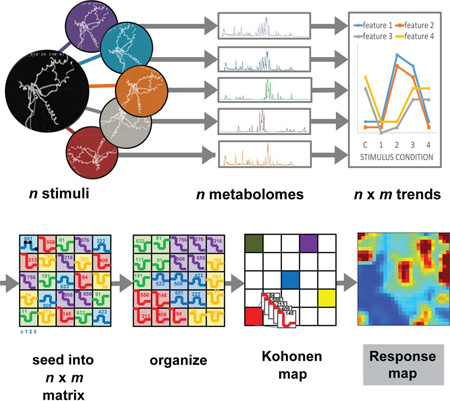
Abstract
Secondary metabolite biosynthesis in microorganisms responds to discrete chemical and biological stimuli; however, untargeted identification of these responses presents a significant challenge. Herein we apply multiplexed stimuli to Streptomyces coelicolor and collect the resulting response metabolomes via ion mobility-mass spectrometric analysis. Self-organizing map (SOM) analytics adapted for metabolomic data demonstrate efficient characterization of the subsets of primary and secondary metabolites that respond similarly across stimuli. Over 60% of all metabolic features inventoried from responses are either not observed under control conditions or produced at greater than two-fold increase in abundance in response to at least one of the multiplexing conditions, reflecting how metabolites encode phenotypic changes in an organism responding to multiplexed challenges. Using abundance as an additional filter, each of sixteen known S. coelicolor secondary metabolites is prioritized via SOM and observed at increased levels (1.2 – 22-fold compared to unperturbed) in response to one or more challenge conditions.
Graphical abstract
Microbial producers of secondary metabolites typically contain gene clusters encoding dozens of secondary metabolite families (Zerikly et al., 2009; Udwary et al., 2007), the expression of which appear to be tightly regulated in response to discrete chemical and/or biological stimulus. For example, exposure of actinomycetes to mixed fermentation conditions has demonstrated that secondary metabolite families are produced selectively via intergeneric (Onaka et al., 2011; Traxler et al., 2013) and interkingdom (Moree et al, 2012) microbial interactions. Similarly, the acquisition of antibiotic resistance via point mutations (Hosaka et al., 2009; Tanaka et al., 2013), exposure to rare earth metals (Tanaka et al., 2010; Ochi et al., 2014), exposure to small molecules (Nodwell et al., 2012); Sayedsayamdost et al, 2014), and the formulation of production media (Bode et al., 2002), have also been linked to gene-cluster specific upregulation of secondary metabolites in actinomycetes. These data are consistent with secondary metabolites governing adaptive organismal responses to environmental stimuli. Identifying secondary metabolites and associating them to gene clusters that are linked to discrete chemical and biological stimuli can provide insight into the chemical ecological role of secondary metabolites. Moreover, the ability to selectively stimulate native expression of secondary metabolic gene clusters via chemical or biological stimuli, and detect their corresponding products without resorting to genetic recombinant methods, would greatly expedite microbial secondary metabolite discovery.
If secondary and primary metabolite regulation has adapted to selectively respond to chemical and biological stimulus, then metabolites possessing selective responses may be identifiable within metabolomes by possessing characteristic abundance trends across multiplexed stimulus conditions. To investigate this hypothesis and enable secondary metabolite discovery, we herein assess the potential for stimulus-mediated production of secondary metabolites in the native microbe by multiplexed chemical and biological stimulation. To access a broad spectrum of responses, a battery of 23 perturbations in a single growth medium was utilized from three reported categories of activating conditions for Streptomyces coelicolor A3(2). The resulting collected sum of detectable metabolomic response inventories was analyzed by ultra performance liquid chromatography-ion mobility-mass spectrometric analysis (UPLC-IM-MS). To structure and categorize the response-specificity of metabolic features within these data, we developed and implemented a self-organizing map (SOM)-based analysis (Goodwin et al., 2014; Eichler et al., 2003) for the identification and prioritization of increased metabolite production resulting from the multiplexed perturbations. SOM analysis converted the collected metabolomes into a navigable topological response phenotype map and efficiently identified specific primary and secondary metabolites that are produced at increased levels in response to stimuli. For example, in primary metabolism, we identified discrete changes in guanosine and phenylalanine pools on lanthanide exposure and evidence of unique adaptive cell wall remodelling in several conditions. Notably, a large fraction (16 total secondary metabolites) of detected secondary metabolites was prioritized via this workflow as the most intense response-specific features, providing insight into the roles secondary metabolism plays in adapting to chemical stimuli and microbial interactions. The combination of multiplexed stimulation of native expression and structuring of the resulting metabolomic responses comprise a generalizable method for activating and detecting products of natively regulated primary and secondary metabolism.
RESULTS
Multiplexing stimuli of secondary metabolism
Streptomyces coelicolor A3(2) was cultivated under a battery of processes known to potentiate secondary metabolism. S. coelicolor was selected as a model microorganism because it has been extensively mined for secondary metabolites (Bentley et al., 2002), methods for native gene cluster activation have been most commonly developed for this organism (Hosaka et al., 2009; Luti and Mavituna, 2011b; Tanaka et al., 2010; Xu et al., 2002), and the majority of secondary metabolites isolated from this strain have been correlated to a gene cluster (Barona-Gomez et al., 2006; Bentley et al., 2002; Challis, 2013; Song et al., 2006).
We selected three known categories of activating stimuli: eliciting spontaneous resistance to transcription or translation-targeting antibiotics, exposure to rare earth elements, and cultivation in the presence of competing microorganisms. Our specific adaptations and standardizations of these reported methods are described in the Electronic Supplementary Information (ESI). In brief, using a single growth medium (International Streptomyces Protocol 2, ISP2), we cultivated (1) liquid cultures in the presence and absence of five separate scandium or five lanthanum concentrations, (2) liquid cultures of ten different spontaneous rifampicin or streptomycin resistance mutants, and (3) agar plate ISP2 co-cultures with three different challenge organisms, Micrococcus luteus, Rhodococcus wratislaviensis, or Tsukamurella pulmonis. Hence, we generated a total of 23 conditions, including controls, spanning these three methodologies.
Total cellular extracts were generated from fermentations via methanol extraction, concentrated, and processed for reverse phase UPLC analysis. Technical triplicates of extracts were analyzed in a randomized sequence using UPLC-IM-MS (Waters Synapt G2, Milford, MA) with lock mass correction to provide accurate mass measurements. During each spectral acquisition, an intact and fragmentation spectrum was taken for all ions present (herein referred to as MSE analysis (Plumb et al., 2006)) (Goodwin et al., 2012; McLean, 2009). Fragmentation was performed subsequent to IM separation, which allowed for the correlation of product ions to precursor origins through matched mobility.
Raw data were converted to distinct mass-to-charge (m/z) and retention time (Rt) pairs, termed “features,” and aligned across all samples (Smith et al., 2006). The resultant data matrix of discrete features, or ions, and associated intensities for each condition were averaged across technical replicates and subjected to multivariate statistical analysis (MVSA) and SOM.
