Summary
Neuronal plasticity helps animals learn from their environment. However, it is challenging to link specific changes in defined neurons to altered behavior. Here we focus on circadian rhythms in the structure of the principal s-LNv clock neurons in Drosophila. By quantifying neuronal architecture, we observed that s-LNv structural plasticity changes the amount of axonal material in addition to cycles of fasciculation and defasciculation. We found that this is controlled by rhythmic Rho1 activity that retracts s-LNv axonal termini by increasing myosin phosphorylation and simultaneously changes the balance of pre-synaptic and dendritic markers. This plasticity is required to change clock network hierarchy and allow seasonal adaptation. Rhythms in Rho1 activity are controlled by clock-regulated transcription of Puratrophin-1-like (Pura), a Rho1 GEF. Since spinocerebellar ataxia is associated with mutations in human Puratrophin-1, our data support the idea that defective actin-related plasticity underlies this ataxia.
Graphical Abstract
Introduction
A plastic nervous system allows organisms to adapt and to learn from their environment. Although many neurons in the brain show plasticity, there are relatively few examples where structural changes in defined adult neurons are understood at the molecular level and have clear-cut behavioral consequences. This is due to difficulties in identifying and manipulating the precise neurons whose structure has changed in a densely packed brain during adulthood (May, 2011). Structural plasticity is thus often studied in vitro (Matsuzaki et al., 2004).
Almost all organisms have circadian rhythms of behavior and disturbances to human circadian rhythms can result in psychiatric disorders (Zelinski et al., 2014). Circadian rhythms are controlled by pacemaker neurons in the brain. These neurons receive external signals such as light to synchronize behavior with the solar day, although circadian rhythms persist in constant darkness (DD). A set of clock genes form a molecular clock that drives 24hr oscillations in RNA and protein levels in clock neurons. In Drosophila, this clock is composed of the transcriptional activators Clock (CLK) and Cycle (CYC), which activate the clock genes period (per) and timeless (tim). After translation, PER and TIM enter the nucleus, where PER represses CLK/CYC to close the negative feedback loop. Similar genes act in a conserved manner in the mammalian clock (reviewed by (Yu and Hardin, 2006)). Additional clock-controlled genes such as K+ channels that change firing properties of clock neurons help transduce molecular clock time into rhythmic electrical activity (Meredith et al., 2006; Ruben et al., 2012).
In addition to rhythms in intrinsic excitability, the structure of both Drosophila s-LNvs and mammalian pacemaker neurons is remodeled daily and, at least in s-LNvs, this is clock-controlled (Becquet et al., 2008; Fernandez et al., 2008; Girardet et al., 2010). However, the behavioral correlates of circadian structural plasticity have not yet been identified. The importance of s-LNvs in circadian behavior (Renn et al., 1999; Stoleru et al., 2004) offers an unusual opportunity to connect structural plasticity to behavior.
s-LNv axonal termini are normally maximally spread at dawn, which coincides with their peak excitability (Cao and Nitabach, 2008; Cao et al., 2013; Fernandez et al., 2008). Although it was recently reported that daily changes in s-LNv termini are a cycle of fasciculation and defasciculation (Sivachenko et al., 2013), we found that s-LNvs add and lose axonal material with a 24hr rhythm.
We speculated that actin rearrangements drive s-LNv growth and retraction and therefore that Rho family GTPases (Rho, Rac and Cdc42) are involved. GTPases act as switches that are active when bound to GTP and inactive when GDP-bound. Guanine nucleotide exchange factors (GEFs) increase GTPase activity while GTPase-activating proteins (GAPs) decrease activity (Van Aelst and D’Souza-Schorey, 1997). Rho GTPases are important in neuronal development: Rac1 and Cdc42 promote axonal elongation and branching while RhoA (Rho1 in Drosophila) promotes axonal retraction (Gonzalez-Billault et al., 2012; Hall, 2012).
We found that transiently over-expressing wild type Rho1 keeps s-LNv termini in a dusk-like retracted state and can also override electrical activity-dependent expansion. We discovered that endogenous Rho1 activity shows circadian rhythms in s-LNvs that retract their axonal termini at dusk via actin contraction and myosin light chain phosphorylation. Rho1 activity is regulated by clock-controlled expression of a previously uncharacterized GEF, which we named Puratrophin-1-like (Pura). Pura and Rho1 also orchestrate daily antiphase rhythms in pre-synaptic and dendritic markers in s-LNv termini that modulate signaling. Finally we showed that Rho1-regulated plasticity in s-LNv termini is required for normal circadian rhythms and controls hierarchy in the clock neuronal network so that flies can adapt to different seasons. Pura is an ortholog of human Puratrophin-1, mutations of which are associated with spinocerebellar ataxia (Ishikawa et al., 2005). Our data support the idea that Puratrophin-1 related spinocerebellar ataxia is a disease of defective actin-mediated neuronal plasticity.
Results
s-LNv projections increase axonal volume at dawn
The approaches previously used to quantify the termini of s-LNv projections detected clear time of day differences (Fernandez et al., 2008; Sivachenko et al., 2013). However, these methods are laborious and limited to two dimensions. They therefore cannot calculate the overall axonal volume and determine whether spreading and retraction change the total amount of axonal material. To address this, we created a Matlab script to automatically reconstruct 3D projections from confocal stacks of s-LNv termini.
To test the script, we reconstructed s-LNv projections from flies fixed at dusk (ZT12) and dawn (ZT24). [ZT = Zeitgeber time, time in a 12:12 Light:Dark cycle]. We used two different markers to test reproducibility: a membrane-tethered GFP expressed in LNvs using the Pdf-Gal4 driver; and the Pigment Dispersing Factor (PDF) neuropeptide, which is required for circadian behavior and has higher levels at dawn than dusk (Park et al., 2000; Renn et al., 1999).
The 3D reconstructions and quantification in Figures 1A and S1B show that s-LNv projections are significantly less spread in each axis at ZT12 than ZT24. This makes the 3D spread of both GFP and PDF at ZT12 ~50% of the spread at ZT24 (Figure 1A). Axonal volume is also significantly reduced at ZT12 compared to ZT24 (Figure 1A) and this is independent of fluorescence levels (Figure S1B). These data indicate that axonal growth and contraction takes place simultaneously with fasciculation and defasciculation and that together these constitute the daily expansion and retraction cycles of s-LNv termini.
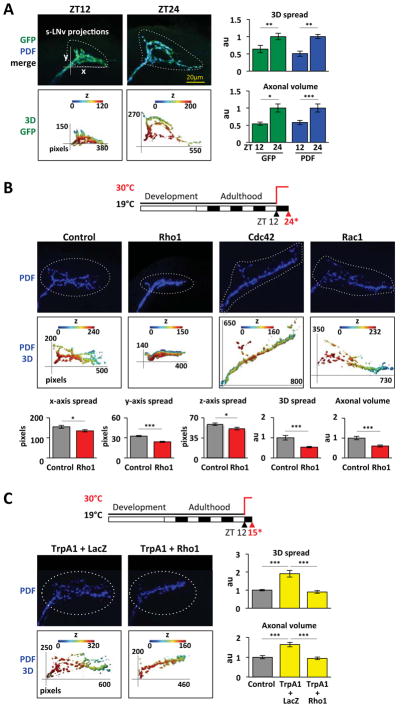
Figure 1. Rho1 prevents s-LNv projections from expanding.
