30AM and 09:30AM by a single direct venipuncture. Upon collection, Vacutainer® (Becton Dickinson, Franklin Lakes, NJ USA) serum separator tubes (SST), were stored upright, at room temperature, for at least 30 min (but no more than 2 hr) until a visible clot formed. Serum was acquired by centrifugation (1000 × g, 25°C for 10 min), after which aliquots (500μL) were generated and immediately transferred to the freezer (-80°C). Blood for the generation of WB samples was collected into tubes containing sodium heparin (Vacutainer®, Becton Dickinson), which were immediately placed in an ice-water bath for 10 min during which aliquots (500μL) were generated; the samples were then promptly transferred to the freezer (-80°C). Blood was also collected on two separate occasions (approximately two weeks apart) from one donor between 0830-0930 via a single venipuncture to generate five WB and serum samples to test the impact of time on sample processing. These samples were processed as described above except that one of each sample was designated as time (T) 0 (immediately processed upon collection), T 30, 60, 120, and 180 minutes. To mimic serum processing conditions, the T30-T180 samples for both WB and serum were kept at room temperature until the time of allocation into separate tubes and centrifugation, respectively, after which aliquots were transferred to the freezer (-80°C).
NMR Spectroscopy Metabolomics
At the time of assay, frozen samples were thawed on ice and macromolecules were removed by the addition of methanol:chloroform (1:1) as previously described (17). Following extraction, the resulting aqueous phase of each sample was assayed by 1H-NMR spectroscopy. The NMR spectra were acquired on a 500 MHz Agilent spectrometer with a 5mm probe housed at the University of Michigan College of Pharmacy's Biochemical NMR core laboratory. The internal standards of known concentration, formate and deuterated 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS-d6; IS-2, Chenomx Internal Standard), were used for quantification of metabolites for serum and WB, respectively. Spectra were processed and compounds were identified and quantified using Chenomx NMR Suite 7.6 (Chenomx, Inc., Edmonton, Alberta, Canada) by two skilled NMR spectroscopists (CM and LY) with Chenomx software experience. In order for a compound to be categorized as detectable, it needed to be present in at least 70% of the samples (WB or serum) at a concentration of greater than or equal to 5μM.
Serum samples from patients with sepsis
Serum samples that were generated from blood collected via indwelling catheters as part of a clinical trial ((18); clinicaltrials.gov NCT 00372502) from patients with sepsis in the emergency department or intensive care unit. The clinical indices for the diagnosis of sepsis were consistent with consensus definitions for severe sepsis or septic shock: confirmed or suspected infection, two or more systemic inflammatory response criteria (19), and hypoperfusion evidenced by either a systolic blood pressure of < 90 mmHg after 20 min of fluid resuscitation or a blood lactate level of at least 36 mg/dL (18). Blood was drawn into a SST, was allowed to clot at least 30 minutes, and was subsequently centrifuged (10 min at 1,800 × g at 15°C) at which time serum was aliquoted in 0.5 mL increments and stored (-80°C) until they were assayed for free hemoglobin.
Free Hemoglobin Assay
Free Hgb levels in serum samples from healthy controls and sepsis patients were measured using a commercially available colorimetric assay kit (700540; Cayman Chemical, Ann Arbor, MI) using a 96-well plate reader (Molecular Devices, Sunnyvale, CA). The assay was performed in accordance with the manufacturer's instructions and sample Hgb concentrations were calculated from a standard curve constructed with known concentration standards provided in the kit using the plate reader's software (Softmax, version 4.1, Molecular Devices).
Data Analysis
Statistical Analysis
The metabolite data set from WB and serum were range scaled and cube root transformed in preparation for parametric statistical analysis. Principal component analysis of the normalized data was performed using SIMCA (SIMCA software, Umetrics, Umeå, Sweden). For quantitative analysis, the mean normalized value of each metabolite common to both WB and serum were compared using a unpaired Student's t-test with Welch's correction for different variances, if applicable, and each resulting p value was corrected for multiple comparisons by calculating a corresponding false discovery rate (FDR) (20). The extent of the association between free Hgb and concentration of metabolites that were common to, but different between serum and WB, was determined by Spearman correlation. To quantitatively compare changes in the total WB and serum metabolomes over time, the median (IQR) concentrations at each time point (0-180 min) of all 32 common metabolites for serum and WB from the single donor experiment were calculated and compared by a Mann Whitney test. All statistics were performed using GraphPad Prism (version 6 for Mac; GraphPad software, La Jolla, CA., www.graphpad.com).
Pathway analysis
Bioinformatics aids in the interpretation and allows for the visualization of metabolomics data. The serum and WB pathway analysis was done with Metscape 2, a plugin for Cytoscape (http://apps.cytoscape.org/apps/metscape) (21). To perform the analysis we uploaded the list of compounds that were significantly underrepresented in serum plus those that were unique to WB and generated metabolic networks. The networks were examined using Metscape pathway filter to identify representative pathways that included input compounds.
RESULTS
The Serum and Whole Blood Metabolomes are Distinct
A total of 20 volunteers were enrolled in the study of which 50% were female and the mean (± S.D.) age was 54.9±8.2 years. Metabolite data were acquired for 19 WB and 20 serum samples; one WB sample did not properly extract and the NMR spectrum was uninterruptable. The urine metabolomics data from this cohort has been previously published (16).