Identifying products of multiplexed stimulation through MVSA
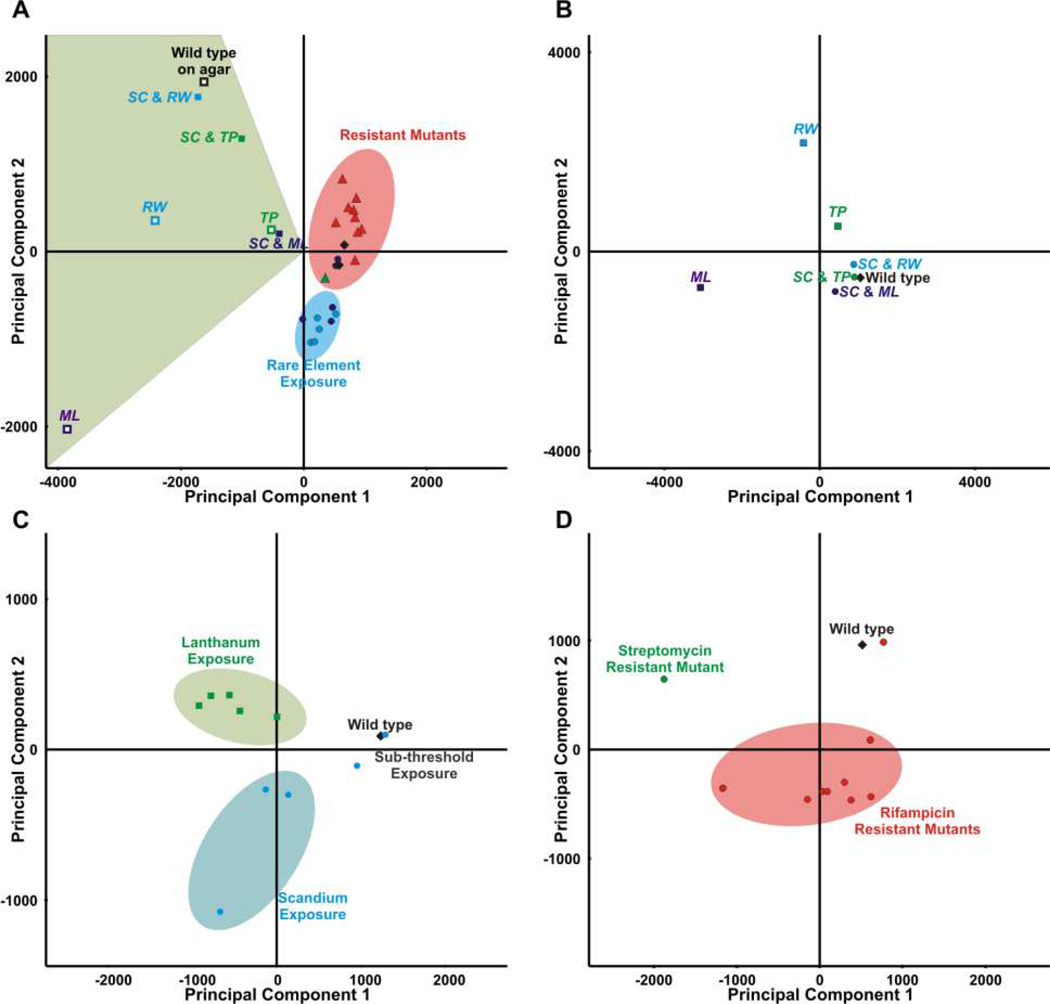
The identification of new metabolites with characteristic responses from multiplexed microbial stimuli requires methods for comparing and classifying covarying ions in the response inventories. Recently, we (Derewacz et al., 2013) and others (Hou et al., 2012; Robinette et al., 2012) have presented MVSA approaches for identifying the most abundant new ions resulting from individual stimulating microbial metabolic perturbations. MVSA methods for data analysis are powerful tools for identifying distinguishing features of small data sets (2 – 3 conditions), or extracting information regarding global sample grouping and which metabolites contribute to coarse trends. However, MVSA methods are not ideal for similar prioritization of metabolites in multiplexed perturbations, as MVSA is inherently biased for the largest differences amongst perturbations and is limited in the ability to reflect multiple stimuli in two- or three-dimensional space. Therefore, it omits the lower abundance or minorly covarying, yet still unique, metabolic reflexes (i.e., changes in metabolism resulting from a stimulus). In applying our previously described MSVA workflow (Derewacz et al., 2013) to the 23 conditions, as shown in Figure 1A, we can visualize the gross distinctions of metabolomic profiles that exist amongst different stimuli. When each perturbation is analyzed in isolation (Figure 1B–D), the distinct differences in global metabolism shifts are seen. A loadings plot analysis can be used to determine which ions contribute to sample distinction, as seen in Figure S3A. For comparison, detected secondary metabolites are annotated, which highlights a significant shortcoming of MVSA-based prioritization for a large number of culturing conditions: the largest contributors to sample differences are highlighted, but specific features of interest can be masked by the covariance of many species in the dataset. To garner conditionally distinct differences, experimental subsets (see Figure 1B – D), or even smaller subsets (e.g., a single co-culture versus monocultures, or orthogonal partial least squares-discriminant analysis approaches) can be analyzed. Thus, MVSA is most suitable for the interrogation of small datasets where the number of species is low (e.g. <50). However, for 25 conditions, pairwise analysis of multiple iterations of stimulus conditions for prioritization purposes becomes a time-intensive method of prioritizing secondary metabolites from extracts. As a result, we have developed and applied SOM-based methods to ion association and filtering. The primary advantage of SOM-based methods is that they are not prone to masking low abundant species of interest in the comparison of large datasets (e.g. >50).
Figure 1. Principal component analyses (PCA) of metabolomic inventories.
A. PCA of all cultures. B. Co-culture of S. coelicolor with M. luteus, R. wratislaviensis, and T. pulmonis. C. Metabolomic profiles in response to rare earth metals. D. Comparison of antibiotic-resistant mutants selected by plating on rifampicin and streptomycin.
Identifying products of multiplexed stimulation by self-organizing maps
Assessing trends in a large number of biochemical or biological conditions requires methods for the rapid visualization and organization of distinct differences in metabolic profiles across many perturbations to sort ions in a response-dependent manner. To address this, we developed a SOM-based approach to sort the complete inventory of ions across all growth conditions into regions based upon similarities in abundance profiles across experiments. This method is particularly well suited for secondary metabolite prioritization, as secondary metabolites are the end products of biochemical pathways and accumulate during fermentation. Differentially expressed, high abundance ions may be ranked subsequent to SOM analysis using the percent contribution of an ion to a region of interest (ROI) on the self- organized map. These ROIs exclusively contain features that respond specifically to a particular perturbation, while other loosely regulated metabolites will cluster outside of these prioritized regions. This method then prioritizes metabolites in a response-specific manner that performs well for the comparison of large datasets. Note that statistically MVSA and SOM are performing similar comparisons, but are representing the resulting comparisons in a different graphical manner. In fact many SOM approaches approximate a similar graphical presentation to MVSA when the datasets decrease to a small number of comparisons (ca. 10–50). Thus, the choice of one approach versus the other directly depends on how a specific query is framed and the corresponding number of features to be compared across datasets.
