Abstract
Purpose: The prognostic value of the expression of STAT3/phosphorylated-STAT3 on survival for cancer patients remains controversial. We performed a meta-analysis of the published literature in this field to identify its impact. Methods: We conducted a meta-analysis of 26 studies (n=3877 patients) that evaluated the relationship between the prognostic value and the expression of STAT3/phosphorylated-STAT3 in 15 different kinds of carcinomas. Studies evaluated the correlation between STAT3/phosphorylated-STAT3, which detected mostly by immunohistochemistry and western blot, and clinical staging, overall survival (OS) and disease free survival (DFS) were included. The impact of STAT3 and phosphorylated-STAT3 was analyzed separately. Results: A total of 26 studies (14 for STAT3 and 16 for phosphorylated-STAT3), comprising 3877 patients, were included for meta-analysis. The expression of STAT3 was strongly associated with a poor impact on overall survival (OS) in all eligible studies [hazard ratio (HR)=2.91, (95% confidence interval (CI), 1.91-4.42)], while a significant association was shown between the expression of phosphorylated-STAT3 and patients’ outcome [HR=1.53, (95% CI, 0.86-2.70)]. No significant effect was shown between the expression of STAT3/phosphorylated-STAT3 and clinical staging, neither with DFS. Conclusion: High expression of STAT3 seems to be associated with poor OS in patients with carcinomas, while phosphorylated-STAT3 does not.
Keywords: Meta-analysis, STAT3/phosphorylated-STAT3, prognosis, tumor
Introduction
Previous studies have shown several potential complex signal transduction systems involved in the process of malignant transformation. The signal transducers and activators of transcription (STAT) protein family has been reported to play vital roles in several oncogenic processes including proliferation, survival, differentiation and angiogenesis [1]. As an important member of STAT family, it is activated by phosphorylation on tyrosine residue in the cytoplasm, after which it translocates into the nucleus to regulate gene expression [2,4]. A large number of studies have focused on the prognostic significance of STAT3 expression in different human carcinomas. However, the formation is still limited.
Phosphorylated-STAT3 (p-STAT3) is the activated form of STAT3 with the ability to be detected in the process of oncogenesis. It dimerizes and translocates into the nucleus, where its occupation of specific DNA-binding sites resulting in the increased transcription of several molecules that are involved directly related to survival, proliferation, self-renewal, and invasion. However, the activation of STAT3 signaling pathway is temporary and in tight control under physical circumstances. If STAT3 is continuously activated to promote the transcription of downstream target genes, malignant transformation will ultimately take place [5]. We cannot figure out the differences between these two indicators in the process of oncogenesis. Therefore, in this study, we performed an analysis of STAT3 and p-STAT3. The meta-analysis is as follows.
Materials and methods
Identification and eligibility of relevant studies
We carried out our meta-analysis according to a predetermined written protocol. To be eligible for our meta-analysis, studies had to deal with expression of STAT3/p-STAT3 factors in different tumors, evaluated the prognostic effect of the expression and survival, and be published in English or Chinese languages. We searched on Pubmed (MEDLINE) 1950-present and EMBASE, with common search terms “Transcription Factor STAT3”, “APRF Transcription Factor”, “IL6-Response Factor”, “Signal Transducer and Activator of Transcription 3”, “STAT3 Protein”, “Acute-Phase Response Factor”, and “Neoplasm”, “Neoplasms”, “Tumors”, “Tumor”, “Cancer”, “Cancers”, and “prognosis”, “prognostic”, “outcome”. The deadline of the eligible articles was August 2013. Reference list from primary identified studies were also searched to prevent missing any studies by the internet searching.
Inclusion criteria for primary studies were as follows: (1) proven clinical diagnosis of different tumors, (2) immunohistochemistry or western blot was applied as a evaluating method for STAT3/p-STAT3, (3) STAT3 detected in the cytoplasm and p-STAT3 in the nuclear were considered suitable positive, and (4) correlation of STAT3/p-STAT3 expression with OS, DFS and TNM staging. Exclusion criteria include: (1) animal experiment, (2) reported the value of STAT3/p-STAT3 other than prognosis, (3) without exploitable survival statistics, (4) evaluate other cell surface markers relate to STAT3/p-STAT3. Full articles were retrieved for further assessment if the eligibility was unclear from the abstracts. Any disagreements were resolved by elaborate discussions. Eligible studies were then carefully examined to ensure the reliability.
Definitions and standardization
We used regular rules to standardize as much as possible the definition of STAT3 positive. The majority of the eligible studies used 10% as a cutoff value [6-13], so we defined STAT3 positive as positive cell stain in at least 10% of tumor cells. As to standard methods used in some articles, we contacted the author for their suggested cutoff value. When it was not retrieved, we just accepted the cutoff value as at least 10% as the majority. Also, as STAT3 and p-STAT3 were not the same form, so we considered them as two different subjects and did the analysis separately.
Data extraction and quality evaluation
Data were carefully extracted from the eligible original studies using a standard information collection form, with the following information: first author, year of publication, nationality of first author, tumor type, number of patients, TNM stage, median age of patients, median follow up year of patients, method of evaluation, antibody used, cutoff value and analysis methods. For different articles based on a same study, we just collected the recently published one or the one with sufficient data for analysis. The Newcastle-Ottawa Scale (NOS) recommended by the Cochrane Collaboration was used, and eight criteria were established to evaluate the quality of the included case-control studies and cohort studies [48]. Such being the case, studies could receive a score of 0-9 points, based on the three criteria of subject selection, comparability between groups and measurement of exposure factors. The main outcomes are shown in Table 1.
Table 1.
