Abstract
Objective: The study aimed to validate the efficacy of detection of fetal cell-free DNA in maternal plasma of trisomy 21, 18 and 13 in a clinical setting. Methods: A total of 2340 women at high risk for Down syndrome based on maternal age, prenatal history or a positive sesum or sonographic screening test were offered prenatal noninvasive aneuploidy test. According to the prenatal noninvasive aneuploidy test, the pregnant women at high risk were offered amniocentesis karyotype analysis and the pregnant at low risk were followed up to make sure the newborn outcome. Results: The prenatal noninvasive aneuploidy test was positive for trisomy 21 in 17 cases, for trisomy 18 in 6 cases and for trisomy 13 in 1 case, which of all were confirmed by karyotype analysis. Newborns of low risk gestational woman detected by prenatal noninvasive aneuploidy for trisomy 21, 18, 13 were followed up and no one was found with trisomy. Conclusions: The prenatal noninvasive aneuploidy test is highly accurate for detection of trisomy 21, 18 and 13, which can be considered as a practical alternative for traditional invasive diagnostic procedures.
Keywords: Down syndrome, fetal cell-free DNA, prenatal noninvasive diagnosis
Introduction
Chromosome aneuploidies are a common kind of genetic defects, which may induce heavy burden to the society and families. The common chromosome aneuploidies among fetuses are trisomy 21 (Down syndrome, T21), trisomy 18 (Edwards syndrome, T18), trisomy 13 (Patau syndrome, T13). The trisomy 21 is the most common aneuploidy with prevalence rate at approximately one in 700 births and the risk of it increases with maternal age. Births with T21 have one more chromosome in chromosome 21. Specific facial features, severe mental retardation and growth retardation are the major clinical manifestations of T21. The morbidity of T13 is estimated at one in 25000. And the births with T13 have one more chromosome in chromosome 13. The malformation and manifestations of T13 are more severe than those of T21. Births with T18 have one more chromosome in chromosome 18 with the clinical features including mental retardation, development disorders and deformity. So far, there is no cure for these chromosomal aneuploidies, nor is there an effective treatment. Therefore, it is highly significant for us to detect these fetuses with chromosomal aneuploidy effectively. Invasive manipulations such as chorionic villi sampling, cord blood sampling and amniocentesis are used to identify those chromosomal aneuploidies. Villus cells and aminotic cells are cultured and used to make a definite diagnose by karyotype analysis which were regarded as golden standards for antenatal diagnosis of T21, T18 and T13 in the past 3 decades [1]. The diagnosis of karyotype analysis is very accurate, but the invasive of which may cause intrauterine infection and abortion, and demands a great experience of techniques and certain technical equipments. Moreover, the accurate results of karyotype analysis may not be obtained due to the bad quality of sampling and cell contamination. It cannot be popularized in all pregnant women using karyotype analysis to confirm chromosomal aneuploidies. Therefore, noninvasive prenatal test should be one of the most important directions in future of obstetrics.
The recently developed noninvasive prenatal test by massively parallel sequencing the cell-free fetal DNA circulating in maternal peripheral blood presents a new era of prenatal screening. The following studies proved cell-free fetal DNA circulating in maternal peripheral blood could be used in prenatal screening for chromosomal aneuploidies [2]. A series of clinical validation studies [3] showed that the detection sensitivity for T21 and T18 was greater than 99% and the detection sensitivity for T13 ranged from 78.6% to 91.7%, with less than 1% false positive rates. However, the above studies were retrospective study and mainly conducted in high-risk and advanced maternal age (≥ 35 years old) populations. The non-invasive prenatal test of T21, T18 and T13 using cell-free fetal DNA circulating in maternal peripheral blood in our follow-up study, which was with cooperation of Huada Genomics Institute and the well diagnostic value were found.
Materials and methods
Study subjects
From March 2012 to May 2013, a total of 2340 pregnant women with fetuses who were diagnosed as high risk of fetal aneuploidies such as T21, T18, T13 were enrolled. The high risks were as follows: over age 35, the histories of abnormal pregnancy including children with Down syndrome and repeated spontaneous abortion, stillbirth in pregnancy periods, abnormal serological screening for Down syndrome at early and mid pregnancy, abnormal screening for fetal nuchal translucency using color duplex ultrasonography between 11-14 weeks of gestation. Fetal cell-free DNA in matemal plasma was used to prenatal diagnosis of fetal aneuploidies for these patients. They had a singleton pregnancy with a gestational age of 12-14 weeks. Among them, 279 women were > 35 years and 2061 women were ≤ 35 years. The basic characteristics of study subjects were shown in Table 1.
