Abstract
Background
Dermatophytes are a scientific label for a group of three genera (Microsporum, Epidermophyton and Trichophyton) of fungus that causes skin disease in animals and humans. Conventional methods for identification of these fungi are rapid and simple but are not accurate comparing to molecular methods.
Objective
This study aimed to isolate human pathogenic dermatophytes which cause dermatophytosis in Riyadh City, Saudi Arabia and to identify these fungi by using conventional and molecular methods.
Methods
The study was conducted in Medical Complex, Riyadh and King Saud University. Samples of infected skin, hairs and nails were collected from 112 patients. Diagnosis of skin infections, direct microscopic test, isolation and identification of dermatophytes by conventional and molecular methods were carried out.
Results
The results indicated that the tinea capitis infection had the highest prevalence among the patients (22.3%) while Tinea barbae had the lowest. In this study the identified dermatophyte isolates belong to nine species as Trichophyton violaceum, Trichophyton verrucosum, Trichophyton rubrum, Trichophyton mentagrophytes, Trichophyton schoenleinii, Trichophyton concentricum, Microsporum canis, Microsporum audouinii and Epidermophyton floccosum which cause skin infections were isolated during this study. Non dermatophyte isolates included 5 isolates from Aspergillus spp. 4 isolates from Acremonium potronii and 15 isolates from Candida spp. M. canis were the most common species (25% of isolated dermatophytes). Out of the 52 dermatophyte isolates identified by conventional methods, there were 45 isolates identified by the molecular method.
Conclusions
The results concluded that approximately M. canis caused a quarter of dermatophyte cases, tinea capitis infection was prevalent and the molecular method was more accurate than conventional methods.
Keywords: Dermatophytes, Microsporum, Epidermophyton, Trichophyton, Tinea
1. Introduction
The dermatophytes are a group of fungi that are able to damage and utilize keratin found in the skin, hair and nails. They are classified into three genera (Microsporum, Trichophyton and Epidermophyton) based on the shape of macroconidia. Dermatophytosis is an infection produced by dermatophytic fungi in the keratinized tissues (Grumbt et al., 2013). Since 1978, it was decided that the dermatophytes contracted half way across the world may become manifest in a country in which the pathogen is not normally found because of the rapid transit, and the increasing mobility of people (Aho, 1988). The dermatophytosis transfers to man from animal (zoophilic dermatophytes) and from soil (geophilic dermatophytes) or through direct infection by personal contact (Aho, 1988; Philpot, 1978).
The infectious phase includes fungal hyphae and arthrospores that have a latency phase that reaches to 24 months inside loose vacuoles keratinocytes of host (Richardson, 1990). The phenotypic characteristics of dermatophyte fungi are changed by many environmental, nutritional and chemical factors, for this reason researchers prefer the molecular methods, genotypic characteristics, to identify the dermatophytes, also the molecular methods are fast and more specific (Faggi et al., 2001; Liu et al., 1997, 2000b). Direct microscopic examination, isolation, cultural features and physiological characteristics are useful to identify the genus and species of dermatophytes but these conventional methods require time and effort (Singh and Beena, 2003). Molecular methods such as random amplified polymorphic DNA (RAPD) or arbitrarily primed PCR (AR-PCR) and Specific nucleotide sequence are used to identify species and sub-species of dermatophytes (Faggi et al., 2001). Recently, multiplex PCR method has been developed to detect dermatophytes in onychomycosis based on chitin synthase and internal transcribed spacer genes (Dhib et al., 2014).
Many studies were carried out in Saudi Arabia to isolate and identify some dermatophytes by conventional methods. Some of those studies found that the onychomycosis (40.3%) was more prevalent than tinea capitis (21.9%) followed by tinea pedis (16%), tinea cruris (15.1%) and tinea corporis (6.7%). In the Riyadh region, Trichophyton mentagrophytes and Microsporum canis were the most common dermatophytes whereas M. canis, Trichophyton rubrum and T. mentagrophytes were the most prevalent in the Eastern Province (Abanmi et al., 2008; Al-Sogair et al., 1991; Al Sogair and Hay, 2000). The aim of this research was to isolate and identify dermatophytes from patients in Riyadh City by a conventional and molecular method.
