Abstract
Objective: To investigate the relationships of resistance mutations of hepatitis B virus (HBV) with replication and genotypes of HBV and to understand the common resistance mutations and mutation pattern. Methods: The mutation patterns related to resistance to nucleoside drugs were analyzed, and the relationships of resistance mutations with HBV genotypes, Ct value, HBeAg, alanine aminotransferase (ALT), age and gender were evaluated. Results: Genotype B was found in 52 patients (73.2%) and genotype C in 19 patients (26.8%). In addition, 32 patients (45.07%) had resistance mutations at different loci, of which single base mutation accounted for 56.25% (18/32) and multi-base mutation for 43.75% (14/32). Of single base mutation, L180M, M204I, M204V and V173L had higher prevalence, and the incidence of L180M was closely related to the genotype of HBV. L180M, M204I and M204V were associated with the resistance to lamivudine and telbivudine; L180M, M204I, M204V and V173L were associated with the resistance to entecavir; A181T, N236T and N/H238T were related to the resistance to adefovir. Of multi-base mutations, L180M combined M204V had a high prevalence and were frequently found in patients with resistance to lamivudine and telbivudine. There was cross-resistance between lamivudine and telbivudine, between lamivudine and entecavir, and between entecavir and telbivudine. The Ct value of HBV DNA, HBeAg, ALT, age and gender were comparable among patients with different resistance mutations and HBV genotypes. Conclusion: Detection of mutations of multiloci resistance genes is helpful for timely identification of HBV resistance and the clinical anti-virus therapy.
Keywords: Hepatitis B virus, genotype, resistance mutation, nucleoside drugs
Introduction
Hepatitis B is a liver inflammatory disease caused by hepatitis B virus (HBV) and may cause injury to multiple organs [1]. Hepatitis B is a public health problem word wide and about 360 million people are infected by HBV. Liver cancer has been the fifth common malignancy worldwide and more than 1 million patients die of hepatitis B related disease or liver cancer. China has a high prevalence of hepatitis B [1,2]. Thus, it is imperative to develop effective strategies for the therapy of hepatitis B.
In the therapy of chronic hepatitis B, anti-virus treatment is crucial. Drugs used for anti-virus treatment are classified as interferons and nucleos(t)ide analogues. Currently, approved and commercially available anti-HBV nucleos(t)ide analogues in China include lamivudine, telbivudine, entecavir, adefovir and tenofovir. Nucleos(t)ide analogues may inhibit the polymerase activity of HBV to exert anti-viral effect [3,4]. The long term and wide application of nucleos(t)ide analogues result in the occurrence of HBV resistance [5]. Hepatitis B patients with resistance to nucleos(t)ide analogues usually develop increase in viral load (an increment of HBV DNA by more than 100 IU/ml), elevation of serum alanine aminotransferase (ALT), persistent serum HBeAg positivity and clinical deterioration [4].
The resistance mutation related to nucleos(t)ide analogues usually found in the polymerase gene of HBV DNA (P region) [6]. Currently, there is a consensus on the nomenclature of resistance mutations: the amino acids from upstream to downstream in the P region is named rt1 to rt344, and P region is also classified as seven sub-regions: A, B, C, D, E, F and G. Mutation loci are mainly found in the A, B, C and D sub-regions. The resistance of HBV is caused by the resistance mutations of bases in the P region of HBV DNA due to long lasting therapy, the subsequent changes in the encoded amino acids and DNA polymerase structure and the nucleos(t)ide analogues failing to bind to DNA polymerase [7]. The presence of drug resistance may significantly compromise the efficacy of anti-viral therapy and deteriorate the disease condition [8,9]. New mutations emerge with the wide and long term application of nucleos(t)ide analogues for HBV. Thus, long lasting, large scale and persistent monitoring of resistance mutations of HBV DNA is necessary [10].
In the present study, sequencing of P region of HBV DNA, viral load detection and genotype analysis were done in 84 patients with hepatitis B in Guangzhou, aiming to investigate the patterns and characteristics of resistance mutations of HBV and explore the relationships of resistance mutations of HBV DNA with clinicopathologic parameters (genotype of HBV DNA, Ct value of HBV, HBeAg, ALT, age and gender). Our findings may be helpful for the understanding of common resistance mutations and mutation patterns of HBV, the identification of meaningful mutations and the rational therapy of hepatitis B, and also provide evidence for the delay or inhibition of drug resistance of HBV.
Materials and methods
Patients
Inpatients and outpatients with chronic hepatitis B who were treated with nucleoside drugs were recruited from the First Affiliated Hospital of Sun Yat-sen University between July 2011 to May 2014, and serum was collected. The chronic hepatitis B was diagnosed according to the Guideline for the Prevention and Therapy of Chronic Hepatitis B (2010) [11], and anti-viral therapy was done with nucleos (t) ide analogues.