A. Confocal images of s-LNv projections from Pdf > CD8::GFP flies stained with antibodies to GFP (green) and PDF (blue) at ZT12 and ZT24. 3D reconstructions (rainbow images) were generated using the Matlab script (see Experimental procedures) with colors indicating depth in the z-axis (blue to red represents dorsal to ventral). White dots show the area quantified. 1pixel = 0.12μm and z-step is 1μm. Graphs on right quantify 3D spread and axonal volume using the Matlab script. B. Top: Induction of Rho GTPase transgenes. Flies were raised at 19°C and entrained in LD cycles at 19°C for at least 3 days before shifting to 30°C at ZT12. Flies were dissected 12hr later (ZT24*) and stained with anti-PDF. Confocal images of s-LNv projections and their 3D reconstructions as above at ZT24* for Control (Pdf, tub-Gal80ts > CD8::GFP), Rho1 (Pdf, tub-Gal80ts > Rho1), Cdc42 (Pdf, tub-Gal80ts > Cdc42CA) and Rac1 (Pdf, tub-Gal80ts > Rac1). Graphs quantify parameters of s-LNv projections from Control and Rho1-induced flies. C. Top: Diagram of induction. Flies were handled and stained as in Figure 1B except dissection was at ZT15*. Confocal images and 3D reconstructions as above for TrpA1 + LacZ (Pdf, tub-Gal80ts > TrpA1, LacZ) and TrpA1 + Rho1 (Pdf, tub-Gal80ts > TrpA1, Rho1). Control flies were Pdf, tub-Gal80ts > myrRFP. Graphs quantify s-LNv projections.
Error bars show SEM. Statistical comparisons are with Student’s t-test. *p<0.05, **p<0.01, ***p<0.001. Significance was also verified with ANOVA. (See also Figure S1)
Rho GTPases dynamically regulate adult s-LNv structure
Given that axonal volume changes between dawn and dusk, we hypothesized that an actin-related pathway underlies s-LNv plasticity and tested if Rho GTPases are involved. Since Rho GTPases affect neuronal development (Gonzalez-Billault et al., 2012), we restricted over-expression to adulthood (Figure 1B). We used tubulin-Gal80ts (McGuire et al., 2003) to repress Pdf-Gal4 activity and raised flies at 19°C when Gal80ts is functional. After entraining to LD cycles at 19°C, Rho GTPase expression was induced in s-LNvs by raising the temperature to 30°C to inactivate Gal80ts. We induced expression of Rho1, Rac1 or Cdc42 for 12hr starting at dusk (ZT12). Brains were dissected, fixed and stained at ZT24* (asterisk indicates prior 12hr of induction), when s-LNvs are normally maximally spread (Fernandez et al., 2008).
We found that inducing wild-type Rho1 significantly reduced the 3D spread of s-LNv projections to ~50% of the 3D spread of control projections at ZT24* (Figure 1B). Inducing constitutively active Cdc42 or wild type Rac1 had the opposite effect (Figure 1B, S1D). Rac1 increased the spread of s-LNv projections in the x- and y-axes, while Cdc42CA increased the spread in x- and y-axes but reduced z-axis spread (Figures 1B, S1D). The rapid response of s-LNv projections to Rho GTPase induction suggests they normally play a role in s-LNv expansion and retraction.
(Sivachenko et al., 2013) proposed that the transcription factor Mef2 regulates expression of the inter-cellular adhesion molecule Fas2 as a mechanism for s-LNv plasticity. However, we found that inducing Fas2 for 12hr did not significantly change the spread in the x- or y-axes, the 3D spread or the axonal volume, although the z-axis spread was reduced (Figure S1E). Given these limited effects of Fas2 induction, some of the defects detected by (Sivachenko et al., 2013) may be due to continuous Fas2 over-expression during development and early adulthood.
(Sivachenko et al., 2013) also found that s-LNv projections can change rapidly. They increased LNv electrical activity at dusk using the heat-sensitive ion channel TrpA1 and found that s-LNv projections expand to a dawn-like state within 2hr. This requires the transcription factor Mef2. We tested whether inducing Rho1 can also affect activity-dependent spreading of s-LNvs. We found that a 3hr shift from 19°C to 30°C starting at ZT12 induces sufficient TrpA1 expression and activity to increase the 3D spread and axonal volume of s-LNvs to a dawn-like state (Figure 1C, S1F). This expansion was blocked by simultaneously inducing Rho1 (Figure 1C). Thus Rho1 can regulate s-LNv plasticity in a highly dynamic manner and the similar phenotypes of Mef2RNAi (Sivachenko et al., 2013) and Rho1 (Figure 1C) suggest they are in the same pathway.
Rho1-induction locks s-LNv projections in a dusk-like state
We decided to focus on Rho1 since it was the only Rho GTPase that blocks the expansion of s-LNvs (Figure 1B, S1D). We next tested if inducing Rho1 expression can reduce s-LNv projections at any time of day. Rho1 was induced starting either at dusk (ZT12) or dawn (ZT24) with projections assayed 12hr later. Figure 2 shows that the 3D spread and axonal volume of s-LNv projections with Rho1 induced are no different to control s-LNv projections fixed at dusk (ZT12*, Figure 2). Thus inducing Rho1 has no effect on s-LNv projections at dusk but only affects s-LNv projections at dawn.
Figure 2. Inducing Rho1 locks s-LNv projections in a dusk-like state.
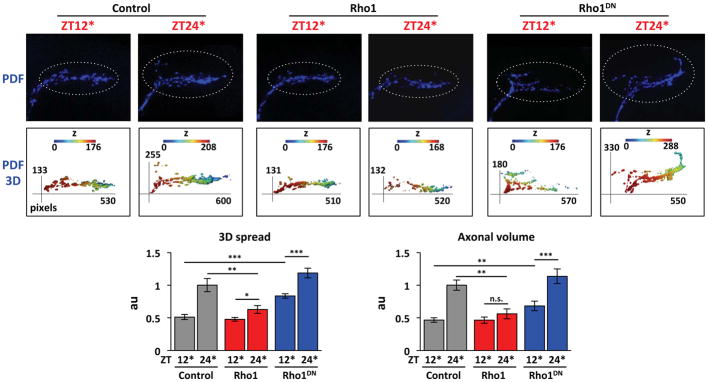
Flies were raised and entrained as in Figure 1B. Rho1 was induced starting either at dawn with brains fixed at ZT12* or starting at dusk with brains fixed at ZT24*. Confocal images of s-LNv projections and their 3D reconstructions as in Figure 1 from Control (Pdf, tub-Gal80ts > myrGFP + myrRFP), Rho1 (Pdf, tub-Gal80ts > Rho1 + myrRFP) and Rho1DN (Pdf, tub-Gal80ts > Rho1DN) flies. s-LNv projections were stained with anti-PDF and quantified as in Figure 1B. 1 pixel = 0.12μm and z-step is 1μm. Statistical comparisons are with Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, n.s.: non-significant. Significance was also verified with ANOVA. (See also Figure S2)
To test how well tubulin-Gal80ts represses Rho1 activity, we raised flies at 19°C and shifted them to 25°C, when Gal80ts is still functional (McGuire et al., 2003). Flies with control or Rho1 transgenes had strong rhythms in the 3D spread and axonal volume of s-LNv projections at 25°C (Figure S2). Thus we conclude that tubulin-Gal80ts represses Rho1 activity at 25°C and lower.