2. Materials and methods
2.1. Reference isolates
Reference dermatophyte isolates were purchased from Assiut University Mycological Center (AUMC), Egypt. (Table 1).
Table 1.
Reference dermatophyte isolates from Assiut University Mycological Center, Egypt.
| No. | AUMC No.∗ | Fungal species |
|---|---|---|
| 1 | 2350 | Microsporum fulvum |
| 2 | 2349 | Microsporum gallinae |
| 3 | 5096 | Microsporum gypseum |
| 4 | 5503 | Trichophyton mentagrophytes |
| 5 | 5488 | Trichophyton rubrum |
| 6 | 2353 | Trichophyton simii |
| 7 | 5097 | Trichophyton violaceum |
| 8 | 2357 | Microsporum praecox |
| 9 | 5448 | Microsporum canis |
| 10 | 5473 | Trichophyton verrucosum |
| 11 | 5495 | Epidermophyton floccosum |
AUMC: Assiut University Mycological Center.
2.2. Specimen collection
The experimental design of current study was approved by the Department of Botany and Microbiology, College of Science, King Saud University (No. 29499001265). This study was performed according to international ethical guidelines for epidemiological studies prepared by the Council for International Organizations of Medical Sciences (CIOMS) in collaboration with the World Health Organization (CIOMS, 2008). The Samples were collected from patients treated in a dermatology clinic-medical complex, Riyadh. The medical diagnosis was done by physicians and consultants as skin diseases. Samples of skin, hair and nails were collected by sterilized scalper, nail clipper, tweezers, toothbrush, vinyl tape strips or moistened cotton swabs depending on nature of infection. The samples were preserved in a sterile Petri dish and were transferred within 6 h to the Medical mycology laboratory in the Department of Botany and Microbiology, College of Science, King Saud University to perform isolation and identification. A portion of each simple was examined by direct microscopic method (KOH 10% with methylene blue) according to Miranda and Silva, 2005.
2.3. Isolation of dermatophytes
Sabouraud dextrose agar (SDA) and dermasel agar (DA) (Oxoid, England) were used to cultivate the dermatophytes. Cycloheximide (50 μg/ml) and Chloramphenicol (50 μg/ml) were added to prepare a selective media. To preserve the isolates, the isolates were cultivated on Potato dextrose (PD) slant agar with additional chloramphenicol (50 μg/ml), Cycloheximide (50 μg/ml), and glycerol (1%), after that they were preserved at −20 °C. The sub-cultivation was performed from the stock isolates to perform subsequent experiments (Hashemi et al., 2010; Khosravi and Mansouri, 2001).
2.4. Identification of dermatophytes by conventional methods
Isolated dermatophytes were identified by study of common dermatophyte identification criteria such as gross colony on sabouraud’s, mycosel agar and corn meal agar (Oxoid, England) with 2% dextrose (front, reverse color and texture), microscopic characteristics (microconidia, macroconidia, chlamydoconidia, chlamydoconidia in chains, and coiled spirals), urease test (on urease dextrose agar ((Oxoid, England)), in vitro hair penetration test, growth on trichophyton agar (No. 1 (casein without vitamin), No. 3 (Thiamin and inositol) and No. 4 (thiamin only)), and growth on boiled rice. Those microscopic and macroscopic morphological characteristics were studied according to Dolenc-Voljc, 2005; Ellabib et al., 2002; Fathi and al-Samarai, 2000; Morales-Cardona et al., 2013. Dermatophytes test medium (DTM) (BBL Company, USA) was used to distinguish between dermatophytes and non dermatophyte fungi. DTM contains phenol red indicator to detect secondary alkaline metabolites that are produced by the dermatophytes whereas the non dermatophytes produce secondary acidic metabolites. The changes of the yellow color medium to red indicate that the isolates are dermatophytes (Jennings and Rinaldi, 2003).
