Abstract
Tuberculosis (TB) is still an infectious disease that greatly threatens human health, and is always refractory to the current therapeutic modalities. Accumulated evidence revealed that microRNAs (miRNAs) are closely with various pathologies, such as TB. The possibilities of miRNAs as diagnostic biomarkers and therapeutic targets have been proved. However, it is still unknown if miRNA is implicated in the TB-associated immunity. This study revealed that miR-155, which has been shown to suppress the activation of natural killer (NK) cells associated with tumors, was downregulated in serum samples of TB patients (n=90), compared with healthy controls (n=31). Cytotoxicity assays indicated that NK cells, which have been demonstrated to promote TB progression, exhibited lower cytotoxicity in high serum miR-155 TB patients (n=37). There is an inverse relationship between serum miR-155 abundance and NK cell cytotoxicity (R=-0.659, P=0.000). Further studies demonstrated that miR-155 level is inversely associated with the concentration of TNFα secreted by NK cells from TB patients (n=37, R=-0.694, P=0.000). Collectively, serum miR-155 level was shown to be negatively associated with the TB-suppressing activity of NK cells, and this miRNA can be used as a potential therapeutic agent for TB treatment.
Keywords: Tuberculosis, miR-155, natural killer cell
Introduction
Although some great advances have been achieved in the field of medical prevention in recent decades, tuberculosis (TB) still risks human public health. There are reported to be more than 30% of population infected with Mycobacterium tuberculosis, the major pathogen for TB [1]. TB is always refractory to the current therapeutic strategies, and therefore, it is highly needed to elucidate the molecular mechanism by which TB progresses, and to develop new therapeutic modalities for TB.
miRNAs are a kind of 20-22 nt non-coding RNA, which are evolutionarily conserved from yeast to mammals. miRNA can suppress the expression of some specific target genes by recognizing and binding their 3’ untranslated region (3’UTR). The binding results in mRNA degradation or the inhibition of their translation [2]. miRNAs have been well documented to play a key role during various physiological processes, such as development and homeostasis [3]. In addition, accumulated evidence also indicates that miRNAs are associated with many diseases, including infectious diseases. For instances, Salmonella infection can reduce the expression of let-7 in macrophages, thereby elevating the secretion of specific immunostimulatory cytokines [4]. This result indicates that miRNA plays an important role in the immune response of host against pathogens.
Similar to its important role in other infectious diseases, immune reaction has also been demonstrated to be closely associated with TB progression [5]. Innate and adaptive immune responses are both implicated in the defense of host against M. tuberculosis. As one of the major immune cells, natural killer (NK) cells can contribute to the progression of TB by secreting some specific cytokines, such as TNF-α [5]. Thus, NK cell activation is essential for the initiation and progression of TB. However, there is still no available molecular marker used to evaluate the activation of NK cells in TB patients.
Recent studies revealed that miRNA has also been proved as a potent regulator for the biology of NK cells [6]. For example, miR-155 is associated with the reduction in the cytolysis capability of NK cells. miR-155-/- NK cells are more prone to be activated, compared with wild type controls. Higher TNF-γ production is also detected in NK cells with miR-155 downregulation. NK cells exhibit high cytotoxicity to cancer cells when miR-155 expression was reduced [7]. However, it is still unclear if miR-155 level can be used to predict the activation of NK cells in TB patients.
To answer the question, we collected serum samples from TB patients to determine the expression profile of miR-155, and to explore the relationship between serum miR-155 levels and the activity of TB-associated NK cells.
Materials and methods
Cell culture
Normal fibroblast cell line, MRC-5, was purchased from Shanghai Cell Culture Center (Shanghai, China). This cell line was cultured with DMEM media containing 10% fetal bovine serum at 37°C in a 5% CO2 humidified condition.
Ethics statement
In our study, peripheral blood was collected from TB patients and healthy volunteers with written informed consent. The samples were collected following the procedures approved by the Ethics Committee of The First Affiliated Hospital of Xinxiang Medical University.
Study design and patients
90 confirmed TB patients and 31 healthy individuals were recruited for this study. TB was diagnosed by clinical manifestation, chest X-ray examination, and sputum test. The blood samples were collected from these TB patients prior to any treatments at the Departments of Tuberculosis medicine, the First Affiliated Hospital of Xinxiang Medical University.
