Abstract
Carboxyl-terminal binding protein-1 (CtBP1) is a transcriptional co-repressor with multiple in vitro targets, but its in vivo functions are largely unknown. We generated keratinocyte-specific CtBP1 transgenic mice with a keratin 5 promoter (K5.CtBP1) to probe the pathological roles of CtBP1. At transgene expression levels comparable with endogenous CtBP1 in acute skin wounds, K5.CtBP1 epidermis displayed hyperproliferation, loss of E-cadherin, and failed terminal differentiation. Known CtBP1 target genes associated with these processes, e.g., p21, Brca1, and E-cadherin were down-regulated in K5.CtBP1 skin. Surprisingly, K5.CtBP1 pups also exhibited a hair loss phenotype. We found that expression of the Distal-less 3 (Dlx3), a critical regulator of hair follicle differentiation and cycling, was decreased in K5.CtBP1 mice. Molecular studies revealed that CtBP1 directly suppressed Dlx3 transcription. Consistently, K5.CtBP1 mice displayed abnormal hair follicles with decreased expression of Dlx3 downstream targets Gata3, Hoxc13, and hair keratins. In sum, this first CtBP1 transgenic model provides in vivo evidence for certain CtBP1 functions predicted from in vitro studies, reveals to our knowledge previously unreported functions and transcriptional activities of CtBP1 in the context of epithelial-mesenchymal interplay, and suggest CtBP1 has a pathogenesis role in hair follicle morphogenesis and differentiation.
Introduction
CtBP was originally identified based on its ability to bind the carboxyl terminus of the E1A oncoprotein (Boyd et al., 1993; Schaeper et al., 1995). Subsequently, CtBP was found as a transcriptional co-repressor involved in a variety of biological processes including proliferation and anti-apoptosis (Chinnadurai, 2002). CtBP indirectly binds DNA with various DNA binding partners at multiple DNA sequences thus CtBP-mediated transcriptional repression is context-specific. For instance, CtBP represses E-cadherin in epithelial cells (Grooteclaes et al., 2003; Grooteclaes and Frisch, 2000; Zhang et al., 2006), IL-4 in human T cells (Kitamura et al., 2009), and dll4, sprouty, and ve-cadherin for endothelial sprouting (Roukens et al., 2010). Recently, we and others found that CtBP down-regulates DNA damage repair by directly suppressing the transcription of breast cancer type 1 susceptibility protein (Brca1) in cancer cells (Deng et al., 2010; Di et al., 2010).
In mammals, there are two isoforms, CtBP1 and CtBP2. Both isoforms are expressed in the wildtype mouse embryo and play overlapping and unique roles (Hildebrand and Soriano, 2002). CtBP1 knockout mice are small and 23% die of an unknown cause by 20 days postpartum. CtBP2-null mice are small in size, have axial truncations, delayed neural, muscular and skeletal development, and defects in heart morphogenesis; they die by E10.5 due to defects in both yolk sac and placental vascularization. In most human and mouse adult tissues, CtBP expression is low. Re-activation of CtBP expression has been shown in pathological conditions, e.g, cancer (Deng et al., 2010; Nadauld et al., 2006), but its in vivo role in adult tissue is virtually unknown.
To determine the effects of aberrant CtBP1 expression in keratinocytes in vivo, we targeted CtBP1 over-expression to the basal layer of the epidermis and hair follicle using a keratin 5 promoter (K5.CtBP1) (He et al., 2002). K5.CtBP1-transgenic epidermis displayed loss of E-cadherin, hyperproliferation and decreased differentiation. Unexpectedly, K5.CtBP1 mice also exhibited defective hair morphogenesis starting at the postnatal stage. Molecular analyses revealed that CtBP1 directly repressed transcription of the Dlx3 gene, a homeobox transcription factor that plays a critical role in hair development by orchestrating the differentiation of the inner root sheath and hair shaft (Hwang et al., 2008). Supporting this link, we found a hair follicle formation defect in the K5.CtBP1 mice, with decreased expression of Dlx3 and its target genes. Our study provides in vivo model for CtBP1 over-expression and reveals that CtBP1 over-expression perturbs epidermal and hair follicle homeostasis.
Results
Generation of the CtBP1 transgenic mice
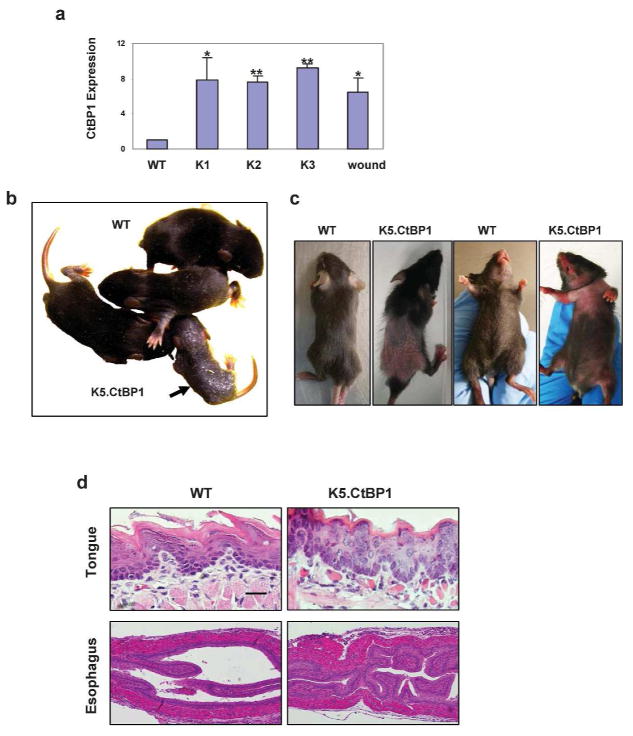
In normal mouse skin, CtBP1 is barely detectable. Acute skin wound by punch biopsy induced CtBP1 expression ~6 fold higher than in non-wounded skin (Fig. 1A). To evaluate the role of CtBP1 over-expression in the skin, we generated K5.CtBP1 transgenic mice by inserting human CtBP1 cDNA (99% amino acid homology to mouse CtBP1 protein) into a K5 vector (He et al., 2002). Three K5.CtBP1 transgenic founders (K1, K2, and K3) were generated. Their CtBP1 transgene expression levels were 7–10 fold higher than endogenous CtBP1, but comparable to wounds at the mRNA level (Fig. 1A). Overall, K5.CtBP1 phenotype severity correlated with transgene expression levels, suggesting CtBP1 over-expression causes the phenotype. Results from the representative line K2 were shown in this study. K5.CtBP1 pups were born without gross abnormality (not shown), but began to exhibit thickened skin at 1 week postpartum when wildtype mice developed their first coat of hair and K5.CtBP1 pups had no hair growth (Fig. 1B). After weaning, K5.CtBP1 mice exhibited hair loss, on their dorsal and ventricle sides (Fig. 1C). These transgenic mice died between 3 to 6 weeks of age due to severe hyperplasia in the esophagus (Fig. 1D) and forestomach (not shown) where CtBP1 transgene was also expressed, compromising food intake. Compared to the wildtype control, these K5.CtBP1 mice also displayed abnormal epithelium in their tongues (Fig. 1D).