Instruments and reagents
Reagents
HBV and resistance mutation assay kit (Shanghai Shenyou Company), HBeAg assay kit (Zhongshan Biotech Co., Ltd), and ALT detection kit (Sekisui Medical Technology, Japan) were used in the present study.
Instruments
7500 fluorescence quantitative PCR instrument, 3730 XL sequencer (ABI, USA), ML-FAME24/20 microplate reader (AusBio Co., Ltd) and AEROSET 2000 automatic biochemistry analyzer (Abbott) were used in the present study.
Methods
Detections of resistance mutations, genotypes and Ct values of HBV
Fluorescence quantitative PCR and Sanger sequencing were performed according to the manufacturers’ instructions.
Analysis of mutations
Results from Sanger sequencing were compared with the HBV DNA sequence obtained from GenBank, and the resistance mutations and mutation patterns were analyzed.
Detection of ALT
Lactate dehydrogenase conjugate UA method was used for the detection of ALT according to the manufacturer’s instructions.
Detection of HBeAg
ELISA was employed for the detection of HBeAg according to the manufacturer’s instructions.
Collection of clinical information
The clinical information was collected from the electronic medical records system between July 2011 and May 2014.
Statistical analysis
Statistical analysis was performed with SPSS version 19.0. Quantitative data (HBV DNA Ct value, ALT) were compared with one sample K-S test. When data showed normal distribution, homogeneity of variance test was done; when homogeneity of variance was present, t test was employed. Qualitative data (resistance mutation, genotype, HBeAg) were compared with chi square test. Binary logistic regression analysis was employed for the evaluation of relationships of resistance mutations with genotypes, age and gender. A value of P<0.05 was considered statistically significant.
Results
Clinical information
Of 84 patients with chronic hepatitis B, there were 67 males and 17 females with the male to female ration of 4:1. The mean age was 35.86 ± 11.58 years (range: 15-68 years). All the patients were treated with nucleos (t) ide analogues. In 13 patients, the HBV DNA was lower than the detection limit; HBV DNA was detectable in 71 patients, of whom HBV resistant mutants were found in 32 patients and wild-type HBV in 39. Detections of ALT and HBeAg were done in 39 patients.
Genotypes of HBV
Of 71 patients, genotype B was found in 52 patients (73.2%), genotype C in 19 and other genotypes or mixed genotypes were not observed.
Genotypes, resistance mutation, HBV DNA Ct value, ALT and HBeAg
Of 71 patients, the resistance mutation had an incidence of 40.4% (21/52) and the mean Ct value was 27.31 ± 7.01 in patients with genotype B; the resistance mutation had an incidence of 57.9% (11/19) and the mean Ct value was 25.63 ± 6.52 in patients with genotype C. Of 39 patients receiving detections of ALT and HBeAg, the mean ALT was 88.25 ± 45.77U/L and 14 patients were positive for HBeAg (50%) in 28 patients with genotype B; the mean ALT was 66.27 ± 63.20 U/L and 7 patients were positive for HBeAg (63.6%) in 11 patients with genotype C. Genotypes had no significant relationship with resistance mutations (P=0.189), HBV DNA Ct value (P=0.367), ALT (P=0.607) and HBeAg (P=0.442) (Table 1).
Table 1.
Genotypes, resistance mutations, Ct value, HBeAg and ALT in patients with chronic hepatitis B
| Variables | Genotype | P value | |
|---|---|---|---|
|
| |||
| B (n) | C (n) | ||
| Resistance mutation (%) | 40.4 (21/52) | 57.9 (11/19) | 0.189 |
| Ct value (x ± SD) | 27.31 ± 7.01 (52) | 25.63 ± 6.52 (19) | 0.367 |
| HBeAg (%) | 50.0 (14/28) | 63.6 (7/11) | 0.442 |
| ALT (x ± SD) | 88.25 ± 134.02 (28) | 66.27 ± 63.20 (11) | 0.607 |
Resistance mutations, HBV DNA Ct value, ALT and HBeAg
Of 71 patients, the overall incidence of HBV resistance mutation was 45.07%. The mean Ct value was 25.88 ± 6.12 in 32 patients with resistance mutations and 27.67 ± 7.42 in 39 patients without resistance mutations.
Of 39 patients receiving detections of ALT and HBeAg, 14 patients had resistance mutations, of whom the mean ALT was 67.5 ± 60.79 U/L and 6 were positive for HBeAg (42.86%); 25 had no resistance mutations, of whom the mean ALT was 90.2 ± 50.63 U/L and 15 were positive for HBeAg (60.00%). Resistance mutations were no associated with Ct value (P=0.278), ALT (P=0.571) and HBeAg (Table 2).
Table 2.