Serum collection
We collected serum samples following the procedures previously described. Briefly, blood were collected from vena and transferred into serum collection tubes. The samples were centrifuged at 820 g for 10 min at 4°C, after 1 h coagulation at room temperature. Then, the serum samples were transferred into new tubes for centrifugation at 16000 g for 10 min at 4°C.
Quantitative PCR (qPCR) assay
Total RNA was extracted the serum samples with the Trizol Solution (Invitrogen, Carlsbad, CA, USA) according to the routine protocols. The obtained RNA was subjected to reverse transcription with TaKaRa microRNA transcription kit (Takara, Japan), in order to produce cDNA. The resulting cDNA was subjected to quantitative PCR (qPCR) assays using SYBR Premix Ex Taq II kit (Takara, Japan) on an ABI-7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) supplied with analytic software. The primers used for miR-155 detection are as follows: Forward: GGTATCGAATGACCGACT; Reverse: TATTGACCTATGCAGGCT. U6 was used as endogenous references in this study.
NK cell sorting
Blood samples were collected from vena, followed by elimination of erythrocytes with Red Blood Cell Lysis Buffer (Beyotime Institute of Biotechnology, China). CD56+CD16- cells, which represented NK cell population (Figure 4), were sorted using Human NK Cell Isolation Kit (Miltenyi Biotec, Germany) following the procedures provided by the manufacturer. The purity and recovery of NK cell were both higher than 95% after sorting. The obtained NK cells were cultured with Alpha Minimum Essential Medium without ribonucleosides and deoxyribonucleosides but with 2 mM L-glutamine and 1.5 g/l sodium bicarbonate, and supplemented with 0.2 mM inositol; 0.1 mM 2-mercaptoethanol; 0.02 mM folic acid; 100-200 U/ml recombinant IL-2; adjust to a final concentration of 12.5% horse serum and 12.5% fetal bovine serum. The cells were cultured at 37°C in a 5% CO2 humidified condition.
Figure 4.
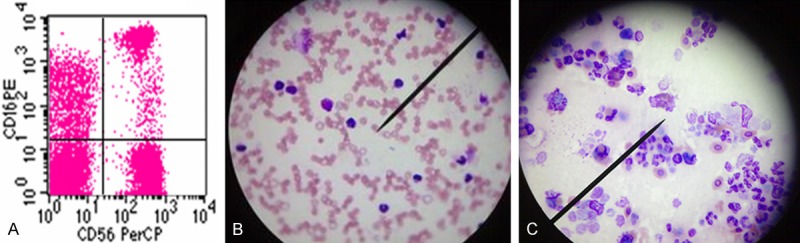
A. The cell population co-expressing low CD16 and high CD56 molecules was determined as NK cells by flow cytometry. B. NK cells derived from TB patients with high serum miR-155 level display low phagocytosis-associated cytotoxicity. C. NK cells derived from TB patients with low serum miR-155 level display high phagocytosis-associated cytotoxicity.
MTT assays
2×106 NK cells and 2×105 MRC-5 cells were planted in each well of 96-well plates as test groups. While 2×106 NK cells or 2×105 MRC-5 cells were planted as effector control group or target control group, respectively. After 4 h coculture, cells were treated with 10 µl MTT (5 mg/ml). 4 h later, MTT was removed and 150 µl DMSO was replaced. A model 550 microplate reader was used to measure the spectrophotometric absorbance at 570 nm with a reference wavelength of 655 nm. The cytotoxicity of NK cells to MRC-5=100%-[(absorbance of test group-absorbance of effector control group)/absorbance of target control group].
ELISA assay
We used ELISA assay to evaluate TNFα secreted into the media. 3.5×105 cultured NK cells were planted in each well of a 6-well plate. 48 h later, two-antibody sandwich ELISA was applied to determine human TNFα expression level in the supernatant of cells. The involved antibodies are monoclonal mouse anti-human TNFα antibody (R&D Systems), peroxidase-conjugated rabbit anti-goat IgG (H&L) and goat anti-human TNFα antibody (R&D Systems). The absorbance was assessed at a 450 nm wavelength.
Statistical analysis
The difference in miR-155 serum level between TB patients and healthy controls was determined by two-tailed student’s test. The associations between serum miR-155 level and the cytotoxicity of NK cells as well as TNFα level were determined by Pearson analysis. Differences were considered as statistically significant (*) when P<0.05, statistically very significant (**) when P<0.01, and statistically extremely significant when P<0.001 (***).