Basic characteristics of eligible studies
| First Author | Year | Country | Tumor Type | No.of patient (M/F) | Stage I/II (III/IV) | Median age (y) | Median follow-up (m) | Method | Antibody | Cut-off (positive) | Analysis | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Andreas W. | 2002 | Austria | Breast cancer | 73 (0/73) | Sep-63 | 56 | 81.6 | W-B | R-P-A | NR | OS/RFS | 8 |
| Mingzhen Y. | 2010 | China | Breast cancer | 367 (0/367) | 212/155 | 52.8 | 43.3 | IHC/W-B | R-P-A/Tyr705 | ≥10% | DFS | 6 |
| Amir S. | 2013 | Israel | Breast cancer | 375 (0/375) | 188/138 | 50 | >5 | IHC | R-P-A | ≥10% | OS | 6 |
| Yujuan C. | 2013 | China | Breast cancer | 140 (0/140) | 108/32 | 48.79 | 54 | IHC | R-P-A | >25% | OS | 7 |
| Sung-Im D. | 2008 | Korea | Osteosarcoma | 47 (25/22) | Aug-35 | 25 | 76.9 | IHC/RT-PCR | Try705 | >10% | Survival curve | 7 |
| Yucai W. | 2010 | China | Osteosarcoma | 76 (25/51) | NR | NR | NR | IHC/PT-PCR/W-B | R-P-A | ≥5% | OS/DFS | 7 |
| Keinosuke R. | 2010 | USA | Osteosarcoma | 51 (38/13) | NR | 20.2 | 137.6 | IHC/I-B | NR | ≥10% | Survival curve | 6 |
| Weida G. | 2005 | USA | Gastric cancer | 86 (56/30) | 42/44 | 62 | 25.7 | IHC/W-B | G-P-A | ≥10% | OS | 8 |
| Yuichi Y. | 2006 | Japan | Gastric cancer | 111 (63/48) | NR | 68.9 | NR | IHC/I-B | G-P-A | >10% | Survival curve | 7 |
| Sungmin W. | 2011 | Korea | Gastric cancer | 103 (36/67) | 168/117 | 54.5 | 51 | IHC | Tyr705 | >1% | Survival curve | 7 |
| Yanfei J. | 2013 | China | Gastric cancer | 48 (34/14) | 22/26 | NR | NR | IHC/W-B | R-P-A | NR | Survival curve | 6 |
| Bin Z. | 2010 | China | Hepatocellular carcinoma | 196 (136/60) | 100/96 | 48.1±12.6 | NR | IHC | M-M-A | NR | OS | 4 |
| Wenyong W. | 2011 | China | Hepatocellular carcinoma | 113 (93/20) | 86/27 | NR | NR | IHC/RT-PCR | Tyr705 | >25% | OS | 6 |
| Eric B.H. | 2005 | USA | NSCLC | 176 (97/79) | NR | 69 | 37 | IHC | Tyr705 | NR | OS/DFS | 8 |
| Mei Z. | 2011 | China | NSCLC | 68 (38/30) | 27/41 | 59.44 | NR | IHC/W-B | R-P-A/Tyr705 | ≥20% | OS | 7 |
| Chang K-C. | 2005 | Taiwan, China | Thymic epithelial tumor | 118 (65/53) | 49/58 | 52.7 | >9 | IHC | NR | ≥10% | Survival curve | 5 |
| Chao L. | 2013 | China | Thymic epithelial tumor | 80 (47/33) | 43/33 | 46.5 | 61.5 | IHC | NR | >10% | OS | 5 |
| Mustafa B. | 2012 | USA | Acute myeloid leukemia | 63 (32/31) | NR | 64 | 40 | W-B | Y705/C-20 | NR | OS/DFS | 8 |
| Christina B. | 2011 | Greece | Astrocytomas | 97 (60/37) | 20/77 | 59 | 13 | IHC | R-M-A | >5% | Survival curve | 7 |
| Teppei M. | 2011 | USA | Colorectal cancers | 724 (266/458) | 384/301 | NR | 129 | IHC | R-P-A | NR | OS | 8 |
| Chel-Hun C. | 2009 | Korea | Cervical carcinoma | 29 (0/29) | 20he9 | 47 | NR | IHC | G-P-A | >51% | DFS | 6 |
| Sibastian-F S. | 2012 | Austria | Esophagealcarcimoma | 324 (252/72) | 101/78 | 63+10 | NR | IHC | R-M-A | NR | OS/DFS | 5 |
| Yanyang T. | 2010 | China | Gliomas | 96 (61/35) | 18/78 | 50.9 | 42 | IHC/W-B | A-M-IgG | ≥5% | Survival curve | 5 |
| Yan Z. | 2012 | China | LSCC | 163 | 112/51 | NR | 60 | IHC | A-STAT3 | ≥10% | OS | 5 |
| Chih-cheng C. | 2009 | Taiwan, China | Pharyngeal cancer | 95 | 15/80 | 53 | Till death | IHC | NR | >10% | OS/OR | 6 |
| Lijuan Z. | 2013 | China | Wilma’ tumor | 58 (20/38) | 44/14 | 31 months | NR | IHC | R-P-A | ≥51% | OS/DFS | 7 |
*W-B: Western blot; IHC: Immunohistochemistry; NR: Not reported; OS: Over survival; DFS: Disease free survival; NSCLC: None small cell lung cancer; LSCC: lingual squamous cell carcinoma; R-P-A: Rabbit poly antigen; G-P-A: Goat polycolonal antigen; M-M-A: Mouse monoclonal antigen.
Statistical analyses
Eligible studies were first divided into 2 groups for analysis: those detected the expression of STAT3 and others with p-STAT3 expression. Then we performed further statistic analysis in each of the two groups regarding OS, DFS, and TNM stage. The result of the study was labeled ‘positive’ when a high expression of STAT3/p-STAT3 predicted a poor patient survival, and labeled ‘negative’ when a high STAT3/p-STAT3 predicted a good patient survival, and labeled ‘intermediate’ when no significant relationship between the expression of STAT3/p-STAT3 and survival.
The survival result in each individual study was estimated by HRs with 95% confidence interval values. We first extracted HRs and their 95% CI from the original articles. If the data were not available, we used the method described by Parmer et al [13] to calculated HRs. This method would require published data from numbers of patients at risk and total number of events in each group. When data were only available in the form of figures, we interpreted Kaplan-Meier curves by Engauge Digitizer version 4.1 (free software downloaded from http://sourceforge.net) and extracted survival data from each curve of OS, DFS and TNM stage, then we used the survival data to reconstructed HRs and their 95% CI. If the HR>1, we considered it a worse outcome with STAT3/p-STAT3 expression while statistical significance when 95% CI surpass 1 and P<0.05 the same time. Heterogeneity across the studies was evaluated with Cochrane Q test and the I2 statistic. We regarded P>0.10 or I2<50% as indicators of no heterogeneity using a fixed-effect model. Otherwise, P≤0.10 or I2>50% were regarded as indicators of heterogeneity using a random effect model [49]. We performed subgroup analysis regarding with tumor type, region, study methods as well in order to identify the sources heterogeneity across studies. The effect of publication bias on the outcomes was assessed by Begg’s funnel plot and Egger’s linear regression method. (P < 0.05 was considered statistically significant publication bias) [14]. Sensitivity analysis was conducted to evaluate the stability of the results by sequentially omitting one study at each turn with the metaninf algorithm in Stata. version 11.0 (Stata Corporation, College Station, TX, USA). P<0.05 was considered to be statistically significant. Meta-analyses were carried out by Stata version 11.0 (Stata Corporation, College Station, TX, USA).
Results
Studies selection and characteristics
A total of 1068 studies were selected from databases and we evaluated 582 candidate studies in full text. By further review, 556 articles were excluded for animal experiments, no exploitable survival statistics or so (Figure 1). Finally we selected 26 articles (n=3877) with STAT3/p-STAT3 measurements in patients with 15 types of tumor.
Figure 1.