Resistance mutations, HBV DNA Ct value, ALT and HBeAg
| Variables | Mutation | P value | |
|---|---|---|---|
|
| |||
| Yes | No | ||
| HBV DNA Ct value (x ± SD) | 25.88 ± 6.12 (32) | 27.67 ± 7.42 (39) | 0.278 |
| ALT (x ± SD) | 67.5 ± 60.79U/L (14/38) | 90.2 ± 140.83U/L (25/39) | 0.571 |
| HBeAg (%) | 42.86 (6/14) | 60.00 (15/25) | 0.303 |
Genotypes, resistance mutations, age and gender
Binary logistic regression was employed to analyze the relationships of genotypes and resistance mutations with age and gender. Results showed no significant relationships among them (P>0.05) (Table 3).
Table 3.
Binary logistic regression analysis of genotypes, resistance mutations, age and gender
| Variable | Total | Genotype B | Genotype C | P | Mutation | Mutation free | P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| N=71 | n | % | n | % | n | % | n | % | |||
| Age | |||||||||||
| 20~40 | 49 | 38 | 77.6 | 11 | 22.4 | 0.225 | 20 | 40.8 | 29 | 59.2 | 0.284 |
| <20, >40 | 22 | 14 | 63.6 | 8 | 36.4 | 12 | 54.5 | 10 | 45.5 | ||
| Sex | |||||||||||
| Male | 55 | 41 | 74.5 | 14 | 25.5 | 0.646 | 25 | 45.5 | 30 | 54.5 | 0.904 |
| Female | 16 | 11 | 68.8 | 5 | 31.2 | 7 | 43.8 | 9 | 56.2 | ||
Notes: Age from 20 to 40 is the relative high incident age of hepatitis B.
Loci of resistance mutations
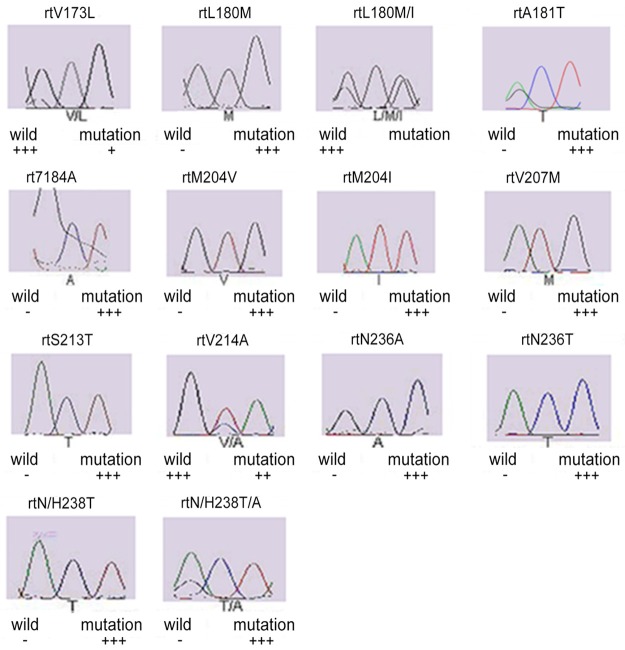
In 32 patients with chronic hepatitis B, single base mutations (n=18; 56.25%) and multi-base mutations (n=14; 43.75%) were found in the P region of HBV DNA. Single base mutations included L180M, M204I, M204V, V207M, S213T, N/H238T, T184A, V214A, N236A, V173L, L180M/I, A181T, N236T and N/H238T/A. Of these mutations, M204I had the highest detection rate, followed by L180M, M204V and V173L (Figure 1).
Figure 1.
Genotyping pictures of the mutation by sequencing.
Multi-base mutations were as follows: (1) L180M+M204I; (2) L180M+M204V; (3) V173L+M204I; (4) L180M+M204V/I; (5) V173L+L180M+M204V; (6) L180M+T184A+M204V; (7) L180M+M204I/L; (8) L180M+M204V+S213T+N/H238T+T184A; (9) L180M+M204I+V207M; (10) V214A+N/H238T; (11) M204I+N236A; (12) V173L+L180M/I+M204I+A181T+N236T. Of these mutations, L180M+M204V had the highest detection rate (Table 4).
Table 4.