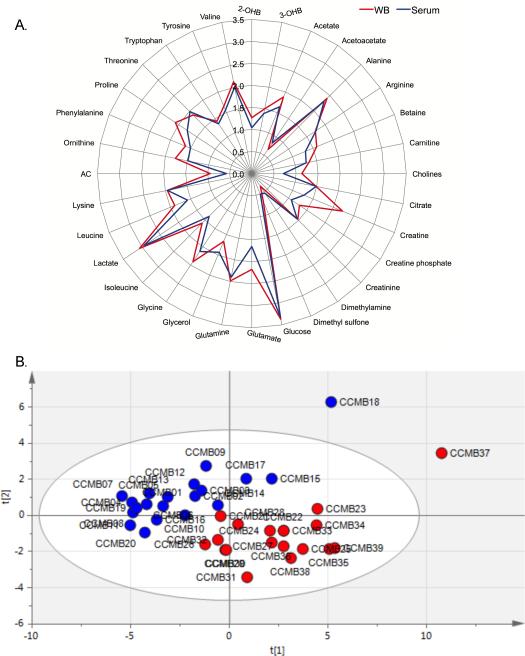
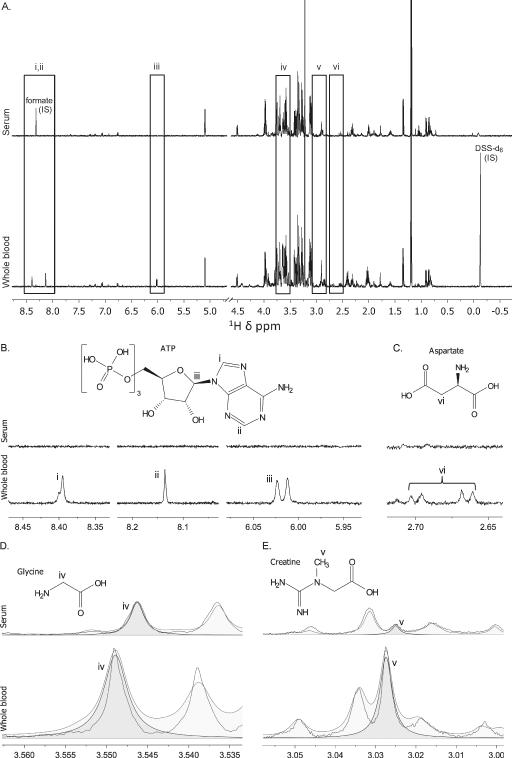
A total of 34 and 50 aqueous metabolites were detected and quantified in serum and WB, respectively. All metabolites that were detected in serum were detected in WB with the exception of acetoacetate, which was only detectable in 45% of the WB samples, and propylene glycol, which may be a contaminant from the SST blood collection tube but has been reported as a human serum metabolite (22), resulting in a total of 32 common metabolites between the two biofluids (Figure 1A). Principal component analysis showed that the metabolomes of the two sample types were distinct (Figure 1B). The 18 unique WB metabolites are shown in Table 1. Representative 1H-NMR spectra of examples of metabolites present in WB and absent in serum or at different concentrations in the two sample types are shown in Figure 2.
Figure 1.
Differences in serum (blue) and whole blood (WB, red) metabolomes as detected by 1H-NMR (500mHz) spectroscopy are illustrated by (A) a radar plot of the 32 common aqueous metabolites of serum and WB and (B) principal component analysis of these metabolites. Samples CCMB 18 and CCMB 37 were acquired from the same study volunteer. The first two principal components (t[1] and t[2]) explain 54% of the variability in the data. Data were generated from 20 serum and 19 WB samples and in the radar plot, data are the median Log10 concentration (μM). 2-; 3-OHB = hydroxybutyrate; AC = acetylcarnitine.
Table 1.
Whole Blood (WB) Metabolites
| Metabolite | KEGGa ID | WB Concentration (μM)b | Reported in the Human Serum Metabolomec |
|---|---|---|---|
| 2-Oxoisocaproate | C00233 | 10.3, 2.7 | No |
| 3-Hydroxyisobutyrate | C06001 | 8.7, 4.9 | No |
| 3-Methyl-2-oxovalerate | C03465 | 11.2, 1.5 | No |
| Adenosine | C00212 | 6.2, 5.0 | No |
| Adenosine diphosphate (ADP) | C00008 | 75.9, 22.7 | No |
| Adenosine monophosphate (AMP) | C00020 | 45.6, 48.3 | No |
| Adenosine triphosphate (ATP) | C00002 | 249.5, 88.0 | No |
| Asparagine | C00152/C01905 | 78.7, 17.0 | Yes |
| Aspartate | C00049 | 69, 73.1 | Yes |
| Formate | C00058 | 31.9, 22.4 | Yesd |
| Glutathione | C00051 | 324.9, 172.7 | No |
| Histidine | C00135 | 55.7, 8.8 | Yes |
| Inosinic acid | C00130 | 30.1, 17.0 | No |
| Malonate | C00383 | 17.4, 8.2 | Yese |
| Acetylcarnitine | C02571 | 8.8, 4.5 | No |
| Pyruvate | C00022 | 23.4, 29.6 | Yesf |
| Succinate | C00042 | 7.0, 2.6 | No |
| Trimethylamine N-oxide | C01104 | 33.3, 12.6 | No |
Kyoto Encyclopedia Genes and Genomes (KEGG) identification (ID) numbers (http://www.genome.jp/kegg/)
concentration data are median, interquartile range of 19 and 20 healthy volunteers for whole blood (WB) and serum, respectively
reported as an aqueous metabolite detected by NMR in reference (30) at an occurrence of 100% of samples unless otherwise noted
reported as occurring in 48% of samples and is removed by MeOH:CHCl3 extraction (24)
reported as occurring in 14% of samples
reported as occurring in 81% of samples
Figure 2.
Differences in the 1H-NMR spectra of whole blood and serum. (A) A representative serum and whole blood spectrum showing regional differences (i-vi) in the spectra that are illustrated by two metabolites, (B) adenosine tri-phosphate (ATP) and (C) aspartate both of which are absent in the 1H-NMR spectrum of serum but present in that of whole blood. The spectral peaks of the associated protons of ATP (i-iii) and aspartate (vi) are shown in the whole blood spectrum. (D) Glycine and (E) creatine are examples of metabolites that are common to serum and whole blood but are higher in concentrations in whole blood than in serum. The spectral peaks of the associated protons of glycine (iv) and creatine (v), as visualized in Chenomx software that is used for metabolite identification and quantification, are shown to illustrate concentration differences. The spectra are not aligned because of the use of different internal standards (chemical shift references), formate for serum and deuterated 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS-d6; IS-2, Chenomx Internal Standard).