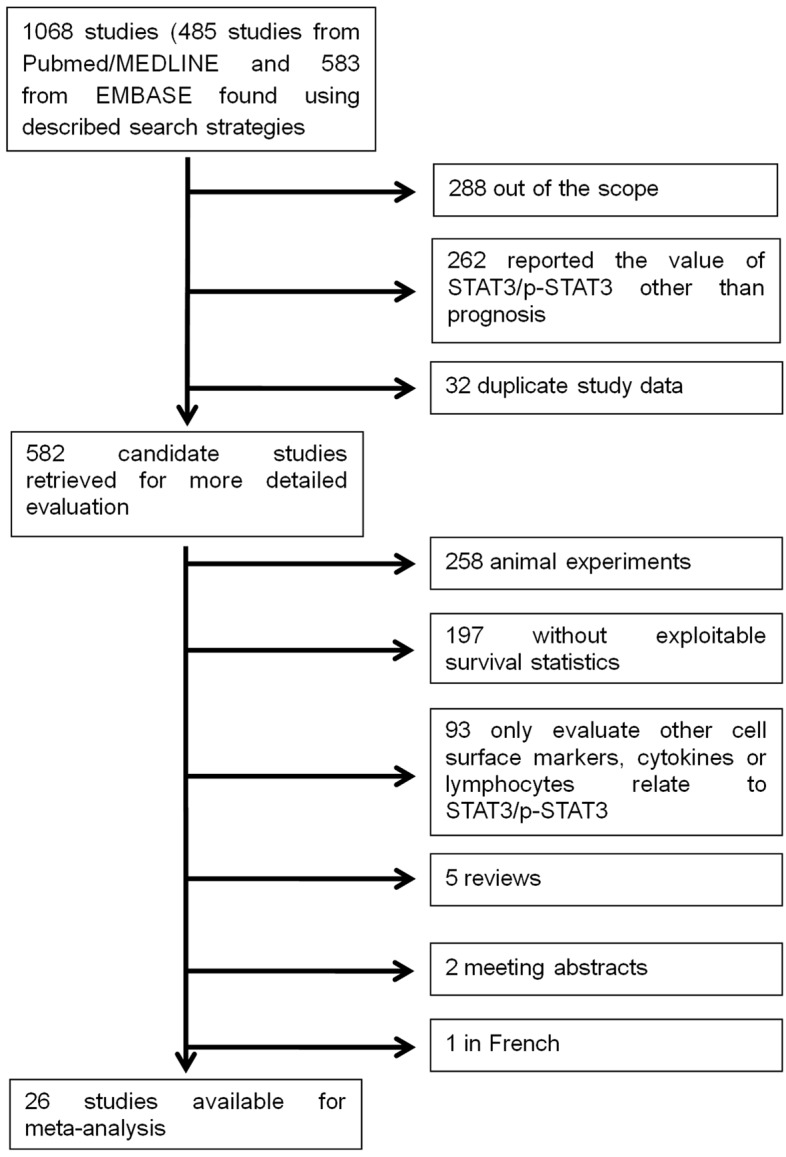
Flow chart of the literature search and selection of included studies.
The characteristics of eligible articles were listed in Table 1. The median age of the patients was 50.2 years ranging from 2.5 to 69. The median follow-up time was 60.9 months from 13 to 137.6. Twenty four articles were evaluated in the level of STAT3/p-STAT3 expression by IHC staining while the other 2 by western blot. The total proportion of patients in TNM grade I/II was 54.6%. The cut-off value was informed in 19 of the retrieved articles and over half of the articles provided the original HRs of OS or DFS or Stage. Twenty four out of 26 can do the meta-analysis OS, in which 14 articles were evaluated the expression of STAT3 and 12 evaluated the expression of p-STAT3. Seven out of the 26 studies could be included into the meta-analysis of DFS while 11 could be included for staging analysis.
Among the total 26 articles, including 4 studies for breast cancer, 3 for osteosarcoma, 4 for gastric cancer, 2 for hepatocellular carcinoma, non-small-cell carcinoma, thymic epithelial tumor each. The remaining studies included acute myeloid leukemia (N=1), astrocytomas (N=1), colorectal cancers (N=1), cervical carcinoma (N=1), esophageal-carcinoma (N=1), gliomas (N=1), lingual squamous cell carcinoma (N=1), pharyngeal cancer (N=1), and Wilms’ tumor (N=1).
All of the total 26 articles were with considerable NOS scores (≥4) that could ensure convincing results.
Data synthesis in two groups: overall survival of STAT3 and p-STAT3
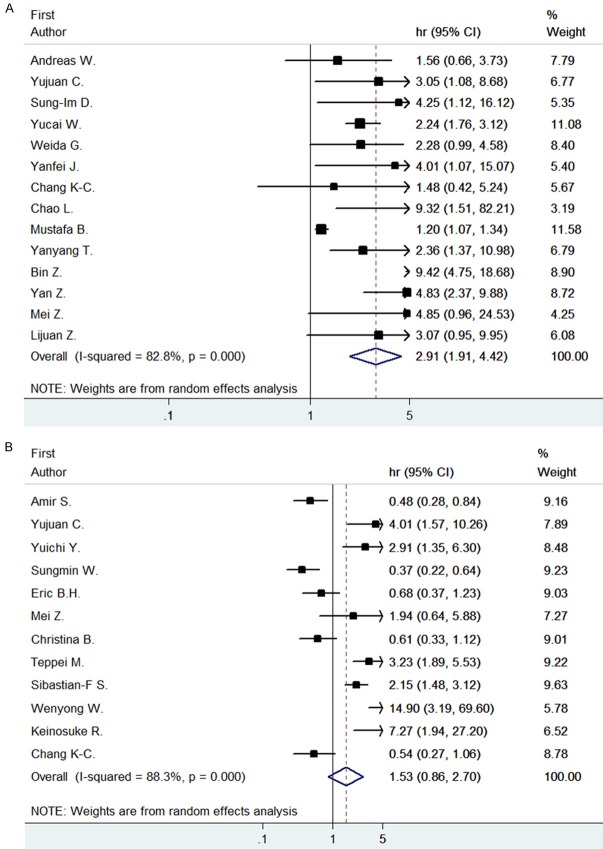
The genotype of STAT3 is evaluated in two different types, STAT3 and p-STAT3, of which they show different results on overall survivals. Statistics for each HR of overall survival are listed in Table 2. Heterogeneity analysis showed P=0.000, I2=75.36, so we conducted the random model. The overall HR for STAT3 is 2.91, (95% CI: 1.91-4.42), while for p-STAT3 is 1.53 (95% CI: 0.86-2.70), which indicate statistical significant only in the OS of STAT3 (Figure 2).
Table 2.