Single-base mutation and multi-base mutation
| Mutation patterns | n | Detection rate (%) | Drugs |
|---|---|---|---|
| Single-base mutation | |||
| L180M | 10 | 31.25 | Lamivudine, telbivudine |
| M204I | 23 | 71.88 | Lamivudine, telbivudine, entecavir |
| M204V | 4 | 12.50 | Lamivudine, telbivudine, entecavir |
| V207M | 2 | 6.25 | Lamivudine |
| S213T | 1 | 3.13 | Lamivudine |
| N/H238T | 2 | 6.25 | Adefovir |
| T184A | 2 | 6.25 | Entecavir |
| V214A | 1 | 3.13 | Adefovir |
| N236A | 1 | 3.13 | Adefovir |
| V173L | 3 | 9.38 | Lamivudine |
| L180M/I | 1 | 3.13 | Lamivudine, telbivudine |
| A181T | 2 | 6.25 | Telbivudine, adefovir |
| N236T | 2 | 6.25 | Adefovir |
| N/H238T/A | 1 | 3.13 | Adefovir |
| Multi-base mutation | n | Ratio | |
| L180M+M204I | 1 | 1/32 | Lamivudine, telbivudine |
| L180M+M204V | 3 | 3/32 | Lamivudine, telbivudine |
| V173L+M204I | 1 | 1/32 | Lamivudine, telbivudine |
| L180M+M204V/I | 1 | 1/32 | Lamivudine, telbivudine |
| V173L+L180M+M204V | 1 | 1/32 | Lamivudine, telbivudine, entecavir |
| L180M+T184A+M204V | 1 | 1/32 | Lamivudine, telbivudine, entecavir |
| L180M+M204I/L | 1 | 1/32 | Lamivudine, telbivudine |
| L180M+M204V+S213T+N/H238T+T184A | 1 | 1/32 | Lamivudine, telbivudine, entecavir, adefovir |
| L180M+M204I+V207M | 1 | 1/32 | Lamivudine, telbivudine, entecavir |
| V214A+N/H238T | 1 | 1/32 | Adefovir |
| M204I+N236A | 1 | 1/32 | Lamivudine, telbivudine, entecavir, adefovir |
| V173L+L180M/I+M204I+A181T+N236T | 1 | 1/32 | Lamivudine, telbivudine, entecavir, adefovir |
Of 4 single-base mutations with high detection rate, the incidence of L180M was 5.77% (3/52) in patients with genotype B and 36.84 (7/19) in those with genotype C; the incidence of M204I was 32.69% (17/52) in patients with genotype B and 31.59% (6/19) in those with genotype C; the incidence of M204V was 1.92% (1/52) in patients with genotype B and 18.85% (3/16) in those with genotype C; the incidence of V173L was 1.92% (1/52) in patients with genotype B and 10.53% (2/19) in those with genotype C. Chi square test showed L180M resistance mutation was related to genotype (P=0.003), but M204I, M204V and V173L resistance mutations had no relationship with genotypes (P>0.05) (Table 5).
Table 5.
Four common single-base mutations in patients with different genotypes
| Mutations | Total | Genotypes | P | |||
|---|---|---|---|---|---|---|
|
| ||||||
| n=40 | B (N=52) | C (N=19) | ||||
| L180M | 10 | 3 | 5.77 | 7 | 36.84 | 0.003 |
| M204I | 23 | 17 | 32.69 | 6 | 31.58 | 0.929 |
| M204V | 4 | 1 | 1.92 | 3 | 15.79 | 0.096 |
| V173L | 3 | 1 | 1.92 | 2 | 10.53 | 0.173 |
Notes: N denotes the total number of the mutations for every gene type. n denotes for the total number of the mutations for four single bases.
Resistance mutations and nucleoside analogues
In 71 patients, 43 (60.56%) were sensitive to lamivudine and telbivudine and 28 were resistant to lamivudine and telbivudine; 61 (85.92%) were sensitive to entecavir and 10 were resistant to entecavir; 64 (90.14%) were sensitive to adefovir and 7 were resistant to adefovir; all the patients were sensitive to tenofovir. Of 14 single-base mutations, L180M, M204I and M204V were related to the resistance to lamivudine, telbivudine and entecavir (P<0.05); V173L was related to the resistance to entecavir (P=0.05); A181T, N236T and N/H238T were related to the resistance to adefovir (P<0.05) (Table 6).
Table 6.
Single-base mutations and sensitivity to 4 anti-viral drugs
| Mutations | Lamivudine/telbivudine | Entecavir | Mutations | Adefovir | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Resistant (n=28) | Sensitive (n=43) | P | Resistant (n=10) | Sensitive (n=61) | P | Resistant (n=7) | Sensitive (n=64) | P | ||||
| L180M | No | 18 | 43 | 0.000 | 5 | 56 | 0.002 | A181T | No | 5 | 64 | 0.008 |
| Yes | 10 | 0 | 5 | 5 | Yes | 2 | 0 | |||||
| M204I | No | 5 | 43 | 0.000 | 3 | 45 | 0.017 | N236T | No | 5 | 64 | 0.008 |
| Yes | 23 | 0 | 7 | 16 | Yes | 2 | 0 | |||||
| M204V | No | 24 | 43 | 0.043 | 7 | 60 | 0.008 | N/H238T | No | 5 | 64 | 0.008 |
| Yes | 4 | 0 | 3 | 1 | Yes | 2 | 0 | |||||
| V173L | No | 25 | 43 | 0.112 | 8 | 60 | 0.050 | |||||
| Yes | 3 | 0 | 2 | 1 | ||||||||
Cross-resistance between lamivudine and telbivudine, between lamivudine and entecavir, between telbivudine and entecavir, adefovir and telbivudine as well as between adefovir and entecavir was detected in 71 patients. Results showed there was cross-resistance between lamivudine and telbivudine, between lamivudine and entecavir as well as between telbivudine and entecavir (P<0.05) (Table 7).