Results
The serum from TB patients contains lower concentration of miR-155
A group of TB patients were recruited for our study, as well as healthy volunteers. We employed qPCR assays to detect the expression level of miR-155 in the serum sample collected from patients with TB (n=90) and healthy volunteers (n=31). The data revealed that miR-155 level was significantly reduced in the serum samples of TB patients, compared with healthy volunteers (P<0.001) (Figure 1).
Figure 1.
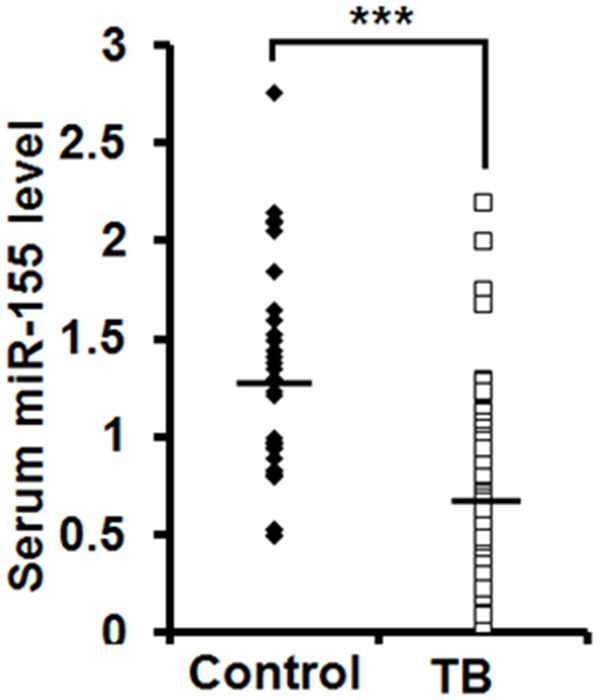
Serum miR-155 abundance is reduced in TB patients. qPCR assay was used to examine the levels of serum miR-155 from TB patients (n=90) and healthy volunteers (Control, n=31). The lines indicated the average value of miR-155 levels in TB patients and healthy group.
The level of serum miR-155 is inversely associated with the cytotoxicity of NK cells
Subsequently, we applied MTT assays to examine the viability of normal fibroblast cell line MRC-5 co-cultured with TB patients-derived NK cells, in order to determine the association between serum miR-155 abundance and the cytotoxicity of NK cells. Pearson analysis revealed that the level of serum miR-155 is inversely associated with the cytotoxicity of NK cells isolated from TB patients (n=37, R=-0.659, P=0.000) (Figure 2).
Figure 2.
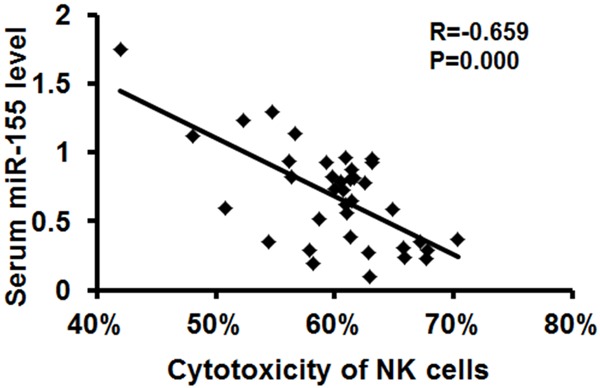
Serum miR-155 level was inversely associated with the cytotoxicity of NK cells. The cytotoxicity of NK cells isolated from TB patients (n=37) were evaluated by MTT assays after co-culturing with MRC-5 cells. The association between serum miR-155 level and the cytotoxicity of NK cells was determined with Pearson analysis.
TNFα expression by NK cells is negatively associated with abundance of serum miR-155
TNFα mediated the effect of NK cells on the progression of TB, since that this cytokine has been well demonstrated to be the major immunostimulatory protein that contribute to TB. Therefore, we employed ELISA assays to detect the concentration of TNFα that was secreted into the media of NK cell culture. The data indicated that NK cells with low serum miR-155 level had higher concentration of TNFα, than high miR-155 group. There is an inverse association between miR-155 level and TNFα from NK cells (n=37, R=-0.694, P=0.000) (Figure 3).
Figure 3.
