Abstract
Background: The extraordinary invasive capability is a major cause of treatment failure and tumor recurrence in lung cancer. Evidence in other cell systems has implicated the regulatory role of microRNA-145 in cell motility and invasion, which promotes us to investigate the biological functions of miR-145 in lung cancer in this regard. Results: We have found that miR-145 is dramatically down-regulated in clinical specimen of lung cancer and is negatively correlated with the tumor pathological grading in the current study. The cells transfected by miR-145 expression vector have demonstrated retarded cell mobility. Using a bioinformatics analysis approach, fascin homolog 1 (FSCN1), actin-binding protein, has been identified as the target of miR-145. Over-expression of miR-145 mimics enhanced protein levels of E-cadherin and fibronectin, indicative of its inhibitory role in EMT occurrence. Mechanistic studies showed that miR-145 mimics inhibited FSCN1 expression and miR-145 inhibitor enhanced it. Over-expression of FSCN1 reversed miR-145-regulated expression of EMT markers, suggesting that FSCN1 mediated the inhibitory effects of miR-145. Our results revealed a novel mechanism that miR-145 inhibits lung cancer cells migration and invasion via FSCN1 downregulation. Conclusions: These results suggest that miR-145 may function as anti-migration and anti-invasion influence in lung cancer and provides a potential approach for developing miR-145-based therapeutic strategies for malignant lung cancer therapy.
Keywords: miR-145, FSCN1, lung cancer
Introduction
Lung cancer is one of the leading causes of cancer death worldwide. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer [1]. The 5 year survival rate for lung cancer remains poor. In order to develop more effective therapies, it is important to obtain a better understanding of the molecular biology of lung cancer. miRNAs are an abundant group of endogenous small non-coding RNAs that function as regulators of protein encoding genes through translational repression and/or degradation of their target mRNAs [2]. Extensive miRNA research has revealed that miRNAs are involved in the regulation of numerous key cellular functions such as metabolism, cell proliferation, tumorigenesis, apoptosis, development and differentiation [3]. To regulate target mRNAs, mature miRNAs are bound to AGO proteins and guide the AGO-associated RNA induced silencing complex (RISC) to mRNA targets through imperfect base pairing between the miRNA and the target. This often involves perfect base paring between the 5’ end of the miRNA strand and its target, also termed the seed site. Bioinformatics analyses suggest that each miRNA can control hundreds of target genes in humans and it has recently been reported that over 60% of protein encoding genes are under selective pressure to maintain pairing with miRNAs, indicating that miRNAs have the potential to regulate the majority of protein encoding genes [4]. Given the overwhelming evidence that miRNAs are important regulators of tumor genesis [5], identification of miRNA targets is necessary in order to understand the mechanistic basis for the involvement of miRNAs in cancer.
MiR-145 has frequently been reported as down-regulated in cancers, including prostate cancer [6], bladder cancer [7], colon cancer [8], ovarian cancer [9] as well as B-cell malignancies [10]. Accordingly, miR-145 over expression has been demonstrated to have a growth inhibitory effect and to suppress anchor age independent growth. It has furthermore been demonstrated that miR-145 expression induced by p53. Moreover, miR-145 inhibits cell growth by targeting c-Myc and IRS-1 [11], suppresses the pluripotent potential of embryonic and cancer stem cells by targeting OCT4, SOX2 and NANOG [11,12] and regulates cell migration, invasion and metastasis by targeting ADAM17 [13], mucin1 [14], FSCN1 [15].
In this study we examined the expression of miR-145 in lung cancer and its effects on lung cancer cell functions. We found that miR-145 was downregulated in lung tumors as compared to normal lung tissues and that overexpression of miR-145 in A549 cells and H460 cells decreased cell migration. MiR-145 has tumor-suppressive function and directly controls the oncogenic actin-binding protein, Fascin homolog1 gene (FSCN1). Furthermore, FSCN1 expression is inversely correlated with miR-145 expression levels in clinical lung cancer specimens. Insight into the association between miRNAs and their target gene net works could enhance our understanding of the molecular mechanism of lung cancer carcinogenesis.