2.5. Identification of dermatophytes by molecular methods
Random short primers [OPU15 (5′-ACGGGCCAGT-3′), and OPD18 (5′-GAGAGCCAAC-3′)] were purchased from Qiagen company, USA. Finger prints for isolated dermatophytes and reference dermatophytes isolates were carried out by arbitrarily primed polymerase chain reaction (AP-PCR) according to Liu et al., 1997, 2000b; Lopandic et al., 2006. Genomic DNA was extracted and purified by DNeasy plant mini Kit (Qiagen, USA). Concentration and purity of DNA were determined by a spectrophotometer according to the instructions of the manufacturer (Qiagen, USA). PuReTaqTM Ready_To-GoTM Polymerase Chain Reaction (PCR) Beads (GE Healthcare Company, UK Limited) were used in amplification of the DNA segments consisting of a bacterial Tag DNA polymerase, Molecular bases (200 μM dATP, 200 μM dCTP, 200 μM dGTP and 200 μM dTTP), reaction buffer (10 μM Tris–HCl, pH 9.0 at room Temperature), 50 μM KCl and 1.5 μM MgCl2) and RNase (100 mg/ml), the amplification was done according to the instructions of the manufacturer (GE Healthcare, UK). Polymerase chain reaction (PCR) was performed in a Primus 96 plus, (MWG-AG, Biotech) programed for 39 cycles at 93 °C for 1 min; 50 °C for 1 min and 72 °C for 10 min. The electrophoresis on agarose gel with ethidium bromide was carried out then the gel images were photographed and stored in a Compact or UVP gel documentation Image analysis system.
3. Statistical analysis
The experiment was designed as completed random design (CRD). The data were expressed as percentages. Statistical analysis was performed by analysis of variance (SAS, 2002).
4. Results
4.1. Diagnosis of skin infections
In this study, 112 skin infections were diagnosed and were distributed as follows: tinea capitis (22.3%), tinea corporis (21.4%), tinea unguium (17.9%), tinea pedis (17%), tinea manuum (11.6%), tinea cruris (4.5%), tinea faces (2.7%), tinea favosa (1.8%) and Tinea barbae (0.9%).
4.2. Identification of dermatophytes by conventional methods
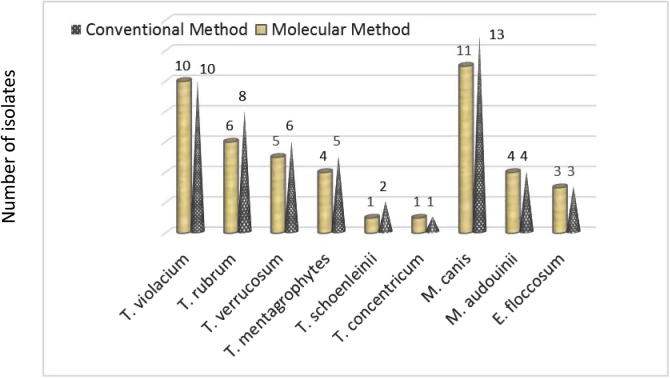
The fungal structures were directly observed in samples by KOH 10% with methylene blue. In Fig. 1, it can be seen that among the 112 isolates, there were 37 (33%) dermatophyte samples, 19 (17%) non-dermatophyte samples and 56 (50%) negative samples (the absence of the fungal elements). The results indicated that there are statistically significant differences (P < 0.05) between percentage of dermatophytes, non-dermatophytes and negative samples. The results of microscopic and macroscopic characteristics showed that out of the 112 specimens, 52 (46.4%) isolates were identified as dermatophytes while 24 (21.4%) samples were identified as non dermatophytes. The negative samples were 36 samples which had not shown any growth on several media in this research (Fig. 2). There were remarkable significant differences (P < 0.05) in number of identified isolates as dermatophytes between the direct microscopic test (37 samples) and cultivation method (52 samples). Nine species of dermatophytes were identified by microscopic and macroscopic morphological characteristics. These species belong to three genera: Trichophyton (T. violaceum, T. verrucosum, T. rubrum, T. mentagrophytes, T. schoenleinii and T. concentricum), Microsporum (M. canis and M. audouinii) and Epidermophyton (E. floccosum). The non dermatophyte isolates included 5 isolates from Aspergillus spp., 4 isolates from Acremonium potronii and 15 isolates from Candida spp. The results in Fig. 3 demonstrate that M. canis was the most common species (13 isolates) whereas T. concentricum was the least (1 isolate). There was a significant association (P < 0.05) between the dermatophyte species and skin infection. The most common species (M. canis) were major causative agents for prevailing skin infections (tinea capitis) while the least isolated species (T. concentricum) was isolated from one beard infection (Tinea barbae). A significant correlation (P < 0.05) was observed between M. canis and T. violaceum with tinea capitis and tinea corporis. Also some species correlated with specific skin infections; for example, T. rubrum correlated with tinea capitis, corporis, manuum and pedis while T. verrucosum correlated with tinea capitis, manuum, pedis, faces and barbae (Table 2).