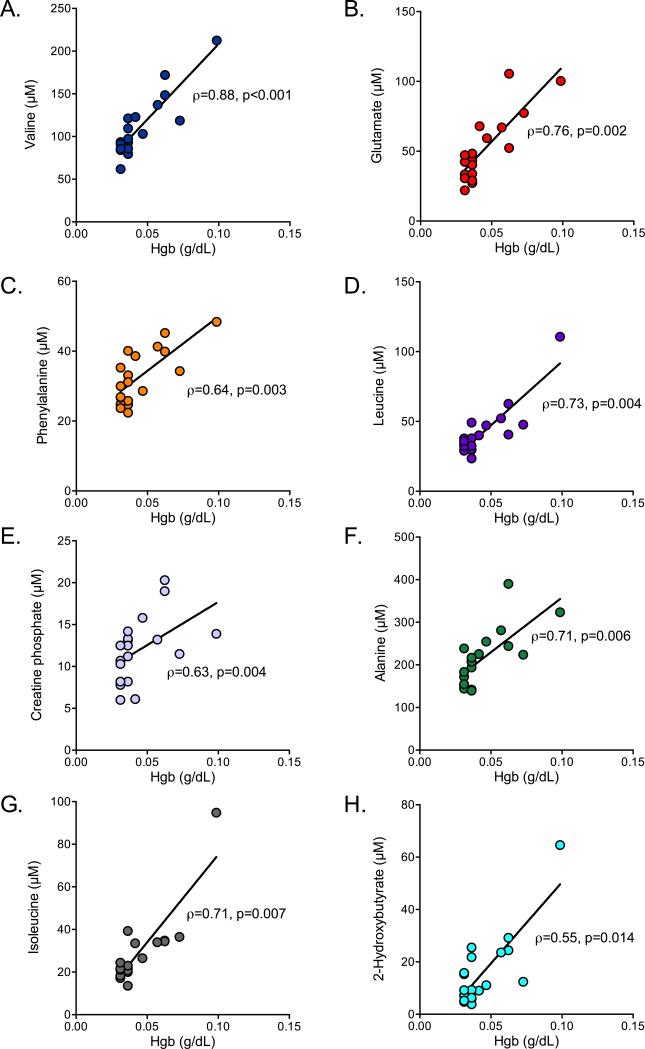
Using a FDR of ≤ 5%, statistical comparison of the normalized concentrations of the 32 metabolites common to both sample types identified 18 metabolites that were different between WB and serum, all of which, with the exception of glycerol, had median concentrations that were higher in WB (Table 2). Of the 18 differentiating metabolites, 8 were associated with free Hgb levels (Figure 3A-H).
Table 2.
Metabolites Common to Whole Blood and Serum as Measured by 1H-NMR Spectroscopy
| WB | Serum | ||||
|---|---|---|---|---|---|
| Metabolite | KEGGa ID | Concentration (μM)b | p value | FDRc (%) | |
| Choline | C00114 | 13.6, 4.3 | 5.2, 5.4 | 0.0001 | 0.14 |
| Creatine | C00300 | 167.2, 60.3 | 19.2, 10.9 | 0.0001 | 0.14 |
| Glutamate | C00217 | 149.7, 27.3 | 45.2, 33.2 | 0.0001 | 0.14 |
| Glycine | C00037 | 254.7, 75.8 | 139.4, 38.2 | 0.0001 | 0.14 |
| Leucine | C00123/C01570 | 77.8, 10.5 | 37.9, 15.4 | 0.0001 | 0.14 |
| Ornithine | C00077/C00515 | 58,8, 19.0 | 30.4, 17.6 | 0.0001 | 0.14 |
| Proline | C00148/C00763 | 121.0, 46.4 | 57.5, 27.8 | 0.0001 | 0.14 |
| Serine | C00065/C007740 | 115.3, 63.4 | 69.1, 29.8 | 0.0001 | 0.14 |
| Taurine | C00245 | 169.3, 48.5 | 79.8, 43.2 | 0.0001 | 0.14 |
| Creatine phosphate | C02305 | 19.8, 10.2 | 12.0, 5.2 | 0.0004 | 0.14 |
| Isoleucine | C00407 | 39.0,8.2 | 24,4, 13.1 | 0.0013 | 0.42 |
| Methionine | C00073 | 24.3, 6.1 | 16.0, 8.5 | 0.0015 | 0.44 |
| Phenylalanine | C00079 | 42.7, 11.8 | 32.2, 13.5 | 0.0021 | 0.56 |
| Betaine | C00719 | 39.2, 21.0 | 20.7, 8.8 | 0.0028 | 0.66 |
| Alanine | C00041/C00133 | 255.8, 77.2 | 208.4, 72.4 | 0.0029 | 0.66 |
| Glycerol | C00116 | 46.4, 18.6 | 85.8, 131.5 | 0.0031 | 0.66 |
| Valine | C00183 | 132.8, 27.8 | 100.2, 38.6 | 0.0230 | 4.50 |
| 2-Hydroxybutyrate | C05984 | 18.5, 8.0 | 11.0, 15.3 | 0.0239 | 4.50 |
| Tyrosine | C00082 | 45.3, 13.6 | 36.2, 15.1 | 0.0342 | 6.08 |
| Ethanol | C00469 | 29.7, 21.2 | 39.2, 31.7 | 0.0463 | 7.80 |
| Glucose | C00031 | 2341.3, 636.4 | 2124.2, 428.1 | 0.0490 | 7.84 |
| Acetate | C00033 | 75.2, 44.3 | 43.6, 19.8 | 0.1027 | 15.27 |
| Threonine | C00188 | 75.8, 42.2 | 96.2, 36.5 | 0.1050 | 15.27 |
| Glutamine | C00819/C00064 | 304.3, 73.3 | 257.2, 48.4 | 0.1101 | 15.32 |
| Carnitine | C00318 | 20.9, 3.2 | 18.0, 5.4 | 0.1203 | 16.04 |
| Lactate | C00186/C00256 | 1142.0, 741.0 | 973.6, 630.0 | 0.2341 | 29.96 |
| Tryptophan | C00078/C00525 | 26.5, 6.6 | 22.7, 9.8 | 0.3058 | 37.64 |
| Arginine | C00062/C00792 | 54.0, 22.8 | 56.2, 26.4 | 0.3982 | 47.19 |
| Lysine | C00047/C00739 | 89.6, 29.3 | 89.3, 29.8 | 0.4538 | 51.86 |
| 3-Hydroxybutyrate | C01089 | 31.5, 31.9 | 24.5, 28.0 | 0.5377 | 59.33 |
| Creatinine | C00791 | 29.6, 8.9 | 28.4, 10.4 | 0.7736 | 82.52 |
| Citrate | C00158 | 30.1, 11.1 | 29.9, 8.4 | 0.9113 | 94.07 |
Kyoto Encyclopedia Genes and Genomes (KEGG) identification (ID) numbers (http://www.genome.jp/kegg/)
concentration data are median, interquartile range of 19 and 20 healthy volunteers for whole blood (WB) and serum, respectively
false discovery rate
Figure 3.