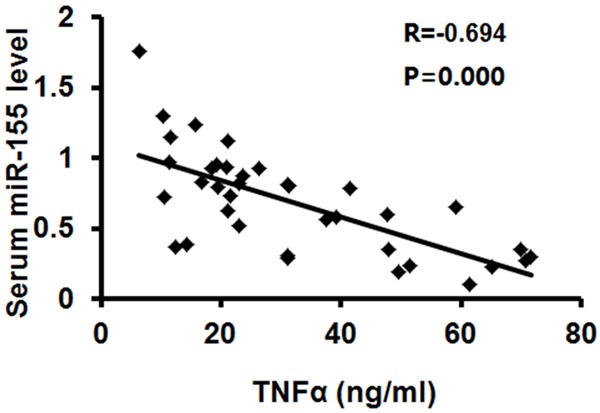
Serum miR-155 level was inversely associated with TNFα production by TB-associated NK cells. The concentration of TNFα in the media of NK cells was examined by ELISA assay (ng/ml). The association between miR-155 level and TNFα concentration was determined by Pearson analysis.
Discussion
Much evidence has shown that miRNA is closely associated with the progression of TB [8]. Although the mechanisms, by which miRNA was secreted from cells, is still unknown, miRNA can be detected not only in cell lysate, but also in the blood sample collected from animals [9]. Therefore, some miRNAs have been used to discriminate TB patients from healthy people [10-12] for diagnostic purposes. In this study, low level of miR-155 was detected in the serum of TB patients, and there is a significant difference in its expression level between TB patients and healthy controls. The data revealed that miR-155 can be used to distinguish TB patients from healthy individuals.
Recently, high miR-155 expression has been identified to account for the decline in the cytotoxicity of NK cells, maybe by suppressing TNF-γ production [7]. The above finding implied that serum miR-155 can be used as a biomarker to predict the activation of NK cells under pathological conditions. However, it is still unknown if miR-155 abundance in blood is associated with the activation of NK cells in TB patients.
This study is aimed to study the relationship between serum miR-155 level and the bioactivity of NK cells. The results showed that the level of serum miR-155 is closely relevant with the activation of NK cells, evidenced by increased cytotoxicity of NK cells and higher production of TNFα in low miR-155 TB patients. This finding demonstrated that serum miR-155 abundance can be used to evaluate the activation of NK cells in TB patients.
In order to elucidate the mechanism by which miR-155 participates in the progression of TB, further studies are still needed on TB patients. These efforts are believed to contribute to our understanding of molecular mechanism of immune response of TB patients against Mycobacterium tuberculosis.
Taken together, we demonstrated that miR-155 is closely associated with the development of TB. The level of serum miR-155 is significantly lower in TB patients than healthy individuals. The level of serum miR-155 is relevant to the activation of NK cells. These results suggested that miR-155 may be an effective agent for TB treatment.
Disclosure of conflict of interest
None.
References
- 1.Dheda K, Gumbo T, Gandhi NR, Murray M, Theron G, Udwadia Z, Migliori GB, Warren R. Global control of tuberculosis: from extensively drug-resistant to untreatable tuberculosis. Lancet Respir Med. 2014;2:321–338. doi: 10.1016/S2213-2600(14)70031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozzano F, Marras F, De Maria A. Immunology of Tuberculosis. Mediterr J Hematol Infect Dis. 2014;6:e2014027. doi: 10.4084/MJHID.2014.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Squadrito ML, Etzrodt M, De Palma M, Pittet MJ. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013;34:350–359. doi: 10.1016/j.it.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan RP, Fogel LA, Leong JW, Schneider SE, Wong R, Romee R, Thai TH, Sexl V, Matkovich SJ, Dorn GW 2nd, French AR, Fehniger TA. MicroRNA-155 tunes both the threshold and extent of NK cell activation via targeting of multiple signaling pathways. J Immunol. 2013;191:5904–5913. doi: 10.4049/jimmunol.1301950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iannaccone M, Dorhoi A, Kaufmann SH. Host-directed therapy of tuberculosis: what is in it for microRNA? Expert Opin Ther Targets. 2014;18:491–494. doi: 10.1517/14728222.2014.897696. [DOI] [PubMed] [Google Scholar]
- 9.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 10.Weiner J, Maertzdorf J, Kaufmann SH. The dual role of biomarkers for understanding basic principles and devising novel intervention strategies in tuberculosis. Ann N Y Acad Sci. 2013;1283:22–29. doi: 10.1111/j.1749-6632.2012.06802.x. [DOI] [PubMed] [Google Scholar]
- 11.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. 2011;49:4246–4251. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One. 2012;7:e43184. doi: 10.1371/journal.pone.0043184. [DOI] [PMC free article] [PubMed] [Google Scholar]