Next we measured the effect of reducing Rho1 activity using a dominant-negative Rho1 transgene (Rho1DN). Figure 2 shows that the 3D spread of s-LNv projections expressing Rho1DN at ZT12* is significantly higher than in control flies, indicating that s-LNv projections need maximal Rho1 activity to fully retract. However, there are still oscillations in the 3D spread and axonal volume between ZT12* and ZT24* in Rho1DN-expressing s-LNvs (Figure 2). This suggests that either 12hr of Rho1DN induction does not completely eliminate Rho1 activity and/or that additional factors drive s-LNv expansion. We favor the latter idea since inducing other Rho GTPases increases s-LNv expansion at dawn (Figure 1B, S1D).
Circadian oscillations in endogenous Rho1 activity in s-LNv axons
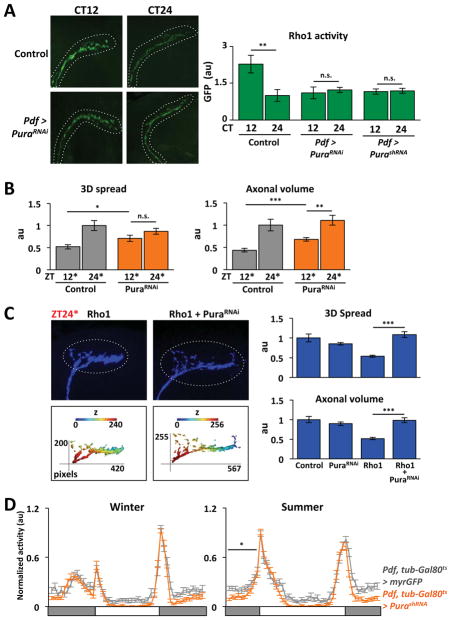
One explanation for the time-specific effect of Rho1 induction is that endogenous Rho1 activity is rhythmic. Thus over-expressing Rho1 would only affect s-LNv projections when endogenous Rho1 activity is low. To test this, we used a Rho1-specific activity sensor which has three Rho1-GTP binding domains fused to eGFP (Simoes et al., 2006). Sensor fluorescence is diffuse in the cytoplasm until endogenous Rho1 activation concentrates the sensor at the membrane to give intense fluorescence (Simoes et al., 2006).
We expressed the sensor in s-LNvs, fixed fly brains at different times of day and used a GFP antibody to quantify endogenous Rho1 activity. The data in Figure 3A show that endogenous Rho1 activity in s-LNv axons is highest around dusk. We detected oscillations in s-LNv axonal termini and the straight portion of the s-LNv axons below their ramifications (Figure 3A) but not in s-LNv cell bodies (Figure 3B). We found no rhythms in cytosolic GFP or membrane-bound RFP (Figure 3A), indicating that Rho1 sensor rhythms are not due to altered s-LNv morphology.
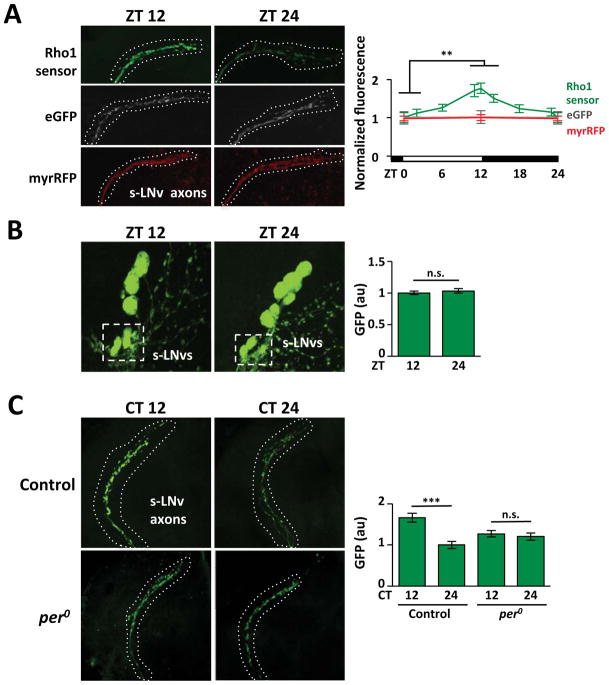
Figure 3. Circadian oscillations in Rho1 activity in s-LNv axons.
A. Confocal images of s-LNv axons from flies expressing Rho1-activity sensor (Pdf > PKNG58AeGFP, green), eGFP (Pdf > eGFP, grey) or myrRFP (Pdf > myrRFP, red) stained with anti-GFP or anti-RFP. Graph shows average fluorescence levels in LD measured with Fiji with ZT0 data replotted at ZT24. B. Confocal images of s-LNv cell bodies from flies expressing Rho1-activity sensor with average GFP levels plotted on the right. C. Confocal images of s-LNv axons and termini from control (Pdf > PKNG58AeGFP) and per0 flies (per0; Pdf > PKNG58AeGFP) with average GFP levels plotted on the right.
Error bars show SEM. Statistical comparisons are with Student’s t-test. **p<0.01, ***p<0.001, n.s.: non-significant.
Next we measured sensor activity in DD to test if Rho1 rhythms are circadian. We found that Rho1 activity was higher at CT12 than CT24 (Figure 3C), in phase with LD rhythms. Rho1 activity rhythms in LD and DD made it likely that these oscillations are clock-dependent, like s-LNv plasticity itself (Fernandez et al., 2008). To test this, we assayed Rho1 sensor levels in per null mutant flies. We found that Rho1-activity oscillations were lost in per0 mutants (Figure 3C), indicating that endogenous Rho1 activity rhythms are normally clock-controlled.
Rho1 regulates an output pathway important for circadian behavior
We next assayed the behavioral consequences of altered s-LNv plasticity by comparing the locomotor activity of control (Pdf, tub-Gal80ts > myrRFP) and Rho1-induced flies (Pdf, tub-Gal80ts > Rho1). Flies were grown at 19°C and their behavioral rhythms assayed in DD at 25°C and 30°C. Flies of both genotypes had similarly strong circadian rhythms at 25°C when Gal80ts is active (Figure 4A, Table S1). However, flies with Rho1 induced by shifting to 30°C were either arrhythmic or weakly rhythmic (Figure 4A, Table S1). We confirmed that maintaining flies at 30°C keeps s-LNv projections in a dusk-like retracted state at CT24* even 2.5 days after inducing Rho1 (data not shown).
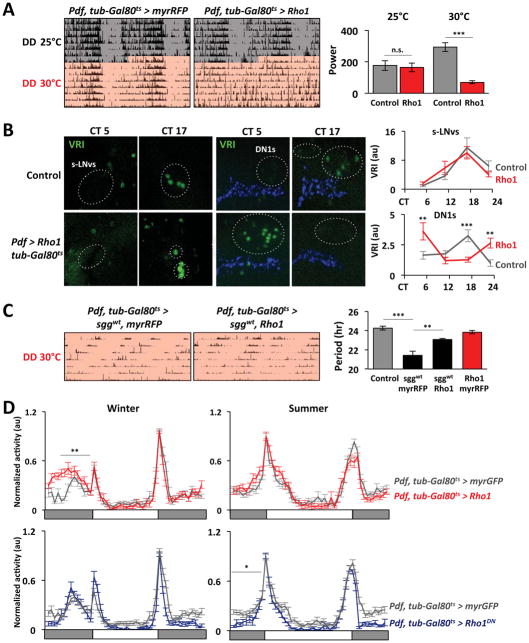
Figure 4. Rho1 regulates an output pathway important for circadian behavior and seasonal adaptation.