Figure 1.
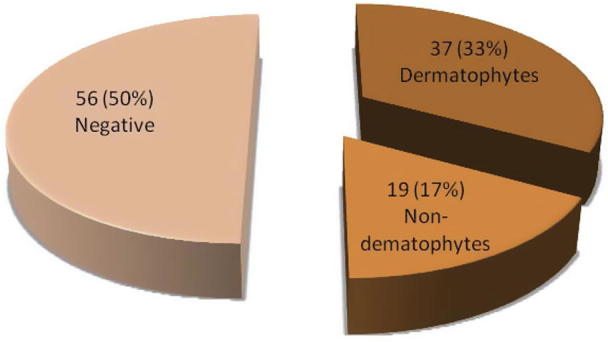
Number and percentage of dermatophytes, non-dermatophytes and negative samples by direct microscopic examination.
Figure 2.
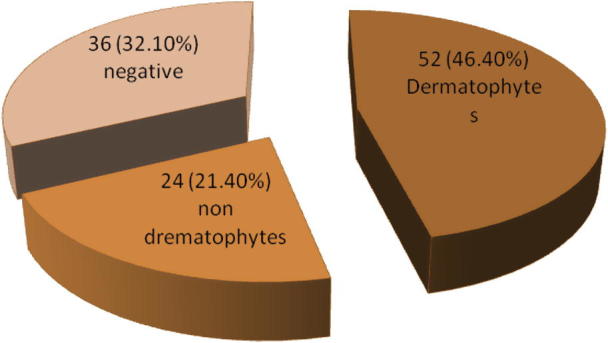
Number and percentage of dermatophytes, non-dermatophytes and negative samples by the microscopic, and macroscopic characteristics.
Figure 3.
Comparison between number of isolates identified by the microscopic, and macroscopic characteristics and the molecular methods.
Table 2.
Percentage of dermatophyte isolates and their correlation with type of skin infections.
| Species of dermatophytes | Types of dermatophytosis (Type of tinea) (%) |
Total (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T. capitis | T. corporis | T. unguium | T. pedis | T. manuum | T. cruris | T. faces | T. favosa | T. barbae | ||
| Microsporum canis | 19.2 | 5.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 |
| Trichophyton violaceum | 9.6 | 9.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19.2 |
| Trichophyton rubrum | 1.9 | 9.6 | 0 | 1.9 | 1.9 | 0 | 0 | 0 | 0 | 15.3 |
| Trichophyton verrucosum | 1.9 | 0 | 0 | 1.9 | 3.8 | 0 | 1.9 | 0 | 1.9 | 11.4 |
| Trichophyton mentagrophytes | 0 | 1.9 | 0 | 0 | 1.9 | 5.8 | 0 | 0 | 0 | 9.6 |
| Microsporum auoduinii | 7.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.7 |
| Epidermophyton floccosum | 0 | 0 | 3.8 | 0 | 1 1.9 | 0 | 0 | 0 | 0 | 5.8 |
| Trichophyton schoenleinii | 1.9 | 0 | 0 | 0 | 0 | 0 | 0 | 1.9 | 0 | 3.8 |
| Trichophyton concentricum | 0 | 1.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.9 |
| Total | 42.3 | 28.8 | 3.8 | 3.8 | 9.6 | 5.8 | 1.9 | 1.9 | 1.9 | 100 |
4.3. Identification by molecular method
The short random primer (OPU 15) and reference dermatophyte isolates were used to identify some isolated dermatophytes. The PCR amplified productions are summarized in Table 3. Six species (T. violaceum. T. rubrum, T. verrucosum, T. mentagrophytes, M. canis and E. floccosum) were identified by this primer. To identify the remaining isolates, the PCR amplified productions of OPD 18 were compared with the study of Liu and his colleagues (Liu et al., 2000a) (Table 4). To calculate the similarity, the following equation was used (Lopandic et al., 2006).