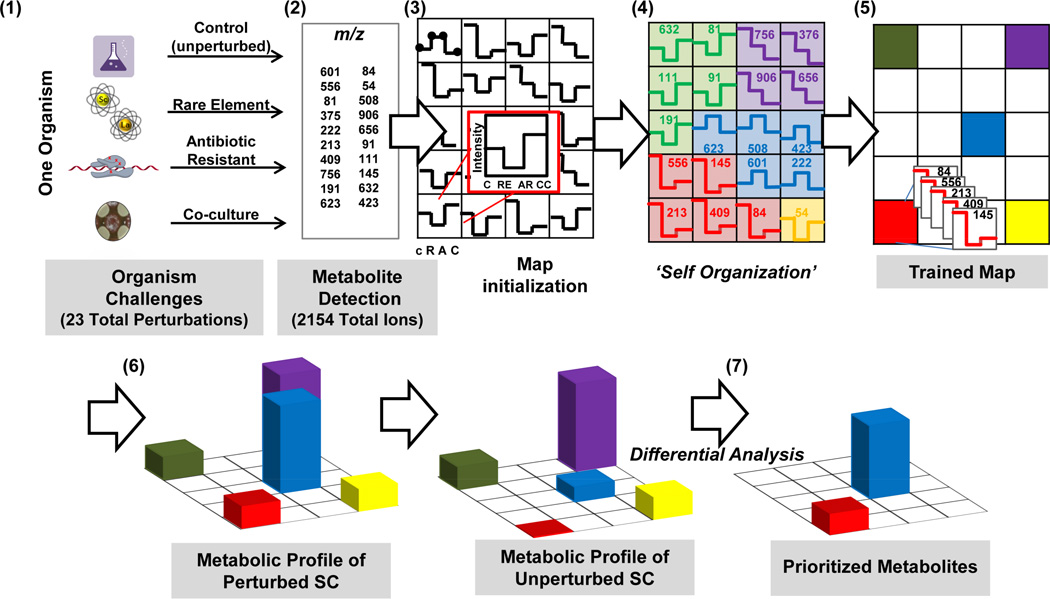
Figure 2 demonstrates the general workflow of the SOM-based approach, as applied to multiple perturbing conditions. Experimental and control conditions are processed by UPLC-IM-MS (Step 1), significant m/z retention time features are identified, and integrated intensity trends lines are generated for each feature. SOM analysis of these feature trends is performed using the Gene Expression Dynamics Investigator (GEDI) software (Step 2) (Eichler et al., 2003). For a more in-depth description, see Figure S2. Conceptually, detected ion abundance trends are first randomly seeded into a user-defined asymmetric grid (Step 3). The coordinates of the grid are only meaningful in relation to other grid locations and have no associated dimensions. Feature intensity trends are then iteratively organized based upon intensity similarities across experiments in a competitive-cooperative process analogous to a tile puzzle (Step 4) (Kohonen et al., 1998). As a result, metabolites that are produced as similar responses to the experimental conditions occupy the same or close coordinates in the grid. This sorts features in a data-driven manner into regions of correlated feature response (Step 5).
Figure 2. The general self-organizing map (SOM)-based approach to feature prioritization.
(1) This method begins with extracts from cultures of an organism cultivated under a battery of perturbing conditions. (2) Extracts are analyzed using UPLC-IM-MSE (or other feature-producing methodology) and converted into a matrix of discrete, aligned peaks with associated intensities for each culturing condition. (3–5) These features are then organized based upon intensity trends across culturing conditions. (6) Subsequently, extracts are represented as heat maps based upon the sum abundance of each organized metabolite in a region. (7) Differential analysis comparing data from perturbed cultures to controls allows generation of regions of interest.
The presumed correspondence of secondary metabolite expression profiles to responses is premised on the hypothesis that microorganisms use secondary metabolites to respond to discrete external stimuli (e.g., antibiotic challenge, competition, and metal exposure). The metabolic profile of each sample or experimental condition is then depicted as a topological heat map, which is a function of the intensity of each ion in that sample (Step 6). Features occupying the same coordinates in the SOM are summed. These heat maps (cf. Figure 3), or metabolic profiles, are then differentially compared with unperturbed metabolic profiles, resulting in heat maps with prioritized ROIs, indicating metabolic responses to experimental conditions (Step 7). Each pixel or node within the heat maps contains m/z-Rt feature lists, which are used for subsequent feature identifications. We selected six ROIs based on visual comparison of the differential phenotype heat maps, generated tables of covarying features via summing islands of high intensity within the heat maps, and ranked features by percentage (for more details, see ESI). The species occupying these regions of interest are then prioritized for further identification using accurate mass and fragmentation data acquired using MSE technology. A given ROI may comprise only one or several species. To ascertain rank within a given ROI, percent contributions of each species to the total ROI intensity may be determined (see ESI). Through self-organization, features corresponding to fragment ions, adducts, and isotopes are also all clustered for rapid triage. The determination of molecular identity of features is facilitated by the untargeted fragmentation acquisition, accurate mass measurements, retention time, ion mobility drift time, and other fragmentation interpretation afforded by the ion mobility separation dimension, as described in Figure S2.
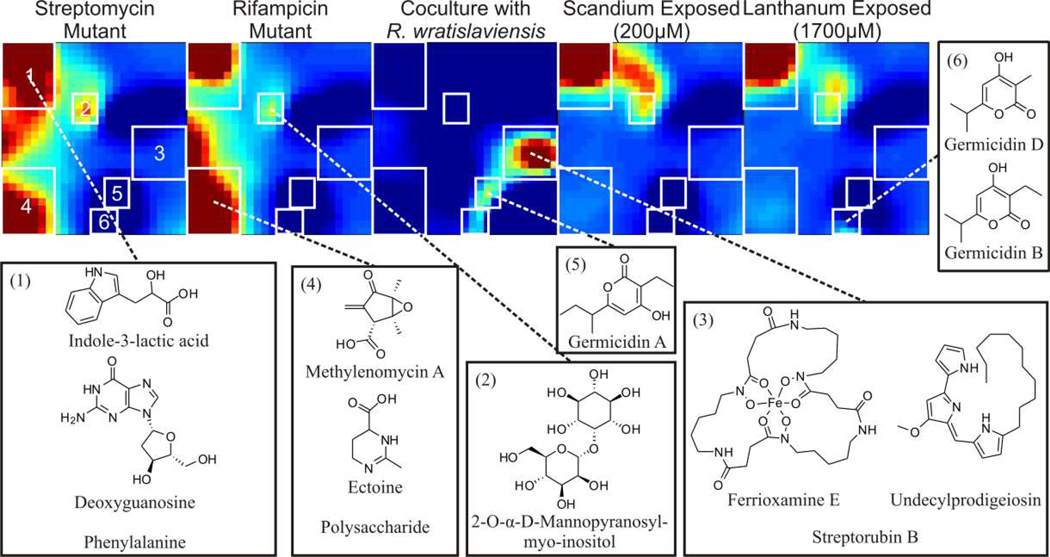
Figure 3. Differential metabolic phenotype heat maps representing increased production/decreased consumption of molecules using a single growth medium.