Figure 1.
Generation of K5.CtBP1 mice and phenotypes. (A) CtBP1 mRNA expression in skin of wildtype mice (WT) and K5.CtBP1 transgenic mice (K1, K2, K3), and acutely wounded WT skin (wound). The mRNA level in WT skin was arbitrarily set as “1”. Error bars indicate s.d. (n=3), significance was determined using Student’s t test. **, p<0.01; *, p<0.05. (B) Hyperplasia/hyperkeratotic gross appearance of a K5.CtBP1 transgenic pup 8 days postpartum, in comparison with WT littermates. (C) Thin and patchy hair in K5.CtBP1 mice 3 weeks postpartum, in comparison with WT littermates. (D) H&E staining of WT and K5.CtBP1 tongue (top panels) and esophagus (bottom panels). Scale bar: 15 μm (top panels) and 40 μm (bottom panels).
K5.CtBP1 mice displayed epidermal hyperproliferation, with decreased expression of p21 and Brca1
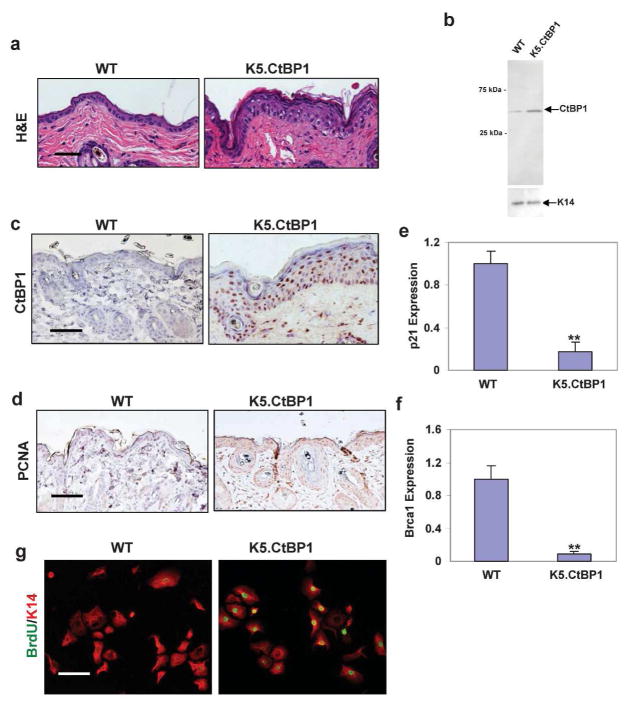
Re-activation of CtBP1 expression has been shown in cancer (Deng et al., 2010; Nadauld et al., 2006) and during wound healing (Fig. 1A), presumably contributing to the hyper-proliferative process in these conditions. We biopsied the K5.CtBP1 skin at day 21 postpartum to examine morphology changes (Fig. 2A) and expression of CtBP1 (Fig. 2B and 2C). Hyperplasia in the epidermis of transgenic mice became obvious at the microscopic level (Fig. 2A). Although the K5 promoter targets CtBP1 expression to basal keratinocytes, nuclear CtBP1 staining was apparent throughout the entire epidermis and hair follicles in K5.CtBP1 transgenic mice (Fig. 2C), possibly due to protein retention in differentiated layers. In contrast, weak CtBP1 staining was detected in wildtype skin (Fig. 2B and 2C). Interestingly, although CtBP1 transgene is targeted to keratinocytes, CtBP1-positive cells were also observed in the K5.CtBP1 dermis but not in wildtype dermis (Fig. 2C). This is likely increased endogenous CtBP1 in stromal cells in response to changes in the epidermis and hair follicles. The origin of the cells over-expressing endogenous CtBP1 in the transgenic dermis remains to be determined.
Figure 2.
Microscopic characteristics of K5.CtBP1 skin. (A) H&E staining of 21 day-old mice shows hyperplasia of epidermis in K5.CtBP1 skin compared with WT skin. (B) Western blotting of CtBP1 in WT and K5.CtBP1 skin. Keratin 14 (K14) was used as loading control. Immunohistochemistry using antibodies specific for CtBP1 (C) and PCNA (D). Scale bar: 40 μm (all panels). qRT-PCR analysis shows down-regulation of CtBP1 target genes p21 (E) and Brca1 (F) in K5.CtBP1 skins. p21 mRNA was decreased to 0.17±0.09 and Brca1 mRNA was decreased to 0.09±0.03 in transgenic skin. (G) Keratinocytes from K5.CtBP1 mice displayed an increased BrdU index. Keratin 14 (red) was used as a counterstain. Scale bar: 15 μm.