A. Left: Actograms show locomotor activity in DD of Control (Pdf, tub-Gal80ts > myrRFP) and Rho1-inducible flies (Pdf, tub-Gal80ts > Rho1) for 7 days at 25°C (grey) or 30°C (pink). Right: Average rhythm power at 25°C and at 30°C in DD (also see Table S1). B. Flies were entrained to LD cycles at 19°C and then transferred to DD at 30°C. Confocal images of s-LNv cell bodies (left panels) and DN1 clock neurons (right panels) from Control (Pdf, tub-Gal80ts > +) and Rho1-induced flies (Pdf, tub-Gal80ts > Rho1) stained with antibodies to VRI (green) and PDF (blue) at CT5 and CT17 on day 3 in DD. Graphs show average VRI fluorescence. The phase of the oscillation in DN1s was significantly different between genotypes (p<0.01, ANOVA). C. Left: Actograms of Pdf, tub-Gal80ts > sggwt,myrRFP and Pdf, tub-Gal80ts > sggwt,Rho1 flies at 30°C in DD. Right: Graph shows average period of rhythmic flies (also see Table S1). D. Locomotor activity of Control (Pdf, tub-Gal80ts > myrGFP, grey), Rho1- (Pdf, tub-Gal80ts > Rho1, red) and Rho1DN-induced flies (Pdf, tub-Gal80ts > Rho1DN, blue) in winter (10L:14D) or summer (14L:10D) light conditions.
Error bars show SEM. Statistical comparisons are with Student’s t-test (A–C) and ANOVA (D). *p<0.05, **p<0.01, ***p<0.001, n.s.: non-significant. (See also Figure S3)
The arrhythmicity in Pdf, tub-Gal80ts > Rho1 flies could be associated with defects in the s-LNv molecular clock and/or s-LNv signaling. To test the first possibility, we measured levels of the core clock proteins TIM and Vrille (VRI) on day 3 in DD at 30°C. We found that VRI and TIM protein oscillations were indistinguishable when comparing s-LNvs in control and Rho1-induced flies (Figure 4B, S3A). Thus the cause of arrhythmic behavior must lie downstream of the s-LNv molecular clock.
To test if inducing Rho1 in s-LNvs alters their ability to signal, we measured the molecular clocks in DN1 clock neurons whose phase is set by the s-LNvs (Stoleru et al., 2005; Yao and Shafer, 2014; Yoshii et al., 2009). We found that the phase of the clock proteins VRI and PAR-Domain Protein 1 (PDP1) in DN1 clocks is shifted by ~6hr between control and Rho1-induced flies after 3 days in DD (Figures 4B, S3B). Rhythmic DN1 clocks differ from their arrhythmicity in Pdf null mutants (Yoshii et al., 2009). Thus keeping s-LNv projections in a retracted state seems to change rather than abolish s-LNv signaling, as is presumably the case in Pdf mutants.
To test this idea, we built on the work of (Stoleru et al., 2005). They had found that overexpressing the clock kinase Shaggy (Sgg) / GSK3 only in LNvs speeds up the molecular clock in many other clock neurons, leading to short period behavioral rhythms and indicating that s-LNvs determine the period of the entire clock network in DD. We reasoned that if retracting s-LNv projections alters their signaling, this should alter the period length of flies overexpressing sgg.
We measured the period of flies co-expressing sgg and either Rho1 or a control myrRFP transgene, again using Pdf-Gal4, tubulin-Gal80ts to restrict expression to adults. Figure 4C shows that rhythmic flies expressing sgg and myrRFP have a short period of 21.4hr, similar to expressing sgg throughout development. More sgg-expressing flies were arrhythmic at 30°C than at 25°C, probably due to higher Gal4 and/or Sgg activity at 30°C. In contrast, flies co-expressing sgg and Rho1 have a 23.1hr period (Figure 4C, Table S1). Thus we conclude that constitutively retracting s-LNv projections reduces their signaling strength and their ability to set the pace of the clock network.
LNv structural plasticity is important for seasonal adaptation
The circadian system is plastic with different neuronal groups taking the role of main oscillators as day length changes with the seasons (Stoleru et al., 2007; Zhang et al., 2010). s-LNvs are the dominant oscillators on short winter days while the LNds, 5th s-LNv and some DN1s assume control on long summer days (Stoleru et al., 2007). However, it is unclear how different oscillators take control of the network. Since retracting s-LNv projections reduces their ability to control the network (Figure 4B–C), we tested whether s-LNv plasticity is important for seasonal adaptation.
Flies were raised at 19°C and then transferred to 30°C and either winter (10:14) or summer (14:10) LD cycles. We compared the behavior of control flies (Pdf, tub-Gal80ts > myrGFP) and experimental flies with induced expression of either Rho1 or Rho1DN. Figure 4D shows that control flies shift their morning and evening activity peaks to align with seasonal dawn and dusk. However, flies with Rho1 induced in LNvs are active earlier before dawn in winter conditions than control flies, although their activity during summer is the same as control flies. Conversely, Rho1DN flies increase their activity later than control flies on summer mornings but behave like controls in winter (Figure 4D) and this is independent of activity levels (Figure S3C). We interpret these phenotypes as follows: high Rho1 activity constitutively retracts s-LNv projections, preventing s-LNvs from controlling the network in winter. In contrast, when s-LNvs cannot fully retract in Rho1DN-induced flies, s-LNvs cannot cede control of the network in summer.
We also measured the behavior of Pdf null mutants in our winter and summer conditions. Pdf null flies show defective morning behavior in both conditions (Figure S3D) as in (Renn et al., 1999; Yoshii et al., 2009). Since the behavior of Pdf mutants differs from flies expressing Rho1 transgenes, this supports the idea that s-LNvs with altered plasticity still release PDF. Thus structural plasticity likely modulates s-LNv signaling strength, with retracted s-LNv projections reducing signaling from s-LNvs to downstream cells. Reduced s-LNv signaling with Rho1 induced in LNvs probably explains why DN1 molecular clock oscillations do not stop as in Pdf mutants (Yoshii et al., 2009), but persist albeit with altered phase (Figure 4B).
Rho1 retracts s-LNvs via myosin phosphorylation
(Billuart et al., 2001) showed that Rho1 retracts axonal branches during Drosophila mushroom body development by phosphorylating myosin light chain (MLC) to contract actin filaments. To understand how Rho1 regulates s-LNv plasticity, we asked if phosphorylated MLC (P-MLC) levels change between dusk and dawn in s-LNv axons using a P-MLC specific antibody (Lee and Treisman, 2004). We found that P-MLC levels in s-LNv axons were two-fold higher at ZT12 than ZT24 (Figure 5A). These data are consistent with higher Rho1 activity at dusk and serve as an independent marker of endogenous Rho1 activity rhythms. P-MLC oscillations were blocked in per0 flies (Figure S4A), indicating that MLC phosphorylation is clock-controlled in s-LNv axons, like Rho1 activity. To test if Rho1 is responsible for MLC phosphorylation, we induced Rho1 for 12hr. This increased P-MLC levels at ZT24* ~2-fold over controls (Figure 5B). Thus we conclude that high Rho1 activity in s-LNv axons around dusk increases MLC phosphorylation.
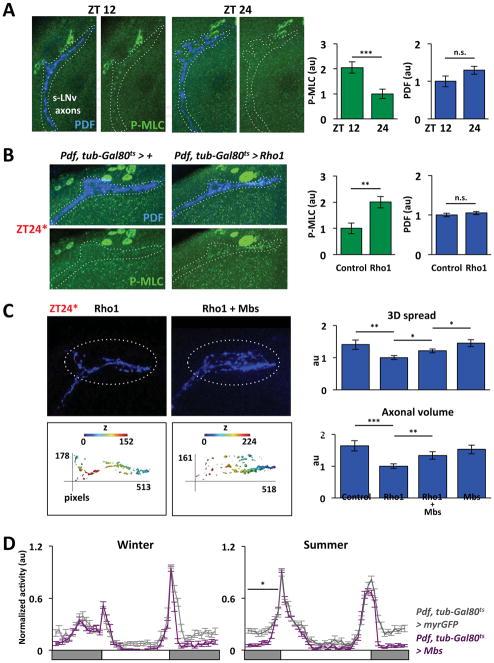
Figure 5. Rho1 controls rhythmic myosin light chain phosphorylation in s-LNv axons.