Table 1.
General data of 2340 pregnant women
| Clinical features | Cases |
|---|---|
| Maternal age | |
| < 35 years | 2061 |
| ≥ 35 years | 279 |
| Gestation age at blood sampling | |
| 12-14 weeks | 80 |
| 15-20 weeks | 2239 |
| ≥ 24 weeks | 21 |
| Miscarriage history | |
| 0 time | 2172 |
| 1 time | 150 |
| 2 times | 14 |
| ≥3 times | 4 |
| Pregnant women(≥ 35 years) not screening for T21 | 147 |
| Down’s syndrome screening (high risk of T21) | 1189 |
| High risk of T18 | 18 |
| Only abnormal results of HCG marker | 558 |
| Only abnormal results of AFP marker | 319 |
| Only abnormal results of uE3 marker | 109 |
| Abnormal results of ultrasound (NT > 2.5 mm) | 72 |
HCG: Human Chorionic Gonadotropin; AFP: Alpha-fetoprotein; uE3: unconjugated estrio; NT: Nuchal Translucency.
All women provided written informed consent prior to participation in the current study and were told that this examine had the limitations of diagnose for chromosomal chimaera and microdeletions. They were provided with insurance plans on behalf of Shenzhen Huada Genomics Institute. Positive results of noninvasive prenatal test between 16-24 weeks of gestation were in 24 pregnant women. And then they were scheduled to receive invasive diagnostic testing (Amniocentesis). The amnioreduction and karyotyping were performed in these hosptials such as Shenzhen Maternal and Child Health Hospital which had the qualification of prenatal diagnosis. Fetuses from 2316 pregnant women with negative results of noninvasive prenatal test were followed up.
Methods
A total of 5 ml peripheral blood was drawn into an EDTA tube. Each tube was made a mark and kept at 4 degrees. Free DNA in matemal plasma was extracted using QIAamp Circulation Nucleic Acid Kit from Qiagen (Hilden, Germany). The resulting plasma DNA was sequenced using new high-throughput sequencing platforms. Compared with base sequence, the chromosome of free DNA was identified. The number and percentage of Unique read in each chromosome was calculated. At last, z-score of chromosome in samples was analyzed, and the cases of fetal chromosome aneuploidies (T21, T18, and T13) were identified. The flow chart was seen in Figure 1.
Figure 1.
The flow chart of noninvasive prenatal testing for T21, T18 and T13 using DNA.
Statistical analysis
The software package SPSS 13.0 was conducted for statistical analysis. Kappa statistic was employed to evaluate concordance between the two methods. P<0.05 was regarded as significance.
Results
General data of pregnant women
A total of 2340 pregnant women were enrolled in this study. General data including gestational age, conceptional age at time of blood drawing, screening result for Down syndrome and nuchal translucency were seen in Table 1. The testing results of fetal free DNA in plasma from all pregnant women for T21, T18 and T13 were seen in Figure 2. The comparison results of T-score between pregnant women with high risk of fetal aneuploidies such as T21, T18, T13 and normal pregnant women were seen in Table 2.
Figure 2.
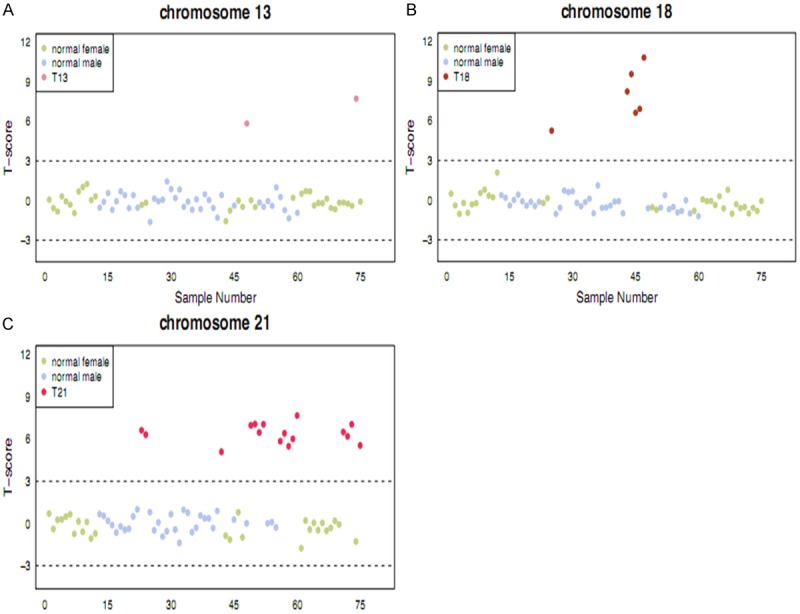
The results of noninvasive prenatal testing using fetal cell-free fetal DNA in 2340 pregnant women. A: Trisomy 13; B: Trisomy 18; C: Trisomy 21. The yellow and purple points represents T-score of pregnant women, red point indicates T-score of pregnant women with high risk of Trisomy 13, 18, 21.