In vitro studies show that CtBP1 has both anti-apoptotic and proliferative effects (Grooteclaes et al., 2003; Mroz et al., 2008), potentially causing epidermal hyperplasia. Apoptosis, as determined by cleaved Caspase-3, was not changed in K5.CtBP1 skin (not shown). Proliferative cells, identified by PCNA staining, were sporadic in the basal layer of the epidermis and hair follicles of wildtype skin, but were expanded to suprabasal layers of K5.CtBP1 transgenic epidermis and hair follicles (Fig. 2D). Furthermore, we found mRNA levels of p21 and Brca1 were decreased in skin with CtBP1 over-expression, with 5- and 11-fold reduction in the K5.CtBP1 skin compared to that of the control skin (Fig. 2E and 2F), supporting the notion that CtBP1 positively contributes to proliferation in keratinocytes. We cultured keratinocytes from K5.CtBP1 mice. Consistent with the hyperplasic phenotype observed in the K5.CtBP1 mice, keratinocytes derived from these animals displayed a higher BrdU index than wildtype controls (Fig. 2G), suggesting that the proliferative property is cell autonomous. No major difference was detected in the cell cycle analysis and migration assay (not shown).
CtBP1 transgene expression reduced differentiation and caused loss of E-cadherin in the epidermis and hair follicles
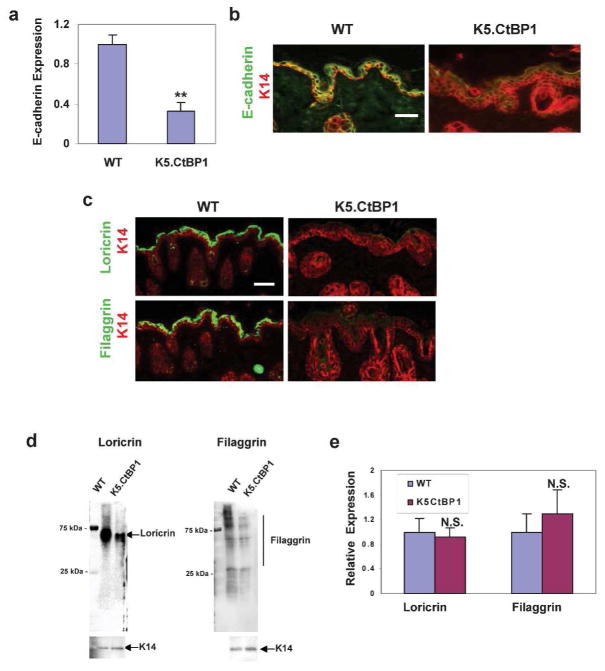
Previously, we and others demonstrated that CtBP1 represses the transcription of E-cadherin by directly targeting the E-cadherin promoter (Grooteclaes et al., 2003; Grooteclaes and Frisch, 2000; Zhang et al., 2006). To determine if CtBP1 expression in epithelial cells leads to E-cadherin reduction, we performed qRT-PCR on the K5.CtBP1 skin at day 21 postpartum and found that E-cadherin mRNA was decreased compared to wildtype control littermates (Fig. 3A). Consistent with the mRNA decrease, E-cadherin protein was largely lost in K5.CtBP1 epidermis and hair follicles as seen by immunostaining (Fig. 3B). The E-cadherin loss in K5.CtBP1 mice is progressive, as only a small decrease of E-cadherin was detected in day one pup skin compared to wildtype littermates (not shown).
Figure 3.
Aberrant differentiation in the epidermis and hair follicles of K5.CtBP1 skin. (A) Down-regulation of CtBP1 target gene E-cadherin in K5.CtBP1 skin. **: p<0.01. (B) Immunofluorescence staining for E-cadherin (green) demonstrates significant loss of E-cadherin in K5.CtBP1 epidermis and hair follicles. Keratin 14 (red) was used as a counterstain. Scale bar: 40 μm. (C) Immunofluorescence staining (green) for differentiation markers loricrin and filaggrin demonstrates loss of these proteins in K5.CtBP1 epidermis. Keratin 14 (red) was used as a counterstain. Scale bar: 40 μm. (D) Western blotting of loricrin and filaggrin in WT and K5.CtBP1 skin. Keratin 14 (K14) serves as a loading control. (E) qRT-PCR analysis of the differentiation marker genes loricrin and filaggrin in K5.CtBP1 skins. N.S., no statistical significance by Student’s t test.
Next, we examined the epidermal differentiation of K5.CtBP1 mice. Early epidermal differentiation markers, keratins K1 and K10, were not altered (not shown), but epidermal terminal differentiation markers loricrin and filaggrin were largely diminished by immunostaining in the K5.CtBP1 epidermis (Fig. 3C) and significantly reduced by western blotting (Fig. 3D). Because CtBP1 is a classic transcriptional co-repressor and potentially contributes to the decreased differentiation by transcriptional repression of the terminal differentiation players, we assayed the mRNA levels of loricrin and filaggrin, important physical barrier components in the epidermis against the environment (Candi et al., 2005; Kalinin et al., 2001). Different from the protein loss observed in the K5.CtBP1 epidermis, neither loricrin nor filaggrin showed a decrease in their mRNA levels (Fig. 3E), suggesting the defect in epidermal terminal differentiation is a secondary effect.