Representative extracts from each culturing condition are shown above, with regions of interest boxed and labelled. Corresponding putative identifications and structures for each ROI are labelled, and comprehensive catalogues of inhabiting features for each region, including relative abundance and percent contribution to total ROI intensity, are presented in the ESI.
Figure 3 demonstrates the utility of the SOM-based approach for molecular prioritization using this workflow across the multiplexed inducing conditions reported for enhanced secondary metabolite production. For heat maps of all tested conditions, please refer to Figure S3. Each heat map is representative of 2154 detected features (including detected isotopologs) observed in S. coelicolor grown with a unique perturbation or condition, following subtraction of the unperturbed culture extract. In the case of monoclonal cultures (i.e., streptomycin- and rifampicin-selected point mutations, rare element exposure), this baseline subtraction is simply subtraction of the ISP2 unperturbed culture metabolic profile. In co-culturing conditions, metabolic profiles from both wild type S. coelicolor and competing organism monoculture were subtracted, resulting in a map of feature inventories that are produced at increased levels in each mixed culture in comparison to its constituent monocultures. Hence, caveats in interpreting mixed culture data are that increased feature production can be a result of either organism, and the output of the mixed culture is likely more than the sum of its parts. Six dominant ROIs are indicated as boxed regions in Figure 3, and identified ions that occupy these regions are annotated (for a full list of all features occupying these regions, see ESI (Tables S2, S3). A majority of the annotated ions correspond to secondary metabolites S. coelicolor is known to produce (Challis, 2013). However, we gain additional biological insight into the microbial response to the various stimuli by observing the other biochemical results which are sorted with these secondary metabolites (e.g., deoxyguanosine, phenylalanine).
Measuring and structuring metabolic perturbations
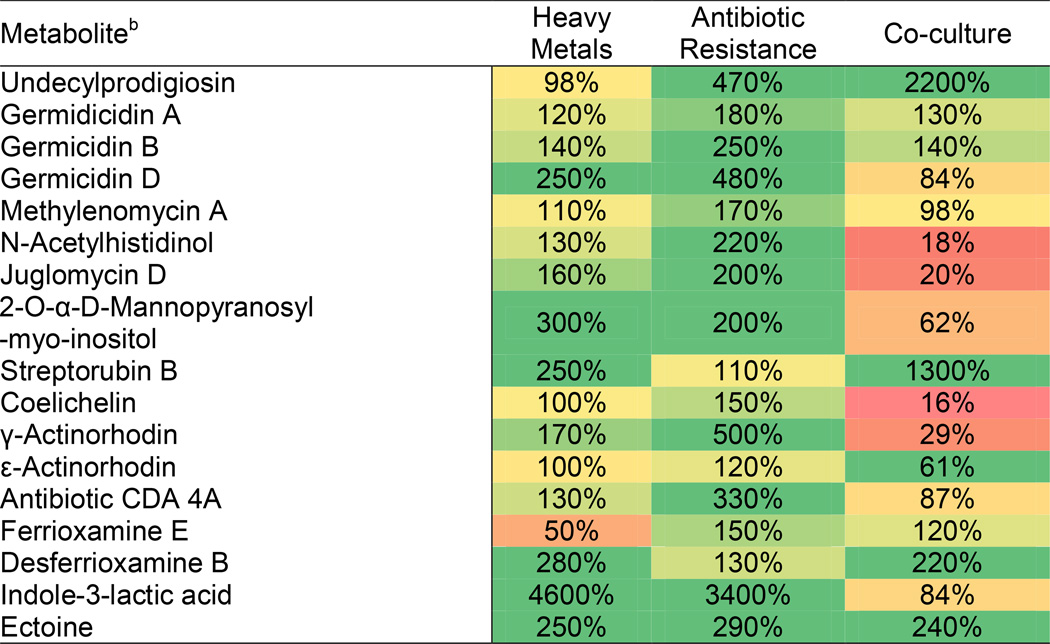
Each inducing condition provoked unique metabolic responses, as observed in the differential profiles in Figure 3. In total, of the 2154 significant features detected, 1318 were found to be either previously undetected or produced in at least two-fold abundance in at least one perturbed system relative to control (see ESI). This corresponds to induced overproduction of ~61% of all detected species. For the subset of known secondary metabolites, 16 were observed in at least one S. coelicolor expression condition (<20 ppm mass accuracy), including monoclonal culturing in liquid culture or agar. Molecular ions, corresponding mass accuracies, and comparative relative intensities appear in the ESI (Table S4, Figures S4). We found increased production of each of the 16 detected metabolites in at least one perturbed culture when compared to the matched unperturbed control. The magnitude of amplification is shown in Figure 4. In certain cases, a nearly 22-fold increase in production was found (i.e., undecylprodigiosin).
Figure 4.
Maximum resultant metabolite production abundances compared to matched control cultures in ISP2 medium.a
a Color scale: 0% (red); 100% (yellow); 200% (green). Heavy metal and antibiotic resistance performed in liquid cultures and co-culture performed on agar medium.
b Putative metabolite identification of metabolites reported to be produced by S. coelicolor based on accurate mass measurement, and fragmentation pattern, when available.
Response profile analysis
Secondary metabolic gene clusters in microorganisms are often organized into operons, and within a given organism, gene clusters share common regulatory elements programmed to respond to specific cellular states (e.g., pleiotropic signals) (Bibb, 2005). Correspondingly, we hypothesize that secondary metabolite features that are differentially produced as a result of multiplexed chemical and biological stimuli will structure into grouped regions in a SOM of metabolites based on similarity of production response profiles. Herein, we describe and demonstrate the application of this approach, which ameliorates the limitations of an MVSA-based analytics for multiplexed stimuli data interpretation. One practical advantage of the SOM approach is that dozens of chemical, biochemical, or genetic perturbations may be analyzed using a single computation (in this case 25 × 3 analyses, comprising >780,000 spectra, in excess of 58 gigabytes of data, spanning three classes of stimuli), resulting in the generation of sets of simple and easily navigable metabolic phenotype graphical representations that still retain the attributes of MVSA-based analytics. Additionally, though features within an ROI can be ranked by abundance, SOM organizes features by intensity trends, so low intensity features can also be identified even in the presence of large datasets.
