Abstract
There is a need to develop a colorectal cancer (CRC) screening test that is noninvasive, cost effective, and sensitive enough to detect preneoplastic lesions. This case-control study examined the feasibility of using circulating extracellular miRNAs to differentiate a spectrum of colorectal neoplasia of various severity and hence for early detection of colorectal neoplasia.
Archived serum samples of 10 normal controls and 31 cases, including 10 with non-advanced adenoma, 10 with advanced adenoma, and 11 with CRC, were profiled for circulating miRNAs using next-generation sequencing. Multiple linear regression, adjusting for age, gender, and smoking status, compared controls and the three case groups for levels of 175 miRNAs that met stringent criteria for miRNA sequencing analysis. Of the 175 miRNAs, 106 miRNAs were down-regulated according to severity of neoplasia and showed a relative decrease in expression from controls to non-advanced adenoma to advanced adenoma to CRC (Ptrend < 0.05). Pair-wise group comparisons showed that 39 and 80 miRNAs were differentially expressed in the advanced adenoma and CRC groups, as compared to the controls, respectively. Differences in miRNA levels between the non-advanced adenoma group and controls were modest.
Our study found that expression of many miRNAs in serum was inversely correlated with the severity of colorectal neoplasia, and differential miRNA profiles were apparent in preneoplastic cases with advanced lesions, suggesting circulating miRNAs could serve as potential biomarkers for CRC screening.
INTRODUCTION
Screening for colorectal cancer (CRC) by fecal occult blood test (FOBT), fecal immunochemical test (FIT), flexible sigmoidoscopy (FSG), and colonoscopy can lower the incidence and mortality of colorectal cancer.1-5 However, these recommended screening methods have shortcomings; FOBT and FIT have a low sensitivity (~40%) for detecting preneoplastic lesions,4,5 while FSG and colonoscopy are limited by their high costs, the requirement of bowel preparation, and their invasive nature.5,6 Therefore, compliance remains an issue for CRC screening. In 2012, only 65% of adults aged 50-75 years were up-to-date with the U.S. Preventive Services Task Force recommendations for CRC screening.7 There is a pressing need to develop a screening test that is sensitive, noninvasive, and cost effective to detect biomarkers for both CRC and advanced preneoplastic lesions.
MiRNAs, small non-coding RNAs of ~22 nucleotides in length, mediate gene silencing by binding to specific mRNA targets and repressing translation of mRNA through degradation and/or translational inhibition.8,9 As such, they have profound effects on regulating gene expression, including genes that are involved in tumorigenesis.10 Extracellular miRNAs have the potential to serve as cancer biomarkers because they are derived from cells, show tissue specific expression,11,12 and present in peripheral blood in a remarkably stable form.13-15 In addition, tumors can be a source of circulating miRNAs.13,16-18
Because of these properties, a noninvasive blood test for assessment of extracellular miRNA markers in circulation could potentially be useful as a screening test for CRC. This is supported by observations that differential miRNA expression profiles were found between colorectal tumor and normal tissues,19-22 that miRNAs dysregulated in tumor tissues also had aberrant levels in plasma of the same colorectal cancer patients,22,23 and that circulating levels of specific miRNAs decreased after tumor resection.22,23 Epidemiological studies have also shown differential circulating miRNA profiles between normal controls and individuals with either advanced adenoma or CRC.22-33
However, none of the previous epidemiological studies have examined whether circulating miRNA concentrations vary across the spectrum of neoplasia (i.e., from normal to non-advanced adenoma to advanced adenoma to CRC). If so, a screening test for miRNA markers may be feasible for prevention and early detection of CRC. This is the first case-control study of colorectal neoplasia to comprehensively profile circulating miRNAs in serum using next-generation sequencing and to correlate miRNA levels with severity of colorectal neoplasia.