Table 7.
Cross-resistance among 4 anti-viral nucleoside analogues
| Lamivudine | Adefovir | Entecavir | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Resistant (n=28) | Sensitive (n=43) | P | Resistant (n=7) | Sensitive (n=64) | P | Resistant (n=10) | Sensitive (n=61) | P | ||
| Telbivudine | Resistant | 27 | 1 | 0.000 | 4 | 24 | 0.547 | 10 | 18 | 0.000 |
| Sensitive | 1 | 42 | 3 | 40 | 0 | 43 | ||||
| Entecavir | Resistant | 10 | 0 | 0.000 | 3 | 7 | 0.053 | |||
| Sensitive | 18 | 43 | 4 | 57 | ||||||
| Adefovir | Resistant | 3 | 4 | 1.000 | ||||||
| Sensitive | 25 | 39 | ||||||||
Discussion
HBV infection has geographic regional distribution and HBV in Chinese patients with chronic hepatitis B is characterized by genotypes A, B, C and D [12,13] although the dominant genotype varies among regions. In the present study, fluorescence quantitative PCR and Sanger sequencing were performed to detect the genotypes of HBV in 71 patients with chronic hepatitis B. Results showed genotype B was dominant genotype of HBV in Guangzhou and accounted for 73.2% (52/71), genotype C accounted for 26.8% (19/71), and other genotypes were not found, which were consistent with previously reported [12,13]. Serum HBV DNA Ct value, ALT and HBeAg were comparable between patients with genotype B and those with genotype C, and age and gender had no relationship with HBV genotypes. Our study did not reveal the association between HBV genotype and resistance mutation, which might be ascribed to the small number of patients with genotype C.
With the increase in the number of patients with hepatitis B and the wide application of nucleoside analogues, the incidence of resistance mutations is changing. Our results showed the incidence of nucleoside analogues induced resistance mutations was at a high level and as high as 45.1% (32/71). These mutations could be classified as single-base mutation and multi-base mutation. Single-base mutation was dominant and accounted for 56.25% (18/32) and multi-base mutation accounted for 43.75% (14/32). Of single-base mutations, M204I had the highest incidence (71.88%), followed by L180M (31.25%), M204V (12.5%) and V173L (9.38%). Moreover, L180M resistance mutation was significantly related to HBV genotype: patients with genotype C had a higher risk for L180M as compared to those with genotype B. However, M204V, M404I and V173L were not associated with HBV genotype. Of multi-base mutations, L180M+M204V had the highest incidence 9.4% (3/32). The detection rate of mutations at rt207, rt213, rt214, rt237 and rt238 reduced, but that at rt169, rt194 and rt233 increased from August 2012 to now as compared to that from July 2011 t0 July 2012, which might be ascribed to the dynamic process of nucleoside analogues induced resistance mutations. Serum HBV DNA Ct value, ALT and HBeAg were comparable between patients with resistance mutations and those without resistance mutations, suggesting that the presence of resistance mutation may not immediately cause increase in viral load and elevation of ALT. In addition, age and gender had no associated with resistance mutations of HBV [14].
Our results showed all the patients with chronic hepatitis B in Guangzhou were sensitive to tenofovir, which may be related to more recent use of tenofovir in clinical practice. In addition, 90.14% and 85.92% of patients were sensitive to adefovir and entecavir, respectively. Of note, these patients had a consistently low sensitivity to lamivudine and telbivudine, which may be explained that both lamivudine and telbivudine are levo-nucleoside analogues and were approved for anti-viral therapy earlier than entecavir, adefovir and tenofovir. There is cross-resistance among nucleoside analogues [15-18]. This study also investigated the cross-resistance among 5 nucleoside analogues, aiming to provide evidence for the rational clinical use of nucleoside analogues. Results showed cross-resistance between lamivudine and telbivudine, between entecavir and lamivudine, and between entecavir and telbivudine in Guangzhou, indicating that combined therapy is necessary once resistance mutation is found in hepatitis B patients [19]. Clinicians should pay attention to the cross-resistance in hepatitis B patients [20].