Table 2.
The results of T value in pregnant women with high risk of T21, T18 and T13 using noninvasive prenatal testing of cell-free fetal DNA in maternal blood
| Case NO. | Karyotype | T-score |
|---|---|---|
| S1 | Trisomy 21 | 6.618500358 |
| S2 | Trisomy 21 | 6.310893329 |
| S3 | Trisomy 21 | 5.092127056 |
| S4 | Trisomy 21 | 6.968898206 |
| S5 | Trisomy 21 | 7.059833972 |
| S6 | Trisomy 21 | 6.463319739 |
| S7 | Trisomy 21 | 7.042486975 |
| S8 | Trisomy 21 | 5.840891226 |
| S9 | Trisomy 21 | 6.403097077 |
| S10 | Trisomy 21 | 5.483233799 |
| S11 | Trisomy 21 | 6.010570766 |
| S12 | Trisomy 21 | 7.666579033 |
| S13 | Trisomy 21 | 6.494635916 |
| S14 | Trisomy 21 | 6.191701823 |
| S15 | Trisomy 21 | 7.041483212 |
| S16 | Trisomy 21 | 5.539834097 |
| S17 | Trisomy 21 | 5.2479369 |
| S18 | Trisomy 18 | 6.595397725 |
| S19 | Trisomy 18 | 6.893230399 |
| S20 | Trisomy 18 | 8.205994041 |
| S21 | Trisomy 18 | 9.522978803 |
| S22 | Trisomy 18 | 10.76841774 |
| S23 | Trisomy 18 | 7.715531248 |
| S24 | Trisomy 13 | 5.837859312 |
Results of prenatal diagnosis from fetal free DNA in maternal plasma
Once fetuses were regarded as high risk of T21, T18 and T13 through detection of fetal free DNA in maternal plasma, karyotyping were employed to make a definite diagnosis with fetal aneuploidies. According to the detection results of fetal free DNA in maternal plasma, all fetuses with low risk of T21, T18 and T13 were followed up and there was no case of fetal aneuploidies among them, seen in Table 3.
Table 3.
The comparison results between noninvasive prenatal testing and amniocentesis testing
| noninvasive prenatal testing (cases) | karyotype analysis (cases) | |
|---|---|---|
| Trisomy 21 | 17 | 17 |
| Trisomy 18 | 6 | 6 |
| Trisomy 13 | 1 | 1 |
| Low risk of Trisomy 21, 18, 13 | 2316 | 0 |
Discussion
Down syndrome, Patau syndrome and Edwards syndrome are the most common chromosomal aneuploidies in prenatal testing and the major cause of congenital mental retardation. Among them, 90% Down syndrome occurs due to nondisjunciton of chromosome 21 during the period of meiosis [4]. These diseases with chromosomal aneuploidies were mainly intervened by prenatal screening and diagnose. At present, the pregnant women firstly are conducted prenatal screening, and then invasive operations are performed for women with high risk of chromosomal aneuploidies according to the result of prenatal screening. Invasive prenatal testing are directly employed to pregnant women with advanced maternal age (≥ 35 years old) and history of Down syndrome in births. Serum markers such as AFP, HCG and uE3 are detected in Down syndrome screening at early (7-12 weeks) and mid (15-20 weeks) pregnancy. The possibility of the fetus developing into a genetic disease are evaluated in combination with Down syndrome screening results, ultrasonic parameters, age, gestational weeks and weight.