Serum metabolites associated with free hemoglobin levels. (A) Valine, a branched chain amino acid; (B) glutamate, which is integral in glucose metabolism and amino acid transport and utilization, particularly in skeletal muscle; (C) phenylalanine, which is transported by RBC; (D) leucine, a branched chain amino acid; (E) creatine phosphate (see text); (F) alanine, which is transported by RBC; (G) isoleucine, a branched chain amino acid; and (H) 2-hydroxybutyrate, a by-product in the pathway of glutathione synthesis were all associated with free Hgb levels in serum. Metabolite and free Hgb levels were available from 19 of 20 serum samples obtained from healthy subjects. The Spearman correlation coefficients (ρ) and the associated p values are reported.
Free Hemoglobin Levels in Sepsis Patients are Highly Variable
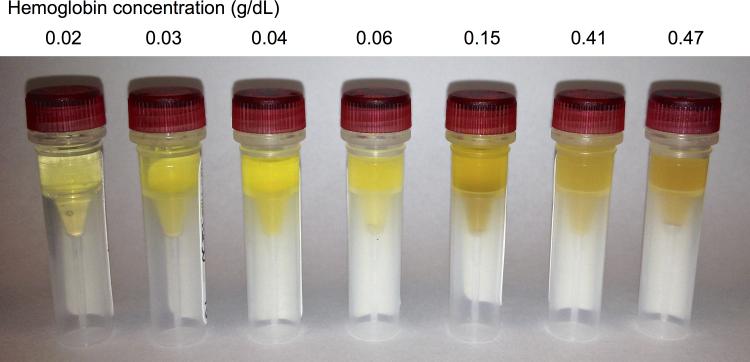
In the serum of sepsis patients (n=18; mean±S.D. age: 54.7±17.3 years), the median Hgb level was 0.05 g/dL (range: 0.02-0.46 g/dL), which was, as expected, associated with a range of visible but subtle differences in hemolysis (Figure 4). Serum free Hgb levels were available from 19 of the 20 healthy control serum samples (median, range: 0.04, 0.03-0.10 g/dL). but there was no difference between them and those of sepsis patients (Mann Whitney test, p = 0.138), suggesting that this level of hemolysis is common to sample processing and not necessarily a result of underlying pathology.
Figure 4.
A range of subtle visual differences in red blood cell hemolysis in seven representative serum samples acquired from patients with sepsis. Samples are ordered from left to right in ascending order of free hemoglobin (Hgb) levels acquired from each sample, which reflects over a 20-fold difference in Hgb concentration.
Both the Serum and Whole Blood Metabolomes Change Over Time
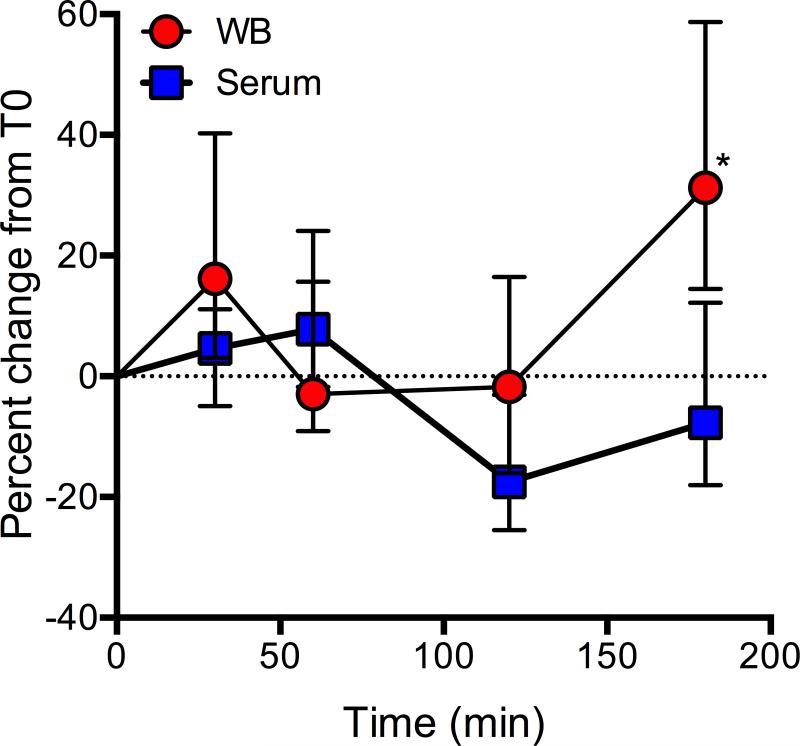
Temporal changes in serum and WB were assessed using blood from a single donor. During the time course, WB was left at room temperature to mimic the conditions of serum processing. All metabolites in both serum and WB changed over time (Figure 5), but the difference between serum and WB metabolomes was not different until T180 (Sidak's p value < 0.001). The median (IQR) percent change at T30 was 4.6% (16.1%) and 16.2% (37%) for serum and WB, respectively. The metabolite with the greatest percent decline in serum was creatine (72% at T120) and the metabolite with the greatest percent increase was taurine (152%) at T180. In WB, arginine increased the most (271% at T180) and glycerol declined the most (51% at T30). Over the course of time, glycerol increased in serum by an average of 52% and in WB, decreased by an average of 28% (data not shown). This may contribute to the higher detected glycerol levels in serum compared with WB.