Table 3.
Summary of PCR amplified products from isolated dermatophytes DNA by OPU 15 primer.
| Dermatophytes species | PCR products (bp) |
|---|---|
| Trichophyton violaceum | 2700, 2200, 1900, 800 |
| Trichophyton rubrum | 2000, 1800, 800,500, 2600 |
| Trichophyton verrucosum | 2600, 2000, 1800, 800, 500. |
| Trichophyton mentagrophytes | 2500, 2000, 1400, 800 |
| Microsporum canis | 600, 1200 |
| Epidermophyton floccosum | 800 |
PCR: polymerase chain reaction, bp: base pairs.
Table 4.
Summary of PCR amplified products from isolated dermatophytes DNA by OPD 18 primer and comparing them with results’ Liu et al. (2000a).
| PCR amplified products (bp) by OPD18 primer |
|||
|---|---|---|---|
| In this research | Results’ Liu et al. (2000a) | Similarity % | |
| Trichophyton schoenleinii | 3200, 2000 | 3200, 2000 | 100 |
| Trichophyton concentricum | 3400, 2100, 1200 | 3400, 2100, | 85.7 |
| 2000, 1200 | |||
| Microsporum audouinii | 3600, 3400, 2800, | 2800, 1600, | 80 |
| 1600, 1400, 1100 | 1400, 1100 | ||
PCR: polymerase chain reaction, bp: base pairs.
The comparison indicated that the isolates that were not identified by POU 15 primer were T. schoenleinii, T. concentricum and M. audouinii. The results in Fig. 3 show that some isolates were identified by the conventional method but they were not identified by the molecular method (two T. rubrum isolates, one T. verrucosum isolates, one T. schoenleinii isolates and two M. canis isolates).
5. Discussion
In most cases, the mycological analysis is required because the clinical diagnosis is not obvious. The dermatophytes exist and are endemic in most regions but the species vary from one environment to another (Gupta et al., 2005). In this study, the distribution of skin infections agreed somewhat with some results obtained from some studies (Abanmi et al., 2008; Al-Sogair et al., 1991; Al Sogair and Hay, 2000) in Riyadh City, Saudi Arabia but tinea imbricate was not observed in this research. Also the tinea capitis was the most common skin infection in this study while another research (Abanmi et al., 2008) found that onychomycosis was the most frequent infection (40.3%). In this occasion, it should be noted that the term onychomycosis refers to fungal infection of the nail that is caused by non dermatophyte fungi such as Scopulariopsis brevicaulis and Candida albicans whereas the term tinea unguium means fungal infection of nail that is caused by dermatophyte fungi (Elewski, 1998). In this research, the results agreed with studies that were carried out in Alexandria – Egypt and in Brazil that found that 50% of positive cultures were in samples from toenails (Costa-Orlandi et al., 2012; Omar, 2000). Although dermatophytosis is found throughout the world, the developed countries have high rates of tinea pedis and onychomycosis, while developing countries have high rates of tinea capitis (Achterman and White, 2012; Seebacher et al., 2008). Many studies in other countries (Tripoli, Libya and Sana’a, Yemen) did not agree with this research. Those studies indicated that tinea corporis was the most common in Libya and Yemen, respectively (Ellabib et al., 2002; Mahmoud, 2002). The dermatophyte isolates were more than the non-dermatophytes, this result was approved by another research that was performed in Eastern Province of Saudi Arabia that examined 4294 clinically suspected cases of dermatomycoses and concluded that the non dermatophyte fungi were dominant (Al-Sogair et al., 1991). Generally in developing countries, the tinea capitis is present in more than 19.7% of the general population (Achterman and White, 2012).