Previous studies documented that the detection accuracy for T21 was greater than 90% [5], with a high false positive rate. It was usually hard to get ideal detection rate due to the difference in health care, economic level and screening strategy among countries and areas. Children with Down syndrome were usually born on account of leak detection. In this study, only 24 cases (1.026%) were diagnosed with T21, T18 or T13 in 2340 pregnant women with abnormal screening results or demanding prenatal testing. Meanwhile, because of false position existence in these techniques, pregnant women with normal fetuses may receive invasive prenatal diagnose, which may brought risks of abortion and intrauterine infection for normal pregnant women. One normal fetus in all three T21 cases miscarried when pregnant women with high risk received invasive prenatal testings [6,7]. The shortcomings of these prenatal screening and diagnoses were as follows: they were only suitable for pregnant women with 10-20 gestational weeks and the screening markers were strictly accordance with duration of pregnancy.
Lo et al. firstly discovered that fetal cell-free DNA floated in maternal blood in 1997 [8]. The results of Iiianes et al. showed cell-free fetal DNA in maternal blood could be detected and obtained reliable result at 4 and 7 weeks of gestation respectively [9]. The cell-free fetal DNA in maternal blood mainly from apoptotic trophoblastic cells and fetal cells existed in the form of small fragments. The cell-free fetal DNA accounted for 3-6% of total DNA in maternal plasma [8,10]. The cell-free fetal DNA in maternal blood is the basic of non-invasive prenatal diagnosis. Lo et al. [11] demonstrated that PLAC4 gene located in chromosome 21, only specially expressed in placenta and not in maternal tissues, which provided scientific foundation for clinical application of cell-free fetal DNA in non-invasive prenatal diagnose of chromosomal aneuploidies. The features of cell-free fetal DNA including content-rich, small fragments, only 163 bp size, simple sampling and analysis techniques, and detection at early pregnancy leaded cell-free fetal DNA to easily apply to the high-throughput detective method of clinical large samples [12].
Fan et al. [13] reported that Down syndrome was firstly detected by high-throughput shotgun method using plasma samples at 14 weeks pregnancy. In this method, millions of short DNA fragments from fetus and mother were sequenced and the sequence tag density of certain chromosome was calculated. Then, copy-number change of fetal chromosomal aneuploidy or fetal smaller chromosomal imbalances was examined. Cell-free fetal DNA in maternal blood was applied to diagnose with T21, T18 and T13 by high-throughput sequencing platforms with cooperation of Huada Genomics Institute. This method could reduce pregnant women’s anxiety caused by waiting results for it can be firstly employed for detection of plasma samples at 12 weeks pregnancy. Non-invasive pregnant diagnose using cell-free fetal DNA in maternal blood can reduce physical and psychological burden of pregnant women for its advantages ofsafe, reliability and no damage to maternal and child. This method was particularly suitable for pregnant women with advanced maternal age, because they need prenatal diagnose for increased risk of chromosomal aneuploidies more frequently. Watt et al. [14] reported that a woman’s fertility declines over time, particularly for women with age over 35 years. Most of these women usually became pregnant with the help of assisted reproductive technology, thus the safety of pregnancy was great important for them. And invasive pregnant diagnose may give them a very heavy burden. In this study, 279 pregnant women with over age 35 years directly took non-invasive prenatal diagnose due to the shortcoming of invasive prenatal diagnose. The final results showed that only 15 cases (5.38%) had abnormalities and other 264 cases had normal results. non-invasive prenatal diagnose undoubtedly were beneficial for them. Lee et al. suggested that fetal DNA content in plasma of pregnant women with T21 fetus were 1.7 times of that of normal pregnant women [15], which also indicated these women were suitable for this non-invasive prenatal diagnose. Copy-number analysis of fetal chromosome using cell-free fetal DNA in maternal blood brought a breakthrough for clinical technology of non-invasive prenatal diagnose.