Figure 5.
Percent change over time in the whole blood (WB) and serum metabolomes. Data represent the median value (± IQR) of the concentrations of the 32 common metabolites of WB and serum at each time that were collected from a single donor. Both the WB and serum sample were kept at room temperature until the time of centrifugation (serum) or transfer into the freezer (-80°C). This graph is meant to illustrate the variance that may be introduced into metabolomics data due to prolonged (30-180 min) sample processing times. ANOVA Sidak post-test *p < 0.0001.
Pathway Analysis Reveals Underrepresented Metabolism in Serum Samples
Since our NMR analysis found marked differences in the WB and serum metabolomes, we used Metscape 2 (21) to conduct pathway analysis of metabolites that differentiated the two biofluids. This type of analysis aids in the understanding of the metabolic significance of the found differences between WB and serum. The metabolic pathways containing WB-specific metabolites and the most significant metabolites (FDR < 5%) are shown in Table 3. These include amino acid metabolism, bile acid biosynthesis and the urea cycle. These results underscore the importance of the differences we found such that these pathways will be underrepresented in metabolomics data derived from serum samples.
Table 3.
Pathways involving metabolites underrepresented or absent from the serum metabolome
| Pathways | Metabolites |
|---|---|
| Bile acid biosynthesis | Taurine |
| Glycine, serine, alanine and threonine metabolism | Creatine, Choline, Creatine phosphate, Glycine, Pyruvate |
| Glycolysis and Gluconeogenesis | Pyruvate, ADP, ATP |
| Purine metabolism | Adenosine, AMP, ADP, ATP, IMP |
| TCA cycle | Succinate |
| Urea cycle and metabolism of arginine, proline, glutamate, aspartate and asparagine | Glutamate, Ornithine, Glycine, Aspartate, Asparagine, Formate, Glutathione, Pyruvate, O-Acetylcarnitine, Succinate |
| Valine, leucine and isoleucine degradation | Leucine, 3-Hydroxyisobutyrate, 2-Oxoisocaproate |
| Vitamin B9 (folate) metabolism | Glycine, Formate |
DISCUSSION
Metabolomics is a rapidly evolving field of discovery science for which the generation of reliable data hinges on the proper collection and processing of samples as well as accurate interpretation. The aim of this study was to determine the extent of the differences between the serum and WB metabolomes and influence of hemolysis on metabolite concentrations. The latter was accomplished by assessing the extent of the associations between free Hgb levels and serum metabolites. Here we demonstrate that the WB metabolome is distinct compared to that of serum when measured by 1H-NMR spectroscopy. This is supported by two primary pieces of evidence: 1) a greater number of metabolites were detected in WB than in serum; and 2) there were significant differences in metabolite concentrations between WB and serum samples. Specifically, of the 32 common metabolites of WB and serum, 17 of them had higher concentrations in WB. In addition, we showed that variability is introduced into the serum metabolome through prolonged (30-180 min) sample processing (Figure 5) and by RBC hemolysis, as the concentrations of multiple metabolites (valine, glutamate, phenylalanine, leucine, creatine phosphate, alanine, isoleucine and 2-hydroxybutyrate) were associated with free hemoglobin levels (Figure 3A-H). Collectively, the impact of using serum rather than WB for metabolomics studies (regardless of the presence of disease) is demonstrated by our pathway analysis, which showed that the absence or underrepresentation of metabolites in serum samples introduces potential bias into the biological interpretation of metabolomics data. A number of recently published tools in addition to pathway mapping calculate statistical significance of pathways or other predefined groups of metabolites based on whether they are enriched with differentiating metabolites (23, 24). Application of such methods may further compound this bias.
Implications for Sepsis Metabolomics
The application of untargeted metabolomics to critical care is a rapidly emerging field that holds great promise for lending insight to the mechanistic underpinnings of complex critical illnesses such as sepsis (25, 26). Studying the metabolome in this group is complicated by a number of pathophysiological and logistical hurdles. These include the fact that metabolism in critically ill patients is rapidly dynamic, and its measurement, as demonstrated in this study, is highly likely to be confounded by RBC and leukocyte lysis in circulating blood, which is well known to be part of the illness. In addition, a major logistical challenge in studying critical ill patients is their unscheduled presentation into relatively poorly controlled areas of the health care systems (e.g., the emergency department, the operating room, the ICU), which may contribute to delays, and technical variation inherent in sample handling and processing even in a clinical research setting. Nevertheless, metabolomics holds promise to inform of underlying etiology and may be a more sensitive gauge of illness severity and outcome than traditional biomarkers (e.g., SOFA scores) (17). This makes the implications of our findings particularly relevant to sepsis because the employment of sound metabolomics approaches are critical for the generation of reliable metabolomics data, though these findings are also likely to be relevant in any number of disease states notable for alterations in blood rheology. In this regard, WB will be more informative than serum because it can be processed more quickly, minimizing time-dependent changes in metabolite levels and it consistently reflects the contribution of RBC. The use of serum, with its lengthy processing time and highly variable free Hgb levels, which is indicative of the presence of the RBC metabolome, will introduce bias into the metabolomics analysis.