Materials and methods
Tissue samples and cell lines
A total of 30 NSCLC tissues and matched normal tissues were obtained from untreated patients undergoing primary surgical treatment. The specimens were snap frozen in liquid nitrogen and stored at -80°C. This project had the informed consents from all the patients and was approved by the Ethics Committee of Jiangxi Provincial People’s Hospital, and informed consent was taken from all subjects. Three NSCLC cell lines, A549, H460, and H1299, and a normal lung bronchus epithelial cell line HBE were obtained from the American Type Culture Collection (ATCC; Manassas, VA USA). All the cell lines were in DMEM media (Invitrogen, Carlsbad, CA, USA) supplemented 10% FBS at 37°C with 5% CO2, at the normal conditions according to the protocol from ATCC.
RNA isolation reverse transcription PCR and real-time PCR assay
Tissues and cells were treated with TRIzol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions, for total RNA extraction. RNA concentration was determined spectrophotometrically, and integrity was checked by gel electrophoresis. RNA quality was confirmed in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Real-time PCR was carried out by using miScript SYBR Green PCR kit (Qiagen). For transcription factor detection, 2 µg of total RNA was used to perform reverse transcription by using TransScript First-strand cDNA Synthesis SuperMix kit. The real-time PCR was performed by using STBR Premix Ex Taq™ kit (TaKaRa Bio, Otsu, Japan). GAPDH was used as internal control. The primers of miR-145 and U6 are acquired from miScript Primer assays kit (Qiagen). The primers for the detection of transcription factors were list in Table 1.
Table 1.
Primer sequences used for quantitative RT-PCR
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| FSCN1 | 5’-TCA GAG CTC TTC CTC ATG AAG CT-3’ | 5’-GTC CAG TAT TTG CCT GTG GAG TC-3’ |
| U6 | 5’-CTC AAC TGG TGT CGT GGA GTC GGC AAT TGA CAA GTT GAA ATA TG-3’ | 5’-ACA CTC CAA GGG CTG TAA CGG GTG CCG GAA-3’ |
| miR-145 | 5’-TCC ACT AGT CAG AGG GTT TCC GGT AC-3’ | 5’-TCG GCT AGC GAT GGA AAG AAA AGC AA-3’ |
| GAPDH | 5’-TCT CCT CTG ACT TCA ACA GCG A-3’ | 5’-GTC CAC CAC CCT GTT GCT GT-3’ |
Reagents and antibodies
miR-145 mimic/inhibitor and the controls were obtained from RiboBio (Guangzhou, China). The 3’-UTR of FSCN1 mRNA was amplified using the following primers: sense 5’-CGA TCG CTC GAG TCT GGC ACC TCT TTC TTC TGA-3’ and antisense 5’-GGC CGC TCT AGG TTT AAA CGA CAT GTG CCC AGC TCT CT-3’. The PCR product was inserted into psiCHECK-2 vector within XhoI and NotI restriction sites (Promega, Madison, WI, USA). Mutation experiment was performed using a fast mutation kit (NEB, Ipswich, Canada).
MTT cell proliferation assay
Approximately 3×103 cells were seeded in each well of a 96-well plate after 24 h incubation at the end of transfection. At various times following treatment, MTT (20ll of 5 mg/ml) was added to each well and incubated at 37°C for 4 h. Then, the cells were harvested and centrifuged and 150 μl of DMSO was added to the formazan precipitates. The absorbance values at 490 nm were detected using a MRX II absorbance reader (DYNEX Technologies, Chantilly, VA, USA) Colony formation assay 500 transfected cells were plated into a 6-well plate and cultured in DMEM containing 10% FBS for 14 days. Colonies were fixed and stained with absolute methanol for 15 min, followed by 0.5% crystal violet for 20 min. Colonies were quantified using Olympus INT-2 inverted microscope (Tokyo, Japan).