MATERIAL AND METHODS
Study population
Thirty-one cases with various degree of colorectal neoplasia and 10 controls were identified from the Digestive Diseases Tissue Resource at the University of Pittsburgh. This program routinely collects and archives tissue and blood samples from individuals who participated in colonoscopy screening as well as from those who were diagnosed with CRC in doctors' offices. With informed consent, blood samples were collected before colonoscopy from screening participants or before treatment for colorectal cancer cases, processed within 1-2 hours of collection, and serum samples were stored at -80°C. The 31 cases in this study included 10 with non-advanced adenoma, 10 with advanced adenoma, and 11 colorectal cancer patients. Of the cancer patients, all had adenocarcinoma, 5 had stage I/II cancer, and 6 had stage III/IV cancer. The 10 controls were individuals who were screen negative by colonoscopy.
In addition to the 41 samples from cases and controls, duplicate aliquots from the same blood sample from six subjects were included to assess reproducibility of miRNA sequencing. All 47 samples were processed blindly without knowledge of the case-control status or identity of the duplicate samples. The study protocol was approved by the institutional review board of the two institutes involved in this study; it was carried out according to The Code of Ethics of the World Medical Association (Declaration of Helsinki), and informed consent was obtained.
Lab procedures
Total RNA was purified from 300 μL of serum using the miRNeasy serum/plasma kit (Qiagen). The median total RNA yield was 32 ng/10 L (range:15 - 61). From this, 10 ng of total RNA was used for small RNA cDNA library preparation. Quality of the library constructs was examined using High Sensitivity DNA Chip Kit (Agilent Technologies) for number of peaks, size distribution, and concentration of fragments. All samples showed a library peak around 145 bp, which corresponds to the 5’-3’ adapter ligated miRNA constructs derived from the 20-30 nucleotides small RNAs.
MiRNA sequencing was done by Illumina Hi-Seq2000. To analyze sequencing data, we developed an automated analytical pipeline. Briefly, data provided from the HiSeq2000 sequencer in a standard fastq format were trimmed of adapter sequences and low quality reads (more than 3 low quality base-calls) through a C++ program.34 These sequences were then collapsed to remove redundancy using the Galaxy Genome Browser tool fastx,35 followed by alignment to the known human miRNA/small RNA database, miRBase (http://www.mirbase.org/), release 19 (August 2012).36 Before statistical analysis, miRNA read counts were normalized using the reads per million (rpm) method, which divided read count of a given miRNA by the total number of miRNA reads in that sample and multiplied by a million.37 Following normalization, we applied stringent criteria for a miRNA to be considered for statistical analysis, namely to be present in at least 50% of study samples in greater than 10 copy numbers. MiRNAs with less than 10 reads were not included due to the error rate of Illumina sequencing and stochastic variation in gene expression.38 Of the 822 unique miRNAs that were detected, 175 were retained for data analysis. The top two miRNAs that showed differential expression between cases and controls were verified by qRT-PCR.
Statistical analysis
We first assessed reproducibility of miRNA sequencing. Six individuals were sequenced twice. For each person, we calculated Spearman rank correlation coefficient between the duplicate values of 175 miRNAs. For these six subjects, one of the duplicate samples was randomly selected for further statistical analysis described below.
The effects of serum storage on miRNA sequencing were evaluated by Spearman rank correlations between serum storage duration (months between blood draw and RNA extraction) and total miRNA reads in 41 samples. In addition, for each of the175 miRNAs, we examined the Spearman rank correlation between its read count and serum storage time.
The four study groups (colorectal cancer, advanced adenoma, non-advanced adenoma, and controls) were first examined for differences in the established risk factors for colorectal neoplasia, namely age, gender, race/ethnicity, smoking status, and body mass index (BMI), by Mantel-Haenszel chi-square test for trend for categorical variables and by Kruskal-Wallis test for continuous variables. We then compared values of each miRNA among the four study groups using multiple linear regression adjusting for covariates significantly associated with colorectal neoplasia in this study population, including age, gender, and ever smoking. The outcome variable was the log2 transformed normalized miRNA read counts. The regression model generated three β coefficients; each represented the difference in mean miRNA values between a case group and the controls. As read counts were log2 transformed, the β coefficient was equivalent to the log ratio of geometric mean miRNA values of a case group versus that of the control group (i.e., log2 of fold-change). P values for global tests (evaluating significance of all 3 βs simultaneously) and 2-group comparisons (evaluating significance of each β) were obtained.