Previous studies showed the resistance mutations were different in patients treated with different nucleoside analogues [3,21-24]. Our results showed, of 14 common single-base mutations in Guangzhou, L180M, M204I and M204V resistance mutations were related to resistance to lamivudine and telbivudine [25], and the presence of L180M resistance mutation was usually accompanied by M204V resistance mutation. Lamivudine and telbivudine have a similar structure and belong to levo-nucleoside analogues, and thus lamivudine and telbivudine induced resistance mutations are similar. L180M, M204I, M204V and V173L are the major resistance mutations caused by entecavir. Previous studies reported the entecavir-induced resistance mutations were divided into two types: (1) rtM250V+rtI169T+M204V+L180M and (2) rtT184G+rtS202I+rtM204V+rtL180M. The entecavir induced resistance mutations include mutations of one or more amino acids at rtTl84, rtS202, rtM250 and rtIl69 in the presence of mutations of rtM204+rtL180 [3,21-24]. In the present study, results showed a new entecavir induced resistance mutation, V173L, but its clinical importance is required to be further elucidated. The adefovir induced resistance mutations mainly include A181T/S, N236T and N/H238T1 [26]. No resistance mutations were found to be associated with tenofovir [27,28], which may be ascribed to more recent use of tenofovir in anti-viral therapy.
V207M, S213T, T184A, V214A, N236A, L180M/I and N/H238T/A may be the secondary single-base mutations caused by lamivudine, telbivudine, entecavir, adefovir and tenofovir [3,21-24,29,30]. A single mutation of them might has little influence on the resistance to nucleoside analogues, but the specific important is warranted to be studied in future investigations [31]. Of 12 common multi-base mutations in Guangzhou, L180M+M204V were mainly found in patients with resistance to lamivudine and telbivudine and accounted for 14.29% (4/28). A variety of studies revealed that M204V/I+L180M resistance mutations account for as high as 60% in patients with resistance to lamivudine [32]. Our results showed the incidence of M204V/I+L180M in patients with resistance to lamivudine was lower than previously reported, which may be attributed to the discrepancies in geographic regions and clinical use of nucleoside analogues. We also analyzed the correlations of L180M, M204I, M204V and L180M+M204V resistance mutations with the use of lamivudine, telbivudine and entecavir. Our results showed above 4 mutations in Guangzhou were related to the use of lamivudine, telbivudine and entecavir, which was consistent with the cross-resistance between lamivudine and telbivudine, between entecavir and lamivudine, and between entecavir and telbivudine.
In the present study, we also found 4 new resistance mutations including N236A (N236T as its common mutation), N/H238T/A (N/H238T/D as its common mutation), V207M (V207I/L/G as its common mutation) and V173L. Studies have shown that the presence of N236A resistance mutation is accompanied by M204I, suggesting that N236A resistance mutation might be related to the resistance to lamivudine, telbivudine and entecavir. V207M resistance mutation also could cause clinical resistance to lamivudine, suggesting that V207M is a new resistance mutation related to the resistance to lamivudine. N/H238T/A resistance mutation could cause clinical resistance to adefovir, indicating that N/H238T/A is a new resistance mutation related to the resistance to adefovir; the resistance to entecavir was usually caused by multi-base mutations, and the common rtM204+rtL180 mutations were frequently found at one or more amino acids among rtTl84, rtS202, rtM250 and rtIl69, suggesting that V173L is a new resistance mutation related to the resistance to entecavir. Thus, we speculate that N236A mutation may be associated with the resistance to lamivudine, telbivudine and entecavir, V207M mutation with the resistance to lamivudine, N/H238T/A mutation with the resistance to adefovir and V173L mutation may be a new multi-base mutation related to the resistance to entecavir. Further investigations are required to confirm the relationships between resistance mutations and phenotypic resistance.
The large scale, long lasting clinical use of nucleoside analogues results in increase in resistance mutations over years. Thus, it is necessary to persistently monitor the resistance mutations of HBV. In future studies, the sample size will be enlarged, detection of resistance mutations will be done once resistance to nucleoside analogues is suspected in patients with hepatitis B, and patients will be followed up for a long time. Once new resistance mutations are found, in vitro experiments and bioinformatics will be employed to analyze the mutations and phenotypic resistance.
Disclosure of conflict of interest
None.