CtBP1 transgene suppressed Dlx3, a critical regulator of hair follicle differentiation
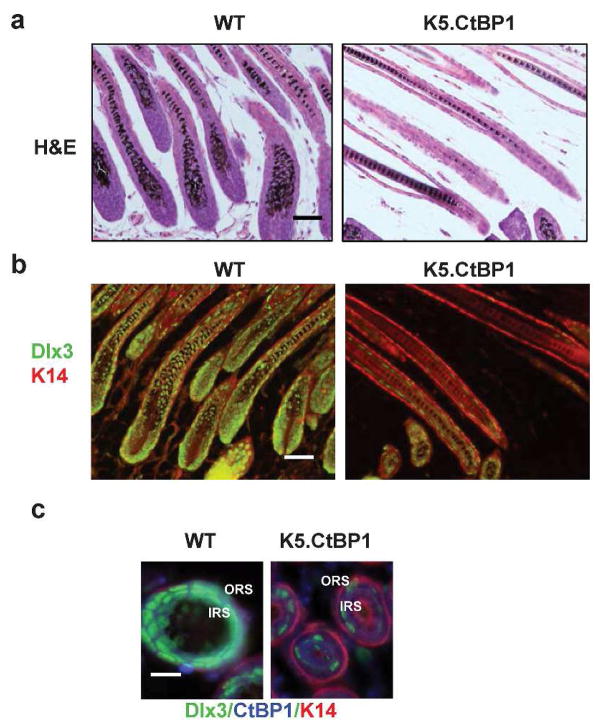
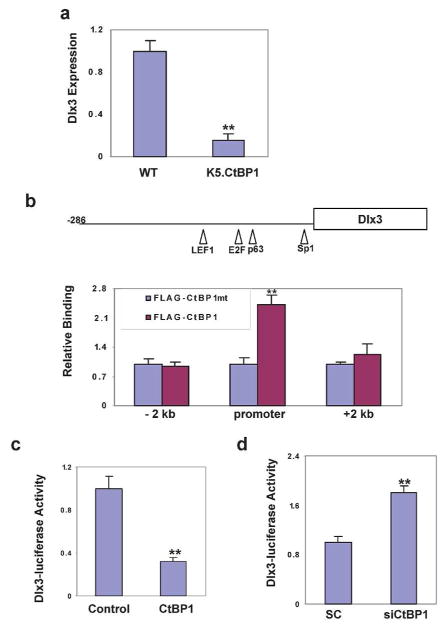
We biopsied neonatal skin and found no obvious hair follicle changes at birth (not shown), suggesting that CtBP1 did not affect hair development. Consistently, similar expression of LEF-1, β-catenin, and pSmad1/5/8 were detected in the CtBP1 transgenic skin hair follicles (not shown), implying that Wnt and BMP signaling pathways are not affected by CtBP1 over-expression in the epidermis. Structural hair follicle abnormalities were observed by day 9 postpartum. Hair follicles had formed large hair bulbs and differentiated hair shafts in wildtype mice. In contrast, CtBP1 transgenic hair follicles displayed smaller hair bulbs, with reduced keratinized medulla and defects in the inner root sheath and hair shaft (Fig. 4A), phenotypes reminiscent of hair follicle abnormities observed in the Dlx3-null mice (Hwang et al., 2008). Therefore, we used the Dlx3 hair follicle differentiation marker to explore the molecular mechanism associated with hair loss in K5.CtBP1 mice. At day 9 postpartum, Dlx3 was expressed in hair matrix cells, inner root sheath, as well as in hair-forming compartments such as the cortex, medulla and cuticle in hair follicles from the wildtype mice (Fig. 4B, left panel). However, expression of Dlx3 was largely reduced in hair follicles from the K5.CtBP1 mice (Fig. 4B, right panel), suggesting that CtBP1 over-expression in hair follicle down-regulates Dlx3 to induce the differentiation defect of the inner root sheath and hair shaft. Cross-sectional co-immunofluorescence staining of Dlx3 and CtBP1 revealed that in wildtype hair follicles the Dlx3 expression is in the inner root sheath, with CtBP1 expression mainly in the outer root sheath (Fig. 4C, left panel). In K5.CtBP1 hair follicles, expression of Dlx3 was decreased, with CtBP1 expansion to the inner root sheath (Fig. 4C, right panel). To evaluate the Dlx3 changes at the mRNA level, in situ hybridization analysis was performed. Dlx3 mRNA was detected in the wildtype mice hair follicles (Supplemental Figure, left panel). In contrast, there was very little Dlx3 detected in the K5.CtBP1 hair follicles (Supplemental Figure, right panel).
Figure 4.
CtBP1 suppresses Dlx3 gene expression in K5.CtBP1 skins. (A) H&E staining of WT and K5.CtBP1 hair shaft and bulb. Scale bar: 15 μm. Note the reduced hair bulbs in the transgenic mice. (B) Immunofluorescence staining of Dlx3 (green) demonstrates its aberrant expression pattern in K5.CtBP1 hair follicles. Keratin 14 (red) was used as a counterstain. Scale bar: 15 μm. (C) Cross-sectional immunofluorescence staining of Dlx3 (green) and CtBP1 (blue) demonstrates the decreased Dlx3 expression with CtBP1 expansion to the inner root sheath (IRS) in K5.CtBP1 hair follicles. ORS, outer root sheath. Keratin 14 (red) was used as a counterstain. Scale bar: 5 μm.
Next, we assayed the mRNA level of Dlx3 in K5.CtBP1 skin by qRT-PCR and compared it to that in wildtype littermates. A significant decrease in Dlx3 mRNA was detected in the CtBP1 transgenic skin (Fig. 5A). To determine if CtBP1 plays a direct role in regulation of the Dlx3 gene, we performed chromatin immunoprecipitation (ChIP) to see if CtBP1 is recruited to the Dlx3 promoter in mouse keratinocytes. Mouse keratinocytes were transfected with a vector expressing CtBP1, either wildtype or the PLDLX-binding deficient mutant tagged with the FLAG epitope, and the cross-linked chromatin was immunoprecipitated with an anti-FLAG antibody. Wildtype CtBP1 bound the Dlx3 promoter region while the PLDLX-binding deficient mutant did not (Fig. 5B). This binding is limited to the promoter region, as no signal was detected with PCR amplification of the ChIP material using primers either 5′ or 3′ 2 kb to the Dlx3 promoter (Fig. 5B). Consistent with mRNA changes observed during CtBP1 knockdown, the luciferase activity of the Dlx3 promoter decreased by 60% with CtBP1 over-expression in mouse keratinocytes (Fig. 5C), indicating that CtBP1 regulates Dlx3 transcription, at least partially, via binding to its promoter. To investigate if the CtBP1-mediated repression of Dlx3 gene occurs in human cells, we knocked down CtBP1 in Fadu cells, a human SCC cell line exhibiting high endogenous CtBP1 (Deng et al., 2010), and assayed the Dlx3-luciferase reporter. CtBP1 knockdown increased the Dlx3-luc reporter activity (Fig. 5D), suggesting that CtBP1’s repressive role in Dlx3 transcription is conserved between mouse and human.