A. Confocal images of s-LNv axons at ZT12 and ZT24 stained with antibodies against P-MLC (green) and PDF (blue). Fluorescent intensity was measured with Fiji and the normalized average plotted on the right. B. Confocal images of s-LNv axons at ZT24* from Pdf, tub-Gal80ts > + and Pdf, tub-Gal80ts > Rho1 flies stained and analyzed as in A. C. Confocal images of s-LNv projections stained with anti-PDF (blue) and their 3D reconstructions as in Figure 1 from flies with Rho1-induced (Rho1) or both Rho1 and Mbs-induced (Rho1 + Mbs). Graphs show average 3D spread and axonal volume for Control (Pdf, tub-Gal80ts > myrRFP + myrGFP), Rho1 (Pdf, tub-Gal80ts > Rho1 + myrRFP), Rho1 + Mbs (Pdf, tub-Gal80ts > Rho1 + Mbs) and Mbs (Pdf, tub-Gal80ts > Mbs + myrRFP) induced flies. 1pixel = 0.12μm and z-step is 1μm. D. Locomotor activity of Control (Pdf, tub-Gal80ts > myrGFP, grey) and Mbs-induced (Pdf, tub-Gal80ts > Mbs, purple) flies as in Figure 4D.
Error bars show SEM. Statistical comparisons are with Student’s t-test (A–C) and ANOVA (D). *p<0.05, **p<0.01, ***p<0.001, n.s.: non-significant. (See also Figure S4 and Table S1)
Rho1 regulates MLC phosphorylation through ROCK (Rho-associated protein kinase) and Mbs (Myosin binding subunit, (Kimura et al., 1996). When Rho1 binds ROCK and Mbs, ROCK phosphorylates and inhibits Mbs, an MLC Phosphatase (MLCP) subunit (Hartshorne et al., 1998). Phosphorylated Mbs decreases MLCP activity, which increases MLC phosphorylation and actomyosin contraction (Kawano et al., 1999; Kimura et al., 1996). If the myosin pathway is downstream of Rho1 in s-LNvs, we hypothesized that over-expressing Mbs would rescue Rho1-induced retraction. We found that the axonal volume and 3D spread of s-LNv projections at ZT24* were higher when Mbs was co-induced with Rho1 than with Rho1 alone (Figure 5C). Similarly, the behavioral arrhythmicity of flies over-expressing Rho1 from birth was rescued by over-expressing Mbs (Figure S4B, Table S1).
Next we tested if MLC phosphorylation is required for s-LNv projections to retract at dusk. We induced Mbs to trigger MLC dephosphorylation for 12hr starting either at dawn or dusk and assayed s-LNv morphology. We found that s-LNv projections in Mbs-induced flies had significantly higher 3D spread and axonal volume at ZT12* than control flies (Figure S4C). This suggests that MLC phosphorylation is required for retraction and that Mbs activity is normally rhythmic and low at dusk when Rho1 activity is high.
We also tested how Mbs-induction affects seasonal adaptation. We found that Mbs-induced flies adapt normally to winter light conditions but less well to summer (Figure 5D), like Rho1DN (Figure 4D). Given that s-LNv projections can never fully retract after inducing Mbs or Rho1DN (Figure 2, S4C), these results strengthen the argument that seasonal adaptation requires structural plasticity. We propose that Rho1 promotes retraction of s-LNv projections at dusk by opposing Mbs activity and promoting MLC phosphorylation.
Pura is a clock-regulated Rho1 GEF that regulates rhythmic Rho1 activity in s-LNvs
Clock-regulated Rho1 activity in s-LNv axons could be explained by rhythms in a Rho1 GEF peaking at dusk and/or a Rho1 GAP peaking at dawn. Our expression profiles of larval LNvs revealed that CG33275, a previously uncharacterized and predicted Rho GEF, is clock-regulated and rhythmically expressed in LD and DD, with RNA levels peaking around dusk (Mizrak et al., 2012; Ruben et al., 2012). CG33275 is also rhythmically expressed in adult s-LNvs, with higher expression at ZT12 than ZT24 (Kula-Eversole et al., 2010). Phylogenetic tree analysis (using ensembl.org) and reciprocal BLAST showed that CG33275 is orthologous to human Puratrophin-1 (Purkinje cell atrophy associated protein-1 / Plekhg4), which functions as a Rho family GEF in vitro (Gupta et al., 2013; Ishikawa et al., 2005). Puratrophin-1 is also rhythmically expressed in the mouse pituitary (CircaDB), suggesting that it has a circadian function. Thus we named CG33275 Puratrophin-1-like (Pura). Pura is 56% identical and 72% homologous to human Puratrophin-1 over the adjoining Dibble-homology and Pleckstrin-like homology domains that are a signature of Rho family GTPase GEFs (Schmidt and Hall, 2002).
To test if rhythmic Pura expression results from transcriptional regulation, we used a Pura-Gal4 line inserted 45bp upstream of the predicted start site of the Pura-B transcript. We crossed Pura-Gal4 flies to a nuclear-localized UAS-destabilized-GFP transgene and found that GFP levels in s-LNvs were higher at ZT14 than ZT2 (Figure S5A), in phase with Pura RNA. This rhythm is specific to Pura-Gal4 since Pdf-Gal4 showed no GFP rhythms (Figure S5A). Pura may be a direct CLK/CYC target since there are 2 E-boxes in the first 1.5kb upstream of Pura-Gal4.
We used genetic interactions to test if Pura can act as a Rho1 GEF in LNvs. We first confirmed that UAS-PuraRNAi and UAS-PurashRNA transgenes that target different Pura sequences reduce Pura RNA levels (Figure S5B). We found that the behavioral arrhythmicity of flies over-expressing Rho1 from birth was rescued by co-expressing either PuraRNAi or PurashRNA (Figure S5C, Table S1). This effect is specific for Rho1: First, we found that the arrhythmic phenotype of flies expressing a constitutively-active Rho1 (Rho1CA) in LNvs was unaltered by co-expressing PuraRNAi (Figure S5C, Table S1). Since Rho1CA binds GTP independently of GEFs, we conclude that Pura only regulates Rho1 activity when Rho1 is GEF-dependent. Second, we found that Rac1 over-expression in LNvs lengthens period to 25hr and that this was unaltered by PuraRNAi (Table S1). Thus, Pura acts on Rho1 but not Rac1.
To test if Pura is the Rho1 GEF responsible for circadian Rho1 activity in s-LNv axons, we used the Rho1 sensor to measure Rho1 activity. The data in Figure 6A show that PuraRNAi and PurashRNA block rhythmic Rho1 activity and reduce Rho1 activity at CT12 to levels comparable to control s-LNv axons at CT24. Reducing endogenous Pura levels did not alter Rho1 activity in s-LNv cell bodies (data not shown), indicating that Pura only regulates Rho1 activity in axons. Taken together, we conclude that Pura regulates the timing and localization of Rho1 activity that connects the core clock to s-LNv outputs.
Figure 6. Pura is a clock-regulated Rho1 GEF that activates Rho1 in s-LNv axons at dusk and is required for seasonal adaptation.