The negative samples reached half (50%) of tested samples by the direct microscopic method and third (32%) of samples by cultivation methods. In direct microscopic test, most studies were based on observing some structural fungi such as septate hyphae, conidia, chlamydospores and arthrospores as preliminary evidences on the presence of dermatophytes in sample while dichotomously branched, fronded hyphae and budding yeast cells as evidences on the non dermatophytes (Singh and Beena, 2003). In direct microscopic test, the fungal elements were not accurate to differ between dermatophyte and non dermatophyte fungi, also the absence of structural fungal elements in samples did not mean the absence of dermatophyte fungi. Some samples did not grow on artificial media that were used in this study, may be a major reason for this is absence of fungi in samples or residual antifungal drugs still exist inside the samples collected from patients.
In this work, candida species and the non dermatophytes (such as A. potronii and Aspergillus spp) were most frequent than other non dermatophyte fungi. Same results were reported by Costa-Orlandi and his colleagues (Costa-Orlandi et al., 2012). M. canis and T. violaceum were the most prevalent among dermatophyte isolates, these results agreed with some studies (Dias et al., 2003; Dolenc-Voljc, 2005) but did not agree with other researches (Costa-Orlandi et al., 2012; Metin et al., 2002) which found that the Trichophyton genus represented over 80% of the isolates (T. rubrum (64.29%), T. mentagrophytes (21.43%)) while Microsporum gypseum represented 14.29%. The differences between this study and previous studies were due to the variations in environment and lifestyle (Nweze and Okafor, 2005).
The correlation between the type of tinea and dermatophytes depends on the kind of keratin (Keratin in hair, keratin in skin, keratin in live tissue or keratin in dead tissue), for example M. canis prefers keratin in scalp and skin while E. floccosum prefers keratin in dead tissue (groin, feet and nails). In this research, M. canis and T. violaceum correlated with tinea capitis and corporis while E. floccosum correlated with tinea unguium and manuum.
Although 52 dermatophyte isolates were identified by the culture method only 45 of 52 isolates were identified by molecular methods; this means that the agreement between two methods was 86.5%. Probably, there are two reasons for reducing the isolates identified by molecular methods, the first: the molecular method is more accurate than cultivation assay and the second: in conventional methods, the great similarity between some dermatophytes in microscopic and macroscopic characteristics leads to a misidentification.
This research concluded that the major types of tinea were tinea capitis and tinea corporis. M. canis, M. audouinii, T. violaceum, T. verrucosum, T. rubrum, T. mentagrophytes, T. schoenleinii, T. concentricum and E. floccosum were responsible for dermatophytosis. M. canis and T. violaceum were the most prevalent among the dermatophyte species.
Acknowledgments
This work was supported by King Saud University, Deanship of Scientific Research, College of Science Research Center.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abanmi A., Bakheshwain S., El Khizzi N., Zouman A.R., Hantirah S., Al Harthi F., Al Jamal M., Rizvi S.S., Ahmad M., Tariq M. Characteristics of superficial fungal infections in the Riyadh region of Saudi Arabia. Int J Dermatol. 2008;47:229–235. doi: 10.1111/j.1365-4632.2008.03563.x. [DOI] [PubMed] [Google Scholar]
- Achterman R.R., White T.C. A foot in the door for dermatophyte research. PLoS Pathog. 2012;8:e1002564. doi: 10.1371/journal.ppat.1002564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho R. Mycological studies on zoophilic dermatophyte isolates of Finish and Swedish origin. Mycoses. 1988;31:295–302. [PubMed] [Google Scholar]
- Al-Sogair S.M., Moawad M.K., al-Humaidan Y.M. Fungal infection as a cause of skin disease in the eastern province of Saudi Arabia: cutaneous candidosis. Mycoses. 1991;34:429–431. doi: 10.1111/j.1439-0507.1991.tb00808.x. [DOI] [PubMed] [Google Scholar]
- Al Sogair S., Hay R.J. Fungal infection in children: tinea capitis. Clin. Dermatol. 2000;18:679–685. doi: 10.1016/s0738-081x(00)00160-7. [DOI] [PubMed] [Google Scholar]
- CIOMS, 2008 International Ethical Guidelines for Epidemiological Studies. Prepared by the Council for International Organizations of Medical Sciences (CIOMS) in collaboration with the World Health Organization (WHO). CIOMS, Geneva (Provisional text – pending printed version).