Figure 5.
CtBP1 suppresses the Dlx3 gene transcription. (A) Down-regulation of the Dlx3 gene in K5.CtBP1 skins from 9 day-old mice. (B) CtBP1 binding to the Dlx3 promoter. ChIP assay was performed in keratinocytes after transfection with FLAG-tagged CtBP1-expressing vector, either wild type or the PLDLX-binding deficient mutant. ** p<0.01 vs. the PLDLX-binding deficient mutant (mt). (C) CtBP1 transfection represses the Dlx3 reporter. The pGL4.26 Dlx3 promoter reporter was generated by cloning the −286 to 0 bp fragment of the Dlx3 promoter. (D) siCtBP1 increases activity of the Dlx3 reporter. Fadu cells were transfected with scrambled siRNA (SC) or siRNA to CtBP1 (siCtBP1) and luciferase activity was measured.
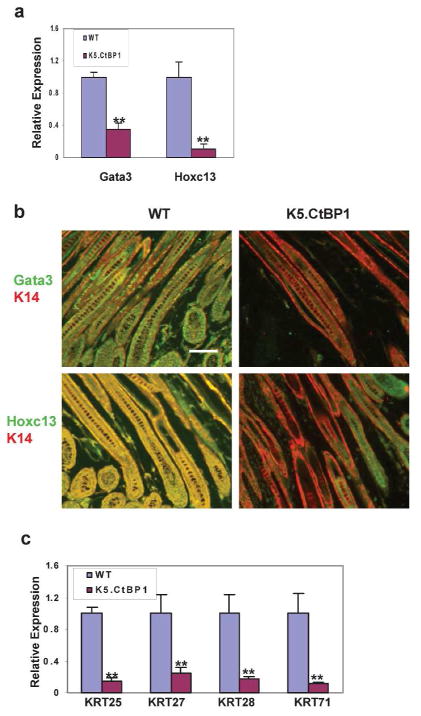
To further study the functional consequence of CtBP1-mediated repression of the Dlx3 gene, we examined mRNA levels of Dlx3 transcriptional targets in K5.CtBP1 skin. Gata3 and Hoxc13 are transcription factors affecting hair differentiation (Godwin and Capecchi, 1998; Kurek et al., 2007). Decreased Gata3 and Hoxc13 expression was detected in genetically engineered K14-Dlx3−/− mice (Hwang et al., 2008). As shown in Fig. 6A, CtBP1 transgene expression decreased mRNA levels of Gata3 to 60% of that seen in control skin; small but significant changes. On the contrary, expression of Hoxc13 was decreased to 10% of that seen in control skin by CtBP1 transgene expression (Fig. 6A). Fig. 6B illustrates the decrease of Gata3 and Hoxc13 expression in hair follicles of K5.CtBP1 mice.
Figure 6.
Down-regulation of Dlx3 target genes in K5.CtBP1 skins. (A) WT and K5.CtBP1 skins were used for qRT-PCR. Gata3 mRNA decreased to 0.35±0.07 and Hoxc13 mRNA decreased to 0.11±0.06 in transgenic skin. **: p<0.01. (B) Immunofluorescence staining (green) of Gata3 and Hoxc13 demonstrates decreased expression in K5.CtBP1 hair follicles compared with WT follicles. Keratin 14 (red) was used as a counterstain. Scale bar: 15 μm. (C) WT and K5.CtBP1 skins were used for qRT-PCR Keratin 25 (KRT25), keratin 27 (KRT27), keratin 28 (KRT28), and keratin 71 (KRT71) mRNA expression levels. Error bars indicate s.d. (n=3), and significance was determined using Student’s t test. **: p<0.01.
A recent study shows Dlx3 up-regulates expression of the inner root sheath forming keratins (Kim et al., 2012). Therefore, we measured the levels of Type I inner root sheath keratin genes. Krt25, Krt27, and Krt28, and Type II inner root sheath keratin gene Krt71 in wildtype and K5.CtBP1 skin. Similar to the decrease of Hoxc13 expression observed in K5.CtBP1 skin, the mRNA levels of Krt25, Krt27, and Krt28 decreased significantly when compared to control skin (Fig. 6C). The expression of Krt71 was decreased by 4-fold as well (Fig. 6C). Taken together, these findings indicate that CtBP1 over-expression triggered a down-regulation of transcription factors and hair keratins critical for the maintenance of hair follicle homeostasis.
Discussion
Epidermal E-cadherin loss, hyperproliferation and poor differentiation are caused by keratinocyte-specific CtBP1 over-expression
We and others have shown that CtBP1 expression can be re-initiated in cancers (Deng et al., 2010; Nadauld et al., 2006), thus we studied if pathologically induced CtBP1 over-expression perturbs skin homeostasis. Consistent with previous in vitro studies, we found hyperproliferation, down-regulated differentiation, and loss of E-cadherin in K5.CtBP1 mouse skin. Among potential CtBP1 targets associated with proliferation, we found down-regulation of p21 and Brca1. Increased proliferation can be, but is not always, associated with reduced differentiation. Both p21 and Brca1 inhibit cell cycle progression and induce differentiation in the epidermis (Berton et al., 2003; Missero et al., 1996). Therefore, reduced p21 and Brca1 could contribute to reduced differentiation of K5.CtBP1 epidermis. We also found gradual E-cadherin loss in the transgenic epidermis and hair follicles. Keratinocyte-specific E-cadherin knockout mice do not show epidermal blisters but display epidermal hyperproliferation and poor differentiation in both the epidermis and hair follicles (Tinkle et al., 2004; Young et al., 2003). Thus, E-cadherin loss in K5.CtBP1 transgenic mice could significantly contribute to progressive hyperplasia and poor terminal differentiation.