A. Confocal images of s-LNv axons at CT12 and CT24 on day 1 in DD from Control (Pdf > PKNG58AeGFP + corazoninRNAi) and Pdf > PuraRNAi flies (Pdf > PKNG58AeGFP + PuraRNAi) expressing the Rho1-activity sensor (green) as in Figure 3. Graph shows the average fluorescence intensity of the Rho1-sensor in s-LNv axons from the above genotypes and from Pdf > PurashRNA flies (Pdf > PKNG58AeGFP + PurashRNA). B. s-LNv projections were stained with anti-PDF and quantified as in Figure 1 from Control (Pdf, tub-Gal80ts > myrRFP) and PuraRNAi flies (Pdf, tub-Gal80ts > PuraRNAi) and entrained and shifted to 30°C for 12hr as in Figure 2. C. Confocal images of s-LNv projections stained with PDF (blue) and their 3D reconstructions from flies with either Rho1 (Pdf, tub-Gal80ts > Rho1 + myrRFP) or Rho1 and PuraRNAi (Pdf, tub-Gal80ts > Rho1 + PuraRNAi) induced for 12hr and fixed at ZT24*. Graphs show the average 3D spread and axonal volume. 1pixel = 0.12μm and z-step is 1μm. D. Locomotor activity of Control (Pdf, tub-Gal80ts > myrGFP, grey) and PurashRNA-induced flies (Pdf, tub-Gal80ts > PurashRNA, orange) as in Figure 4D.
Error bars show SEM. Statistical comparisons are with Student’s t-test (A–C) and ANOVA (D). *p<0.05, **p<0.01, ***p<0.001, n.s.: non-significant. (See also Figure S5 and Table S1)
Pura is required for s-LNv structural plasticity and seasonal adaptation
We also tested whether normal Pura levels are required for s-LNv structural plasticity. We induced PuraRNAi starting at dusk or dawn and assayed s-LNv projections 12hr later. We found that PuraRNAi expression significantly increased the 3D spread and axonal volume at ZT12* compared to control flies (Figure 6B). Thus maximal Pura expression is required for full retraction at dusk. In contrast, s-LNv projections in PuraRNAi flies were similar to control flies at ZT24*. Pura’s time-specific effect on Rho1 activity and LNv projections is consistent with the timing of both Pura RNA and Rho1 activity rhythms.
We also tested whether reducing Pura expression can override Rho1-induced retractions in s-LNv projections. We induced wild type Rho1 for 12hr as in Figure 1B either with or without PuraRNAi. Figure 6C shows that inducing PuraRNAi along with Rho1 (Pdf, tub-Gal80ts > Rho1 + PuraRNAi) restored the axonal volume and 3D spread of s-LNv projections to control ZT24* levels. In addition, inducing PuraRNAi also prevented over-expressed Rho1 from increasing MLC phosphorylation at ZT24* (Figure S5D). Thus we conclude that Rho1 requires sufficient Pura expression to retract s-LNv axons via actin-related structural changes and that rhythmic Pura expression normally limits the timing of Rho1 activity.
We also tested Pura’s importance in seasonal adaptation. We found that inducing PurashRNA reduced morning adaptation in summer conditions, but did not affect winter behavior (Figure 6D), just like Rho1DN and Mbs flies (Figure 4D, 5D).
Flies overexpressing PuraRNAi, PurashRNA, Mbs or Rho1DN have normal locomotor activity in DD, in contrast to Rho1 induction which promotes arrhythmicity (Table S1). We propose that Rho1 activity is normally limited in DD and winter since s-LNvs need to dominate the clock network and thus be in an expanded or permissive state for communication (Stoleru et al., 2005). However, when s-LNvs cede control of the network to other clock neurons during summer (Stoleru et al., 2007), Rho1 activity needs to be maximal, to maintain s-LNv projections in a retracted state that reduces signaling.
Even though Pdf, tub-Gal80ts > PurashRNA flies are rhythmic in DD (Table S1), we tested their molecular clock after 3 days at 30°C. We found that VRI oscillations were similar to control flies in s-LNvs and DN1s (Figure S5E and data not shown), but had slightly altered rhythms in LNds (Figure S5E), whose connectivity with s-LNvs is rhythmic (Gorostiza et al., 2014). Thus s-LNv plasticity is required for optimal signaling with downstream neurons.
s-LNv plasticity changes clock network communication
(Gorostiza et al., 2014) recently showed that rhythms in s-LNv projection morphology are accompanied by rhythms in the number of pre-synaptic active zones. We used a fluorescently-tagged Bruchpilot-short (BRPshort-strawberry) transgene to detect active zones and confirmed that s-LNv projections have more BRP puncta at ZT2* than ZT14* (Figure 7A and S6). We then tested the role of Rho1 and Pura in this rhythm. The data in Figure 7A show that inducing Rho1 for 14hr reduces the number of active zones at ZT2* to control levels at ZT14*, but does not change the low levels at ZT14*. The complementary experiment with PuraRNAi gave the opposite results: active zone numbers were now constitutively high.
Figure 7. Pura and Rho1 regulate synaptic plasticity.
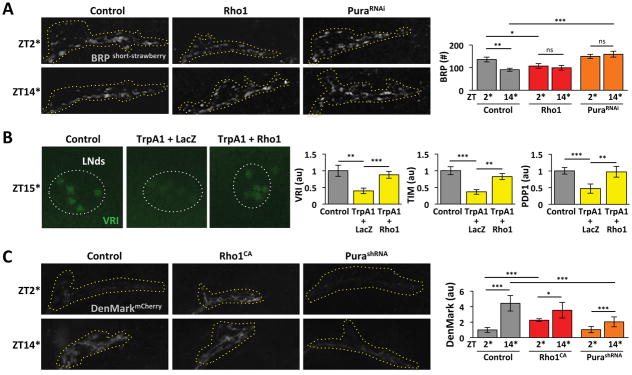
A. Confocal images of s-LNv projections at ZT2* and ZT14* from Control (Pdf, tub-Gal80ts > brpshort-strawberry + LacZ), Rho1 (Pdf, tub-Gal80ts > brpshort-strawberry + Rho1) and PuraRNAi (Pdf, tub-Gal80ts > brpshort-strawberry + PuraRNAi) flies. Flies were raised at 19°C and entrained in LD cycles at 19°C for at least 3 days before shifting to 30°C, starting at ZT12 or ZT24. Flies were dissected 14hr later at ZT2* and ZT14* respectively and brains stained with αDsRed (grey) to visualize Brp. Graph shows average numbers of active zones. B. Confocal images of LNd clock neurons from Control (Pdf, tub-Gal80ts > myrRFP), TrpA1 + LacZ (Pdf, tub-Gal80ts > TrpA1, LacZ) and TrpA1 + Rho1 (Pdf, tub-Gal80ts > TrpA1, Rho1) flies. Flies were raised and entrained in LD at 19°C before shifting to 30°C for 3hr at ZT12 and dissecting 3hr later at ZT15*. Brains were stained for VRI (green), TIM and PDP1. Quantification was as in Figure 4B. C. Confocal images of s-LNv projections at ZT2* (top) and ZT14* (bottom) from Control (Pdf, tub-Gal80ts > DenMarK + LacZ), Rho1 (Pdf, tub-Gal80ts > DenMarK + Rho1CA) and PurashRNA (Pdf, tub-Gal80ts > DenMarK + PurashRNA) flies. Flies were handled as in 7A and brains stained with αDsRed (grey). Quantification of average DenMark fluorescence levels was performed with Fiji.