- Costa-Orlandi C.B., Magalhaes G.M., Oliveira M.B., Taylor E.L., Marques C.R., de Resende-Stoianoff M.A. Prevalence of dermatomycosis in a Brazilian tertiary care hospital. Mycopathologia. 2012;174:489–497. doi: 10.1007/s11046-012-9576-1. [DOI] [PubMed] [Google Scholar]
- Dhib I., Fathallah A., Yaacoub A., Hadj Slama F., Said M.B., Zemni R. Multiplex PCR assay for the detection of common dermatophyte nail infections. Mycoses. 2014;57:19–26. doi: 10.1111/myc.12096. [DOI] [PubMed] [Google Scholar]
- Dias T., Fernandes Ode F., Soares A.J., Passos X.S., Costa M., Hasimoto e Souza L.K., Silva Mdo R. Tinea capitis in children from Goiania, Brazil. Rev. Soc. Bras. Med. Trop. 2003;36:653–655. doi: 10.1590/s0037-86822003000600002. [DOI] [PubMed] [Google Scholar]
- Dolenc-Voljc M. Dermatophyte infections in the Ljubljana region, Slovenia, 1995–2002. Mycoses. 2005;48:181–186. doi: 10.1111/j.1439-0507.2005.01122.x. [DOI] [PubMed] [Google Scholar]
- Elewski B.E. Onychomycosis: pathogenesis, diagnosis, and management. Clin. Microbiol. Rev. 1998;11:415–429. doi: 10.1128/cmr.11.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellabib M.S., Khalifa Z., Kavanagh K. Dermatophytes and other fungi associated with skin mycoses in Tripoli, Libya. Mycoses. 2002;45:101–104. doi: 10.1046/j.1439-0507.2002.00731.x. [DOI] [PubMed] [Google Scholar]
- Faggi E., Pini G., Campisi E., Bertellini C., Difonzo E., Mancianti F. Application of PCR to distinguish common species of dermatophytes. J. Clin. Microbiol. 2001;39:3382–3385. doi: 10.1128/JCM.39.9.3382-3385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi H.I., al-Samarai A.M. Tinea capitis in Iraq: laboratory results. East Mediterr. Health J. 2000;6:138–148. [PubMed] [Google Scholar]
- Grumbt M., Monod M., Yamada T., Hertweck C., Kunert J., Staib P. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J. Invest. Dermatol. 2013;133:1550–1555. doi: 10.1038/jid.2013.41. [DOI] [PubMed] [Google Scholar]
- Gupta A.K., Kohli Y., Batra R. In vitro activities of posaconazole, ravuconazole, terbinafine, itraconazole and fluconazole against dermatophyte, yeast and non-dermatophyte species. Med. Mycol. 2005;43:179–185. doi: 10.1080/13693780410001731583. [DOI] [PubMed] [Google Scholar]
- Hashemi S.J., Gerami M., Zibafar E., Daei M., Moazeni M., Nasrollahi A. Onychomycosis in Tehran: mycological study of 504 patients. Mycoses. 2010;53:251–255. doi: 10.1111/j.1439-0507.2009.01703.x. [DOI] [PubMed] [Google Scholar]
- Jennings M.B., Rinaldi M.G. Confirmation of dermatophytes in nail specimens using in-office dermatophyte test medium cultures. Insights from a multispecialty survey. J. Am. Podiatr. Med. Assoc. 2003;93:195–202. doi: 10.7547/87507315-93-3-195. [DOI] [PubMed] [Google Scholar]
- Khosravi A.R., Mansouri P. Onychomycosis in Tehran, Iran: prevailing fungi and treatment with itraconazole. Mycopathologia. 2001;150:9–13. doi: 10.1023/a:1011028730323. [DOI] [PubMed] [Google Scholar]