This study validated the efficacy of noninvasive prenatal aneuploidy test of T21, T18 and T13 using cell-free fetal DNA in maternal blood in a clinical setting. Newborns of low risk gestational woman who were detected by noninvasive prenatal aneuploidy for T21, T18, T13 were followed up and no one was found with trisomy. The difference between pregnant women with high risk of fetal aneuploidies such as T21, T18, T13 undertaking the cell-free fetal DNA method and these pregnant women undertaking amniocentesis was compared. The detective sensitivity and specificity of non-invasive prenatal genetic diagnose were 100%. Moreover, controlled multicenter clinical trials are needed to further explore the accuracy of cell-free fetal DNA in maternal blood for detecting the fetal aneuploidies, because there were not enough samples in this study. With the development of biological technologies such as DNA separation and purification, real-time PCR and so on, the detection of cell-free fetal DNA in maternal blood will be much more popularized for prenatal screening and diagnose. The cell-free fetal DNA has many advantages, but some problems are still needed to be resolved. For example, this method is not applied in sex chromosome aneuploidies. With progress of molecular biological technologies, we think the detection of cell-free fetal DNA in maternal blood will play an more important role in screening for chromosome aneuploidies.
Disclosure of conflict of interest
None.
References
- 1.Boormans EM, Birnie E, Wildschut HI, Schuring-Blom HG, Oepkes D, van Oppen CA, Nijhuis JG, Macville MV, Kooper AJ, Huijsdens K, Hoffer MV, Go A, Creemers J, Bhola SL, Bilardo KM, Suijkerbuijk R, Bouman K, Galjaard RJ, Bonsel GJ, van Lith JM. Multiplex ligation-dependent probe amplification versus karyotyping in prenatal diagnosis: the M. A.K.E. Study. BMC Pregnancy Childbirth. 2008;8:18. doi: 10.1186/1471-2393-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP MatErnal BLood IS Source to Accurately diagnose fetal aneuploidy (MELISSA) Study Group. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 3.Sparks AB, Wang ET, Struble CA, Barrett W, Stokowski R, McBride C, Zahn J, Lee K, Shen N, Doshi J, Sun M, Garrison J, Sandler J, Hollemon D, Pattee P, Tomita-Mitchell A, Mitchell M, Stuelpnagel J, Song K, Oliphant A. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn. 2012;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci U S A. 1992;89:5847–51. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weisz B, Pandya P, Chitty L, Jones P, Huttly W, Rodeck C. Practical issues drawn from the implementation of the integrated test for Down syndrome screening into routine clinical practice. BJOG. 2007;114:493–497. doi: 10.1111/j.1471-0528.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- 6.Rosen T, D’Alton ME. Down syndrome screening in the first and second trimesters: what do the data show? Semin Perinatol. 2005;29:367–75. doi: 10.1053/j.semperi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs KM, Peipert JF. First trimester Down syndrome screening: public health implications. Semin Perinatol. 2005;29:267–71. doi: 10.1053/j.semperi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 9.Illanes S, Denbow M, Kailasam C, Finning K, Soothill PW. Early detection of cell-free fetal DNA in maternal plasma. Early Hum Dev. 2007;83:563–6. doi: 10.1016/j.earlhumdev.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218–24. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo YM, Tsui NB, Chiu RW, Lau TK, Leung TN, Heung MM, Gerovassili A, Jin Y, Nicolaides KH, Cantor CR, Ding C. Plasma placental RNA allelic ratio permits noninvasive prenatal chromosomal aneuploidy detection. Nat Med. 2007;13:218–23. doi: 10.1038/nm1530. [DOI] [PubMed] [Google Scholar]
- 12.Hahn S, Zhong XY, Holzgreve W. Recent progress in non-invasive prenatal diagnosis. Semin Fetal Neonatal Med. 2008;13:57–62. doi: 10.1016/j.siny.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Steinborn A, Rebmann V, Scharf A, Sohn C, Grosse-Wilde H. Placental abruption is associated with decreased maternal plasma levels of soluble HLA-G. J Clin Immunol. 2003;23:307–314. doi: 10.1023/a:1024592901663. [DOI] [PubMed] [Google Scholar]
- 14.Watt AH, Legedza AT, Ginsburg ES, Barbieri RL, Clarke RN, Hornstein MD. The prognostic value of age and follicle-stimulating hormone levels in women over forty years of age undergoing in vitro fertilization. J Assist Reprod Genet. 2000;17:264. doi: 10.1023/A:1009458332567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee T, LeShane ES, Messerlian GM, Canick JA, Farina A, Heber WW, Bianchi DW. Down syndrome and cell-free fetal DNA in archived maternal serum. Am J Obstet Gynecol. 2002;187:1217–21. doi: 10.1067/mob.2002.127462. [DOI] [PubMed] [Google Scholar]