P values for trend tests were generated by coding and analyzing the four study groups in one linear variable. P values were adjusted for multiple comparisons. An arbitrary cutoff of 0.05 for false discovery rate (FDR) according to the Benjamini and Hochberg procedure was applied to determine statistical significance.39
RESULTS
Reproducibility of miRNA sequencing
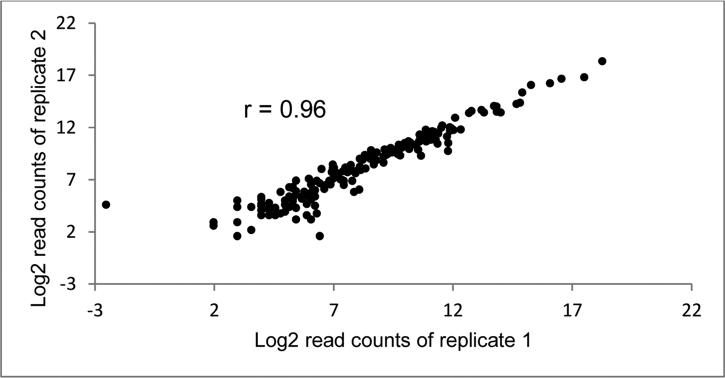
Duplicate aliquots of the same serum sample from six subjects were processed and sequenced twice to assess reproducibility of miRNA sequencing. The correlations between duplicate values of 175 miRNAs in these six individuals ranged from 0.93 to 0.99 (median = 0.97), indicating excellent reproducibility of miRNA sequencing. Figure 1 shows the duplicate miRNA read counts of one subject.
Figure 1.
Replicate read counts of 175 miRNAs in one sample
Effects of serum storage on miRNA levels
Median storage duration of the 41 serum samples was 29.4 months (range 11.1 – 90.2). There was little correlation between serum storage duration and total miRNA reads from sequencing (r = -0.06). Of the175 miRNAs, 166 (95%) of them had a weak to modest correlation (≤ 0.3) between their read counts and storage duration.
Differential miRNA expression according to severity of neoplasia
Median age of the 41 subjects was 61 years old (range 28 - 91), 49% were males, and 95% were Caucasians; 22 (54%) subjects ever smoked cigarettes, but only 4 (10%) were current smokers. Cases, particularly those with advanced adenoma or CRC, tended to be older and were more likely to be a male and an ever-smoker than controls (Table 1).
Table 1.
Characteristics of cases with colorectal neoplasia and controls
| Controls (n=10) | Non-advanced adenoma (n=10) | Advanced adenoma (n=10) | Colorectal cancer (n=11) | P value | |
|---|---|---|---|---|---|
| Median age (IQR)* | 54.5 (52-65) | 59.5 (57-72) | 57.5 (54 – 79) | 68.0 (61 – 87) | 0.093 |
| Median BMI (IQR)* | 30.7 (25.8-31.2) | 29.3 (26.6 – 34.3) | 31.0 (23.8 – 36.5) | 31.5 (27.0 – 37.1) | 0.851 |
| Male, n (%) | 4 (40) | 2 (20) | 6 (60) | 8 (73) | 0.047 |
| Ever smoked, n (%) | 3 (30) | 3 (30) | 8 (80) | 8 (73) | 0.012 |
IQR = Inter-quartile range.