Dlx3 ablation in keratinocytes has been shown to induce hyperplasia of epidermis (Hwang et al., 2008). In the current study, we found CtBP1 suppresses transcription of the Dlx3 gene, which may shift the balance between proliferation and differentiation and contribute to the overly proliferative phenotype of the K5.CtBP1 mice. In addition, the resultant down-regulation of Gata3 may further facilitate over-proliferation because Gata3 ablation in epidermis has been shown to induce epidermal hyperplasia (Kurek et al., 2007). Therefore, keratinocyte proliferation and reduced differentiation of cells in the epidermis and hair follicles appear to be critically regulated by CtBP1, as seen from the synergistic actions of p21, E-cadherin, and terminal differentiation regulators of the epidermis and hair follicles such as Dlx3 and Gata3.
CtBP1 transcriptionally represses the DLX3 gene, a critical regulator of hair follicle differentiation
In our K5.CtBP1 transgenic model, we unexpectedly found defective hair morphogenesis caused by CtBP1 over-expression in keratinocytes. Previous studies have shown that Dlx3 is a transcriptional activator and plays a critical role in the development of epidermis, hair, bone, and placenta (Feledy et al., 1999; Hassan et al., 2004; Hwang et al., 2008; Morasso et al., 1999; Morasso et al., 1996) and Dlx3 mutations are responsible for the defects in hair, teeth and bone development called the Tricho-Dento-Osseous syndrome (Price et al., 1998a; Price et al., 1998b). Our transgenic mice displayed hair follicle differentiation abnormalities similar to the K14.Dlx3−/− mice, with smaller hair bulbs, reduced keratinized medulla and defects in the inner root sheath and hair shaft (Hwang et al., 2008). While the hair loss seems regional, abnormal hair follicles were also observed where hair was retained (not shown), suggesting that mechanical triggers, such as rubbing, facilitate the hair loss. Unlike the genetically engineered mice with Dlx3 ablation in the epidermis in which Wnt and BMP signaling are disturbed, the Wnt and BMP signaling pathways were not perturbed by CtBP1 over-expression in keratinocytes, presumably reflecting an incomplete shutdown of Dlx3 in the K5.CtBP1 mice. The preservation of the BMP and Wnt signaling is consistent with the K5.CtBP1 mice’s ability to regenerate hair despite their abnormal hair formation.
Consistent with Dlx3-mediated effects in hair follicles, keratinocyte-specific over-expression of CtBP1 caused a decrease of Gata3 and Hoxc13, transcription factors involved in hair differentiation (Godwin and Capecchi, 1998; Tkatchenko et al., 2001) and Type I/II keratins. Previous study has shown that HOXC13 regulates human hair keratin gene expression (Jave-Suarez et al., 2002). The inner root sheath forms its structure by obligate heterodimerization of the specified keratins. Type I inner root sheath keratin genes, Krt25, Krt27, and Krt28, and Type II inner root sheath keratin gene Krt71 are specifically expressed in all three layers of the inner root sheath and support the structure (Runkel et al., 2006; Tanaka et al., 2007). We observed that expression of these inner root sheath keratins were down-regulated by CtBP1. As a result of keratin loss, the heterodimer may be reduced, hindering the inner root sheath formation and resulting in further hair abnormality. For instance, mutations in the helix termination motif of mouse Type I inner root sheath keratin genes have been shown to impair the assembly of keratin intermediate filament (Tanaka et al., 2007). The ability to down-regulate both Type I and Type II keratins may further impair the inner root sheath formation.
In addition to the critical role of Dlx3 in hair follicle differentiation, ablation of E-cadherin in keratinocytes has been shown to induce the progressive loss of hair follicle integrity (Tinkle et al., 2004; Young et al., 2003). We reason that CtBP1 contributes to the hair follicle abnormality through multiple targets including Dlx3 and E-cadherin, thus it might become more pronounced with aging. Future studies using an inducible model will help elucidate the impact of CtBP1 over-expression on epidermal and hair follicle differentiation at later stages. This model will also allow us to assess the role of CtBP1 in other pathological processes such as wound healing.
In summary, we report the CtBP1 transgenic mouse model to reveal in vivo CtBP1 transcriptional targets in keratinocytes and functions associated with epidermal hyperproliferation, reduced differentiation, and E-cadherin down-regulation. Further, CtBP1 has a role in hair morphogenesis; which had not been identified previously, and can only be appreciated with CtBP1 over-expression during differentiation of the hair follicle. Our data instigate future studies to determine if pathologically induced CtBP1 over-expression plays a role in skin pathogenesis in human diseases.
Methods
Generation and identification of K5.CtBP1 mice
All animal experiments were performed with the approval of IACUC at University of Colorado Denver. The 1.3 kb full-length wildtype human CtBP1 cDNA was inserted into the K5 expression vector (He et al., 2002). The K5.CtBP1 transgenic mice were generated with the B6D2 strain by microinjection of the transgene into the pronuclei of mouse embryos. Mice were genotyped by PCR analysis of tail DNA utilizing primers specific for BK5 (tctgataggcagcctgcacc) and CtBP1 (atcccagctgctgtggaagg). Throughout this study, all transgenic mice were heterozygous; all wildtype mice were littermates, and at least three independent analyses were performed for each assay, using three to five samples in each group.