Overall survival for STAT3/p-STAT3 expression
| STAT3 OS | ||||||
|
| ||||||
| First Author | Year | Country | Tumor Type | HR (Hazard ratio) | 95% CI | |
|
| ||||||
| Andreas W. | 2002 | Austria | Breast cancer | 1.56 | 0.66 | 3.73 |
| Yujuan C. | 2013 | China | Breast cancer | 3.05 | 0.032 | 0.595 |
| Sung-Im D. | 2008 | Korea | Osteosarcoma | 4.25 | 1.12 | 16.12 |
| Yucai W. | 2010 | China | Osteosarcoma | 2.244 | 1.763 | 3.116 |
| Weida G. | 2005 | USA | Gastric cancer | 2.28 | 0.99 | 4.58 |
| Yanfei J. | 2013 | China | Gastric cancer | 4.01 | 1.07 | 15.07 |
| Chang K-C. | 2005 | Taiwan, China | Thymic epithelial tumor | 1.48 | 0.42 | 5.24 |
| Chao L. | 2013 | China | Thymic epithelial tumor | 9.325 | 1.508 | 82.207 |
| Mustafa B. | 2012 | USA | Acute myeloid leukemia | 1.2 | 1.07 | 1.34 |
| Yanyang T. | 2010 | China | Gliomas | 2.36 | 1.37 | 10.98 |
| Bin Z. | 2010 | China | Hepatocellular carcinoma | 9.416 | 4.747 | 18.676 |
| Yan Z. | 2012 | China | LSCC | 4.83 | 2.37 | 9.88 |
| Mei Z. | 2011 | China | NSCLC | 4.85 | 0.96 | 24.53 |
| Lijuan Z. | 2013 | China | Wilma’ tumor | 3.07 | 0.946 | 9.951 |
|
| ||||||
| p-STAT3 OS | ||||||
|
| ||||||
| First Author | Year | Country | Tumor Type | HR (Hazard ratio) | 95% CI | |
|
| ||||||
| Amir S. | 2013 | Israel | Breast cancer | 0.48 | 0.275 | 0.839 |
| Yujuan C. | 2013 | China | Breast cancer | 4.01 | 1.076 | 8.675 |
| Yuichi Y. | 2006 | Japan | Gastric cancer | 2.91 | 1.35 | 6.3 |
| Sungmin W. | 2011 | Korea | Gastric cancer | 0.37 | 0.22 | 0.64 |
| Eric B.H. | 2005 | USA | NSCLC | 0.68 | 0.37 | 1.23 |
| Mei Z. | 2011 | China | NSCLC | 1.937 | 0.639 | 5.875 |
| Christina B. | 2011 | Greece | Astrocytomas | 0.611 | 0.332 | 1.124 |
| Teppei M. | 2011 | USA | Colorectal cancers | 3.23 | 1.89 | 5.53 |
| Sibastian-F S. | 2012 | Austria | Esophagealcarcimoma | 2.146 | 1.476 | 3.119 |
| Wenyong W. | 2011 | China | Hepatocellular carcinoma | 14.9 | 3.19 | 69.6 |
| Keinosuke R. | 2010 | USA | Osteosarcoma | 7.27 | 1.94 | 27.2 |
| Chang K-C. | 2009 | Taiwan, China | Pharyngeal cancer | 0.5369 | 0.2713 | 1.0627 |
Figure 2.
Forrest plots and meta-analysis of studies evaluating HR of high STAT3 (A) and p-STAT3 (B) counts as compared to low counts. Survival data is reported as oversurvival (OS).
Data synthesis: disease free survival of STAT3 and p-STAT3
The numbers of articles included in the analysis of DFS on STAT3 were 3 and p-STAT3 were 4, because we could not get the exact data from some articles. The overall HR for OS is 2.41 (95% CI: 0.72-8.13) in STAT3 and 1.10 (95% CI: 0.53-2.29) in p-STAT3 analysis, which indicated no statistical significance between the expression of STAT3/p-STAT3 and DFS (Figure 3).
Figure 3.
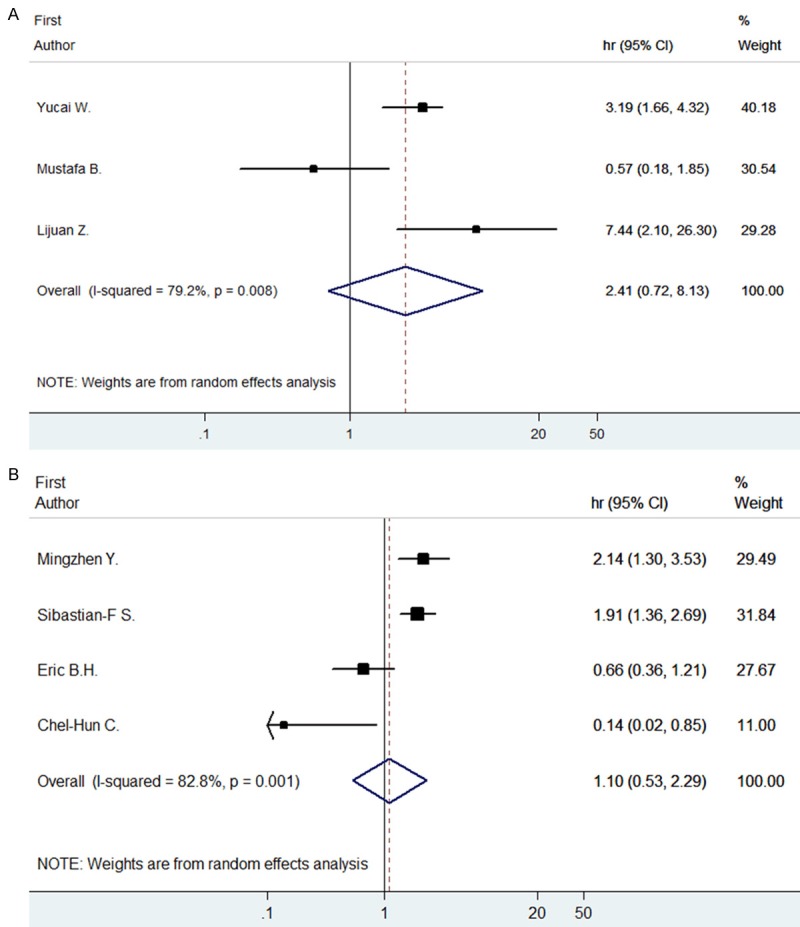
Forrest plots and meta-analysis of studies evaluating HR of high STAT (A) and p-STAT3 (B) counts as compared to low counts. Survival data is reported as disease free survival (DFS).
Data synthesis: clinical stage of STAT3 and p-STAT3
We conducted a meta-analysis to evaluate the clinical stage significance of STAT3/p-STAT3, including 11 out of 26 articles (6 for STAT3 and 5 for p-STAT3). The overall HR for STAT3 is 1.39 (95% CI: 0.30-6.52), indicating an insignificant impact on clinical stage while the overall HR for p-STAT3 is 1.96 (95% CI: 0.93-4.14), which indicated no significant result as well (Figure 4).
Figure 4.