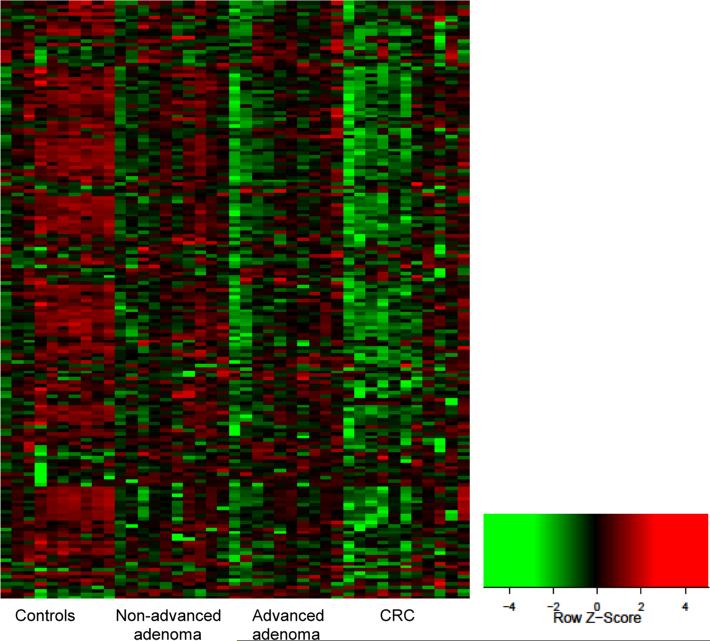
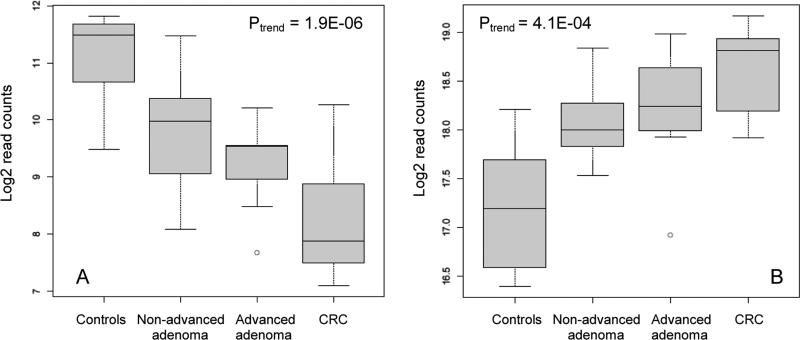
The geometric mean counts of 175 miRNAs in the cases and controls, their fold changes adjusted for age, gender, and smoking status, and FDR adjusted p values are shown in Supplemental Table 1. Heatmap in Figure 2 shows differential expression of miRNAs in the four study groups. Levels of 109 miRNAs were significantly correlated with the severity of colorectal neoplasia, as indicated by their p values for trend test (Supplemental Table 1). Of these, levels of 106 miRNAs were inversely associated with the extent of neoplasia and showed a relative decrease in miRNA levels from the controls to non-advanced adenoma to advanced adenoma to the CRC groups. As an example, Figure 3A shows boxplot of miRNA-30b (Ptrend = 1.9×10−6), which was the most significant among the 106 miRNAs that were down-regulated in the cases. Three miRNAs were up-regulated in the cases with levels increasing with the severity of neoplasia (miRNA-486, miRNA-25, and miRNA-1180). Boxplot of miRNA-486 (Ptrend = 4.1×10−4), the most significant of the three, is shown in Figure 3B.
Figure 2.
Heatmap of expression of 175 miRNAs (rows) in 41 serum samples (columns) Red=High relative expression. Green= Low relative expression
Figure 3.
Boxplots of miRNA-30b (A) and miRNA-486 (B) read counts in the study groups
Differential miRNA expression in pair-wise group comparisons
Two-group comparisons showed that the smallest differences in miRNA levels were between the non-advanced adenoma group and controls, and none of the miRNAs were significantly different between groups. On the other hand, 39 miRNAs were differentially expressed between the advanced adenoma and control groups, and 80 miRNAs were differentially expressed between the CRC and control groups (Supplemental Table 1). As examples, Table 2 shows the top 2 miRNAs (miRNA-30b and miRNA-30c) that were the most significantly different between the CRC and control groups and the top 2 miRNAs (miRNA-146a and miRNA-30d) that were differentially expressed in the advanced adenoma group. Expression levels of miRNA-30b and miRNA-146a were verified by qRT-PCR. The Spearman rank correlations between qRT-PCR and Illumina sequencing expression levels were 0.61 for miRNA-30b and 0.79 for miRNA-146a; qRT-PCR expression patterns in the study groups were consistent with the Illumina sequencing data (Supplemental Figure 1).
Table 2.