At least 22 gene clusters within the S. coelicolor genome have been assigned involvement in secondary metabolite production (Bentley et al., 2002). Of these 22 clusters, SOM maps prioritized metabolites associated with 8 of the 22 gene clusters, which are listed in Table S1, of which all display elevated secondary metabolite production in some capacity as a result of the introduction of challenges (Figure 4). Putative metabolite feature intensity trends are shown in Tables S2. In all cases, feature intensity profiles map consistently to ROI. In some cases, however, putative features show false positive intensity values due to coincident features of similar mass/retention time. For instance, actinorhodins and desferrioxamines are not produced by the challenge organisms, though features with a similar accurate mass were found. For a full table of relative abundances across all conditions, we direct the reader to Table S3. The analytical strategy presented prioritizes secondary metabolites generated from these gene clusters from within the metabolomic pool, yet this begs the question as to the biological rationale of these lower abundance, yet overproduced, species. Significantly, an increase in the production of germicidins (Figure 3, ROIs 5 & 6) was observed in both mixed-fermentation conditions and selected antibiotic-resistant strains. Metabolomic analysis indicated that the process of culturing S. coelicolor on agar versus the liquid cultures affected the production of germicidins. However, germicidins A and B were present in higher concentrations in the mixed fermentation cultures versus the monoclonal cultures grown on agar, in addition to all germicidins observed in increased abundances in many of the antibiotic-resistant cultures. The production of germicidins inhibits spore germination and is a self-regulatory mechanism in the production response to high population densities (Aoki et al., 2011). Additionally, mixed fermentation resulted in the enhanced production of undecylprodigiosin (22-fold increase when co-cultured) and streptorubin B (13-fold increase when co-cultured) (Figure 3, ROI 3), known secondary metabolites of S. coelicolor with antimicrobial and other clinically relevant properties (Williamson et al., 2006). This is consistent with previous studies, which linked undecylprodigiosin and streptorubin B production to external factors, including mixed fermentation with Bacillus subtilis (Luti and Mavituna, 2011a) and salt-stress (Sevcikova and Kormanec, 2004). Within this same ROI, we observe the enhanced production of siderophores functioning as an iron scavenger for nutrient acquisition in all perturbed conditions (Barona-Gomez et al., 2006). This response is likely a concerted rebuttal to the microbial competition encountered in the mixed fermentation environment. A variant of calcium-dependent antibiotic production was observed to be upregulated (3.3-fold increase) specifically in agar culturing and co-culturing conditions with Tsukamurella pulmonis (Figure S4). This Gram-positive targeting metabolite may be attributed to T. pulmonis production of mycolic acid, which activates secondary metabolite production in once silent clusters (Onaka et al., 2011) and underpins the necessity of multi-conditional culturing. We also observed altered production of potentially exo-polysaccharides (Figure 3, ROI 4) as a result of these persistent resistant mutations.
Furthermore, 2-O-α-D-mannopyranosyl-myo-inositol (3-fold increase in rare element exposure) was observed in increased abundance in mutant- and rare element-exposed cultures (Figure 3, ROI 2), the production of which has been demonstrated previously in liquid culture (Pospisil et al., 2007), supported by the absence in mixed fermentation conditions. Elevated production of ectoine (2.8-fold increase) was observed as a general response to perturbations and is consistent with previous results we have observed within rifampicin- and streptomycin-resistant mutants in Nocardiopis (Derewacz et al., 2013). This osmoprotectant has been shown to provide enzyme activity stabilizing effects (Lippert and Galinski, 1992) and stimulate growth in osmotically inhibitory environments (Jebbar et al., 1992).
DISCUSSION
Microbial genome sequencing has revealed a vast reservoir of secondary metabolite-encoding gene clusters, suggesting largely untapped molecular diversity with potential biomedical application. Advances in sequencing have outpaced the developments of the requisite steps to produce, study, and ultimately purify the encoded metabolites: gene cluster expression and translation, identification and purification of the resulting produced metabolites, and structure elucidation. Two complementary strategies for addressing the expression component of these processes consist of refactoring targeted gene clusters for increased expression, typically in heterologous hosts (Medema et al., 2011; Wilkinson and Micklefield, 2007; Yamanaka et al., 2014), or expression of gene clusters in their native hosts using non-recombinant chemical or biochemical methods to stimulate native expression (Walsh and Fischbach, 2010; Zerikly and Challis, 2009), or via systematic modification of cultivation parameters, also called OSMAC (One Strain-Many Compounds) (Bode et al,, 2002). In either case, the analysis of the resulting metabolomes for upregulated or otherwise perturbed metabolites potentially becomes the next rate-limiting step. Rapid unbiased identification and prioritization of newly produced metabolites is an essential prerequisite for what remain the most labor-intensive steps of secondary metabolite discovery: purification, isolation, and structure elucidation.
This study analyzes three categories of microbial stimulus (antibiotic induced resistance, heavy metal exposure, and co-culture) on a single metabolomic platform. To convert the microbial metabolomic responses from 23 distinct conditions spanning these three perturbations into navigable phenotypic maps, we develop and implement self-organizing map analytics for multiplexed responses to microbial metabolomics. This approach localizes metabolomic features that covary across conditions into regions of interest that can be used to identify metabolic features that are similarly regulated, or that respond similarly to challenge. Secondary metabolites are the end products of metabolic pathways, accumulate, and are slowly degraded. As a result, they are well suited for the application of SOM analytics that not only prioritize features, but also illuminate trends in similarly responding metabolites. Previous studies of biological, biochemical, and chemical microbial challenge are consistent with the hypothesis that the upregulation of secondary metabolism may be an adaptive response to challenge stimuli (Derewacz et al., 2013; Ochi et al., 2014; Tanaka et al., 2009). Moreover, the current study confirms the recent analysis of metabolomic dynamics engendered by a cohort of interspecies interactions (Traxler et al., 2013), and analyzed by NanoDESI. These results provide additional support for the broad reaching effects of chemical and biological stimulus and a new means for identification of important microbial response chemicals.
Strategies using topological clustering of metabolomic data are finding increasing application in secondary metabolite discovery. For example, a molecular network analysis tool has recently been developed and applied to aid in the organization of exometabolic inventory analysis and to prioritize secondary metabolite discovery (Nguyen et al., 2013; Traxler et al., 2013). Metabolite molecular network analysis uses numerical clustering of tandem mass spectra similarity as an organizing principle and provides a map based on chemical similarity. This method also permits simultaneous graphical visualization of structural relatedness networks for multiple stimuli conditions or organisms and is excellent for dereplication and prioritization by chemical structure. The SOM method described herein is distinct from this method in that the organizing principle is not structure (inferred from fragment data), but rather response trends across more than two dozen conditions. Indeed, because SOM regions of interest can contain hundreds of correlating features, molecular network analysis can potentially be used as a method to prioritize responsive features identified by SOM analytics, underlining the complementary nature of these methods. Other comparative metabolomics methods, such as “bubble plot” visualization, provide a straightforward and easily interpretable tool for determining differences in metabolomic feature production, but are generally only applicable to binary comparison and do not render correlations in ion profiles across many experimental conditions (Patti et al., 2012a). However, meta-XCMS procedures may find significant utility in this type of secondary metabolite prioritization (Patti et al., 2012b). Some unique advantages of the SOM method described here are that it can analyze the response patterns of 25+ stimuli conditions simultaneously, and secondary metabolites can be identified from response trends via simple subtractive analysis using the trained map template as an organizing principle.