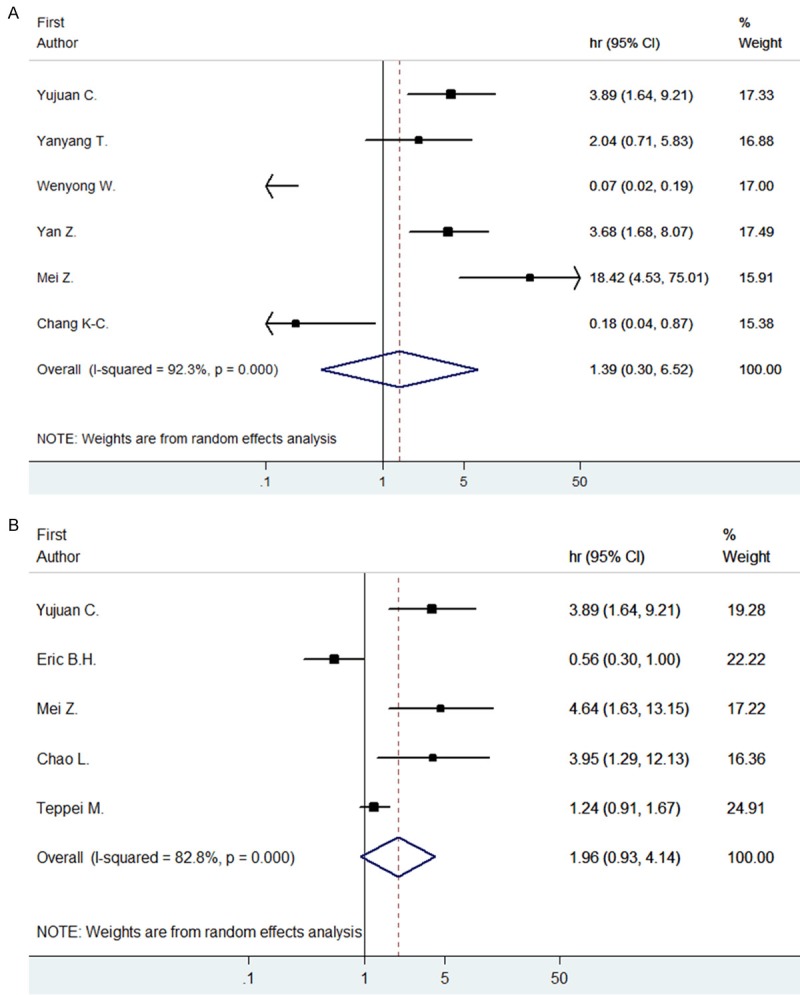
Forrest plots and meta-analysis of studies evaluating HR of high STAT (A) and p-STAT3 (B) counts as compared to low counts. Survival data is reported as clinical staging.
Subgroup analysis
As to identify the heterogeneity of the studies, we first aimed to perform subgroup analysis according to tumor type, but regrettably the data we had was not enough to do this job because we could not mix the HR of STAT3 and p-STAT3 together. So we performed subgroup analysis regard with study method and region, and only found heterogeneity with region in OS of STAT3. The overall HR for OS of STAT3 is 3.64 (95% CI: 2.44-5.43) in Asian countries such as China, Japan and Krea, while in west countries the data is 1.38 (95% CI: 0.97-1.96). These data indicate an significant difference in Asian countries while not in Western countries (Figure 5).
Figure 5.
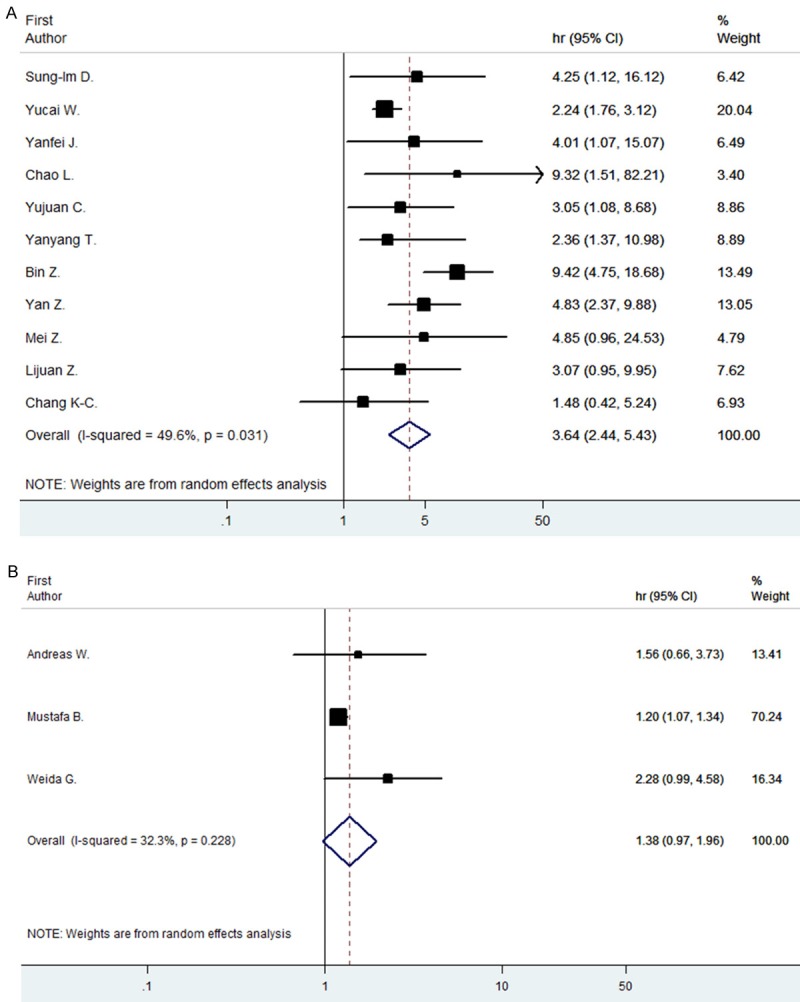
Subgroup analysis for OS by region. Forrest plots and meta-analysis of studies evaluating HR of high STAT3 in Asian country (A) and in Western country (B) compared to low counts.
Evaluation of publication bias
Both Begg’s funnel plot and Egger’s test were performed to assess the publication bias in all eligible studies. Begg’s funnel plot did not reveal any evidence of significant asymmetry in the OS meta-analysis of STAT3 (P=0.622), p-STAT3 (P=0.217), DFS meta-analysis of STAT3 (P=0.602), p-STAT3 (P=0.174), staging meta-analysis of STAT3 (P=0.573), and p-STAT3 (P=0.624). Egger’s test also showed no significant relevance in OS for p-STAT3 (P=0.362); DFS for STAT3 (P=0.822), p-STAT3 (P=0.186); staging for STAT3 (P=0.702), p-STAT3 (P=0.289). While in OS for STAT3 (P=0.002), the Egger’s test indicated a different result comparing to the Begg’s funnel plot (Figure 6).
Figure 6.