The top miRNAs differentially expressed in cases with advanced adenoma or CRC compared to controls
| Geometric mean count | Fold change | FDR adjusted P value | |||||
|---|---|---|---|---|---|---|---|
| miRNA | Controls | Advanced adenoma | CRC | Advanced adenoma vs controls | CRC vs controls | Advanced adenoma vs controls | CRC vs controls |
| 30b | 2309 | 616 | 288 | 0.34 | 0.17 | 0.028 | 0.4 × 10−3* |
| 30c | 4241 | 1074 | 540 | 0.30 | 0.14 | 0.034 | 1.9 × 10−3* |
| 146a | 6442 | 519 | 973 | 0.09 | 0.13 | 0.7 × 10−3† | 4.9 × 10−3 |
| 30d | 30130 | 12646 | 13104 | 0.41 | 0.39 | 1.7 × 10−2† | 3.1 × 10−3 |
MiRNA-30b and miRNA-30c had the lowest FDR adjusted p values when the CRC group was compared to the controls.
MiRNA-146a and miRNA-30d had the lowest FDR adjusted p values when the advanced adenoma group was compared to the controls.
We also repeated the analyses excluding 6 CRC cases with advanced stage III/IV disease to explore if serum miRNAs had the potential to detect early cancers. The results were similar (data not shown). For example, miRNA-30b and miRNA-30c were still among the top miRNAs with significant differential levels between stage I/II CRC cases and controls (FDR adjusted p values were .004 and .007, respectively).
DISCUSSION
In this case-control study of colorectal neoplasia, we examined the feasibility of sequencing circulating extracellular miRNAs in archived serum samples and demonstrated that next-generation sequencing of miRNA had excellent reproducibility, and that archived serum samples were suitable for this technology. Serum storage duration did not affect quantity of total miRNA or levels of individual miRNAs in serum. Previous studies have also found circulating miRNAs to be present in a remarkably stable form that is resilient to freeze-thawing (as many as 10 cycles), RNase degradation, extended storage, and extreme temperature and pH.13-15 These observations suggest that archived serum samples from existing cohort and case-control studies could provide valuable resources for efficient evaluation of circulating miRNAs as cancer biomarkers.
We also assessed the feasibility of using miRNAs, profiled by sequencing, to distinguish normal controls from cases with various degree of colorectal neoplasia. We found levels of many miRNAs to be inversely correlated with the extent of neoplasia. About 12 case-control studies in the literature compared circulating miRNAs between normal controls and cases with either CRC or advanced adenoma.22-33 Most studies found up-regulation of miRNAs in cases, although specific miRNAs were rarely replicated except for miRNA-21 22,31,33 and miRNA-29a 25,29,31 that were found to be over-expressed in CRC cases in three studies. Our results suggesting that serum concentrations of many miRNAs, including miRNA-21 and miRNA-29a, were down-regulated in the cases were inconsistent with those from previous studies. Nevertheless, down-regulation of miRNA expression globally in tumors as compared to normal tissues had been reported previously.12
This discrepancy could be due to the differences in the miRNA detection platform and type of blood sample. While other studies of colorectal neoplasia used qRT-PCR for a few specific miRNAs or microarrays, our study was the first to use miRNA next-generation sequencing. Moreover, almost all previous studies used plasma samples, but we used serum. It has been shown that for some miRNAs, their levels in plasma are different from those in serum.40,41 MiRNA levels in plasma can be affected by centrifugation conditions, hence the amount of residual platelets and cellular debris that contribute miRNAs, and also by certain anticoagulants (e.g., citrate or heparin) that inhibit qRT-PCR.9,40,42,43 Nevertheless, our results were unlikely to be attributed to systematic biases. This is because, in our study, miRNAs were not only down-regulated in cases but also decreased with the severity of colorectal neoplasia in a linear trend; blood samples of the adenoma cases and controls were collected before colonoscopy when neoplastic outcome was not known, and all serum samples were processed and sequenced without knowledge of the case-control status.
Our finding that many miRNAs had significantly lower circulating levels in the cases than controls raises a practical concern of whether down-regulated miRNAs could be utilized as cancer biomarkers in a clinical setting, if their low levels are difficult to be detected by qRT-PCR, a technology that is commonly available in clinical diagnostic laboratories, but less sensitive than next-generation sequencing. In our study, the top 2 miRNAs, namely miRNA-30b and miRNA-146a, that were down-regulated in CRC and advanced adenoma cases, respectively, were verified by qRT-PCR, demonstrating in principle that qRT-PCR could amplify and detect cancer biomarkers that circulate in a below normal level.