Tissue histology, immunofluorescence, immunohistochemistry, western blotting, and in situ hybridization
Skin histology was visualized with hematoxylin and eosin (H&E) staining. Immunofluorescence, immunohistochemistry, and western blotting were performed on frozen and paraffin-embedded sections as previously described (Wang et al., 1999). Immunofluorescence and western blotting were performed using antibodies against CtBP1 (Millipore), loricrin, filaggrin (Covance), Hoxc13 (Abnova); Gata3 (Santa Cruz); E-cadherin (BD Biosciences); Dlx3 (Abcam); and Keratin 14 (Fitzgerald). The antibodies used in immunohistochemistry included CtBP1 (Millipore) and PCNA (Santa Cruz). For immunofluorescence, secondary antibodies to different species IgG were Alexa Fluor® 594 (red) or 488 (green) conjugated (1:200 for all, Invitrogen). For immunohistochemistry, we used secondary biotinylated antibodies to different species IgG (1:300, Vector Labs) and developed using Vectastain ABC kit (Vector Labs). In situ hybridization with antisense digoxigenin-UTP-labeled RNA probes on sections of skin samples was performed as described (Han et al., 2006).
qRT-PCR
Total RNA was isolated using TRIzol (Invitrogen) as previously described (Zhang et al., 2006). One hundred nanograms of RNA from each sample were subjected to qRT-PCR (ThermoFisher). An 18S probe was used as an internal control. Each sample was examined in triplicate. The relative RNA expression levels were determined by normalizing with internal controls, the values of which were calculated using the comparative Ct method.
Cell culture and transfections
Mouse keratinocytes were isolated from neonatal mouse skin as previously described (Han et al., 2011), and cultured in PCT medium (CELLnTEC). Fadu, a human HNSCC line, was purchased from ATCC and cultured in DMEM with 10% FBS. To knock down CtBP1, cells were treated with siRNA against CtBP1 from Dharmacon using Lipofectamine 2000 (Invitrogen) for 48 hours, then harvested. Keratinocytes were treated with BrdU; cells that incorporated BrdU were stained by FITC-anti-BrdU.
Chromatin immunoprecipitation (ChIP) and luciferase reporter assay
Keratinocytes were transfected with a CtBP1-expressing vector, either wildtype or the PLDLX-binding deficient mutant tagged with FLAG epitope. ChIP assay was performed with an anti-FLAG antibody as described previously (Zhang et al., 2006). Primer sets spanning the Dlx3 promoter were used to q-PCR-amplify the ChIP sample. The pGL4.26 DLX3 promoter luciferase reporter plasmid was generated by cloning a PCR-amplified −286 to 0 bp fragment of the Dlx3 promoter into the XhoI and NheI sites of pGL4.26 vector (Promega). Dlx3 promoter–specific primers used were 5′-TATCTCGAGCCGCACAGCCAAC-3′ (forward) and 5′-AATGCTAGCGCCAGCTCCGCCC-3′ (reverse). An empty renilla luciferase vector (pGL4.79) was used for normalization. Mouse keratinocytes were co-transfected with the reporters and CtBP1-expressing plasmids for 48 hr and the luciferase activity was measured (Zhang et al., 2002). Human Fadu cells were co-transfected with the reporters and siRNA to CtBP1 for 48 hr and the luciferase activity was measured (Zhang et al., 2002). Scrambled siRNA or empty plasmid was used for controls.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH, R01CA87949 (to X. J. W.) and R01CA115468 and R03DA033982 (to Q.Z.), pilot grants from P30CA046934 and P30 AR057212 (to Q.Z.), and seed grants from Cancer League of Colorado and CCTSI (to Q. Z.). Hui Deng was supported by grants from the Sixth People’s Hospital of Shanghai. Fulun Li was supported by grants (No. 81102596) from the National Science Foundation of China (NSFC), The Shanghai Rising-Star Program (NO.12QA1403300), and The Innovation Program of Shanghai Municipal Education Commission (NO.12YZ067). We thank Laura Hoaglin and Pamela Fernandez for excellent technical support, Jiang Chen, Maranke Koster, Peter Koch, and Denis Roop for helpful comments, and Pamela J. Garl for editing the manuscript.
Footnotes
Conflict of Interests: The authors declare no conflict of interests.