Cell proliferation and invasion assays in NSCLC cell lines
Mature miRNA molecules, Pre-miR TM miRNA precursors (hsa-miR-145, and negative control miRNA) (Applied Biosystems) and si-FSCN1 (LU-019576-00 and J-019576-08) and negative control siRNA (D-001810-10) (Thermo Fisher Scientific, Waltham, MA) were incubated with LipofectamineTM RNAiMAX (Invitrogen, Tokyo, Japan) before transfection. As recommended by the manufacturer, we first tested the transfection efficacy of miRNA in ESCC cell lines based on the downregulation of protein tyrosine kinase 9 (PTK9) mRNA after transfection with miR-1 (This method was recommended by the manufacture). Cells were reverse transfected with 10 nM miRNA or siFSCN1 and plated in 96-well plates at 3×103 cells per well. After 72 hr, cell viability was determined by XTT assay using the Cell Proliferation Kit II (Roche Molecular Biochemicals, Mannheim, Germany) as previously described [7]. In each treatment group, 3 wells were assayed for cell viability. A cell invasion assay was carried out using modified Boyden Chambers containing transwell-precoated matrigel membrane filter inserts with 8-lm pores in 24-well tissue culture plates (BD Biosciences, Bedford, MA). All experiments were performed in triplicate.
Luciferase reporter assays
HEK293 cells were cultured in 24-well plate for 24 h and co-transfected with 150 nM of miR-145 or control mimics and WT or Mut 30-UTR of FSCN1 using Lipofectamine 2000. 48 h after transfection, HEK293 cells were collected, and the relative luciferase activity was assayed using DualLuciferase Reporter Assay System (Promega, Wisconsin, WI, USA).
Western blotting
Cells were lysed and quantified by a BCA kit (Thermo, Waltham, MA, USA). Proteins were separated by 10% SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes. Then the membranes were blocked with 3% non-fat milk and incubated with antibodies against monoclonal FSCN1 antibody (1:100, ab49815, Abcam, UK) or GAPDH (1:2000, Sangon Biotech, Shanghai, China) overnight. The membranes were incubated with HRP-conjugated secondary antibody for 1 h and detected by ECL system (Pierce, Rockford, IL, USA).
Statistical analysis
All data are presented as mean ± SD and analyzed by SPSS 16.0. One-way analysis of variance (ANOVA) or two-tailed Student’s t test was used to determine the statistical significance of differences. P<0.05 was considered to be statistically significant.
Results
miR-145 expression is downregulated in NSCLC tumors
miR-145 has been reported to be down-expressed in various types of tumors; however, its expression in NSCLC tumors has not been reported. Expression of miR-145 in 30 NSCLC and matched nontumoral normal tissue samples was measured by qRT-PCR. miR-145 was significantly decreased in NSCLC tissues compared to that in the non-tumor normal tissues (Figure 1A). Expression of miR-145 in three NSCLC cell lines, A549, H460, and H1299, was significantly decreased compared to that in the 16HBE (Human bronchial epithelial) cells (Figure 1B).
Figure 1.
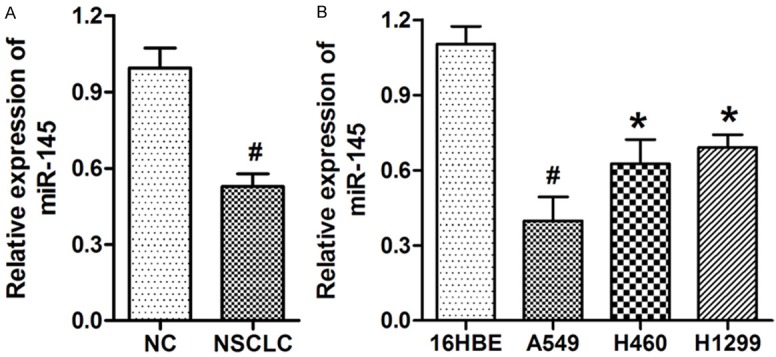
miR-145 was elevated in NSCLC tissues and cell lines. A. miR-145 was significantly decreased in 30 NSCLC tissues compared to that in the non-tumoral tissues (NC). B. miR-145 was remarkably decreased in three NSCLC cell lines compared to that in the normal bronchial epithelial cell line HBE. U6 was used as an internal control. *P<0.05, *P<0.01 versus the control group.