As accumulating dead ends of metabolic pathways, secondary metabolites are ideal candidates for the application of SOM analytics; however, the utility of this approach extends beyond secondary metabolism. For instance, we have recently applied the SOM metabolomics to the analysis of mouse serum and used it to identify diagnostic features in cocaine addicted mice, demonstrating the general utility of this method for both abundant and nonabundant comparative metabolomics (Goodwin et al 2014). Additionally, Kohonen's self-organizing map analytics has been used to understand time dependent metabolite changes in rice plants, identifying synchronously fluctuating metabolites (Sato et al 2008). Importantly, as datasets incorporate ever increasing amounts of data (e.g. time, perturbation, etc.) the corresponding ROIs provide correspondingly higher specificity.
From a “genome mining” perspective, we observed substantial metabolomic expansion of the biomolecular inventory of S. coelicolor grown in a single medium using multiplexed chemical and biochemical induction methods. Of the nearly 2200 total detected molecular features, 61% were found to be either undetected in control cultures or produced in at least two-fold greater amounts, relative to control, in at least one culture challenge. Indeed, using these methodologically simple and rapid non-recombinant techniques, we have observed the increased production of all 16 of the detected secondary metabolites, comprising products of up to 8 out of 22 annotated gene clusters in at least one unique culturing condition, and prioritizing eight natural products within ROIs. These results challenge the notion of “silent” gene clusters in native hosts and support the potential of systematic induction of native secondary metabolism as a method of accessing the hidden reservoirs of secondary metabolic diversity in microorganisms. Indeed, with a comparatively small set of stimuli, which can be generated and analyzed in less than a month, the majority of known secondary metabolism was activated. Future studies, combining analysis of transcriptional and metabolomic covariance with stimuli, offer the potential to target the activation of specifically regulated gene clusters with a rationally selected set of challenges. In this way, chemistry and biology may be rationally manipulated in the future to quickly elicit the expression of cryptic or silent gene clusters in cultivatable organisms, or alternatively in the assessment of heterologously or endogenously refactored gene clusters in microorganism.
EXPERIMENTAL PROCEDURES
Materials and Methods
All reagents were obtained from the Sigma-Aldrich chemical company unless otherwise specified. Streptomyces coelicolor A3(2) was obtained from the John Innes Center, Tsukamurella pulmonis from the American Type Culture Collection (ATCC 700081), and Rhodococcus wratislaviensis was obtained via dilution plating from hypogean sediments.
Eliciting antibiotic resistance and fermentations
To generate antibiotic-resistant mutants, the spore inoculum of S. coelicolor was uniformly spread on GYM (glucose 0.4 %, yeast extract 0.4 %, malt extract 1 %, peptone 0.1 %, sodium chloride 0.2 %, agar 2 %) agar plates containing streptomycin at one of two concentrations (100 µg/mL, 300 µg/mL) or rifampicin at either 200 µg/mL or 400 µg/mL (concentrations of antibiotics were chosen so they exceed the minimum inhibitory concentration for S. coelicolor on GYM medium). After two weeks of incubation at 30 °C, the agar plates were inspected for the presence of resistant colonies, which were then aseptically transferred to antibiotic-free ISP2 (glucose 0.4%, yeast extract 0.4 %, malt extract 1 %, agar 2 %) plates. Each S. coelicolor mutant was then inoculated to 20 mL ISP2 liquid seed culture, incubated for 7 days, and from seed culture to 50 mL liquid ISP2 fermentation culture for 7 days of incubation at 30 °C. Progenitor S. coelicolor was incubated under the same conditions to generate the control culture.
Rare earth element fermentations
For rare element additives, the spore suspension of S. coelicolor was inoculated on ISP2 agar plates for incubation at 30 °C for 7 days, then inoculated from plates int o 20 mL liquid seed culture and from seed culture to 50 mL liquid ISP2 production cultures containing various concentrations of scandium chloride (20 µM, 50 µM, 100 µM, 200 µM, 500 µM) or lanthanum chloride (1500 µM, 1700 µM, 1900 µM, 2100 µM, 2500 µM) for 7 day incubations at 30 °C. To generate a control, S. coelicolor was incubated in 50 mL additives-free ISP2 medium under the same conditions.
Extraction of liquid fermentations
Total culture metabolite extracts from liquid cultures were generated by adding 50 mL of methanol to each fermentation flask and shaking the flasks on a rotary shaker for 1 h. Mycelia were then separated on a centrifuge, and supernatants were dried in vacuo to yield crude extracts.
Co-culture
Co-culture plates were prepared by addition of 40 mL of sterile ISP2 medium to a one well OmniTray plate. Cryogenic spore suspensions of S. ceolicolor were cultivated on agar plates (100 × 15 mm) containing 30 mL of ISP2 medium and incubated at 30 °C until the production of spores occurred. The spores were removed from the surface of the plate using a sterile loop and suspended in 25 mL of ISP2 liquid medium at a concentration of approximately 108 spores/mL as determined via hemocytometer. This suspension was homogenized and decanted into a one well plate as a reservoir. The pins of a 96 well replicator were submerged into the spore solution and applied to the surface of the solid support within the previously prepared one well OmniTray plate without puncturing the surface (Figure S1). The plates were incubated for 24 hours at 30 °C. Cryogenically stored M. luteus was inoculated into 5 mL of sterile ISP2 medium 8 h prior to application to the co-culture plate. Rhodococcus wratislaviensis stock was inoculated into 5 mL of sterile ISP2 medium 24 h prior to application to the co-culture plate. Cryogenically stored T. pulmonis stock was inoculated into 5 mL of sterile heart infusion medium 24 h prior to application to the co-culture plate. For all competing organisms, once an OD600 of ~1 was achieved, the 5 mL sample was diluted into 30 mL of medium in separate one well plate reservoirs. The pins of a 96 well replicator were submerged into the solution and applied to the surface of the solid support within the one well OmniTray plate without puncturing the surface in an offset manner relative to the previously inoculated actinomycete. After 7 days, co-cultures plates were cut into 1 × 1 cm segments and extracted with equal volumes of methanol by shaking for 3 h at 170 rpm and 30 °C.