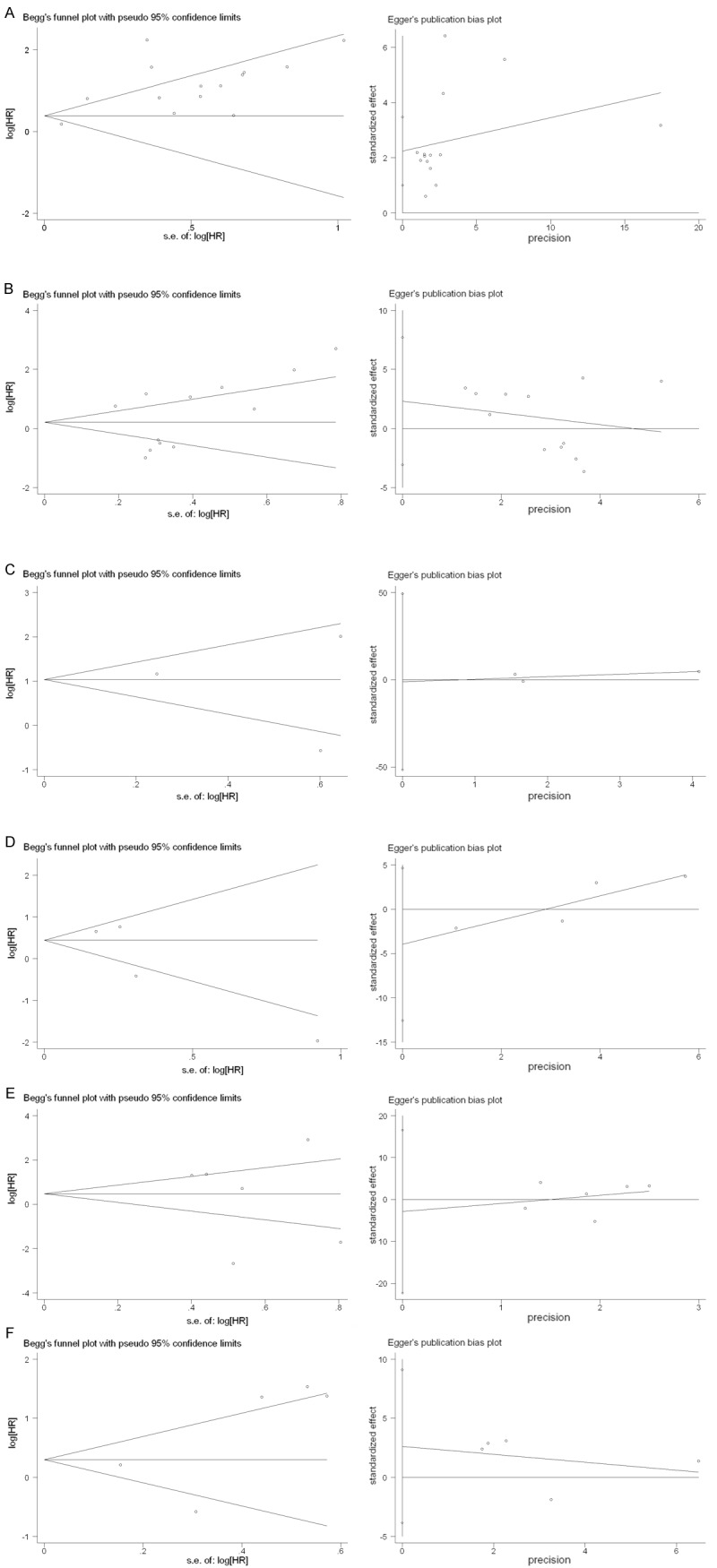
Begg’s funnel graph and Egger’s test for assessment of potential publication bias in studies of STAT3/p-STAT3 in patients with tumors. A. OS for STAT3. B. OS for p-STAT3. C. DFS for STAT3. D. DFS for p-STAT3. E. Stage for STAT3. F. Stage for p-STAT3. The funnel graph plots log HR against the standard error of the log HR.
Sensitivity analysis
Sensitivity analysis was performed based on OS of STAT3 and p-STAT3 and the results demonstrated the robustness of analysis (Data not shown).
Discussion
At present, our study showed insignificant HRs with DFS and clinical stage. However we found that high STAT3 expression level, as detected by immunohistochemistry or western blot, was significantly associated with poor overall survival in different kinds of tumors.
As potential bias exists between studies on STAT3 and p-STAT3, relevant analysis were performed respectively. The patients’ OS of STAT3 was significant, with a HR=2.91 (95% CI: 1.91-4.42) while p-STAT3 with a HR=1.53 (95% CI: 0.86-2.70), which indicated insignificant prognostic effects on tumors. We then compared the two factors and several different key factors.
As we mentioned above, the articles we included covered 15 kinds of tumors. The ones we analyzed for STAT3 were breast cancer [15,16], osteosarcoma [8,17], gastric cancer [9,18], thymic epithelial tumor [12,19], acute myeloid leukemia [20], glioma [21], hepatocellular carcinoma [22], LSCC [23], NSCLC [24], and Wilms’ tumor [25]. And the ones for p-STAT3 were breast cancer [7,16], gastric cancer [10,11], NSCLC [24,26], astrocytoma [27], colorectal cancer [28], esophageal carcinoma [29], hepatocellular carcinoma [30], osteosarcoma [31], and pharyngeal cancer [32]. Through the types of tumors we discovered, only four of them were the same (breast cancer, gastric cancer, NSCLC and hepotocellular carcinoma). By comparison, we found that both STAT3 and p-STAT3 expression in NSCLC both played a poor role in patients’ OS. This result was in line with Y.H Xu and S. Lu in their meta- analysis with NSCLC only [33]. And in gastric cancer we found that both STAT3 and p-STAT3 expression showed bad outcome in patients’ OS as well. While in breast cancer, STAT3 and p-STAT3 seemed to play a controversial role, even subtypes of tumors do influence the outcomes. Thus we suggested that more same type of tumors and subtype of one tumor studies studied to find out the final precise results.
In addition, the factors that have co-influence with STAT3 and p-STAT3 were not the same [34-37]. Aberrantly active STAT3 promotes uncontrolled growth and survival through dysregulating the expression of downstream targeted genes including survivin, Bcl-xL, Bcl-2, Mcl-1, c-Myc and cyclin D1 et al [38,39]. And the factors co-effect p-STAT3 including SOCS3, EGFR, IL-6 and IL-8 et al [6,26,27,32]. Take IL-6 and SOCS3 for example, it has been shown that STAT3 rapidly induced transcriptional activation of the STAT3 gene through an IL-6 response element located in the STAT3 gene promoter. This finding suggests the existence of autoregulatory mechanisms in the IL-6 signal-transducing system at the level of the signal transducing transcription factor. Thus, an increase in SOCS3 can block IL-6 signal transduction, causing a decrease in STAT3 protein expression [40]. What’s more, the degradation of STAT3 has been shown to be involved with the ubiquitin (Ub)-proteosome pathway. This pathway is responsible for selective degradation of short-lived cellular proteins and is critical for the regulation of many cellular processes [41]. As the interaction between factors were not revealed as much as we need, we cannot come to a conclusion that whether the factors influenced STAT3 expression had the same effects on the p-STAT3 expression or not. We then suggested the mechanism of STAT3/p-STAT3 influencing factors been conducted actively.
In the Begg’s funnel plot, the p value for OS of STAT3 is 0.622, while Egger’s test is 0.002. The two numbers showed opposite results for the publication bias in the analysis. Begg’s funnel plot is the most common used method in testing publication bias in meta-analysis [42], but it still has limitations, unless there are more eligible articles [43-45]. Since the number of original articles was limited, this method would be restrictive. So we used Egger’s test [14] for a supplementary detection, finding contradictions. When the number of articles for analysis was 14, we considered the Egger’s results suitable [14,44,46], which revealed an apparent asymmetry that suggested the presence of a potential publication bias. Therefore we suggested the relevant articles been reliable enough not only in types but in numbers and quantities.