References
- 1.Santantonio TA, Fasano M. Chronic hepatitis B: Advances in treatment. World J Hepatol. 2014;6:284–292. doi: 10.4254/wjh.v6.i5.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu FM, Zhuang H. Management of hepatitis B in China. Chin Med J (Engl) 2009;122:3–4. [PubMed] [Google Scholar]
- 3.Fung J, Seto WK, Lai CL, Yuen MF. Extrahepatic effects of nucleoside and nucleotide analogues in chronic hepatitis B treatment. J Gastroenterol Hepatol. 2014;29:428–434. doi: 10.1111/jgh.12499. [DOI] [PubMed] [Google Scholar]
- 4.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. e1591–1592. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 5.Mailliard ME, Gollan JL. Emerging therapeutics for chronic hepatitis B. Annu Rev Med. 2006;57:155–166. doi: 10.1146/annurev.med.57.121304.131422. [DOI] [PubMed] [Google Scholar]
- 6.Suppiah J, Mohd Zain R, Haji Nawi S, Bahari N, Saat Z. Drug-resistance associated mutations in polymerase (p) gene of hepatitis B virus isolated from malaysian HBV carriers. Hepat Mon. 2014;14:e13173. doi: 10.5812/hepatmon.13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoulim F. Mechanism of viral persistence and resistance to nucleoside and nucleotide analogs in chronic hepatitis B virus infection. Antiviral Res. 2004;64:1–15. doi: 10.1016/j.antiviral.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Locarnini S, Mason WS. Cellular and virological mechanisms of HBV drug resistance. J Hepatol. 2006;44:422–431. doi: 10.1016/j.jhep.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 9.Locarnini S, Hatzakis A, Heathcote J, Keeffe EB, Liang TJ, Mutimer D, Pawlotsky JM, Zoulim F. Management of antiviral resistance in patients with chronic hepatitis B. Antivir Ther. 2004;9:679–693. [PubMed] [Google Scholar]
- 10.Hsiao CC, Chang J, Wu JY, Liu WH, Han SY, Chen PJ, Yeh SH. High-resolution melting and real-time PCR for quantification and detection of drug-resistant HBV mutants in a single amplicon. Antivir Ther. 2012;17:291–303. doi: 10.3851/IMP2022. [DOI] [PubMed] [Google Scholar]
- 11.Hinese Society of Hepatology and Chinese Society of Infectious Diseases Chinese Medical Association. The guideline of prevention and treatment for chronic hepatitis B (2010 version) J Clin Hepatol. 2011;27:1–16. doi: 10.3760/cma.j.issn.1007-3418.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Yuan J, Zhou B, Tanaka Y, Kurbanov F, Orito E, Gong Z, Xu L, Lu J, Jiang X, Lai W, Mizokami M. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core and their clinical implications. J Clin Virol. 2007;39:87–93. doi: 10.1016/j.jcv.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Zeng G, Wang Z, Wen S, Jiang J, Wang L, Cheng J, Tan D, Xiao F, Ma S, Li W, Luo K, Naoumov NV, Hou J. Geographic distribution, virologic and clinical characteristics of hepatitis B virus genotypes in China. J Viral Hepat. 2005;12:609–617. doi: 10.1111/j.1365-2893.2005.00657.x. [DOI] [PubMed] [Google Scholar]
- 14.Rong HY, ZHang ZX. Associations of resistance mutations with HBV DNA load and HBeAg in patients with hepatitis B. J Prac Med. 2013;29:404–406. [Google Scholar]
- 15.Lada O, Benhamou Y, Cahour A, Katlama C, Poynard T, Thibault V. In vitro susceptibility of lamivudine-resistant hepatitis B virus to adefovir and tenofovir. Antivir Ther. 2004;9:353–363. [PubMed] [Google Scholar]
- 16.Brunelle MN, Jacquard AC, Pichoud C, Durantel D, Carrouee-Durantel S, Villeneuve JP, Trepo C, Zoulim F. Susceptibility to antivirals of a human HBV strain with mutations conferring resistance to both lamivudine and adefovir. Hepatology. 2005;41:1391–1398. doi: 10.1002/hep.20723. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Qi X, Sabogal A, Miller M, Xiong S, Delaney WEt. Cross-resistance testing of next-generation nucleoside and nucleotide analogues against lamivudine-resistant HBV. Antivir Ther. 2005;10:625–633. [PubMed] [Google Scholar]