UPLC-IM-MS Data Acquisition and Processing
Extract samples were resuspended in methanol at a concentration of 200 mg/mL. UPLC-IM-MSE data acquisition was performed with a 25 min gradient. Mobile phase A consisted of H2O with 0.1 % formic acid, and mobile phase B consisted of acetonitrile with 0.1 % formic acid. A 1×100 mm 1.7 µm particle BEH-T3 C18 column (Waters Co.) was used for chromatographic separations with a flow rate of 75 µL/min and a column temperature of 40 °C. An autosampler with a loop size of 5 µL held at 4 °C was used for sample injection. The i nitial solvent composition was 100 % A, which was held for 1 min and ramped to 0 % A over the next 11 min, held at 0 % A for 2 min, and returned to 100 % A over a 0.1 min period. The gradient was held at 100 % A for the next 10.9 min for equilibration. Prior to analysis of the sample queue, ten sequential column-load injections were performed with 5 µL of the quality control. This protocol increases retention time stability and is critical to reproducible analyses. Quality control injections were then performed after every 10 sample injections to ensure instrument stability. Quality controls were comprised of pooled equal aliquots of all samples analyzed.
IM-MSE spectra were acquired at a rate of 2 Hz from 50–2000 Da in positive ion mode for the duration of each sample analysis on a Synapt G2 HDMS platform (Waters, Milford, MA). The instrument was calibrated to less than 1 ppm mass accuracy using sodium formate clusters prior to analysis. A two-point internal standard of leucine enkephalin was infused in parallel to the sample at a flow rate of 7 µL/min, and data were acquired every 10 s. The source capillary was held at 110 °C and 3.0 kV, with a desolvation gas flow of 400 L/h and a temperature of 150 °C. The sampling cone was held at a setting of 35.0, with the extraction cone at a setting of 5.0. In the MSE configuration, low and high energy spectra were acquired for each scan. High energy data provided a collision energy profile from 10–30 eV in the trapping region, providing post-mobility fragmentation. Ion mobility separations were performed with a wave velocity of 550 m/s, a wave height of 40.0 V, and a nitrogen gas flow of 90 mL/min, with the helium cell flow rate at 180 mL/min. Internal calibrant correction was performed in real time.
Data were converted to mzXML format using the msconvert tool from the ProteoWizard package (Kessner et al., 2008). Peak picking and alignment were performed using XCMS in R (Smith et al., 2006). See Figure S2 for details and package locations. The resulting data matrix contained 2154 detected features and was formatted for analysis using both GEDI and Umetrics. Formatting for GEDI is outlined below; formatting for Umetrics was performed by extracting and transposing the sample-feature intensity matrix generated from XCMS and importing it into Umetrics software. Prior to GEDI and MVSA, analytical triplicates were averaged. For GEDI analysis, a grid of 25 × 26 was generated. Software specific parameters include: 100 first phase training iterations with an initial training radius of 10.0, a learning factor of 0.5, a neighborhood block size of 20, and a conscience of 5.0, and 160 second phase training iterations with a neighborhood radius of 1.0, learning factor of 0.05, neighborhood block size of 2, and conscience of 2.0. A random seed of 10 with a Pearson’s correlation distance metric and random selection initialization was used.
Metabolite identifications were performed using accurate mass measurements and fragmentation spectra extracted from IM-MSE data. Utilizing drift time correlations, product ions were correlated appropriately to precursors for extraction of high energy spectra.
Supplementary Material
SIGNIFICANCE.
We hypothesize that the inventory of metabolic features resulting from a microorganism’s exposure to multiple chemical and biological stimuli can be used to identify induced expression of secondary metabolites. Central to this approach is the premise that microbial secondary metabolites are produced to respond to environmental stimuli. It follows that their production can be revealed by examining the patterns of metabolomic feature responses across multiple stimuli. This ‘responsomics’ approach has been applied here to the well-characterized actinomycete Streptomyces coelicolor, revealing that production of the majority of secondary metabolites in this strain can be induced by simple stimuli and subsequently identified by comparative metabolomics analysis via self-organizing maps. Regions of interest within the response maps reveal those metabolites that are characteristically modulated by multiplexed stimuli and ranking these by abundance provides a means of prioritizing compounds for isolation studies. Advantages of this self-organizing maps analytics are that it ranks features via response profile, not by intensity, permitting the identification of low intensity features contributing to a response phenotype, that it carries out the comparison of large numbers of datasets (up to 25 in this study) in a single computation, and provides easy to navigate heat maps of metabolic response phenotypes. In addition to providing a work-flow for the identification of secondary metabolites, the ability to inventory metabolites that are modulated consistently via multiplexed stimuli may be used to identify features relevant to microbial physiology, development, and chemical ecology.
Highlights.
Secondary metabolite expression is triggered by environmental stimuli
Using stimuli and self-organizing maps, we identify a ‘response metabolome’
Mapping responses to multiplexed stimuli reveals secondary metabolites
In S. coelicolor, this revealed a large fraction of its biosynthetic potential
ACKNOWLEDGEMENTS
We acknowledge support from National Institutes of Health grant 1R01GM92218, the Defense Threat Reduction Agency grant HDTRA1-09-1-0013, the Vanderbilt Institute of Chemical Biology, the Vanderbilt Institute for Integrative Biosystems Research and Education, Vanderbilt University, and the Systems Biology and Bioengineering Undergraduate Research Experience (SyBBURE) funded by Gideon Searle at Vanderbilt. We thank Dr. Carol A. Rouzer for her gracious and significant editorial and intellectual contributions and guidance. We also thank Ms. Allison Price for her editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
D.K.D. performed the heavy metal and antibiotic resistance culturing and extraction. C.R.M and B.C.C. performed the co-culturing and extraction. C.R.G. and R.C.M developed the mixed culture apparatus. C.R.G. performed the mass spectrometry acquisition and analytics. D.K.D, B.C.C, C.R.G., J.A.M., J.P.W., and B.O.B. wrote the manuscript.