Over-expression of miR-145 promoted NSCLC cell growth in vitro
To explore the function of miR-145 in the regulation of NSCLC cell growth, A549 cells were transfected with miR-145 mimic. Over-expression of miR-145 remarkably promoted the growth of A549 cells (Figure 2A). Similarly, over-expression of miR-145 significantly promoted the colony formation of A549 cells (Figure 2B). The effect of miR-145 mimic was confirmed by qRT-PCR (Figure 2C).
Figure 2.
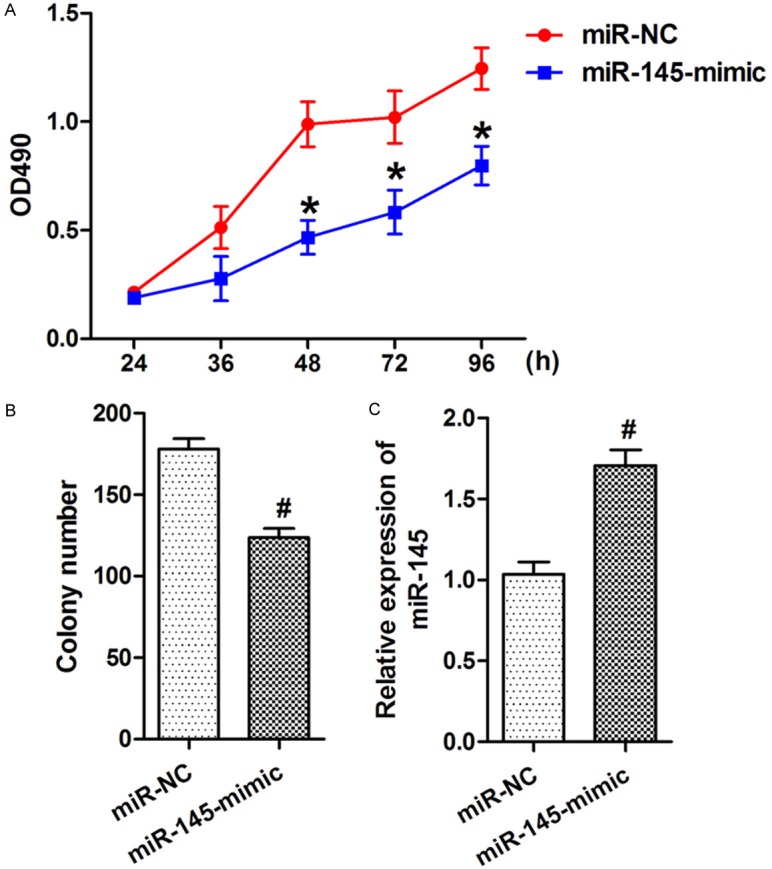
Inhibition of miR-145 suppressed NSCLC cell growth in vitro. A. At 24, 48, 36, 72, or 96 h after transfection, MTT assay was used to determine the proliferation of A549 cells. Anti-miR-145 transfection significantly promoted A549 cell proliferation. B. Anti-miR-145 transfection significantly promoted A549 cells colony formation. C. The expression of miR-145 was substantially decreased after miR-145 inhibitor transfection. Data were drawn from four independent experiments. *P<0.05, *P<0.01 versus the control group.
miR-145 suppresses NSCLC cell migration and invasion
To further investigate the role of miR-145 on motility in NSCLC cells we transfected the A549 and H460 cells with a miR-145 mimic and examined the effect of the miR mimic on cell growth and migration. The expression of synthetic miR-145 did not exert a significant effect on cell proliferation (data not shown). In contrast, we found that the miR-145 mimic significantly decreased cell migration by about 47% in A549 cells (Figure 3A, 3B, 3E) and by about 39% in the H460 cells (Figure 3C, 3D, 3F) as determined by transwell migration assay.
Figure 3.