The contribution of RBC metabolites to the variance of the serum metabolome is particularly relevant because there is growing evidence that in vivo hemolysis contributes to the pathogenesis and severity of sepsis (6, 13, 27). Hemolysis can be a consequence of sepsis that perpetuates inflammation and it has been recently shown that free Hgb is associated with mortality in patients with sepsis (6). We found multiple components of the metabolome to be associated with free Hgb levels, which has implications for the interpretation of data regarding the underlying pathophysiologic changes in metabolism in the setting of sepsis, and ultimately in the design of interventions to affect these processes. For example, the concentration of glutamate (Figure 3B) was more than 3X higher in WB than in serum (Table 2) and is known to be 10X higher in RBC than in plasma (28). Glutamate is integral in glucose metabolism and amino acid transport and utilization, particularly in skeletal muscle, and has also been identified as a potential differentiating metabolite of sepsis (26) as have phenylalanine (Figure 3C) and alanine (Figure 3F), both of which are transported by RBC (10). Also, creatine phosphate has been reported to decline in sepsis (26). The association between creatine phosphate, a low abundant serum metabolite, and free Hgb (Figure 3E) warrants further investigation because even though RBC are known to contain creatine (29), it is generally accepted that RBC lack creatine kinase (30) (EC 2.7.3.2), the enzyme responsible for phosphorylating creatine to creatine phosphate. Of further relevance, RBC possess lactate dehydrogenase (LDH) (31) (EC 1.1.1.27) which catalyzes the conversion of α-ketobutyrate to 2-hydroxybutyrate (Figure 3H), a pathway that may be involved in the human inflammatory response to endotoxin (32). Finally, red cells are also a microcosm of branched chain amino acid (BCAA) metabolism and transport (7, 10) so an association between the concentrations of these amino acids and free Hgb is not surprising (Figure 3A, 3D, 3G). Nevertheless, this finding is relevant to sepsis because studies that have utilized serum or plasma samples have shown that BCAA metabolism is disrupted in critical illness (26, 32, 33). A recent metabolomics study of plasma collected from healthy volunteers that received endotoxin likewise showed a shift in amino acid (including BCAA) metabolism and elevated 2-hydroxybutyrate, the levels of which are all influenced by free Hgb as shown here (32).
The differences between the serum and WB NMR metabolomes however, are not completely explained by RBC hemolysis because only 8 of the 18 differentiating WB metabolites were associated with Hgb (Figure 3A-H). The median creatine concentration was nearly 9 times higher in WB than in serum. Despite the lack of association with Hgb, transport and exchange of creatine is an important function of RBC, particularly in skeletal muscle (29). This points to differences in sample processing between the two sample types as a possible explanation for the higher creatine levels in WB compared with serum (5). As a whole, both the serum and WB metabolomes changed over time but did not differ from each other within the first two hours at room temperature, which mimicked serum processing conditions (Figure 5). However, these sampIes remained metabolically active as evidenced by changes in individual metabolite levels (e.g., creatine, taurine) over time as reported here and by others (34). This illustrates the importance of timely and consistent sample processing for the generation of reliable and consistent metabolomics data.
There were also 18 metabolites that were only detected in WB (Table I). Some of these have been previously reported in serum (22) but in our study, they did not meet our detection criteria. A number of these metabolites, however, have not been reported in serum including adenosine, AMP, ADP and ATP which are known to be abundant in RBC. Also, RBC have a reservoir of glutathione, the most prevalent anti-oxidant in humans and one that is essential for the protection of hemoglobulin (7, 8). The distinct presence of these metabolites in WB further highlights the contribution RBC makes to the NMR detected blood metabolome. Collectively, the implications of these results are illustrated by our pathway analysis which shows that several metabolic pathways including BCAA metabolism, glycolysis, and gluconeogenesis, which are important pathways in sepsis, are underrepresented in the serum metabolome.
Limitations and Conclusions
One limitation of our study is that healthy controls were used to illustrate the differences in WB and serum NMR-based metabolomes. We elected to utilize this approach for this preliminary study because it permitted collection of samples under controlled conditions and the simultaneous acquisition of a WB and serum sample from a single venipuncture of the same subject. To determine whether the changes found in this controlled setting were likely relevant to clinical sepsis research, we measured free Hgb levels in serum from both control and sepsis patients with a representative visible range of hemolysis. In vivo hemolysis can occur under a number of conditions (6, 13-15), though can commonly occur in sepsis and is increasingly recognized as contributing to the pathology of the condition, forming some of the motivation for this investigation. We do acknowledge, however, that in our study, the free Hgb levels in serum collected from sepsis patients were not different from those of healthy controls. This does not necessarily mean that sepsis patients and healthy subjects are equally prone to in vivo hemolysis. We collected a single sample from each healthy control under well-controlled conditions (e.g., single direct venipuncture by a skilled phlebotomist) between 0830AM and 0930AM in the morning. Samples from sepsis patients were collected at all times of the day and at various points during the course of illness. So, while mechanical hemolysis may contribute to the absence of a detected difference between the two groups, we cannot draw confident conclusions about changes in free Hgb that may have occurred over time in sepsis patients because we only collected a single sample from each patient (6). However, this lack of difference between groups does provide reassurance that the findings in healthy controls is highly likely to be relevant in clinical samples and for data interpretation.