† Electronic Supplementary Information (ESI) available, including detailed methods for growth, analysis and computational methods. See DOI: 10.1039/b000000x/
References
- Aoki Y, Matsumoto D, Kawaide H, Natsume M. Physiological role of germicidins in spore germination and hyphal elongation in Streptomyces coelicolor A3 (2) The Journal of Antibiotics. 2011;64:607–611. doi: 10.1038/ja.2011.59. [DOI] [PubMed] [Google Scholar]
- Barona-Gomez F, Lautru S, Francou F-X, Leblond P, Pernodet J-L, Challis GL. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology. 2006;152:3355–3366. doi: 10.1099/mic.0.29161-0. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Bibb MJ. Regulation of secondary metabolism in streptomycetes. Current Opinion in Microbiology. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Challis GL. Exploitation of the Streptomyces coelicolor A3 (2) genome sequence for discovery of new natural products and biosynthetic pathways. Journal of Industrial Microbiology & Biotechnology. 2013:1–14. doi: 10.1007/s10295-013-1383-2. [DOI] [PubMed] [Google Scholar]
- Craney A, Ozimock C, Pimentel-lardo SM, Nodwell JR. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chemistry and Biology. 2012;24:1020–1027. doi: 10.1016/j.chembiol.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Derewacz DK, Goodwin CR, McNees CR, McLean JA, Bachmann BO. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2336–2341. doi: 10.1073/pnas.1218524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- Goodwin CR, Fenn LS, Derewacz DK, Bachmann BO, McLean JA. Structural Mass Spectrometry: Rapid Methods for Separation and Analysis of Peptide Natural Products. Journal of Natural Products. 2012;75:48–53. doi: 10.1021/np200457r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin CR, Sherrod SD, Marasco CC, Bachmann BO, Schramm SN, Wikswo JP, McLean JA. Phenotypic mapping of metabolic profiles using self-organizing maps of high-dimensional mass spectrometry data. Analytical Chemistry. 2014;86:6563–6571. doi: 10.1021/ac5010794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nature Biotechnology. 2009;27:462–464. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- Hou Y, Braun DR, Michel CR, Klassen JL, Adnani N, Wyche TP, Bugni TS. Microbial Strain Prioritization Using Metabolomics Tools for the Discovery of Natural Products. Analytical Chemistry. 2012;84:4277–4283. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebbar M, Talibart R, Gloux K, Bernard T, Blanco C. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. Journal of Bacteriology. 1992;174:5027–5035. doi: 10.1128/jb.174.15.5027-5035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T. Proceedings of ICANN98, the 8th International Conference on Artificial Neural Networks. Springer; 1998. Self-Organization of Very Large Document Collections: State of the Art; pp. 65–74. [Google Scholar]
- Lippert K, Galinski E. Enzyme stabilization by ectoine-type compatible solutes: protection against heating, freezing and drying. Applied Microbiology and Biotechnology. 1992;37:61–65. [Google Scholar]
- Luti K, Mavituna F. Streptomyces coelicolor increases the production of undecylprodigiosin when interacted with Bacillus subtilis. Biotechnology Letters. 2011a;33:113–118. doi: 10.1007/s10529-010-0401-y. [DOI] [PubMed] [Google Scholar]
- Luti KJK, Mavituna F. Streptomyces coelicolor increases the production of undecylprodigiosin when interacted with Bacillus subtilis. Biotechnology letter. 2011b;33:113–118. doi: 10.1007/s10529-010-0401-y. [DOI] [PubMed] [Google Scholar]
- McLean J. The mass-mobility correlation redux: The conformational landscape of anhydrous biomolecules. Journal of the American Society for Mass Spectrometry. 2009;20:1775–1781. doi: 10.1016/j.jasms.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Medema MH, Breitling R, Bovenberg R, Takano E. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nature Reviews. 2011;9:131–137. doi: 10.1038/nrmicro2478. [DOI] [PubMed] [Google Scholar]
- Moree WJ, Phelan VV, Wu CH, Bandeira N, Cornett DS, Duggan BM, Dorrestein PC. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DD, Wu C-H, Moree WJ, Lamsa A, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K, Tanaka Y, Tojo S. Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. Journal of Industrial Microbiology & Biotechnology. 2014;41:403–414. doi: 10.1007/s10295-013-1349-4. [DOI] [PubMed] [Google Scholar]
- Onaka H, Mori Y, Igarashi Y, Furumai T. Mycolic Acid-Containing Bacteria Induce Natural-Product Biosynthesis in Streptomyces Species. Applied and Environmental Microbiology. 2011;77:400–406. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Tautenhahn R, Rinehart D, Cho K, Shriver LP, Manchester M, Nikolskiy I, Johnson CH, Mahieu NG, Siuzdak G. A view from above: cloud plots to visualize global metabolomic data. Analytical Chemistry. 2012;85:798–804. doi: 10.1021/ac3029745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nature Protocols. 2012;7:508–516. doi: 10.1038/nprot.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. UPLC/MSE; a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Communications in Mass Spectrometry. 2006;20:1989–1994. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- Pospisil S, Sedmera P, Halada P, Petricek M. Extracellular Carbohydrate Metabolites from Streptomyces coelicolor A3(2) Journal of Natural Products. 2007;70:768–771. doi: 10.1021/np0606188. [DOI] [PubMed] [Google Scholar]
- Robinette SL, Bruschweiler R, Schroeder FC, Edison AS. NMR in Metabolomics and Natural Products Research: Two Sides of the Same Coin. Accounts of Chemical Research. 2012;45:288–297. doi: 10.1021/ar2001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost, Mohammad R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proceedings of the National Academy of Sciences 111. 2014;20:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Arita M, Soga T, Nishioka T, Tomita M. Time-resolved metabolomics reveals metabolic modulation in rice foliage. BMC Systems Biolohy. 2008;2:51:1–13. doi: 10.1186/1752-0509-2-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevcikova B, Kormanec J. Differential production of two antibiotics of Streptomyces coelicolor A3(2), actinorhodin and undecylprodigiosin, upon salt stress conditions. Archives of Microbiology. 2004;181:384–389. doi: 10.1007/s00203-004-0669-1. [DOI] [PubMed] [Google Scholar]
- Smith CA, Elizabeth J, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Song L, Barona-Gomez F, Corre C, Xiang L, Udwary DW, Austin MB, Noel JP, Moore BS, Challis GL. Type III Polyketide Synthase B-Ketoacyl-ACP Starter Unit and Ethylmalonyl-CoA Extender Unit Selectivity Discovered by Streptomyces coelicolor Genome Mining. Journal of the American Chemical Society. 2006;128:14754–14755. doi: 10.1021/ja065247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Komatsu M, Okamoto S, Tokuyama S, Kaji A, Ikeda H, Ochi K. Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Applied and Environmental Microbiology. 2009;75:4919–4922. doi: 10.1128/AEM.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Hosaka T, Ochi K. Rare earth elements activate the secondary metabolite biosynthetic gene clusters in Streptomyces coelicolor A3 (2) The Journal of Antibiotics. 2010;63:477–481. doi: 10.1038/ja.2010.53. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kasahara K, Hirose Y, Murakami K, Kugimiya R, Ochi K. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampicin resistance (rpoB) mutations in actinomycetes. J Bacteriol. 2013;195:2959–2970. doi: 10.1128/JB.00147-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies Interactions Stimulate Diversification of the Streptomyces coelicolor Secreted Metabolome. American Society for Microbiology. 2013;4 doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J, Hendricks N, Meehan M, Dorrestein PC. Capturing bacterial metabolic exchange using thin film desorption electrospray ionization-imaging mass spectrometry. Analytical Chemistry. 2010;82:1598–1600. doi: 10.1021/ac9027388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. Journal of the American Chemical Society. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B, Micklefield J. Mining and engineering natural-product biosynthetic pathways. Nature Chemical Biology. 2007;3:379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- Williamson NR, Fineran PC, Leeper FJ, Salmond GPC. The biosynthesis and regulation of bacterial prodiginines. Nature Reviews. 2006;4:887–899. doi: 10.1038/nrmicro1531. [DOI] [PubMed] [Google Scholar]
- Xu J, Tozawa Y, Lai C, Hayashi H, Ochi K. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3 (2) Molecular Genetics and Genomics. 2002;268:179–189. doi: 10.1007/s00438-002-0730-1. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerikly M, Challis GL. Strategies for the Discovery of New Natural Products by Genome Mining. ChemBioChem. 2009;10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.