Error bars show SEM. Statistical comparisons are with Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, n.s.: non-significant. (See also Figure S6)
Changes in the number of active zones supports the idea that s-LNv plasticity changes signaling. To test this idea further and to test how rapidly Rho1 can regulate s-LNv signaling, we built on the finding that s-LNv firing rapidly reduces levels of the core clock protein TIM in LNd clock neurons (Guo et al., 2014). We induced expression and activity of TrpA1 in s-LNvs for 3hr starting at ZT12 and measured levels of VRI, TIM and PDP1 in LNds at ZT15*. Inducing LNv firing reduced levels of all three proteins (Figure 7B). However, co-inducing Rho1 along with TrpA1 in LNvs blocked these rapid effects of LNvs on the LNd molecular clock (Figure 7B). Thus Rho1 activity can modulate the effect of LNv firing on downstream neurons.
Inputs from other clock neurons help synchronize individual s-LNv molecular clocks and regulate their neuronal activity (Collins et al., 2012; Collins et al., 2014). Since EM studies of s-LNv axonal projections had revealed they have input synapses (Yasuyama and Meinertzhagen, 2010), we tested if s-LNv plasticity is also associated with altered accumulation of post-synaptic components. We used DenMark to mark dendrites (Nicolai et al., 2010) and found that DenMark levels at ZT14* are ~4-fold higher than at ZT2* (Figure 7C). Increasing Rho1 activity by inducing Rho1CA increased the normally low DenMark levels at ZT2*, whereas reducing Pura levels using PurashRNA reduced the high DenMark levels at ZT14*, although both manipulations left reduced amplitude DenMark oscillations. The antiphase rhythms of DenMark and BRP in wild type s-LNvs suggest that s-LNv morphological plasticity is accompanied by a switch from predominantly receiving signals around dusk to predominantly sending signals around dawn. This synaptic plasticity is also regulated by Pura and Rho1.
Discussion
Directly linking plasticity to behavior is challenging in the adult brain. We took advantage of predictable and quantifiable changes in s-LNv structure and the precision of Drosophila genetics for spatial and temporal manipulation. We identified circadian rhythms in Rho1 GTPase activity in s-LNv axons that are regulated by rhythmic transcription of Pura, a Rho1 GEF. Pura activates Rho1 to retract s-LNv axons, decrease active zone numbers and reduce s-LNvs’ influence on the clock network. Thus we make strong links between transcription, plasticity, network hierarchy and behavior.
Mammalian SCN pacemaker neurons show rhythms in post-synaptic densities and neuron-glia connections (Becquet et al., 2008; Girardet et al., 2010). Even liver cells show daily rhythms in actin dynamics and cell size (Gerber et al., 2013). It will be interesting to test if SCN synapses show Rho-regulated plasticity as a potentially conserved mechanism to regulate clock neuron communication.
SCA and Pura
Mutations in human Puratrophin-1 are linked to Spinocerebellar ataxia (SCA), a neurodegenerative disease affecting cerebellar Purkinje cells (Ishikawa et al., 2005). Atrophic Purkinje cells from these SCA patients have cytoplasmic aggregates containing Puratrophin-1 and the actin-binding protein Spectrin (Ishikawa et al., 2005), consistent with fly and mammalian proteins both having actin-related functions.
The SCA-associated mutations reduce Puratrophin-1 RNA levels (Amino et al., 2007; Ishikawa et al., 2005). Thus cerebellar Puratrophin-1 expression seems to be tightly regulated like Pura in s-LNvs. We speculate that low Puratrophin-1 expression reduces activity of RhoA, the Rho1 ortholog. This could misregulate the ROCK/myosin pathway, reducing plasticity and neuronal connectivity and lead to Purkinje cell atrophy.
Rho GTPases and neuronal signaling
Rho GTPases are regulated by Rho GEFs and GAPs (Van Aelst and D’SouzaSchorey, 1997). Pura seems to provide spatiotemporal specificity for Rho1 in s-LNvs: Pura protein localization likely restricts Rho1 activity to axons and rhythmic Pura expression limits the timing of Rho1 activity.
(Gorostiza et al., 2014) added two levels of s-LNv plasticity beyond morphology: s-LNvs change connections with other neurons over 24hr and show rhythms in the numbers of pre-synaptic sites. We found rhythms in post-synaptic markers at s-LNv termini in antiphase to pre-synaptic markers. Since Rho1 activity regulates s-LNv structural and synaptic plasticity, Rho1 activity profoundly influences the effectiveness of s-LNv signaling and can modulate the intrinsic excitability of LNvs.
We propose that external cues can control Rho1 activity. (Yuan et al., 2011) showed that long days reduce larval LNv dendrite length. We propose that long days keep adult s-LNv axons retracted by increasing Rho1 activity, while s-LNv firing around dawn decreases Rho1 activity to allow axonal expansion. We recently showed that increased LNv activity reduces Pura RNA levels (Mizrak et al., 2012). Thus Pura expression may integrate clock state and electrical activity.
s-LNv plasticity may also involve Rho GTPases such as Rac1 and Cdc42 at dawn. It will be interesting to test if s-LNv plasticity is regulated by the opposing dynamics of different Rho GTPases. These neurons regulating innate behavior are surprisingly plastic and their transcriptionally programmed dynamics make s-LNvs an exciting system to understand the role of actin dynamics in structural and synaptic plasticity.
Experimental Procedures
Adult locomotor activity
For locomotor activity experiments that included temperature induction, adult flies were entrained for 3 days in 12:12 LD cycles at 19°C and then transferred to DD at 25°C or 30°C. For other experiments, adults were entrained for 3 days in 12:12 LD cycles at 25°C before transfer to DD. Locomotor activity was recorded using the DAM system (TriKinetics, Waltham, MA) and we used χ2 analysis in ClockLab (Actimetrics, Wilmette, IL) to calculate the power above the significance line (p<0.01) for each fly. Flies with a power <100 were considered arrhythmic and excluded from period calculations but included in average power calculations. Flies were kept for 5 days at 30°C in 10:14 and 14:10 LD cycles for winter and summer conditions respectively. Activity data from the last 2 days were averaged, normalized and plotted using a bin size of 30min. Morning anticipation was measured as the activity 6hrs before dawn (ZT19–ZT24).
Immunocytochemistry
Immunocytochemistry was carried out as in (Collins et al., 2012; Collins et al., 2014). Antibodies are described in the supplementary information. Images were scanned on the 20x lens of a Leica SP5 confocal microscope with 4–6.1x digital zoom. For P-MLC staining, brains were cleared through an isopropanol series and mounted in Murray Clear (1:2 benzyl alcohol: benzyl benzoate) as in (Veeman and Smith, 2013). Mean staining intensity was quantified using Fiji (http://pacific.mpi-cbg.de/wiki/index.php/Main_Page), with background staining levels subtracted. BRP puncta were quantified as in (Gorostiza et al., 2014).
Quantification of structural plasticity
127 x 127 μm confocal stack jpg images with 1024 x 1024 pixel resolution were imported into Matlab and s-LNv projections starting from where axons turn dorsally were selected for quantification. The Matlab script generates a 3D surface contour over regions (pixels) with >70% of the maximum fluorescent intensity to identify s-LNv projections. A 3D curve that runs through the pixels that form the s-LNv projections is then computed along with the average spread in x, y- and z-axes (Figure S1A). We mutliply these values to obtain the 3D spread. The script also calculates the total amount of pixels, which is the axonal volume. See supplementary experimental procedures for a detailed description.