In summary, our results highlight the metabolic relevance of the contribution of RBC to the blood metabolome. These cells survey all regions of the human body but lack of internal membrane bound organelles has left them with a reputation of mere oxygen transporters. These cells are pivotal to the transport and tissue exchange of amino acids and are a reservoir of anti-oxidants like glutathione and a number of enzymes such as LDH. The metabolic capabilities of RBC, which remain underappreciated, extend beyond glycolysis. Exclusion of RBCs from blood metabolomics studies causes important information about a number of metabolites to be lost, many of which are involved in metabolic pathways relevant to sepsis. Although some work has been done in RBC metabolomics (6, 35), to the best of our knowledge, this is the first study to directly compare the WB and serum metabolomes. The use of serum or plasma without accounting for free Hgb levels or sample processing time likely enhances metabolomics data variance and may lead to misinterpretation of concentration data, both of which have implications in sepsis metabolomics. Given our findings, we believe WB metabolomics may have unique advantages as the choice biofluid, particularly in disease states characterized by changes in RBC fragility. As such, we recommend that when serum or plasma is chosen for metabolomics studies, free Hgb levels be measured in order to gauge the potential contribution of RBC hemolysis to metabolite measurements. Furthering knowledge of the metabolic importance of RBC in critical illness will lend new insight into the molecular mechanisms that underlie sepsis and sepsis severity and expand the scope of drug target opportunities.
Acknowledgements
Sources of support This study was supported by a National Institutes of Health Common Fund Metabolomics Supplement (KAS, AK) to R01 GM069438-07 (JGY), a pilot study award from the University of Michigan's School of Medicine's Department of Bioinformatics and Computational Medicine, and a University of Michigan College of Pharmacy Upjohn-Valteich Award (LY). Dr. Puskarich's effort was supported by a K23 award (GM113041) from the National Institute of General Medical Sciences (NIGMS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Portions of this work were presented at the American Thoracic Society Meeting, May 15-20th, 2015, in Denver, CO.
References
- 1.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;184(6):647–55. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, Rom W, Sanda M, Sorbara L, Stass S, Wang W, Brenner DE. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res. 2009;8(1):113–7. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sowemimo-Coker SO. Red blood cell hemolysis during processing. Transfus Med Rev. 2002;16(1):46–60. doi: 10.1053/tmrv.2002.29404. [DOI] [PubMed] [Google Scholar]
- 4.Yucel D, Dalva K. Effect of in vitro hemolysis on 25 common biochemical tests. Clin Chem. 1992;38(4):575–7. [PubMed] [Google Scholar]
- 5.Yin P, Peter A, Franken H, Zhao X, Neukamm SS, Rosenbaum L, Lucio M, Zell A, Haring HU, Xu G, Lehmann R. Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clin Chem. 2013;59(5):833–45. doi: 10.1373/clinchem.2012.199257. [DOI] [PubMed] [Google Scholar]
- 6.Adamzik M, Hamburger T, Petrat F, Peters J, de Groot H, Hartmann M. Free hemoglobin concentration in severe sepsis: methods of measurement and prediction of outcome. Crit Care. 2012;16(4):R125. doi: 10.1186/cc11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darghouth D, Koehl B, Madalinski G, Heilier JF, Bovee P, Xu Y, Olivier MF, Bartolucci P, Benkerrou M, Pissard S, Colin Y, Galacteros F, Bosman G, Junot C, Romeo PH. Pathophysiology of sickle cell disease is mirrored by the red blood cell metabolome. Blood. 2011;117(6):e57–66. doi: 10.1182/blood-2010-07-299636. [DOI] [PubMed] [Google Scholar]
- 8.Pasini EM, Lutz HU, Mann M, Thomas AW. Red blood cell (RBC) membrane proteomics--Part I: Proteomics and RBC physiology. J Proteomics. 2010;73(3):403–20. doi: 10.1016/j.jprot.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Castagnola M, Messana I, Sanna MT, Giardina B. Oxygen-linked modulation of erythrocyte metabolism: state of the art. Blood Transfus. 2010;8(Suppl 3):s53–8. doi: 10.2450/2010.009S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tunnicliff G. Amino acid transport by human erythrocyte membranes. Comp Biochem Physiol Comp Physiol. 1994;108(4):471–8. doi: 10.1016/0300-9629(94)90329-8. [DOI] [PubMed] [Google Scholar]
- 11.Purtle SW, Moromizato T, McKane CK, Gibbons FK, Christopher KB. The association of red cell distribution width at hospital discharge and out-of-hospital mortality following critical illness*. Crit Care Med. 2014;42(4):918–29. doi: 10.1097/CCM.0000000000000118. [DOI] [PubMed] [Google Scholar]
- 12.Jo YH, Kim K, Lee JH, Kang C, Kim T, Park HM, Kang KW, Kim J, Rhee JE. Red cell distribution width is a prognostic factor in severe sepsis and septic shock. Am J Emerg Med. 2013;31(3):545–8. doi: 10.1016/j.ajem.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 13.Meinders AJ, Dijkstra I. Massive hemolysis and erythrophagocytosis in severe sepsis. Blood. 2014;124(6):841. doi: 10.1182/blood-2014-04-565663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrnthaller C, Ignatius A, Gebhard F, Huber-Lang M. New insights of an old defense system: structure, function, and clinical relevance of the complement system. Mol Med. 2011;17(3-4):317–29. doi: 10.2119/molmed.2010.