In this study, several members in the miRNA-30 family (miRNA-30b, -30c, and -30d) were among the top miRNAs down-regulated in cases with either advanced adenoma or CRC. It is intriguing that low expression of the miRNA-30 family members have been reported in breast, lung, and colorectal cancers.44-48 MiRNA-30b, the top miRNA down-regulated in CRC cases in our study, may function as a tumor suppressor – it had lower expression in CRC tissue than normal tissues, suppressed growth of CRC cell lines, and repressed expression of KRAS, PIK3CD, and BCL2 genes.47 MiRNA-30d was one of the top miRNAs down-regulated in cases with advanced adenoma in our study. MiRNA-30d targets GRP78, a protein that is commonly over-expressed in cancers as a major endoplasmic reticulum chaperone and signaling regulator, and has also been found to be down-regulated in colon tumors and a colon cancer cell line.48
MiRNA-146a was another top miRNA that was significantly down-regulated in cases with advanced adenoma. It is involved in innate immunity by inhibiting Toll-like receptor signaling and regulating cytokine responses. MiRNA-146a may function as a key negative regulator of inflammation 49-51 and could be involved in the pathogenesis of colorectal neoplasia, an inflammation-associated disease.52-54 Of the miRNAs that were up-regulated among cases with advanced adenoma or CRC, MiRNA-486 was the most significant. Its role in carcinogenesis, however, is not quite clear. It was reported to suppress NF-κB-negative regulators, resulting in sustained NF-κB activity, in one study,55 but shown to inhibit tumor growth in others.56,57 Nevertheless, one study found up-regulation of miRNA-486 in colorectal cancer tissue to be associated with KRAS mutation.58
With a small sample size, this study was not designed to identify and validate diagnostic or early detection miRNA markers for colorectal neoplasia, but rather a proof of concept to demonstrate differential expression of circulating miRNAs according to severity of disease using next-generation sequencing, which, to the best of our knowledge, has not been studied previously. To further pursue the potential of measuring circulating miRNAs for CRC screening, future studies need to focus on several areas that are neglected by existing studies so far: (1) to discover differentially expressed circulating miRNA markers in cases targeted by screening, specifically a large sample size of individuals with early stage I-II CRC or advanced adenoma; (2) to identify and validate promising miRNA markers while adjusting for possible confounding factors, such as presence of inflammatory bowel disease or other inflammatory conditions, that could affect miRNA expression in study groups; (3) to evaluate whether serum levels of promising markers correlate with those in tissue and are specific for colorectal neoplasia; (4) to examine variation and latency of these promising miRNAs over time in serial blood samples; and (5) to assess the intended clinical utility of qRT-PCR in detecting promising miRNA markers in a target population for CRC screening. Given that most significant markers were down-regulated in individuals with disease in our study, it would be crucial for future studies to pinpoint the markers that, despite their levels in diseased individuals are below normal, can be detected reliably with high sensitivity by a clinical diagnostic technology.
In summary, in this study of circulating miRNAs in colorectal neoplasia, we found that levels of circulating miRNAs correlated with severity of colorectal neoplasia. Moreover, cases with neoplastic lesions, as early as advanced adenoma and stage I/II CRC, had differential miRNA profiles compared to normal controls. These results, if confirmed and validated, suggest that miRNAs have the potential to be screening biomarkers for prevention and early detection of CRC.
Supplementary Material
Translational Significance.
Our study found that levels of circulating miRNAs, determined by sequencing, correlated with severity of colorectal neoplasia. Cases with neoplastic lesions, as early as advanced adenoma and stage I/II cancer, had differential miRNA profiles compared to controls, suggesting that miRNAs have the potential to be screening biomarkers for prevention and early detection of colorectal cancer.