- 18.Tenney DJ, Levine SM, Rose RE, Walsh AW, Weinheimer SP, Discotto L, Plym M, Pokornowski K, Yu CF, Angus P, Ayres A, Bartholomeusz A, Sievert W, Thompson G, Warner N, Locarnini S, Colonno RJ. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498–3507. doi: 10.1128/AAC.48.9.3498-3507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatsuji H, Suzuki F, Sezaki H, Akuta N, Suzuki Y, Kawamura Y, Hosaka T, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Watahiki S, Iwasaki S, Kobayashi M, Kumada H. Low risk of adefovir resistance in lamivudine-resistant chronic hepatitis B patients treated with adefovir plus lamivudine combination therapy: two-year follow-up. J Hepatol. 2008;48:923–931. doi: 10.1016/j.jhep.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Kim SS, Cho SW, Kim SO, Hong SP, Cheong JY. Multidrug-resistant hepatitis B virus resulting from sequential monotherapy with lamivudine, adefovir, and entecavir: clonal evolution during lamivudine plus adefovir therapy. J Med Virol. 2013;85:55–64. doi: 10.1002/jmv.23440. [DOI] [PubMed] [Google Scholar]
- 21.Inoue J, Ueno Y, Wakui Y, Niitsuma H, Fukushima K, Yamagiwa Y, Shiina M, Kondo Y, Kakazu E, Tamai K, Obara N, Iwasaki T, Shimosegawa T. Four-year study of lamivudine and adefovir combination therapy in lamivudine-resistant hepatitis B patients: influence of hepatitis B virus genotype and resistance mutation pattern. J Viral Hepat. 2011;18:206–215. doi: 10.1111/j.1365-2893.2010.01301.x. [DOI] [PubMed] [Google Scholar]
- 22.Sheng YJ, Liu JY, Tong SW, Hu HD, Zhang DZ, Hu P, Ren H. Lamivudine plus adefovir combination therapy versus entecavir monotherapy for lamivudine-resistant chronic hepatitis B: a systematic review and meta-analysis. Virol J. 2011;8:393. doi: 10.1186/1743-422X-8-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang JX, Liu BM, Li XG, Yan CH, Xu J, Sun XW, Wang YH, Jiao XJ, Yan L, Dong JP, Hou CS, Abuduheilili X, Li T, Zhuang H. Profile of HBV antiviral resistance mutations with distinct evolutionary pathways against nucleoside/ nucleotide analogue treatment among Chinese chronic hepatitis B patients. Antivir Ther. 2010;15:1171–1178. doi: 10.3851/IMP1677. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Liu Y, Zhao P, Wang Y, Chen L, Xin S, Zhang XX, Xu D. Investigation into drug-resistant mutations of HBV from 845 nucleos(t)ide analoguenaive Chinese patients with chronic HBV infection. Antivir Ther. 2015;20:141–7. doi: 10.3851/IMP2813. [DOI] [PubMed] [Google Scholar]
- 25.Li MW, Hou W, Wo JE, Liu KZ. Character of HBV (hepatitis B virus) polymerase gene rtM204V/I and rtL180M mutation in patients with lamivudine resistance. J Zhejiang Univ Sci B. 2005;6:664–667. doi: 10.1631/jzus.2005.B0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Li X, Xin S, Xu Z, Chen R, Yang J, Liu L, Wong VW, Yang D, Chan HL, Xu D. The rtA181S mutation of hepatitis B virus primarily confers resistance to adefovir dipivoxil. J Viral Hepat. 2015;22:328–334. doi: 10.1111/jvh.12298. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Wang CM, Cheng J, Liang ZL, Zhong YW, Ren XQ, Xu ZH, Zoulim F, Xu DP. Hepatitis B virus in tenofovir-naive Chinese patients with chronic hepatitis B contains no mutation of rtA194T conferring a reduced tenofovir susceptibility. Chin Med J (Engl) 2009;122:1585–1586. [PubMed] [Google Scholar]
- 28.Hann HW, Chae HB, Dunn SR. Tenofovir (TDF) has stronger antiviral effect than adefovir (ADV) against lamivudine (LAM)-resistant hepatitis B virus (HBV) Hepatol Int. 2008;2:244–249. doi: 10.1007/s12072-008-9045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, Chung YH, Kim JA, Jin YJ, Park WH, Kim SE, Lee D, Shim JH, Kim KM, Lim YS, Lee HC, Lee YS, Suh DJ. rtL180M mutation of hepatitis B virus is closely associated with frequent virological resistance to adefovir dipivoxil therapy. J Gastroenterol Hepatol. 2012;27:300–305. doi: 10.1111/j.1440-1746.2011.06853.x. [DOI] [PubMed] [Google Scholar]
- 30.Lin CL, Chien RN, Hu CC, Lai MW, Yeh CT. Identification of hepatitis B virus rtS117F substitution as a compensatory mutation for rtM204I during lamivudine therapy. J Antimicrob Chemother. 2012;67:39–48. doi: 10.1093/jac/dkr416. [DOI] [PubMed] [Google Scholar]
- 31.Locarnini S. Primary resistance, multidrug resistance, and cross-resistance pathways in HBV as a consequence of treatment failure. Hepatol Int. 2008;2:147–151. doi: 10.1007/s12072-008-9048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westland CE, Yang H, Delaney WEt, Wulfsohn M, Lama N, Gibbs CS, Miller MD, Fry J, Brosgart CL, Schiff ER, Xiong S. Activity of adefovir dipivoxil against all patterns of lamivudine-resistant hepatitis B viruses in patients. J Viral Hepat. 2005;12:67–73. doi: 10.1111/j.1365-2893.2005.00578.x. [DOI] [PubMed] [Google Scholar]