References
- Berton TR, Matsumoto T, Page A, Conti CJ, Deng CX, Jorcano JL, Johnson DG. Tumor formation in mice with conditional inactivation of Brca1 in epithelial tissues. Oncogene. 2003;22:5415–5426. doi: 10.1038/sj.onc.1206825. [DOI] [PubMed] [Google Scholar]
- Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. Embo J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- Deng Y, Liu J, Han G, Lu SL, Wang SY, Malkoski S, Tan AC, Deng C, Wang XJ, Zhang Q. Redox-dependent Brca1 transcriptional regulation by an NADH-sensor CtBP1. Oncogene. 2010;29:6603–6608. doi: 10.1038/onc.2010.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di LJ, Fernandez AG, De Siervi A, Longo DL, Gardner K. Transcriptional regulation of BRCA1 expression by a metabolic switch. Nat Struct Mol Biol. 2010;17:1406–1413. doi: 10.1038/nsmb.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feledy JA, Morasso MI, Jang SI, Sargent TD. Transcriptional activation by the homeodomain protein distal-less 3. Nucleic Acids Res. 1999;27:764–770. doi: 10.1093/nar/27.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci U S A. 2003;100:4568–4573. doi: 10.1073/pnas.0830998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, Ten Dijke P, Lin X, Wang XJ. Smad7-induced beta-catenin degradation alters epidermal appendage development. Dev Cell. 2006;11:301–312. doi: 10.1016/j.devcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Han G, Li F, Ten Dijke P, Wang XJ. Temporal smad7 transgene induction in mouse epidermis accelerates skin wound healing. Am J Pathol. 2011;179:1768–1779. doi: 10.1016/j.ajpath.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Li AG, Wang D, Han S, Zheng B, Goumans MJ, Ten Dijke P, Wang XJ. Overexpression of Smad7 results in severe pathological alterations in multiple epithelial tissues. Embo J. 2002;21:2580–2590. doi: 10.1093/emboj/21.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Mehrani T, Millar SE, Morasso MI. Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development. 2008;135:3149–3159. doi: 10.1242/dev.022202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jave-Suarez LF, Winter H, Langbein L, Rogers MA, Schweizer J. HOXC13 is involved in the regulation of human hair keratin gene expression. J Biol Chem. 2002;277:3718–3726. doi: 10.1074/jbc.M101616200. [DOI] [PubMed] [Google Scholar]
- Kalinin A, Marekov LN, Steinert PM. Assembly of the epidermal cornified cell envelope. J Cell Sci. 2001;114:3069–3070. doi: 10.1242/jcs.114.17.3069. [DOI] [PubMed] [Google Scholar]
- Kim BK, Lee HY, Choi JH, Kim JK, Yoon JB, Yoon SK. Hairless plays a role in formation of inner root sheath via regulation of Dlx3 gene. J Biol Chem. 2012;287:16681–16688. doi: 10.1074/jbc.M111.320770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N, Motoi Y, Mori A, Tatsumi H, Nemoto S, Miyoshi H, Kitamura F, Miyatake S, Hiroi T, Kaminuma O. Suppressive role of C-terminal binding protein 1 in IL-4 synthesis in human T cells. Biochem Biophys Res Commun. 2009;382:326–330. doi: 10.1016/j.bbrc.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Kurek D, Garinis GA, van Doorninck JH, van der Wees J, Grosveld FG. Transcriptome and phenotypic analysis reveals Gata3-dependent signalling pathways in murine hair follicles. Development. 2007;134:261–272. doi: 10.1242/dev.02721. [DOI] [PubMed] [Google Scholar]
- Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto GP. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 1996;10:3065–3075. doi: 10.1101/gad.10.23.3065. [DOI] [PubMed] [Google Scholar]
- Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci U S A. 1999;96:162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morasso MI, Markova NG, Sargent TD. Regulation of epidermal differentiation by a Distal-less homeodomain gene. J Cell Biol. 1996;135:1879–1887. doi: 10.1083/jcb.135.6.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz EA, Baird AH, Michaud WA, Rocco JW. COOH-terminal binding protein regulates expression of the p16INK4A tumor suppressor and senescence in primary human cells. Cancer Res. 2008;68:6049–6053. doi: 10.1158/0008-5472.CAN-08-1279. [DOI] [PubMed] [Google Scholar]
- Nadauld LD, Phelps R, Moore BC, Eisinger A, Sandoval IT, Chidester S, Peterson PW, Manos EJ, Sklow B, Burt RW, Jones DA. Adenomatous polyposis coli control of C-terminal binding protein-1 stability regulates expression of intestinal retinol dehydrogenases. J Biol Chem. 2006;281:37828–37835. doi: 10.1074/jbc.M602119200. [DOI] [PubMed] [Google Scholar]
- Price JA, Bowden DW, Wright JT, Pettenati MJ, Hart TC. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum Mol Genet. 1998a;7:563–569. doi: 10.1093/hmg/7.3.563. [DOI] [PubMed] [Google Scholar]
- Price JA, Wright JT, Kula K, Bowden DW, Hart TC. A common DLX3 gene mutation is responsible for tricho-dento-osseous syndrome in Virginia and North Carolina families. J Med Genet. 1998b;35:825–828. doi: 10.1136/jmg.35.10.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukens MG, Alloul-Ramdhani M, Baan B, Kobayashi K, Peterson-Maduro J, van Dam H, Schulte-Merker S, Baker DA. Control of endothelial sprouting by a Tel-CtBP complex. Nat Cell Biol. 2010;12:933–942. doi: 10.1038/ncb2096. [DOI] [PubMed] [Google Scholar]
- Runkel F, Klaften M, Koch K, Bohnert V, Bussow H, Fuchs H, Franz T, Hrabe de Angelis M. Morphologic and molecular characterization of two novel Krt71 (Krt2-6g) mutations: Krt71rco12 and Krt71rco13. Mamm Genome. 2006;17:1172–1182. doi: 10.1007/s00335-006-0084-9. [DOI] [PubMed] [Google Scholar]
- Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci U S A. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Miura I, Yoshiki A, Kato Y, Yokoyama H, Shinogi A, Masuya H, Wakana S, Tamura M, Shiroishi T. Mutations in the helix termination motif of mouse type I IRS keratin genes impair the assembly of keratin intermediate filament. Genomics. 2007;90:703–711. doi: 10.1016/j.ygeno.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Tinkle CL, Lechler T, Pasolli HA, Fuchs E. Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc Natl Acad Sci U S A. 2004;101:552–557. doi: 10.1073/pnas.0307437100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkatchenko AV, Visconti RP, Shang L, Papenbrock T, Pruett ND, Ito T, Ogawa M, Awgulewitsch A. Overexpression of Hoxc13 in differentiating keratinocytes results in downregulation of a novel hair keratin gene cluster and alopecia. Development. 2001;128:1547–1558. doi: 10.1242/dev.128.9.1547. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Liefer KM, Tsai S, O’Malley BW, Roop DR. Development of gene-switch transgenic mice that inducibly express transforming growth factor beta1 in the epidermis. Proc Natl Acad Sci U S A. 1999;96:8483–8488. doi: 10.1073/pnas.96.15.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Boussadia O, Halfter H, Grose R, Berger P, Leone DP, Robenek H, Charnay P, Kemler R, Suter U. E-cadherin controls adherens junctions in the epidermis and the renewal of hair follicles. Embo J. 2003;22:5723–5733. doi: 10.1093/emboj/cdg560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103:9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.