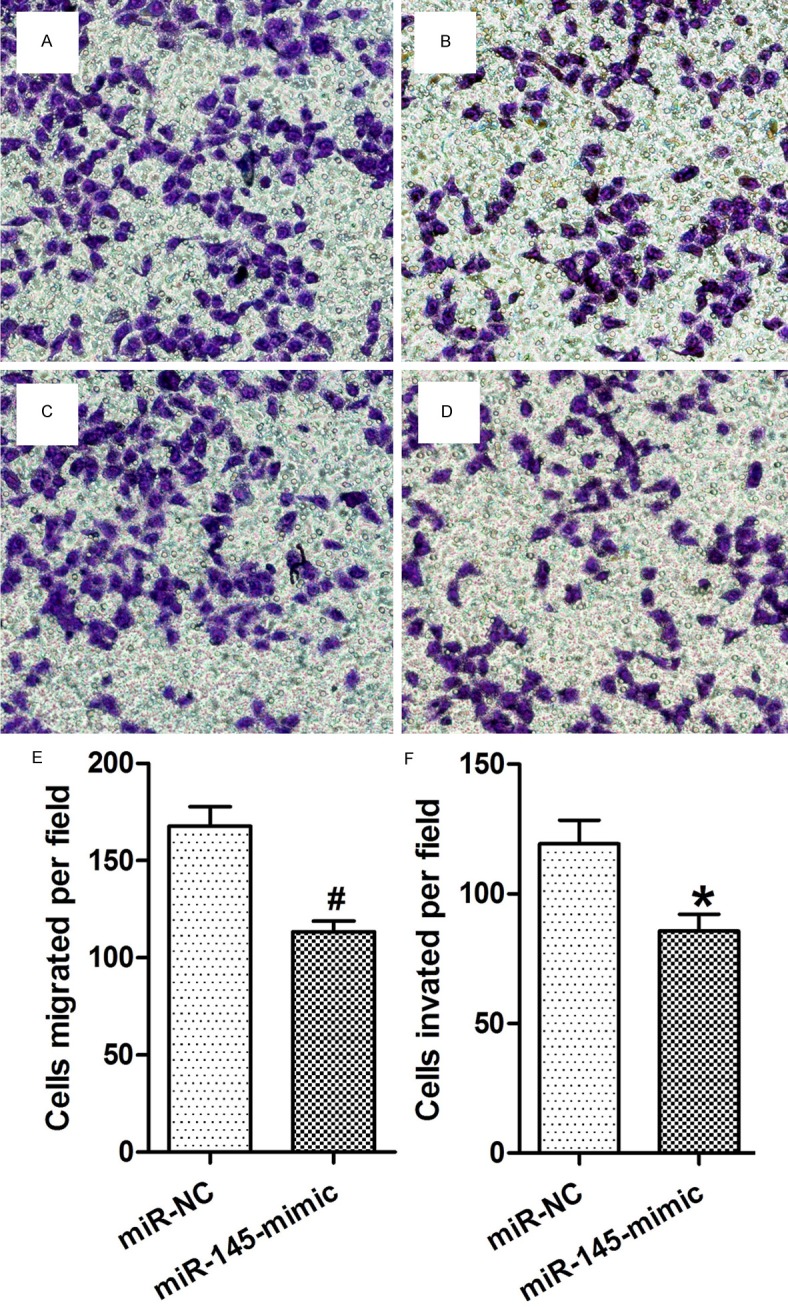
Inhibition of miR-145 promoted NSCLC cell migration and invasion. A, B. Migration assay of A549 cells transfected with miR-NC or miR-145-mimic. Inhibition of miR-145 significantly suppressed the migration of A549 cells. C, D. Invasion assay of A549 cells transfected with miR-NC or miR-145-mimic. Overexpression of miR-145 significantly suppressed the invasion of A549 cells. E, F. Statistical analysis of migrated or invaded cells after miR-145 transfection. Data were drawn from four independent experiments. *P<0.05, *P<0.01 versus the control group.