- Liu D., Coloe S., Baird R., Pedersen J. PCR identification of Trichophyton mentagrophytes var. interdigitale and T. mentagrophytes var. mentagrophytes dermatophytes with a random primer. J. Med. Microbiol. 1997;46:1043–1046. doi: 10.1099/00222615-46-12-1043. [DOI] [PubMed] [Google Scholar]
- Liu D., Coloe S., Baird R., Pedersen J. Application of PCR to the identification of dermatophyte fungi. J. Med. Microbiol. 2000;49:493–497. doi: 10.1099/0022-1317-49-6-493. [DOI] [PubMed] [Google Scholar]
- Liu D., Coloe S., Baird R., Pederson J. Rapid mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 2000;38:471. doi: 10.1128/jcm.38.1.471-471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopandic K., Zelger S., Banszky L.K., Eliskases-Lechner F., Prillinger H. Identification of yeasts associated with milk products using traditional and molecular techniques. Food Microbiol. 2006;23:341–350. doi: 10.1016/j.fm.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Mahmoud A.L. A study of dermatophytoses in Sana’a, Yemen Republic. Mycoses. 2002;45:105–108. doi: 10.1046/j.1439-0507.2002.00729.x. [DOI] [PubMed] [Google Scholar]
- Metin A., Subasi S., Bozkurt H., Calka O. Tinea capitis in Van, Turkey. Mycoses. 2002;45:492–495. doi: 10.1046/j.1439-0507.2002.00802.x. [DOI] [PubMed] [Google Scholar]
- Miranda M.F., Silva A.J. Vinyl adhesive tape also effective for direct microscopy diagnosis of chromomycosis, lobomycosis, and paracoccidioidomycosis. Diagn. Microbiol. Infect. Dis. 2005;52:39–43. doi: 10.1016/j.diagmicrobio.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Morales-Cardona C.A., Valbuena-Mesa M.C., Alvarado Z., Solorzano-Amador A. Non-dermatophyte mould onychomycosis: a clinical and epidemiological study at a dermatology referral centre in Bogota, Colombia. Mycoses. 2013;57:284–293. doi: 10.1111/myc.12157. [DOI] [PubMed] [Google Scholar]
- Nweze E.I., Okafor J.I. Prevalence of dermatophytic fungal infections in children: a recent study in Anambra state, Nigeria. Mycopathologia. 2005;160:239–243. doi: 10.1007/s11046-005-0124-0. [DOI] [PubMed] [Google Scholar]
- Omar A.A. Ringworm of the scalp in primary-school children in Alexandria: infection and carriage. East Mediterr. Health J. 2000;6:961–967. [PubMed] [Google Scholar]
- Philpot C.M. Geographical distribution of the dermatophytes: a review. J. Hyg. (Lond) 1978;80:301–313. doi: 10.1017/s0022172400053663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M.D. Diagnosis and pathogenesis of dermatophyte infections. Br. J. Clin. Pract. Suppl. 1990;71:98–102. [PubMed] [Google Scholar]
- SAS . SAS Inst. Inc.; Cary, NC: 2002. SAS User’s Guide: Statistics. [Google Scholar]
- Seebacher C., Bouchara J.P., Mignon B. Updates on the epidemiology of dermatophyte infections. Mycopathologia. 2008;166:335–352. doi: 10.1007/s11046-008-9100-9. [DOI] [PubMed] [Google Scholar]
- Singh S., Beena P.M. Comparative study of different microscopic techniques and culture media for the isolation of dermatophytes. Indian J. Med. Microbiol. 2003;21:21–24. [PubMed] [Google Scholar]