Supplementary Material
Acknowledgments
We thank Vivian Budnik, Paul Hardin, Brian McCabe, Tim Mosca, Jae Park, Michael Rosbash, Amita Sehgal, Stephan Sigrist, Sergio Simoes, Jessica Treisman, the DSHB and the VDRC, Kyoto and Bloomington Stock Centers for flies and antibodies. We thank Anita Burgos and Samantha Raymond for their contributions and Hsaio-Yun Liu for advice on shRNA. We thank Rich Bonneau, Lionel Christiaen, Claude Desplan, Erik Herzog, Adrian Rothenfluh, Chris Rushlow and Mark Siegal for invaluable discussions, Claire Bertet, Matthieu Cavey, Ben Collins, Chris Hackley and Zhonghua Zhu for comments on the manuscript and Erin Meekhof for making the graphical abstract. We also thank Herman Wijnen for communicating unpublished data. This investigation was conducted in facilities constructed with support from Research Facilities Improvement Grant Number C06 RR-15518-01 from the NCRR, NIH. Imaging was performed at NYU’s Center for Genomics & Systems Biology. AP was partly supported by NYU’s GSAS MacCracken Program and a Dean’s Dissertation Fellowship. TPS was partly supported by an NYU Courant Institute postdoctoral fellowship. This work was supported by NIH grants GM063911 and NS077156 to JB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amino T, Ishikawa K, Toru S, Ishiguro T, Sato N, Tsunemi T, Murata M, Kobayashi K, Inazawa J, Toda T, et al. Redefining the disease locus of 16q22.1-linked autosomal dominant cerebellar ataxia. J Hum Genet. 2007;52:643–649. doi: 10.1007/s10038-007-0154-1. [DOI] [PubMed] [Google Scholar]
- Becquet D, Girardet C, Guillaumond F, Francois-Bellan AM, Bosler O. Ultrastructural plasticity in the rat suprachiasmatic nucleus. Possible involvement in clock entrainment. Glia. 2008;56:294–305. doi: 10.1002/glia.20613. [DOI] [PubMed] [Google Scholar]
- Billuart P, Winter CG, Maresh A, Zhao X, Luo L. Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell. 2001;107:195–207. doi: 10.1016/s0092-8674(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CircaDB Circadian expression profiles Data Base. http://bioinf.itmat.upenn.edu/circa.
- Collins B, Kane EA, Reeves DC, Akabas MH, Blau J. Balance of activity between LNvs and glutamatergic dorsal clock neurons promotes robust circadian rhythms in Drosophila. Neuron. 2012;74:706–718. doi: 10.1016/j.neuron.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B, Kaplan HS, Cavey M, Lelito KR, Bahle AH, Zhu Z, Macara AM, Roman G, Shafer OT, Blau J. Differentially timed extracellular signals synchronize pacemaker neuron clocks. PLOS Biology. 2014;12:e1001959. doi: 10.1371/journal.pbio.1001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MP, Berni J, Ceriani MF. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLOS Biology. 2008;6:e69. doi: 10.1371/journal.pbio.0060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A, Esnault C, Aubert G, Treisman R, Pralong F, Schibler U. Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell. 2013;152:492–503. doi: 10.1016/j.cell.2012.12.027. [DOI] [PubMed] [Google Scholar]
- Girardet C, Blanchard MP, Ferracci G, Leveque C, Moreno M, Francois-Bellan AM, Becquet D, Bosler O. Daily changes in synaptic innervation of VIP neurons in the rat suprachiasmatic nucleus: contribution of glutamatergic afferents. Eur J Neurosci. 2010;31:359–370. doi: 10.1111/j.1460-9568.2009.07071.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Billault C, Munoz-Llancao P, Henriquez DR, Wojnacki J, Conde C, Caceres A. The role of small GTPases in neuronal morphogenesis and polarity. Cytoskeleton. 2012;69:464–485. doi: 10.1002/cm.21034. [DOI] [PubMed] [Google Scholar]
- Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pirez N, Ceriani MF. Circadian pacemaker neurons change synaptic contacts across the day. Current Biology. 2014;24:2161–2167. doi: 10.1016/j.cub.2014.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife. 2014;3:e02780. doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Kamynina E, Morley S, Chung S, Muakkassa N, Wang H, Brathwaite S, Sharma G, Manor D. Plekhg4 is a novel Dbl family guanine nucleotide exchange factor protein for Rho family GTPases. J Biol Chem. 2013;288:14522–14530. doi: 10.1074/jbc.M112.430371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- Hartshorne DJ, Ito M, Erdodi F. Myosin light chain phosphatase: subunit composition, interactions and regulation. J Muscle Res Cell Motil. 1998;19:325–341. doi: 10.1023/a:1005385302064. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Toru S, Tsunemi T, Li M, Kobayashi K, Yokota T, Amino T, Owada K, Fujigasaki H, Sakamoto M, et al. An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5′ untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am J Hum Genetics. 2005;77:280–296. doi: 10.1086/432518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Fukata Y, Oshiro N, Amano M, Nakamura T, Ito M, Matsumura F, Inagaki M, Kaibuchi K. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J Cell Biol. 1999;147:1023–1038. doi: 10.1083/jcb.147.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. PNAS. 2010;107:13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Treisman JE. Excessive Myosin activity in Mbs mutants causes photoreceptor movement out of the Drosophila eye disc epithelium. Mol Biol Cell. 2004;15:3285–3295. doi: 10.1091/mbc.E04-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A. Experience-dependent structural plasticity in the adult human brain. Trends Cogn Sci. 2011;15:475–482. doi: 10.1016/j.tics.2011.08.002. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Meredith AL, Wiler SW, Miller BH, Takahashi JS, Fodor AA, Ruby NF, Aldrich RW. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nature Neuroscience. 2006;9:1041–1049. doi: 10.1038/nn1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrak D, Ruben M, Myers GN, Rhrissorrakrai K, Gunsalus KC, Blau J. Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons. Current Biology. 2012;22:1871–1880. doi: 10.1016/j.cub.2012.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. PNAS. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. PNAS. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Ruben M, Drapeau MD, Mizrak D, Blau J. A mechanism for circadian control of pacemaker neuron excitability. J Biol Rhythms. 2012;27:353–364. doi: 10.1177/0748730412455918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes & Development. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Simoes S, Denholm B, Azevedo D, Sotillos S, Martin P, Skaer H, Hombria JC, Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–4267. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- Sivachenko A, Li Y, Abruzzi KC, Rosbash M. The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron. 2013;79:281–292. doi: 10.1016/j.neuron.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernandez MP, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes & Development. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Smith WC. Whole-organ cell shape analysis reveals the developmental basis of ascidian notochord taper. Developmental Biology. 2013;373:281–289. doi: 10.1016/j.ydbio.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuyama K, Meinertzhagen IA. Synaptic connections of PDF-immunoreactive lateral neurons projecting to the dorsal protocerebrum of Drosophila melanogaster. J Comp Neurol. 2010;518:292–304. doi: 10.1002/cne.22210. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wulbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Forster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Xiang Y, Yan Z, Han C, Jan LY, Jan YN. Light-induced structural and functional plasticity in Drosophila larval visual system. Science. 2011;333:1458–1462. doi: 10.1126/science.1207121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelinski EL, Deibel SH, McDonald RJ. The trouble with circadian clock dysfunction: Multiple deleterious effects on the brain and body. Neurosci Biobehav Rev. 2014;40C:80–101. doi: 10.1016/j.neubiorev.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner RA, Hardin PE, Allada R. DN1p circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Current Biology. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.