Targeting of FSCN1 by miR-145 mediates the inhibitory effect of miR-145 on lung cancer cell migration
To identify the target of miR-145 in NSCLC, TargetScan 6.2 was used. FSCN1 was predicted to be a potential target of miR-145. Relative luciferase activity assay found that miR-145 significantly inhibited the luciferase activity of the 30-UTR of FSCN1 in HEK293 cells. Moreover, overexpression of miR-145 significantly inhibited FSCN1’s expression, while inhibition of miR-145 significantly elevated FSCN1’s expression (Figure 4A, 4B). miR-145 was negatively correlated with FSCN1 in NSCLC tissues. Expression of FSCN1 in 30 NSCLC and non-tumor tissues was measured. Results showed that FSCN1 mRNA level was significantly increased in NSCLC tissues compared to that in the non-tumor tissues (Figure 5A). Moreover, FSCN1 was found to be inversely correlated with miR-145 level in NSCLC tissues (r = -0.633, Figure 5B).
Figure 4.
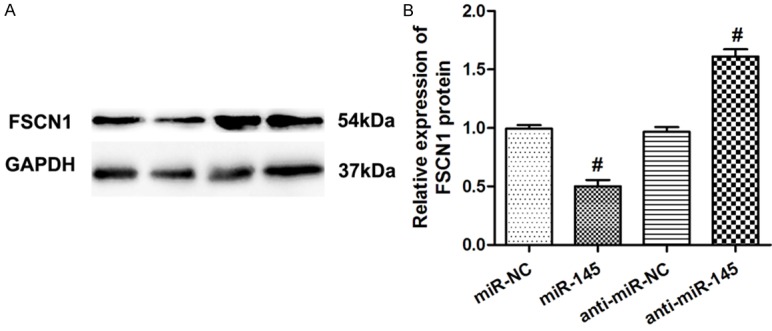
FSCN1 was a direct target of miR-145 in NSCLC cells. A. A549 cells transfected with miR-145/miR-NC, or anti-miR-145/anti-miR-NC, and representative Western blotting images were used to detect the protein levels of FSCN1. GAPDH was used as control. B. Relative expression of FSCN1 protein. Data were drawn from four independent experiments. *P<0.01 versus the control group.
Figure 5.
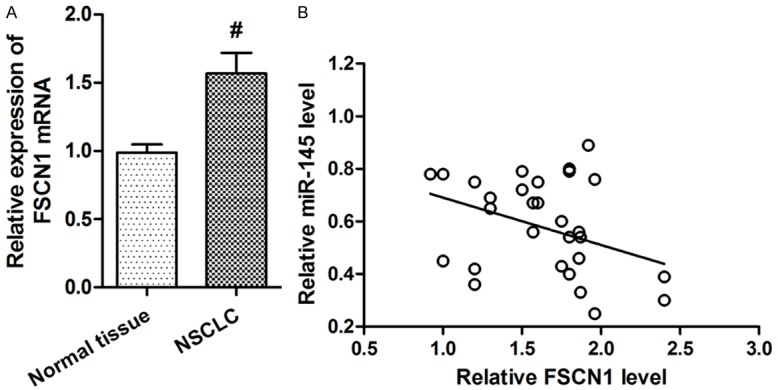
miR-145 was negatively correlated with FSCN1 in NSCLC tissues. A. FSCN1 mRNA level was significantly increased in NSCLC tissues compared to that in the non-tumoral tissues. B. FSCN1 mRNA level was inversely correlated with miR-145 level in NSCLC tissues (Spearman’s correlation analysis, r = -0.633). *P<0.01 versus the control group.
Discussion
Lung cancer is one of the malignancies with the highest incidence in the Western world and despite all advances in diagnosis and therapy one of the leading causes of cancer related death [16]. Nearly 85% of these patients suffer from non-small cell lung cancer, which is subdivided into squamous cell carcinoma, adenocarcinoma, large cell carcinomas and other rare subtypes [17].
miRNAs regulate a variety of cellular pathways by regulating the expression of multiple target genes. Therefore, we carried out genome-wide gene expression analysis to find miR-145 targets. The microarray data obtained after miR-145 transfection of cancer cells led us to focus on FSCN1.