Moreover, variability in definitions, outcomes, measurements, experimental procedure, and even antibody concentration may contribute to heterogeneity between studies [47]. Even though we made efforts to minimize the confounding bias, such as subgroup analysis and sensitivity analysis, the factors controlled were not so much and still differed between studies. We recommended the following criteria in the future study: (1) precisely describe the factors evaluation including cut-off value staining and antibody concentration; (2) the including patients be in an age that not so spread; (3) the follow-up years should define at a same level. Our study still discovered a difference in the cut-off values, for some of the studies set the value at 20%, while others at 10% or even using their own-defined score systems. We just chose the majorities as a final result. Although the using of standardized cut-off value in different studies did not differ greatly for OS in the total population analysis, conclusions need to be reached more cautiously.
In conclusion, we come to a conclusion that STAT3 factors have poor effect on the prognosis of many types of tumors while p-STAT3 do not. As discussed above, the clinical use should be of further concerned and measured more carefully. We anticipate more studies being reliable, revealing more clearly consequences, and leading to develop new therapeutic strategies against certain type of tumors.
Acknowledgements
We thank all authors whose publication could be included in our meta-analys.
Disclosure of conflict of interest
None.
References
- 1.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 2.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an Oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 3.Frank DA. STAT signaling in the pathogenesis and treatment of cancer. Mol Med. 1999;5:432–456. [PMC free article] [PubMed] [Google Scholar]
- 4.Choi JH, Ahn MJ, Park CK, Han HX, Kwon SJ, Lee YY, Kim IS. Phospho-Stat3 expression and correlation with VEGF, p53, and Bcl-2 in gastric carcinoma using tissue microarray. Apmis. 2006;114:619–625. doi: 10.1111/j.1600-0463.2006.apm_401.x. [DOI] [PubMed] [Google Scholar]
- 5.Hao M, Zhao WH. Constitutive activation of signal transducer and activator of transcription 3 in epithelial ovarian carcinoma. J Obstet Gynaecol Res. 2009;35:918–925. doi: 10.1111/j.1447-0756.2009.01045.x. [DOI] [PubMed] [Google Scholar]
- 6.Ying M, Li D, Yang L, Wang M, Wang N, Chen Y, He M, Wang Y. Loss of SOCS3 expression is associated with an increased risk of recurrent disease in breast carcinoma. J Cancer Res Clin Oncol. 2010;136:1617–1626. doi: 10.1007/s00432-010-0819-6. [DOI] [PubMed] [Google Scholar]
- 7.Sonnenblick A, Uziely B, Nechushtan H, Kadouri L, Galun E, Axelrod JH, Katz D, Daum H, Hamburger T, Maly B, Allweis TM, Peretz T. Tumor STAT3 tyrosine phosphorylation status, as a predictor of benefit from adjuvant chemotherapy for breast cancer. Breast Cancer Res Treat. 2013;138:407–413. doi: 10.1007/s10549-013-2453-x. [DOI] [PubMed] [Google Scholar]
- 8.Wang YC, Zheng LH, Ma BA, Zhou Y, Zhang MH, Zhang DZ, Fan QY. Clinical value of signal transducers and activators of transcription 3 (STAT3) gene expression in human osteosarcoma. Acta Histochem. 2011;113:402–408. doi: 10.1016/j.acthis.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Gong W, Wang L, Yao JC, Ajani JA, Wei D, Aldape KD, Xie K, Sawaya R, Huang S. Expression of activated signal transducer and activator of transcription 3 predicts expression of vascular endothelial growth factor in and angiogenic phenotype of human gastric cancer. Clin Cancer Res. 2005;11:1386–1393. doi: 10.1158/1078-0432.CCR-04-0487. [DOI] [PubMed] [Google Scholar]
- 10.Yakata Y, Nakayama T, Yoshizaki A, Kusaba T, Inoue K, Sekine I. Expression of p-STAT3 in human gastric carcinoma: significant correlation in tumour invasion and prognosis. Int J Oncol. 2007;30:437–442. [PubMed] [Google Scholar]
- 11.Woo S, Lee BL, Yoon J, Cho SJ, Baik TK, Chang MS, Lee HE, Park JW, Kim YH, Kim WH. Constitutive activation of signal transducers and activators of transcription 3 correlates with better prognosis, cell proliferation and hypoxia-inducible factor-1alpha in human gastric cancer. Pathobiology. 2011;78:295–301. doi: 10.1159/000321696. [DOI] [PubMed] [Google Scholar]
- 12.Chang KC, Wu MH, Jones D, Chen FF, Tseng YL. Activation of STAT3 in thymic epithelial tumours correlates with tumour type and clinical behaviour. J Pathol. 2006;210:224–233. doi: 10.1002/path.2041. [DOI] [PubMed] [Google Scholar]
- 13.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widschwendter A, Tonko-Geymayer S, Welte T, Daxenbichler G, Marth C, Doppler W. Prognostic significance of signal transducer and activator of transcription 1 activation in breast cancer. Clin Cancer Res. 2002;8:3065–3074. [PubMed] [Google Scholar]
- 16.Chen Y, Wang J, Wang X, Liu X, Li H, Lv Q, Zhu J, Wei B, Tang Y. STAT3, a Poor Survival Predicator, Is Associated with Lymph Node Metastasis from Breast Cancer. J Breast Cancer. 2013;16:40–49. doi: 10.4048/jbc.2013.16.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Do SI, Jung WW, Kim HS, Park YK. The expression of epidermal growth factor receptor and its downstream signaling molecules in osteosarcoma. Int J Oncol. 2009;34:797–803. doi: 10.3892/ijo_00000205. [DOI] [PubMed] [Google Scholar]
- 18.Jia Y, Liu D, Xiao D, Ma X, Han S, Zheng Y, Sun S, Zhang M, Gao H, Cui X, Wang Y. Expression of AFP and STAT3 is involved in arsenic trioxide-induced apoptosis and inhibition of proliferation in AFP-producing gastric cancer cells. PLoS One. 2013;8:e54774. doi: 10.1371/journal.pone.0054774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wang Z, Liu Y, Wang P, Zhang R. STAT3 expression correlates with prognosis of thymic epithelial tumors. J Cardiothorac Surg. 2013;8:92. doi: 10.1186/1749-8090-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benekli M, Xia Z, Donohue KA, Ford LA, Pixley LA, Baer MR, Baumann H, Wetzler M. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood. 2002;99:252–257. doi: 10.1182/blood.v99.1.252. [DOI] [PubMed] [Google Scholar]