ACKNOWLEDGEMENTS
Research reported in this publication was supported in part by (1) the Albert Einstein Cancer Center of Yeshiva University, (2) the National Center for Research Resources (NCRR) and the National Center for Advancing Translational Sciences (NCATS), components of the National Institutes of Health (NIH), through CTSA grant numbers UL1RR025750, TL1RR025748, KL2RR025749, UL1TR000086, TL1TR000087, and KL2TR000088, (3) U01CA152753 from the Early Detection Research Network of the National Cancer Institute (awarded to RES), (4) NIH grants HD068546 and CA179564 (awarded to ZW), and (5) NIH grants CA180126 and AG017242 (awarded to YS).
Abbreviations
- BMI
body mass index
- CRC
colorectal cancer
- FDR
false discovery rate
- FIT
fecal immunochemical test
- FOBT
fecal occult blood test
- FSG
flexible sigmoidoscopy
- miRNA
micro-RNA
- RT
reverse transcription
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors have any financial or personal relationship with organizations that could potentially be perceived as influencing the described research, and all authors have read the journal's policy on disclosure of potential conflicts of interest. All authors have read the journal's authorship agreement and that the manuscript has been reviewed by and approved by all named authors.
REFERENCES
- 1.Atkin WS, Edwards R, Kralj-Hans I, et al. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. 2010;375:1624–33. doi: 10.1016/S0140-6736(10)60551-X. [DOI] [PubMed] [Google Scholar]
- 2.Schoen RE, Pinsky PF, Weissfeld JL, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med. 2012;366:2345–57. doi: 10.1056/NEJMoa1114635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandel JS, Church TR, Bond JH, et al. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000;343:1603–7. doi: 10.1056/NEJM200011303432203. [DOI] [PubMed] [Google Scholar]
- 4.Whitlock EP, Lin J, Liles E, et al. Screening for colorectal cancer: A targeted, updated systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2008;149:638–58. doi: 10.7326/0003-4819-149-9-200811040-00245. [DOI] [PubMed] [Google Scholar]
- 5.Bretthauer M. Colorectal cancer screening. J Intern Med. 2011;270:87–98. doi: 10.1111/j.1365-2796.2011.02399.x. [DOI] [PubMed] [Google Scholar]
- 6.Miller S, Steele S. Novel molecular screening approaches in colorectal cancer. J Surg Oncol. 2012;105:459–67. doi: 10.1002/jso.21704. [DOI] [PubMed] [Google Scholar]
- 7.CDC Vital signs: colorectal cancer screening test use--United States, 2012. MMWR Morb Mortal Wkly Rep. 2013;62:881–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 9.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–69. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 16.Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–48. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 18.LaConti JJ, Shivapurkar N, Preet A, et al. Tissue and serum microRNAs in the Kras(G12D) transgenic animal model and in patients with pancreatic cancer. PLoS ONE. 2011;6:e20687. doi: 10.1371/journal.pone.0020687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20:1272–86. doi: 10.1158/1055-9965.EPI-11-0035. [DOI] [PubMed] [Google Scholar]
- 20.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121:141–58. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 21.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–51. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 22.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a Diagnostic and Prognostic Biomarker in Colorectal Cancer. J Natl Cancer Inst. 2013;105:849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 24.Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674–80. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 25.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 26.Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Huang Z, Ni S, et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS ONE. 2012;7:e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faltejskova P, Bocanek O, Sachlova M, et al. Circulating miR-17-3p, miR-29a, miR-92a and miR-135b in serum: Evidence against their usage as biomarkers in colorectal cancer. Cancer Biomark. 2012;12:199–204. doi: 10.3233/CBM-130308. [DOI] [PubMed] [Google Scholar]
- 29.Giraldez MD, Lozano JJ, Ramirez G, et al. Circulating MicroRNAs as Biomarkers of Colorectal Cancer: Results From a Genome-Wide Profiling and Validation Study. Clin Gastroenterol Hepatol. 2013;11:681–8. e3. doi: 10.1016/j.cgh.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Xiang J, Li Z, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2013;136:152–61. doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 31.Luo X, Stock C, Burwinkel B, Brenner H. Identification and Evaluation of Plasma MicroRNAs for Early Detection of Colorectal Cancer. PLoS ONE. 2013;8:e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanaan Z, Roberts H, Eichenberger MR, et al. A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258:400–8. doi: 10.1097/SLA.0b013e3182a15bcc. [DOI] [PubMed] [Google Scholar]