FSCN1 is a 55-kDa globular protein that organizes F-actin into well-ordered, tightly packed parallel bundles in cells. Vertebrate genomes encode 3 forms of FSCN: FSCN1, which is widely expressed by mesenchymal tissues and in the nervous system; FSCN2, which is expressed by retinal photoreceptor cells; and FSCN3, which is testis specific [18]. FSCN1 contributes to the organization of 2 major forms of actin-based structures: cortical cell protrusions that mediate cell interactions and migration, and cytoplasmic microfilament bundles that contribute to cell architecture and intracellular movements. Many of the studies described above demonstrate how closely the formation of FSCN1-containing protrusions is linked with the activation of cell migration by physiological stimuli. Two new biological contexts where FSCN1 participates in motility and invasion have recently emerged: the movement of intracellular pathogenic bacteria and the clinical aggressiveness of human carcinomas. With the exception of the basal layer of the epidermis, FSCN1 has not been reported in normal adult epithelia. During mouse development, the equivalent epithelia are also FSCN1 negative. (A) recent burst of publications have indicated that upregulated FSCN1 protein was observed in human carcinomas from many body sites. In studies of NSCLC and gastric adenocarcinoma, high FSCN1 expression within the tumor correlated with poor survival. Increased FSCN1 content has also been documented in other invasive tumor types such as high-grade astrocytoma [19]. Our study has indicated a critical role of miR-145 in EMT and revealed its molecular mechanism. Our results suggest that miR145 targets genes essential for EMT, a prerequisite step for migration and invasion during metastasis.
In conclusion, miR-145 was overexpressed in NSCLC, and increased miR-145 could affect various biological processes of NSCLC, including proliferation, colony formation, migration, and invasion, partially by inhibiting FSCN1expression. Due to its multiple roles in tumor formation and metastasis, miR-145 may serve as an effective target for cancer diagnosis and therapy.
Acknowledgements
We thank Mr. Erjia Peng kindly help to our work.
Disclosure of conflict of interest
None.
References
- 1.Blanco R, Maestu I, de la Torre MG, Cassinello A, Nunez I. A review of the management of elderly patients with non-small-cell lung cancer. Ann Oncol. 2015;26:451–463. doi: 10.1093/annonc/mdu268. [DOI] [PubMed] [Google Scholar]
- 2.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 3.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 7.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 8.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 10.Yuan W, Sun W, Yang S, Du J, Zhai CL, Wang ZQ, Zhang J, Zhu TH. Downregulation of microRNA-142 by proto-oncogene LMO2 and its co-factors. Leukemia. 2008;22:1067–1071. doi: 10.1038/sj.leu.2405001. [DOI] [PubMed] [Google Scholar]
- 11.La Rocca G, Badin M, Shi B, Xu SQ, Deangelis T, Sepp-Lorenzinoi L, Baserga R. Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J Cell Physiol. 2009;220:485–491. doi: 10.1002/jcp.21796. [DOI] [PubMed] [Google Scholar]
- 12.Jia Y, Liu H, Zhuang Q, Xu S, Yang Z, Li J, Lou J, Zhang W. Tumorigenicity of cancer stem-like cells derived from hepatocarcinoma is regulated by microRNA-145. Oncol Rep. 2012;27:1865–1872. doi: 10.3892/or.2012.1701. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, Chopp M, Zheng X, Katakowski M, Buller B, Jiang F. MiR-145 reduces ADAM17 expression and inhibits in vitro migration and invasion of glioma cells. Oncol Rep. 2013;29:67–72. doi: 10.3892/or.2012.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sachdeva M, Mo YY. miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res. 2010;2:170–180. [PMC free article] [PubMed] [Google Scholar]
- 15.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 16.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 17.Wakelee HA, Bernardo P, Johnson DH, Schiller JH. Changes in the natural history of nonsmall cell lung cancer (NSCLC)--comparison of outcomes and characteristics in patients with advanced NSCLC entered in Eastern Cooperative Oncology Group trials before and after 1990. Cancer. 2006;106:2208–2217. doi: 10.1002/cncr.21869. [DOI] [PubMed] [Google Scholar]
- 18.Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Peraud A, Mondal S, Hawkins C, Mastronardi M, Bailey K, Rutka JT. Expression of fascin, an actin-bundling protein, in astrocytomas of varying grades. Brain Tumor Pathol. 2003;20:53–58. doi: 10.1007/BF02483447. [DOI] [PubMed] [Google Scholar]