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holst LB, Haase N, Wetterslev J, Wernerman J, Aneman A, Guttormsen AB, Johansson PI, Karlsson S, Klemenzson G, Winding R, Nebrich L, Albeck C, Vang ML, Bulow HH, Elkjaer JM, Nielsen JS, Kirkegaard P, Nibro H, Lindhardt A, Strange D, Thormar K, Poulsen LM, Berezowicz P, Badstolokken PM, Strand K, Cronhjort M, Haunstrup E, Rian O, Oldner A, Bendtsen A, Iversen S, Langva JA, Johansen RB, Nielsen N, Pettila V, Reinikainen M, Keld D, Leivdal S, Breider JM, Tjader I, Reiter N, Gottrup U, White J, Wiis J, Andersen LH, Steensen M, Perner A. Transfusion requirements in septic shock (TRISS) trial - comparing the effects and safety of liberal versus restrictive red blood cell transfusion in septic shock patients in the ICU: protocol for a randomised controlled trial. Trials. 2013;14:150. doi: 10.1186/1745-6215-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacy P, McKay RT, Finkel M, Karnovsky A, Woehler S, Lewis MJ, Chang D, Stringer KA. Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites. PLoS One. 2014;9(1):e85732. doi: 10.1371/journal.pone.0085732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puskarich MA, Finkel MA, Karnovsky A, Jones AE, Trexel J, Harris BN, Stringer KA. Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Ann Am Thorac Soc. 2015;12(1):46–56. doi: 10.1513/AnnalsATS.201409-415OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones AE, Shapiro NI, Trzeciak S, Arnold RC, Claremont HA, Kline JA, I. Emergency Medicine Shock Research Network Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: a randomized clinical trial. JAMA. 2010;303(8):739–46. doi: 10.1001/jama.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bone RC, Sibbald WJ, Sprung CL. The ACCP-SCCM consensus conference on sepsis and organ failure. Chest. 1992;101(6):1481–3. doi: 10.1378/chest.101.6.1481. [DOI] [PubMed] [Google Scholar]
- 20.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Annals of Statistics. 2003;31(6):2013–2035. [Google Scholar]
- 21.Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, Athey B, Omenn GS. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28(3):373–80. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. The human serum metabolome. PLoS One. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chagoyen M, Pazos F. MBRole: enrichment analysis of metabolomic data. Bioinformatics. 2011;27(5):730–1. doi: 10.1093/bioinformatics/btr001. [DOI] [PubMed] [Google Scholar]
- 24.Xia J, Wishart DS. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr Protoc Bioinformatics. 10:2011. doi: 10.1002/0471250953.bi1410s34. Chapter 14:Unit 14. [DOI] [PubMed] [Google Scholar]
- 25.Langley RJ, Tsalik EL, van Velkinburgh JC, Glickman SW, Rice BJ, Wang C, Chen B, Carin L, Suarez A, Mohney RP, Freeman DH, Wang M, You J, Wulff J, Thompson JW, Moseley MA, Reisinger S, Edmonds BT, Grinnell B, Nelson DR, Dinwiddie DL, Miller NA, Saunders CJ, Soden SS, Rogers AJ, Gazourian L, Fredenburgh LE, Massaro AF, Baron RM, Choi AM, Corey GR, Ginsburg GS, Cairns CB, Otero RM, Fowler VG, Jr., Rivers EP, Woods CW, Kingsmore SF. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;5(195):195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mickiewicz B, Tam P, Jenne CN, Leger C, Wong J, Winston BW, Doig C, Kubes P, Vogel HJ. Integration of metabolic and inflammatory mediator profiles as a potential prognostic approach for septic shock in the intensive care unit. Crit Care. 2015;19(1):11. doi: 10.1186/s13054-014-0729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutra FF, Alves LS, Rodrigues D, Fernandez PL, de Oliveira RB, Golenbock DT, Zamboni DS, Bozza MT. Hemolysis-induced lethality involves inflammasome activation by heme. Proc Natl Acad Sci U S A. 2014;111(39):E4110–8. doi: 10.1073/pnas.1405023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Divino Filho JC, Hazel SJ, Furst P, Bergstrom J, Hall K. Glutamate concentration in plasma, erythrocyte and muscle in relation to plasma levels of insulin-like growth factor (IGF)-I, IGF binding protein-1 and insulin in patients on haemodialysis. J Endocrinol. 1998;156(3):519–27. doi: 10.1677/joe.0.1560519. [DOI] [PubMed] [Google Scholar]
- 29.Preen DB, Dawson BT, Goodman C, Beilby J, Ching S. Comparison of erythrocyte and skeletal muscle creatine accumulation following creatine loading. Int J Sport Nutr Exerc Metab. 2005;15(1):84–93. doi: 10.1123/ijsnem.15.1.84. [DOI] [PubMed] [Google Scholar]
- 30.Greenson JK, Farber SJ, Dubin SB. The effect of hemolysis on creatine kinase determination. Arch Pathol Lab Med. 1989;113(2):184–5. [PubMed] [Google Scholar]
- 31.Tanaka KR, Zerez CR. Red cell enzymopathies of the glycolytic pathway. Semin Hematol. 1990;27(2):165–85. [PubMed] [Google Scholar]
- 32.Kamisoglu K, Sleight KE, Calvano SE, Coyle SM, Corbett SA, Androulakis IP. Temporal metabolic profiling of plasma during endotoxemia in humans. Shock. 2013;40(6):519–26. doi: 10.1097/SHK.0000000000000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lusczek ER, Lexcen DR, Witowski NE, Determan C, Jr., Mulier KE, Beilman G. Prolonged induced hypothermia in hemorrhagic shock is associated with decreased muscle metabolism: a nuclear magnetic resonance-based metabolomics study. Shock. 2014;41(1):79–84. doi: 10.1097/SHK.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 34.Breier M, Wahl S, Prehn C, Fugmann M, Ferrari U, Weise M, Banning F, Seissler J, Grallert H, Adamski J, Lechner A. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS One. 2014;9(2):e89728. doi: 10.1371/journal.pone.0089728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabenstein DL. H-1-Nmr Methods for the Noninvasive Study of Metabolism and Other Processes Involving Small Molecules in Intact Erythrocytes. Journal of Biochemical and Biophysical Methods. 1984;9(4):277–306. doi: 10.1016/0165-022x(84)90013-7. [DOI] [PubMed] [Google Scholar]