- 21.Tu Y, Zhong Y, Fu J, Cao Y, Fu G, Tian X, Wang B. Activation of JAK/STAT signal pathway predicts poor prognosis of patients with gliomas. Med Oncol. 2011;28:15–23. doi: 10.1007/s12032-010-9435-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Zhong DW, Wang QW, Miao XY, Dai WD, Liu C, Pan KH. [Study on correlation of JAK/STAT signal pathway with progression and prognosis in hepatocellular carcinoma] . Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2010;26:368–370. [PubMed] [Google Scholar]
- 23.Zhao Y, Zhang J, Xia H, Zhang B, Jiang T, Wang J, Chen X, Wang Y. Stat3 is involved in the motility, metastasis and prognosis in lingual squamous cell carcinoma. Cell Biochem Funct. 2012;30:340–346. doi: 10.1002/cbf.2810. [DOI] [PubMed] [Google Scholar]
- 24.Zhao M, Gao FH, Wang JY, Liu F, Yuan HH, Zhang WY, Jiang B. JAK2/STAT3 signaling pathway activation mediates tumor angiogenesis by upregulation of VEGF and bFGF in non-small-cell lung cancer. Lung Cancer. 2011;73:366–374. doi: 10.1016/j.lungcan.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Zhang LJ, Liu W, Gao YM, Qin YJ, Wu RD. The expression of IL-6 and STAT3 might predict progression and unfavorable prognosis in Wilms’ tumor. Biochem Biophys Res Commun. 2013;435:408–413. doi: 10.1016/j.bbrc.2013.04.102. [DOI] [PubMed] [Google Scholar]
- 26.Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11:8288–8294. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- 27.Piperi C, Samaras V, Levidou G, Kavantzas N, Boviatsis E, Petraki K, Grivas A, Barbatis C, Varsos V, Patsouris E, Korkolopoulou P. Prognostic significance of IL-8-STAT-3 pathway in astrocytomas: correlation with IL-6, VEGF and microvessel morphometry. Cytokine. 2011;55:387–395. doi: 10.1016/j.cyto.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Morikawa T, Baba Y, Yamauchi M, Kuchiba A, Nosho K, Shima K, Tanaka N, Huttenhower C, Frank DA, Fuchs CS, Ogino S. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17:1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoppmann SF, Jesch B, Friedrich J, Jomrich G, Maroske F, Birner P. Phosphorylation of signal transducer and activator of transcription 3 (STAT3) correlates with Her-2 status, carbonic anhydrase 9 expression and prognosis in esophageal cancer. Clin Exp Metastasis. 2012;29:615–624. doi: 10.1007/s10585-012-9475-3. [DOI] [PubMed] [Google Scholar]
- 30.Wu WY, Li J, Wu ZS, Zhang CL, Meng XL. STAT3 activation in monocytes accelerates liver cancer progression. BMC Cancer. 2011;11:506. doi: 10.1186/1471-2407-11-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryu K, Choy E, Yang C, Susa M, Hornicek FJ, Mankin H, Duan Z. Activation of signal transducer and activator of transcription 3 (Stat3) pathway in osteosarcoma cells and overexpression of phosphorylated-Stat3 correlates with poor prognosis. J Orthop Res. 2010;28:971–978. doi: 10.1002/jor.21088. [DOI] [PubMed] [Google Scholar]
- 32.Chen CC, Chen WC, Lu CH, Wang WH, Lin PY, Lee KD, Chen MF. Significance of interleukin-6 signaling in the resistance of pharyngeal cancer to irradiation and the epidermal growth factor receptor inhibitor. Int J Radiat Oncol Biol Phys. 2010;76:1214–1224. doi: 10.1016/j.ijrobp.2009.09.059. [DOI] [PubMed] [Google Scholar]
- 33.Xu YH, Lu S. A meta-analysis of STAT3 and phospho-STAT3 expression and survival of patients with non-small-cell lung cancer. Eur J Surg Oncol. 2014;40:311–317. doi: 10.1016/j.ejso.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Jin L, Yuan RQ, Fuchs A, Yao Y, Joseph A, Schwall R, Schnitt SJ, Guida A, Hastings HM, Andres J, Turkel G, Polverini PJ, Goldberg ID, Rosen EM. Expression of interleukin-1beta in human breast carcinoma. Cancer. 1997;80:421–434. doi: 10.1002/(sici)1097-0142(19970801)80:3<421::aid-cncr10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 35.Sieweke MH, Thompson NL, Sporn MB, Bissell MJ. Mediation of wound-related Rous sarcoma virus tumorigenesis by TGF-beta. Science. 1990;248:1656–1660. doi: 10.1126/science.2163544. [DOI] [PubMed] [Google Scholar]
- 36.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit Rev Oncol Hematol. 2008;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, Markowitz D, Wu W, Liu C, Reisfeld RA, Xiang R. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest. 2006;116:2132–2141. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boucher MJ, Morisset J, Vachon PH, Reed JC, Laine J, Rivard N. MEK/ERK signaling pathway regulates the expression of Bcl-2, Bcl-X(L), and Mcl-1 and promotes survival of human pancreatic cancer cells. J Cell Biochem. 2000;79:355–369. [PubMed] [Google Scholar]
- 39.Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, Tomita K, Komiyama S, Weinstein IB. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
- 40.Schmitz J, Weissenbach M, Haan S, Heinrich PC, Schaper F. SOCS3 exerts its inhibitory function on interleukin-6 signal transduction through the SHP2 recruitment site of gp130. J Biol Chem. 2000;275:12848–12856. doi: 10.1074/jbc.275.17.12848. [DOI] [PubMed] [Google Scholar]
- 41.Kong F, Guo X, Noel JG, Wells DA, Lovell GJ, Ogle CK. Thermal injury-induced increases of hepatocyte SOCS3 lead to decreases in STAT3. Shock. 2002;18:374–379. doi: 10.1097/00024382-200210000-00014. [DOI] [PubMed] [Google Scholar]
- 42.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 43.Biljana M, Jelena M, Branislav J, Milorad R. Bias in meta-analysis and funnel plot asymmetry. Stud Health Technol Inform. 1999;68:323–328. [PubMed] [Google Scholar]
- 44.Xu TC, Li X, Wang WL, Hu P, Du FL. Detection of publication bias in meta-analysis of dichotomous variable-Egger test and Begg test. J Evidence-Based Med. 2009;19:181–184. [Google Scholar]
- 45.Zheng HL, Wang ZX, Wang ZZ. Applying the SAS Program to the Complete Begg’s Test, the Egger’s Test, and the Macaskill’s Test for Publication Bias of Meta-analysis. Chin J Evid-based Med. 2009;9:910–916. [Google Scholar]
- 46.Wang Z, Zhang YH, Xu QQ. Introduction of some methods for publication bias. Chinese Journal of Health Statistics. 2009;29:539–541. [Google Scholar]
- 47.Simon R, Altman DG. Statistical aspects of prognostic factor studies in oncology. Br J Cancer. 1994;69:979–985. doi: 10.1038/bjc.1994.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ownby RL, Crocco E, Acevedo A, John V, Loewenstein D. Depression and risk for Alzheimer disease: systematic review, meta-analysis, and metaregression analysis. Arch Gen Psychiatry. 2006;63:530–538. doi: 10.1001/archpsyc.63.5.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analysis. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]