- 33.Zanutto S, Pizzamiglio S, Ghilotti M, et al. Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer. 2014;110:1001–7. doi: 10.1038/bjc.2013.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–71. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blankenberg D, Von Kuster G, Coraor N, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;89:19.0.1–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedlander MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 37.Motameny S, Wolters S, Nurnberg P, Schumacher B. Next generation sequencing of miRNAs - strategies, resources and methods. Genes. 2010;1:70–84. doi: 10.3390/genes1010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10:490–7. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:238–300. [Google Scholar]
- 40.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833–40. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 41.Yamada A, Cox MA, Gaffney KA, et al. Technical factors involved in the measurement of circulating microRNA biomarkers for the detection of colorectal neoplasia. PLoS ONE. 2014;9:e112481. doi: 10.1371/journal.pone.0112481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moldovan L, Batte KE, Trgovcich J, et al. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014;18:371–90. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng HH, Yi HS, Kim Y, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS ONE. 2013;8:e64795. doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo J, Zhao Q, Zhang W, et al. A novel panel of microRNAs provides a sensitive and specific tool for the diagnosis of breast cancer. Mol Med Rep. 2014;10:785–91. doi: 10.3892/mmr.2014.2274. [DOI] [PubMed] [Google Scholar]
- 45.Ouzounova M, Vuong T, Ancey PB, et al. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics. 2013;14:139. doi: 10.1186/1471-2164-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong Z, Xia Y, Wang P, Liu B, Chen Y. Low expression of microRNA-30c promotes invasion by inducing epithelial mesenchymal transition in non-small cell lung cancer. Mol Med Rep. 2014;10:2575–9. doi: 10.3892/mmr.2014.2494. [DOI] [PubMed] [Google Scholar]
- 47.Liao WT, Ye YP, Zhang NJ, et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232:415–27. doi: 10.1002/path.4309. [DOI] [PubMed] [Google Scholar]
- 48.Su SF, Chang YW, Andreu-Vieyra C, et al. miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene. 2013;32:4694–701. doi: 10.1038/onc.2012.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheedy FJ, O'Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis. 2008;67(Suppl 3):iii50–5. doi: 10.1136/ard.2008.100289. [DOI] [PubMed] [Google Scholar]
- 50.Labbaye C, Testa U. The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J Hematol Oncol. 2012;5:13. doi: 10.1186/1756-8722-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–40. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Pohl C, Hombach A, Kruis W. Chronic inflammatory bowel disease and cancer. Hepatogastroenterol. 2000;47:57–70. [PubMed] [Google Scholar]
- 53.Tsilidis KK, Branchini C, Guallar E, et al. C-reactive protein and colorectal cancer risk: a systematic review of prospective studies. Int J Cancer. 2008;123:1133–40. doi: 10.1002/ijc.23606. [DOI] [PubMed] [Google Scholar]
- 54.Cole BF, Logan RF, Halabi S, et al. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. J Natl Cancer Inst. 2009;101:256–66. doi: 10.1093/jnci/djn485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song L, Lin C, Gong H, et al. miR-486 sustains NF-kappaB activity by disrupting multiple NF-kappaB-negative feedback loops. Cell Res. 2013;23:274–89. doi: 10.1038/cr.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang X, Hou J, Shen X, et al. MicroRNA-486-5p, which is downregulated in hepatocellular carcinoma, suppresses tumor growth by targeting PIK3R1. FEBS J. 2015;282:579–94. doi: 10.1111/febs.13167. [DOI] [PubMed] [Google Scholar]
- 57.Zhang G, Liu Z, Cui G, Wang X, Yang Z. MicroRNA-486-5p targeting PIM-1 suppresses cell proliferation in breast cancer cells. Tumour Biol. 2014;35:11137–45. doi: 10.1007/s13277-014-2412-0. [DOI] [PubMed] [Google Scholar]
- 58.Mosakhani N, Sarhadi VK, Borze I, et al. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51:1–9. doi: 10.1002/gcc.20925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.